Abstract
In this report, we demonstrate that cellular stress regulates expression of IFRD1 by a post-transcriptional control mechanism. IFRD1 mRNA and protein are elevated in tunicamycin-treated human kidney epithelial cells via stabilization of the mRNA. IFRD1 mRNA instability in resting cells requires translation of an upstream open reading frame (ORF) that represses translation of the major ORF. During stress response, the mRNA is stabilized via inhibition of translational initiation mediated by phosphorylated eIF2α. Translation of the major ORF of IFRD1 involves both leaky scanning at the upstream AUG codon and re-initiation at the major AUG codon and is not altered during stress. Finally, the instability mechanism depends upon UPF1, suggesting that it is related to nonsense-mediated decay. Importantly, the sequence and length of the upstream ORF are critical but do not need to code for a specific peptide. Moreover the sequence environment of the upstream ORF termination site is not an essential feature of instability. These features of decay collectively define a distinct upstream ORF-mediated instability mechanism whereby cellular stress can modulate specific gene expression through alteration of mRNA half-life.
Keywords: GENE/Regulation, RNA/Messenger RNA/mRNA, RNA/Translation, RNA/Turnover, Translation Initiation Factors, eIF2α, Endoplasmic Reticulum Stress
Introduction
IFRD1, also known as TIS7 (mouse) or PC4 (rat), was initially identified as an immediate early gene implicated in control of growth and differentiation for neurons, myocytes, and intestinal enterocytes (1–5). The IFRD1 protein appears to function as a co-activator/repressor of gene transcription via interaction with selected transcription factors and histone deacetylase complexes (6–10). Multiple reports have also observed elevated expression of IFRD1 in models of acute tissue injury, including ischemia/reperfusion, stroke, surgical resection, and muscle trauma (4, 11–13). IFRD1 protein contributes to the magnitude of neutrophil inflammatory phenotype and determines the efficacy of muscle repair following traumatic injury (4, 10). Collectively, these findings suggest that the IFRD1 gene product may play an important role in controlling cell and tissue repair following stress or injury by modulating patterns of gene expression. Little is known, however, regarding the regulation of its expression.
Although traumatic injury and infectious challenge are known to initiate inflammatory response, it is becoming increasingly evident that cellular stress mechanisms such as the unfolded protein response (UPR)2 are also commonly engaged at sites of tissue injury (14–16). Such responses serve to limit cellular damage and engage processes requisite to repair and restoration of normal structure and function but may also promote cell death via apoptosis or autophagy when prolonged stress results in irreparable cellular damage. Moreover, cellular stress pathways not only reflect response to detrimental conditions but have also been implicated in the pathogenesis of numerous chronic diseases, including diabetes, heart disease, and cancer (17–20).
The UPR is initiated by at least three major signaling pathways within the endoplasmic reticulum (ER) that individually and collectively act to modulate the pattern of gene expression. Sensor proteins, localized in the ER membrane, include IRE-1, ATF6, and the ER-localized eIF2α kinase PERK, all of which are activated when the level of misfolded protein rises (14–16). There are three additional eIF2α kinases (PKR, GCN2, and HRI), each of which can mediate responses under distinct cellular perturbations (21, 22). While the most prominent effect of eIF2α phosphorylation is the global inhibition of 5′ cap-dependent translation initiation, there is a selection of mRNAs, which contain upstream open reading frames (uORF) and whose translation is increased by the action of phospho-eIF2α (23, 24). Interestingly, there is a small but growing body of evidence demonstrating that such uORFs can also regulate mRNA half-life in response to phosphorylation of eIF2α (25). Several mRNAs from both yeast and mammalian cells have been shown to exhibit uORF translation-dependent instability and, for some of these, the instability mechanism depends upon a protein (UPF1) that is required for the process of nonsense-mediated decay (NMD) (21, 25–27). NMD serves to ensure quality control of mRNA sequence by eliminating messages containing premature termination codons and mRNAs that contain uORFs with a termination codon in the 5′-leader region would be predicted to exhibit instability (28, 29). However, not all mRNAs that contain uORFs are targets for NMD (30–32), and the critical determinants of sensitivity remain to be fully appreciated (33, 34).
We now report that the transcript 1 (TR1) variant of human IFRD1 exhibits Upf-1-dependent instability linked to translation of an uORF. IFRD1 mRNA is stabilized in response to chemical stress via the phosphorylation of eIF2α resulting in a substantial increase in levels of mRNA and protein production. The instability mechanism is, however, distinct from those described previously; whereas the sequence and length of the uORF are critical, it does not need to encode a specific peptide. Furthermore, the environment of the uORF termination codon does not appear critical for instability. These findings identify a post-transcriptional mechanism that causes suppression of IFRD1 expression in resting tissues but elevation of mRNA and protein production in response to the phosphorylation of eIF2α.
EXPERIMENTAL PROCEDURES
Reagents
Dulbecco's modified Eagle's medium, Dulbecco's phosphate-buffered saline, and penicillin/streptomycin were obtained from Central Cell Services of the Lerner Research Institute (Cleveland, OH). Fetal bovine serum was purchased from BioWhittaker (Walkersville, MA). G418, formamide, MOPS, salmon sperm DNA, diethyl pyrocarbonate, and tunicamycin (Tm) were purchased from Sigma-Aldrich. Doxycycline (Dox) and the vector pTRE2 were obtained from Clontech. PolyFect transfection reagent was obtained from Qiagen (Valencia, CA), Tri-Reagent was purchased from Molecular Research Center (Cincinnati, OH), and Nylon transfer membrane was purchased from Micron Separation (Westboro, MA). A KC ELISA kit was purchased from R&D Systems (Minneapolis, MN). PerkinElmer Life Sciences was the source of [α-32P]dCTP. Protogel and related buffers were obtained from National Diagnostics Inc. (Atlanta, GA). Protein assay reagents were purchased from Bio-Rad (Richmond, CA). Mouse monoclonal antibodies specific for IFRD1 and GAPDH were purchased from Sigma or Chemicon International (Temecula, CA), respectively, while a polyclonal goat anti-UPF1 antibody was obtained from abCAM (Cambridge, MA). siRNAs with a sequence specific for hUPF1/Rent1 (35) or unrelated sequence were chemically synthesized by Dharmacon Research Inc.
Plasmids
Radiolabeled cDNA probes for use in Northern hybridization analysis were prepared as described previously (36). Plasmids used to drive expression of different versions of IFRD1 and CXCL1 were prepared in pTRE2 (Clontech Inc.). pTRE2(-RBG) was prepared by excising residues 533–1762 in pTRE2 (corresponding to the sequence from the rabbit β-globin gene) with XbaI and SapI. Transcript 1 (TR1) and transcript 2 (TR2) were created by insertion of full-length IFRD1 TR1 (residues 1–2255) and TR2 (residues 1–2469) cDNA isoforms into MluI/NheI sites of pTRE2(-RBG). The 5′TR1-KC and 5′TR2-KC constructs were obtained by inserting full-length 5′-leader sequences (for TR1, residues 1–229; for TR2, residues 1–443) and the coding region of KC (residues 53–359) into the MluI/NheI sites of pTRE2. Stem loop 5′TR1-KC and stem loop 5′TR2-KC were produced by inserting a fragment (5′-CCCGGAGCGCCCAGATCTGGGCGCTCCGGGGTAC-3′) immediately upstream of the 5′-leader sequence in the 5′TR1-KC and 5′TR2-KC (37). The construct (−46)-5′TR1-KC was made by deleting the first 46 nt (residues 1–46) from the 5′TR1-KC construct. The construct 46only-5′TR1-KC was made by removing residues 47–229 from 5′TR1-KC. The sequential deletions of the TR1 5′-leader fragment contain residues 1–46 (Δ4–5′TR1-KC), 1–92 (Δ3–5′TR1-KC), 1–138 (Δ2–5′TR1-KC), and 1–184 (Δ1–5′TR1-KC). The construct KC was obtained by removing full-length TR1 5′-leader fragment from 5′TR1-KC. Δ5–5′TR1-KC was made from Δ1–5′TR1-KC by deletion of 92 nt (residues 47–138) of TR1 5′-leader. The constructs sub1–5′TR1-KC, sub2–5′TR1-KC, and sub3–5′TR1-KC were made from 5′TR1-KC by replacing residues 31–92, 93–138, and 139–184, respectively, with unrelated sequence derived from the coding region of mouse formyl peptide receptor 2 mRNA. The full-length hUPF1 cDNA clone (Clone ID: 5555509) and EIF2S1 (Clone ID: 3139624) were obtained from Open Biosystems Inc. hUPF1-R844C, a helicase-defective dominant negative mutant of UPF1 (38), eIF2α-S51D, a phosphomimetic of eIF2α, NS39 (rabbit β-globin R39*), and mutant versions (uAUGmut-5′TR1-KC, 2nt-in5′TR1-KC, 3nt-in5′TR1-KC, 2nt-in-re5′TR1-KC, Tx-uORF-5′TR1-KC, 10NTsub5′TR1-KC, uUGAmt5′TR1-KC, OPTuAUG-5′TR1-KC, and newTC5′TR1-KC) of 5′TR1-KC were prepared by oligonucleotide site-directed mutagenesis as described previously (36). All expression constructs containing sequence from IFRD1 TR1 or IFRD1 TR2 were equivalent with respect to transcription start site (in pTRE2) and differed only in the specific sequence regions.
Cell Culture and Transient Transfection
Immortalized normal kidney epithelial (NKE) cells (39) were maintained in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, and streptomycin in humidified 5% CO2. HeLa tet-off cells were obtained from Clontech and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, penicillin, streptomycin, and G418 in humidified 5% CO2. HeLa tet-off cells in 100-mm plastic Petri dishes were transfected with a total of 2 μg of the indicated plasmids using PolyFect according to the manufacturer's protocol. After 3 h the cultures were subdivided into 60-mm dishes and rested for 24 h before individual treatments. In experiments with hUPF-R844C or eIF2α-S51D, cells were transfected with 2 μg of reporter plasmid and up to 6 μg of the indicated transgene. For experiments using siRNA, 15 μg of siRNA oligonucleotide duplex were transfected in serum-free medium (Opti-MEM) using Lipofectamine 2000 (Invitrogen). Total RNA was prepared using Tri reagent and analyzed by Northern blot hybridization as previously described (40). Autoradiographs were quantified by image analysis using the NIH Image software package. IFRD1 and KC mRNA levels were normalized to levels of GAPDH mRNA measured in the same RNA sample. The half-life of specific mRNA was calculated as described previously (40).
RT-PCR
Reverse transcription was carried out on total RNA using Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). First strand cDNA was synthesized from 3 μg of total RNA, which had been treated with TURBO DNase (Ambion), using random hexamer primers (Invitrogen). For semi-quantitative PCR, 3 μl of the RT reaction was used under standard conditions with gene-specific primers in a Thermal cycler (Applied Biosystems). Products were separated on 2% agarose gels and revealed by ethidium bromide staining. Quantitative real-time PCR was used to quantify IFRD1 primary transcript (for both TR1 and TR2, primers span junction of exon 2 and intron 2) and mature IFRD1 TR1 with the AB 7300 Real-time PCR System using SYBR Green PCR Master Mix (Applied Biosystems). The results were normalized with primers specific for GAPDH.
RESULTS
IFRD1 mRNA and Protein Expression Is Induced during the UPR
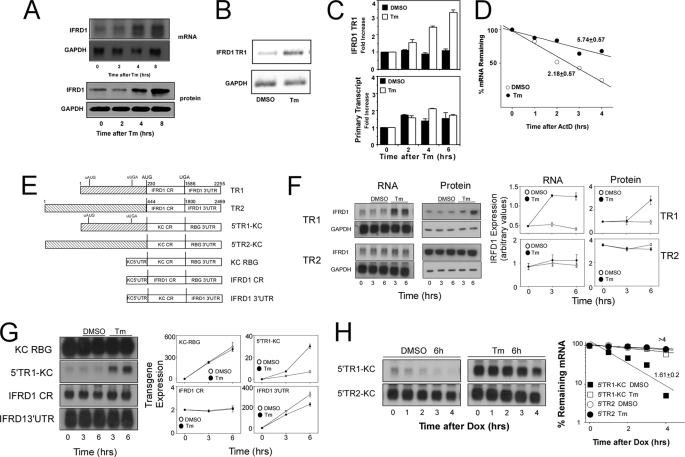
IFRD1 mRNA and protein are inducible in NKE cells following treatment with Tm (Fig. 1A). Tm treatment promoted a substantial increase in IFRD1 mRNA expression by 4 h, which was maintained through 8 h of treatment. IFRD1 protein was detectable in resting cells but was progressively increased after treatment. Two separate transcripts have been reported for the IFRD1 gene that differ only in sequence within the 5′-leader region (see supplemental Fig. 1). TR1 mRNA levels increased in NKE cells treated with Tm, whereas TR2 mRNA was undetectable (not shown) as determined using semi- quantitative RT-PCR with primers specific for each transcript (Fig. 1B).
FIGURE 1.
The 5′-leader sequence of IFRD1 TR1 confers Tm sensitivity and mRNA instability. A, NKE cells in 100-mm Petri dishes were treated with tunicamycin (Tm, 8.5 μm) for the indicated times prior to isolation of total RNA or protein lysate for determination of IFRD1 protein and mRNA levels by Western blot and Northern hybridization, respectively. Blots were stripped and used to assess levels of GAPDH protein and mRNA to control for load. B, RNA from duplicate experiments as described in A were subjected to semi-quantitative RT-PCR with primers specific for IFRD1 TR1 or GAPDH, and the products (25 cycles for IFRD1, 20 cycles for GAPDH) were separated by electrophoresis, the gel was stained with ethidium bromide and photographed with 365 nm UV illumination. C, NKE cells in 100-mm Petri dishes were untreated or treated with DMSO or Tm (8.5 μm) for 2, 4, or 6 h prior to preparation of total RNA. The RNA was digested with RNase free DNase prior to analysis of primary transcript abundance by real time RT-PCR as described under “Experimental Procedures.” Results are presented as mean -fold increase (duplicate determinations) ± 0.5 range. D, NKE cells in 100-mm Petri dishes were treated with DMSO or Tm (8.5 μm) for 6 h prior to the addition of actinomycin D (ActD, 5 μg/ml), and total RNA was isolated after further incubation for the indicated times. IFRD1 TR1 and GAPDH mRNA levels were determined by quantitative real-time RT-PCR. The values for IFRD1 TR1 mRNA were normalized to those for GAPDH in the same samples and are plotted as the % remaining mRNA. The half-life of IFRD1 TR1 mRNA in either untreated or Tm-treated cells was determined from the calculated best fit line, and the mean ± 0.5 range from two separate experiments is presented. E, a schematic representation of reporter constructs. Constructs TR1 and TR2 contain the full-length IFRD1 mRNAs. 5′TR1-KC and 5′TR2-KC contain the corresponding full 5′-leader sequences of IFRD1 TR1 and TR2 linked with the coding region of KC mRNA and the 3′-UTR from rabbit β-globin mRNA as indicated. KC RBG contains the 5′-leader and coding region from KC and the RBG 3′-UTR. IFRD1 CR contains the full coding region from human IFRD1 linked to the 5′-UTR from KC and 3′-UTR from RBG. IFRD1 3′-UTR contains the 5′-UTR and coding region of KC linked with the full 3′-UTR from human IFRD1. F, HeLa tet-off cells were transiently transfected with plasmid constructs TR1 or TR2 (2 μg/dish) and 3 h after transfection were separated into 10 separate dishes and cultured overnight. Cultures were untreated or treated with vehicle or Tm (8.5 μm) for the indicated times prior to determination of IFRD1 and GAPDH mRNA and protein levels by Northern and Western blot analysis, respectively. Data are presented as the mean ± 0.5 range of two separate experiments. G, HeLa tet-off cells were transiently transfected with plasmid constructs KC RBG, 5′TR1-KC, IFRD1 CR, or IFRD1 3′-UTR (2 μg/dish) and 3 h after transfection were separated into separate dishes and cultured overnight. Cultures were untreated or treated with DMSO or Tm (8.5 μm) for the indicated times prior to analysis of KC or IFRD1 mRNA (Northern blot) and protein levels (Western blot for IFRD1, ELISA for KC). Protein levels are in nanograms of KC/106 cells except for the IFRD1 CR panel, which are arbitrary values obtained from quantifying a Western blot and are presented as the mean ± 0.5 range of two separate experiments. H, HeLa tet-off cells were transiently transfected with plasmid constructs 5′TR1-KC or 5′TR2- KC (2 μg/dish) and 3 h after transfection were separated into 10 separate dishes and cultured overnight. Cultures were treated with either vehicle or Tm (8.5 μm) followed by Dox (1 μg/ml) for the indicated times prior to analysis of KC and GAPDH (not shown) mRNA levels by Northern blot. The films were quantified, and the percent remaining mRNA over time is shown. Half lives were calculated and presented as the mean ± 0.5 range of duplicate determinations.
The UPR has been reported to modulate transcription, mRNA translation, and, more recently, mRNA half-life (14, 15, 23–25, 30, 32). As a measure of IFRD1 transcription we determined the abundance of primary transcripts by real-time RT-PCR using primers spanning an intron-exon junction in NKE cells treated with vehicle (DMSO) or Tm for 2, 4, or 6 h. Although the abundance of mature IFRD1 TR1 increased with time following Tm treatment, there was no Tm-dependent increase in the amount of primary transcript (Fig. 1C) indicating that increased transcription was not a major contributing source for the elevated levels of IFRD1 TR1 mRNA. To determine if Tm treatment was acting to alter the half-life of IFRD1 mRNA, NKE cells were treated with DMSO or Tm for 6 h and further transcription was blocked by the addition of actinomycin D prior to determination of residual IFRD1 TR1 and GAPDH mRNA levels by real-time RT-PCR (Fig. 1D). The IFRD1 TR1 mRNA half-life was ∼2 h in resting cells but was increased markedly (to almost 6 h) following treatment with Tm.
To examine expression of IFRD1 mRNA in the absence of transcription from the endogenous gene promoter, we prepared plasmid constructs where IFRD1 TR1 or TR2 sequences were placed under control of a tetracycline-regulated promoter (tet-off). When transfected in HeLa cells expressing the tet-transactivator, the IFRD1 TR1 and TR2 plasmids (see schematic of constructs in Fig. 1E) were expressed well (Fig. 1F). When such cells were treated with Tm, TR1 mRNA and protein were both increased while TR2 mRNA and protein levels showed no sensitivity (Fig. 1F).
To identify the sequence region in IFRD1 TR1 responsible for Tm sensitivity, a series of reporter plasmids was generated in which the 5′-leader, the coding region, or the 3′-UTR sequence from TR1 were independently placed under tet-regulated control (see schematic in Fig. 1E). In all constructs, the transcription start sites were identical in order to ensure that transcription from each was comparable. A plasmid containing the 5′-leader of TR1 in the context of a reporter cDNA that included the mouse CXCL1 or KC coding region and the 3′-UTR from the rabbit β-globin mRNA in pTRE2 (5′TR1-KC) produced appreciably less mRNA and protein (measured as secreted KC protein) as compared with a control containing only KC and the rabbit β-globin 3′-UTR (KC RBG) (Fig. 1G). Furthermore, the levels of reporter mRNA and protein derived from 5′TR1-KC were selectively increased in cells treated with Tm compared with those derived from the KC RBG construct or plasmids containing the full coding region or 3′-UTR of human IFRD1. The half-life of 5′TR1-KC, measured following termination of reporter gene transcription by addition of Dox, was only 1.6 h in untreated cells but increased to >4 h in the presence of Tm, indicating that Tm sensitivity was a result of mRNA stabilization (Fig. 1H). Interestingly, a reporter containing the 5′-leader of TR2 (5′TR2-KC) was stable (t½ > 4 h) both in the presence and absence of Tm. Reporter constructs containing the 5′-leader of TR1 produced markedly less mRNA than did those containing the 5′-leader from TR2 under conditions where the cells were transfected with equal levels of plasmid DNA. When cells were transfected with different plasmid concentrations to achieve comparable levels of mRNA, the same differential sensitivity to Tm was observed (supplemental Fig. 2).
Translation and mRNA Decay Are Regulated by an uORF in the TR1 5′-Leader Region
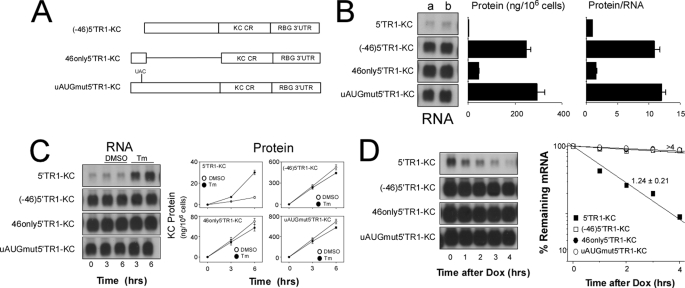
The ratio of protein to RNA for 5′TR1-KC is markedly less than that for 5′TR2-KC suggesting that translation of 5′TR1-KC mRNA is suppressed (supplemental Fig. 3). We hypothesized that the uORF contained within the 5′-leader of TR1 might be suppressing translation and serve as the basis for mRNA instability and Tm sensitivity. To test this possibility three additional plasmid constructs were prepared in which different forms of the TR1 5′-leader were linked with the KC-coding region (Fig. 2A). When HeLa tet-off cells were transfected with a plasmid in which the first 46 nt of the TR1 5′-region (which contains the start site for the uORF) were deleted ((−46)5′TR1-KC), the levels of mRNA and protein were markedly increased as compared with those obtained from intact 5′TR1-KC, and the protein-to-RNA ratio was comparable to that seen in cells transfected with 5′TR2-KC (Fig. 2B and supplemental Fig. 3). Furthermore, mRNA with a 5′ sequence containing only this 46-nt fragment (46only5′TR1-KC), exhibited a reduced protein-to-RNA ratio that was comparable to that seen with full-length 5′TR1-KC. Surprisingly, this construct produced levels of mRNA that were comparable to those obtained with 5′TR2-KC (compare with Fig. 1G). In a third construct the AUG site for the TR1 uORF was mutated to UAC in the context of the full-length TR1 5′-leader region and the protein:RNA ratio of this uAUG mutant construct (uAUGmut5′TR1-KC) was comparable to that seen with the (−46)5′TR1-KC construct (Fig. 2B).
FIGURE 2.
Translational repression and Tm sensitivity for IFRD1 TR1 mRNA depend upon the upstream AUG. A, construct (−46)5′TR1-KC is 5′TR1-KC missing the first 46 nt of the TR1 5′-leader fragment. 46only5′TR1-KC contains only the first 46 nt of the TR1 5′-leader linked with the KC coding region and the RBG 3′-UTR. uAUGmut5′TR1-KC is 5′TR1-KC in which the upstream AUG has been mutated to UAC. All constructs are in the plasmid pTRE2. B, HeLa tet-off cells were co-transfected with the indicated plasmids (2 μg/dish) along with a plasmid encoding firefly luciferase (also in pTRE2) and 3 h after transfection were separated into three individual Petri dishes and cultured overnight. One dish was used to prepare total RNA at t = 0 (a) while a second dish was washed and incubation continued with fresh medium for 3 h. The supernatant was harvested for determination of KC protein secretion by ELISA while the cells were used to prepare total RNA at t = 3 h (b). Levels of KC and GAPDH mRNA were determined by Northern blot hybridization. The third dish was used to prepare whole cell extract for determination of luciferase activity to normalize for transfection efficiency. The ratio of KC protein to mRNA was used as a measure of translation efficiency. Quantified data are presented as the mean ± 0.5 range of two separate experiments. C, HeLa tet-off cells were transfected with the indicated plasmids (each at 2 μg/dish) and 3 h after transfection were separated into individual Petri dishes and cultured overnight. Cultures were either untreated or treated with vehicle or Tm for the indicated time prior to determination of KC and GAPDH (not shown) mRNA and protein levels by Northern blot and ELISA, respectively. RNA levels are representative of two experiments, and protein data are presented as the mean ± 0.5 range of two experiments. D, HeLa tet-off cells were transiently transfected with the indicated plasmids (each at 2 μg/dish) and 3 h after transfection were separated into five separate dishes and cultured overnight. Cultures were treated with Dox (1 μg/ml) for the indicated times prior to analysis of KC and GAPDH (not shown) mRNA levels by Northern hybridization and calculation of half-life.
It is noteworthy that the 5′-leader sequence of TR1 can modulate mRNA half-life and the protein:RNA ratio for the major ORF, although these properties appear to be at least partially independent (see Fig. 2B, 5′TR1-KC as compared with 46only5′TR1-KC). Although all three properties require the 46-nt segment, this sequence is not sufficient for either instability or Tm sensitivity. The full-length TR1 5′-leader conferred marked instability, and deletion of the 46-nt fragment or mutation of the uAUG fully abrogated this property (Fig. 2). The construct containing only the 46-nt fragment produced reduced protein:RNA comparable to that seen with the full-length 5′ sequence, but it did not confer mRNA instability and was insensitive to Tm (Fig. 2, C and D). These findings indicate that the processes by which the TR1 5′-leader sequence controls translation and decay are distinguishable.
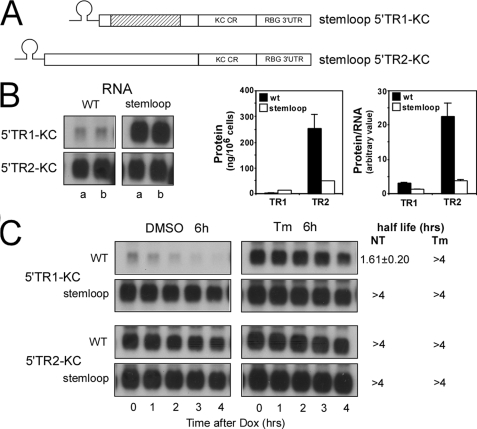
Translation of the uORF Sequence Is Required for Instability/Tm Sensitivity of IFRD1 mRNA and Is Mediated through Phosphorylation of eIF2α
That translation of the uORF was requisite for IFRD1 mRNA instability is inferred from the finding that mutation of the uAUG results in markedly prolonged half-life (Fig. 2). To independently test the translational requirement for instability and Tm sensitivity, constructs were prepared containing a highly stable stem loop structure inserted before the initiation codon in the TR1 5′-leader (stem loop 5′TR1-KC) or the TR2 5′-leader (stem loop 5′TR2-KC) (Fig. 3A). This stem loop structure has been shown to repress translation by restricting ribosome scanning to the translational start site (37). As seen in previous experiments, HeLa tet-off cells transfected with 5′TR1-KC produced less mRNA and protein as compared with 5′TR2-KC. This relationship was altered in cells expressing the stem loop versions, because the stem loop 5′TR1-KC construct produced markedly increased levels of mRNA compared with 5′TR1-KC, whereas the stem loop 5′TR2-KC mRNA levels were equivalent to those obtained with 5′TR2-KC (Fig. 3B). The stem loop addition produced the predicted translational inhibition of 5′TR2-KC as evidenced by an 80% reduction in the amount of KC reporter protein produced and in the protein:RNA ratio (Fig. 3B). The increased mRNA levels seen with the stem loop 5′TR1-KC message results in a substantial increase in protein production even though the protein:RNA ratio was reduced by over 60%. In related experiments, the half-life and Tm sensitivity for mRNAs derived from each version were determined after the addition of Dox (Fig. 3C). Although the 5′TR1-KC mRNA exhibited a half-life of 1.61 h that could be prolonged to >4 h in the presence of Tm, the stem loop 5′TR1-KC version exhibited longer half-life even in untreated cells and showed no response to Tm. 5′TR2-KC and its stem loop version both produced long lived mRNAs (t½ > 4 h) and predictably showed no sensitivity to Tm treatment.
FIGURE 3.
IFRD1 mRNA instability requires translation of the uORF. A, a stem loop sequence was inserted immediately 5′ to the start of the 5′-leader sequence in both 5′TR1-KC and 5′TR2-KC. B, HeLa tet-off cells were transfected with the indicated plasmids (each at 2 μg/dish) and 3 h after transfection were separated into two individual Petri dishes and cultured overnight. One dish was used to prepare total RNA at t = 0 (a) while a second dish was washed and incubation continued with fresh medium for 3 h. The supernatant was harvested for determination of KC protein secretion by ELISA while the cells were used to prepare total RNA at t = 3 h (b). KC and GAPDH (not shown) mRNA levels were determined by Northern blot, and the autoradiographs were quantified as described above. Protein and protein/RNA are presented as the mean ± 0.5 range of duplicate experiments. C, HeLa tet-off cells were transiently transfected with the indicated plasmid constructs (each at 2 μg/100-mm Petri dish) and 3 h after transfection were pooled and separated into 10 dishes and cultured overnight. Cultures were treated with either DMSO or Tm (8.5 μm) for 6 h prior to addition of Dox (1 μg/ml) and analysis of KC and GAPDH (not shown) mRNA levels by Northern hybridization after the indicated times. Half-lives are presented as the mean ± 0.5 range from two separate experiments.
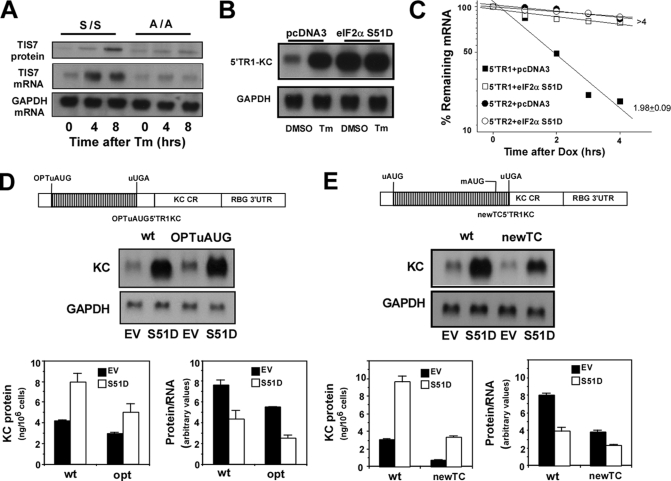
Activation of PERK and phosphorylation of eIF2α blocks 5′ cap-dependent translational initiation (21, 22, 41). Because the instability of IFRD1 mRNA is dependent upon translation of the uORF, we reasoned that phosphorylation of eIF2α would be requisite to Tm-mediated stabilization of IFRD1 mRNA. We first tested this hypothesis by examining the ability of Tm to induce TIS7 expression (the mouse homolog of IFRD1 TR1) in mouse embryo fibroblasts from mice expressing either a wild-type or mutant version of EIF2S1 (eIF2α) in which serine residue 51 has been changed to alanine preventing its phosphorylation (42). Although Tm treatment increased expression of TIS7 mRNA and protein in wild-type mouse embryo fibroblasts (S/S), the response was lost in the A/A mouse embryo fibroblasts (Fig. 4A). This result demonstrates that the TIS7 mRNA is sensitive to Tm and thus behaves similarly to human IFRD1 TR1. In a second experiment we utilized another mutant in which Ser-51 was changed to aspartic acid (S51D) creating a phosphomimetic version of eIF2α (43). Expression of this construct produced a selective increase in co-transfected reporter mRNA derived from 5′TR1-KC very similar to that seen with Tm treatment (Fig. 4B). Furthermore, expression of the S51D version of eIF2α also promoted stabilization of mRNA derived from 5′TR1-KC but did not affect the decay of 5′TR2-KC mRNA (Fig. 4C).
FIGURE 4.
Stabilization of IFRD1 mRNA is involves phosphorylated eIF2α. A, cultures of mouse embryo fibroblasts from either wild-type (S/S) or S51A transgenic (A/A) mice were treated with DMSO or Tm (8.5 μm) for 0, 4, or 8 h prior to preparation of RNA and protein extracts. TIS7 mRNA and protein levels were determined by Northern hybridization and Western blot, respectively, and are representative of two separate experiments. B, HeLa tet-off cells were transiently co-transfected with 5′TR1-KC (2 μg/dish) and either pcDNA3 or eIF2α S51D (2 μg/dish). Separate dishes were treated with vehicle (DMSO) or Tm (8.5 μm) for 6 h prior to determination of KC and GAPDH mRNA levels by Northern hybridization. Results are representative of two separate experiments. C, HeLa tet-off cells were transiently co-transfected with 5′TR1-KC or 5′TR2-KC (each at 2 μg/dish) and either pcDNA3 or eIF2α S51D (0.5 μg/dish). Separate dishes were treated with Dox (1 μg/ml) for the indicated times prior to analysis of KC and GAPDH (not shown) mRNA levels by Northern hybridization. Half-lives are given as mean ± 0.5 range from two experiments. D, HeLa tet-off cells were co-transfected with either 5′TR1-KC or OPTuAUG5′TR1-KC (see schematic) and either pcDNA3 (EV) or eIF2α S51D (each at 2 μg/dish). After overnight culture, the dishes were washed and replenished with fresh medium and after 3 h, the supernatants were harvested to determine KC protein by ELISA, and the cells were used to determine KC and GAPDH mRNA levels by Northern hybridization. The blots were quantified, and protein and protein:RNA ratio are presented as the mean ± 0.5 range from two separate experiments. E, HeLa tet-off cells were co-transfected with 5′TR1-KC or newTC5′TR1-KC and either pcDNA3 (EV) or eIF2α S51D (each at 2 μg/dish). After overnight culture, the dishes were washed and replenished with fresh medium, and after 3 h, the supernatants were harvested to determine KC protein production by ELISA, and the cells were used to determine KC and GAPDH mRNA levels by Northern hybridization. The blots were quantified, and the protein and protein:RNA ratio are presented as the mean ± 0.5 range from two separate experiments.
We also wished to determine if translation of the major ORF in 5′TR1-KC involved leaky scanning at the uORF start site or re-initiation following termination of uORF translation and how either of these might be affected by stress. To examine leaky scanning, we evaluated translation of the major ORF (reporter KC) in constructs containing either the wild-type or an optimal Kozak sequence at the uAUG (OPTuAUG5′TR1-KC, see schematic in Fig. 4D). In HeLa tet-off cells transfected with empty vector (EV) the construct containing the optimal uORF start site exhibited a 30–40% reduction in protein and protein:RNA ratio compared with the construct containing the wild-type IFRD1 uAUG sequence indicating that leaky scanning occurs at this site in the IFRD1 TR1 message (Fig. 4D). Both the wild-type and optimized constructs exhibited increased mRNA and protein accumulation in cells co-transfected with the phosphomimetic eIF2αS51D, but the reduction in translation of the major ORF as a consequence of the inhibition of 5′cap-dependent translation was comparable (Fig. 4D, protein/RNA). To examine the role for re-initiation in translation of the mORF, we prepared a construct in which the uORF termination codon was mutated and a new termination codon created 15 nt downstream of the start site for the mORF (see schematic, Fig. 4E). In this setting re-initiation at the mAUG cannot occur, and any translation of the mORF must be from leaky scanning or an internal ribosome entry site. In cells transfected with this plasmid, protein production and the protein:RNA ratio were both reduced by nearly 60%. This modified uORF still exhibited sensitivity to eIF2α S51D (prolonged half-life, not shown), but there was no selective alteration in any parameter in cells transfected with the eIF2α phosphomimetic as compared with the vector alone (EV). Thus although the translation of the mORF can be accounted for by a combination of leaky scanning at the IFRD1 TR1 uAUG site and re-initiation following translation of the uORF, neither of these features are altered by phosphorylated eIF2α. Hence the effect of stress on expression of IFRD1 appears to be largely dependent upon the change in mRNA half-life and the accompanying increase in mRNA abundance.
Instability of IFRD1 mRNA Is Dependent upon UPF1
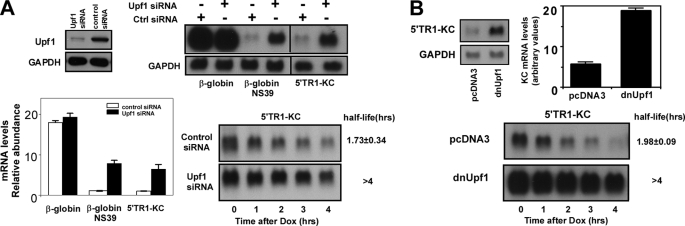
NMD, a mechanism that serves to identify and eliminate mRNAs with a premature termination codon (28, 29), could be involved in the UPR-sensitive instability mechanism governing the levels of IFRD1 mRNA (25). siRNA-mediated depletion of UPF1, a factor necessary for NMD, not only compromises the decay of premature termination codon-containing mRNAs but also increases expression of a subset of normal genes that includes some mRNAs with uORFs (44). To test the role of NMD in control of TIS7/IFRD1 mRNA decay, we depleted UPF1 levels by siRNA in cells transfected with either 5′TR1-KC or with a plasmid encoding either a wild-type or mutant version of rabbit β-globin with a premature termination codon at codon 39 (NS39) known to exhibit sensitivity to NMD (Fig. 5A). Cells transfected with siRNA targeting UPF1 showed a marked reduction in levels of UPF1 protein and a significant increase in levels of the mutant but not wild-type version of the β-globin mRNA. This treatment also increased levels of 5′TR1-KC mRNA to a comparable extent via prolongation of half-life. In a related experiment, a dominant negative mutant version of UPF1 was employed with identical outcome to that seen with the siRNA specific for UPF1 (Fig. 5B).
FIGURE 5.
IFRD1 mRNA instability is dependent on UPF1. A, HeLa tet-off cells were co-transfected with control or UPF1-specific siRNA and either wild-type β-globin, NS39 β-globin, or 5′TR1-KC. 48 h later cultures were harvested for determination of UPF1 or GAPDH protein (by Western blot) or β-globin, KC, or GAPDH mRNA (by Northern blot). Levels of β-globin or KC mRNA were quantified, and data are presented as the mean ± 0.5 range from two experiments. In a second experiment, HeLa tet-off cells were co-transfected with either control or UPF1-specific siRNAs along with 5′TR1-KC. After 48 h, the cultures were treated with Dox, and residual KC and GAPDH (not shown) mRNA levels were determined by Northern hybridization at the indicated times. Half-lives are presented as either stable (>4 h) or as the mean ± 0.5 range from two experiments. B, HeLa tet-off cells were transfected with 5′TR1-KC (2 μg/dish) and either pcDNA3 or hUPF1-R844C (dnUpf1) (each at 6 μg/dish). After 24 h, the cultures were used to determine KC and GAPDH mRNA levels, and the results are presented as mean ± 0.5 range of two experiments. In a second experiment, HeLa tet-off cells were transfected as above and 24 h later were treated with Dox, and residual KC and GAPDH (not shown) mRNA levels were determined by Northern hybridization. Half-lives are presented as above.
Sequence and Length of the IFRD1 uORF Are Critical Determinants of Instability
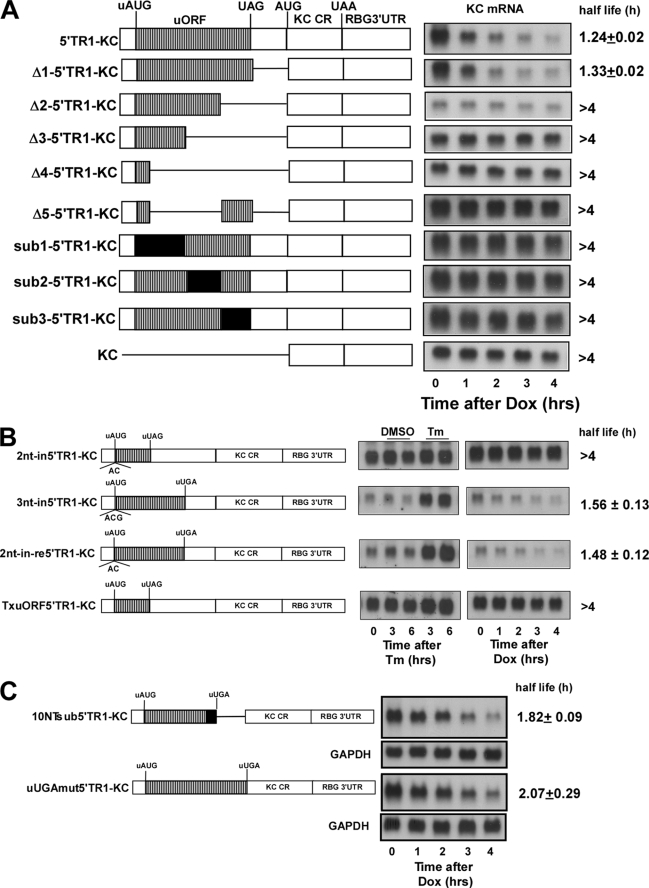
The dependence of IFRD1 mRNA instability on UPF1 suggests that the uORF and particularly the upstream termination codon may be recognized as premature, thereby engaging NMD. Although several mRNAs containing uORFs exhibit short half-lives, the presence of an uORF does not necessarily confer sensitivity to NMD. To explore this experimentally, the half-lives of a series of deletion and substitution mutants of the full-length 5′-leader of TR1 were examined in HeLa tet-off cells following the addition of Dox (Fig. 6A). Previous experiments demonstrated that the first 46-nt segment of the TR1 5′-leader was required for instability and Tm sensitivity (see Fig. 2). Although deletion of the region between the termination codon and the major ORF start codon did not compromise instability (Δ1–5′TR1-KC), all constructs containing a deletion of any portion of the uORF region resulted in loss of the instability function (Fig. 6A). Moreover, substitutions of unrelated sequence in various portions of the uORF, although maintaining the identical length ORF, also produced highly stable mRNA products, suggesting that both the length and specific sequence of the uORF are necessary to confer instability.
FIGURE 6.
Regulatory function of the uORF requires the full sequence and length. A, constructs containing the indicated deletion/substitution mutants of the 5′-leader sequence of IFRD1 TR1 were transiently transfected in HeLa tet-off cells (each at 2 μg/dish). After overnight culture the separate dishes were treated with Dox (1 μg/ml) for the indicated times prior to analysis of KC and GAPDH (not shown) mRNA levels by Northern hybridization. Half-lives are presented as either stable (>4 h) or as the mean ± 1 S.D. from three experiments. B, HeLa tet-off cells were transfected with the indicated plasmid constructs (each at 2 μg/dish) and 3 h after transfection were separated into 10 individual Petri dishes and cultured overnight. Five dishes were either untreated or treated with vehicle or Tm for the indicated times prior to determination of KC and GAPDH (not shown) mRNA levels by Northern hybridization. A separate set of five dishes was treated with Dox (1 μg/ml) for the indicated times prior to analysis of KC and GAPDH (not shown) mRNA levels. Half-lives are presented as either stable (>4 h) or as the mean ± 0.5 range from two experiments. C, the last 10 nt of the uORF were mutated to GAGGUAGCCG, retaining the coding frame and position of the termination codon (10NTsub5′TR1-KC). The UGA termination codon of the uORF was mutated to ACU resulting in a new the termination codon immediately after the AUG of the KC reporter (uUGAmt5′TR1-KC). HeLa tet-off cells were transfected with the indicated plasmid constructs (each at 2 μg/dish) and 3 h after transfection were separated into individual Petri dishes and cultured overnight. Dishes were treated with Dox (1 μg/ml) for the indicated times prior to determination of KC and GAPDH mRNA levels by Northern hybridization. Half-lives are presented as the mean ± 0.5 range from two experiments.
These observations suggested that the uORF might encode a peptide whose translation would be requisite for instability of the mRNA and its sensitivity to UPR, comparable to what has been observed for the yeast CPA1 mRNA (26). Disruption of the coding frame of the original IFRD1 uORF by inclusion of a 2-nt insertion (2nt-in5′TR1-KC) compromised both Tm sensitivity and mRNA instability, whereas a 3-nt insertion (3nt-in5′TR1-KC) had no effect (Fig. 6B). The 2-nt insertion, however, changed not only the coding frame but also produced a new termination codon resulting in a truncation of the uORF. Hence the loss of Tm sensitivity and mRNA instability associated with the 2nt-in5′TR1-KC construct could reflect either a frameshift or an alteration in the length of the uORF. When this plasmid was mutated to restore the original termination codon while maintaining a different coding frame, Tm sensitivity and instability were restored. Finally, a construct was prepared in which a mutation was made to introduce an early termination codon in the original uORF (Tx-uORF5′TR1-KC), thereby truncating the uORF, and this mutation abrogated the regulatory properties of the 5′-leader sequence. The 2nt-in-re5′TR1-KC construct demonstrates that the coding specificity of the uORF is not essential and suggests that either the length of the uORF or the position of the termination codon are critical.
The yeast YAP2 mRNA has an uORF in which the sequence environment at and/or downstream of the termination codon is necessary to promote instability (27). The deletion of the sequence between the termination codon and the major ORF start site in the IFRD1 mRNA 5′-leader did not, however, alter the half-life suggesting that a YAP2-like mechanism is not operative (Fig. 6A). To further assess this possibility, two additional mutations were analyzed. In the first, the last 10 nt of the uORF in IFRD1 TR1 5′-leader were mutated while retaining the same position for the termination codon (10NTsub5′TR1-KC), whereas the second involved mutating the termination codon, which extended the uORF to the start codon of the major ORF. Interestingly, neither of these changes altered the short half-life for the mRNA, demonstrating that the sequence context for termination of the uORF is not a critical feature defining the Upf-1-dependent instability of the IFRD1 TR1 mRNA.
DISCUSSION
Although the IFRD1 gene product has been reported to function in cell development or differentiation, its expression has been reported to be consistently elevated in numerous settings involving tissue injury, conditions that may be linked with accompanying cellular stress (10–13). Indeed, the UPR is increasingly recognized as an important contributing feature of the response to injury in multicellular organisms. The present study was undertaken to test the hypothesis that IFRD1 is a cell stress-inducible gene and to explore the molecular mechanisms through which its induced expression is achieved. The present findings show that the human IFRD1 mRNA is the target of a post-transcriptional mechanism involving stress-induced control of mRNA half-life based upon the following experimental findings. 1) IFRD1 TR1 mRNA and protein levels are increased following treatment with Tm via stabilization of the mRNA. The sensitivity to Tm is dependent upon the uORF, the translation of which leads to both translational repression of the major ORF and selective mRNA instability. 2) Stabilization in response to stress depends upon phosphorylation of eIF2α resulting in reduced translation of the uORF. Although the major ORF is translated either by leaky scanning at the uAUG or re-initiation following termination at the uORF termination codon, these properties are not altered during stress. 3) The instability mechanism depends on UPF1, a factor required for NMD. 4) Finally, the instability mechanism requires the full sequence and length of the uORF but does not need to encode a specific peptide. Moreover, the termination codon and the surrounding sequence environment are not critical for instability. Collectively, these findings identify the IFRD1 instability mechanism as NMD-dependent and demonstrate that it serves as an important target of the integrated stress response through phosphorylated eIF2α. Interestingly, the requirement for specific sequence and length of the uORF distinguish the mechanism from previous reports. Because the sensitivity of uORF-containing mRNAs to NMD and stress-mediated stabilization is not uniform, this feature may represent a broadly applicable mechanism.
Activation of PERK and phosphorylation of eIF2α are known to promote the translation of the major ORF in several uORF-containing mRNAs, the best studied being the transcription factor ATF4 (23, 24). Although the uORF of IFRD1 mRNA does repress the translation of the major ORF, the evidence presented here demonstrates that phospho-eIF2α-mediated inhibition of uORF translation results in marked elevation of IFRD1 mRNA principally by preventing its degradation. Conditions that compromise translation of the uORF (mutation of the uAUG, Tm treatment, expression of eIF2α S51D, or inclusion of a stable stem loop in front of the uAUG) all prolong the half-life of the mRNA and increase mRNA abundance. Under these conditions, translation of the major ORF was also compromised but residual translation was sufficient to elevate levels of protein encoded by the downstream ORF.
uORFs are well known to modulate translation of downstream major ORFs in eukaryotes and are frequently involved in translational control during the UPR (21, 23, 24, 30, 32, 45–48). This raised the issue of how the major ORF in the IFRD1 TR1 mRNA would be translated and whether this would be subject to modulation during cellular stress response. Translation of a major ORF downstream of an uORF results from one of three mechanisms: leaky scanning at the uAUG, re-initiation following termination of the uORF, or the presence of an internal ribosome entry site (31, 49). Several reports suggest that an uORF length of >35 amino acid residues can compromise the capacity for re-initiation at the downstream AUG site (50, 51). The TR1 uORF encodes 52 amino acid residues suggesting that the translation of the major ORF of IFRD1 would not involve a high frequency of re-initiation. However, the results suggest that nearly 60% of the translation from the mORF is a result of re-initiation. The effect of optimizing the uAUG start site reduces downstream ORF protein production by approximately 40% suggesting that leaky scanning is responsible for the residual translation. Phosphorylation of eIF2α is known to change the re-initiation frequency at the major ORF of some mRNAs containing uORFs, although this property is not universal (23, 24, 45, 46, 52, 53). Our findings indicate that neither leaky scanning nor re-initiation exhibit differential sensitivity to stress either for mRNA stabilization or translation of the mORF. Collectively, these findings support the conclusion that the mRNA instability/stabilization mechanism is the primary target for the action of cell stress-mediated induction of IFRD1 protein.
The linkage between translation of uORFs and mRNA degradation via NMD is predicted as a consequence of the presence of a termination codon located within the 5′-leader region (25, 45, 46, 50, 54–56). Furthermore, because NMD requires translation (29), translational suppression during cell stress would be predicted to block the NMD-mediated decay and stabilize the message. Indeed, IFRD1 mRNA levels have been reported to increase in cells depleted of UPF1 through targeting with siRNA, and eIF2α phosphorylation has been linked with stabilizing NMD-dependent decay (25, 44). There are, however, multiple examples of mRNAs containing uORFs that regulate translation of the major ORF without promoting NMD-mediated mRNA decay (30–32, 57–60). For example, the yeast GCN4 mRNA, which contains four uORFs, is not degraded in an UPF1-dependent manner (57, 58). This appears to reflect both the absence of sequence required for NMD as well as the presence of a sequence that can antagonize the NMD process. Because the IFRD1 TR1 5′-leader exhibits NMD-like instability, such stabilizing mechanisms do not seem to be operative. Indeed, although our findings confirm the role of UPF1 in regulating expression of IFRD1, several observations suggest that the mechanism involved in the uORF-dependent decay mechanism exhibits requirements that are distinct for this form of NMD. First, the position of the termination codon for the uORF rather than just its presence is a critical determinant of message instability. More importantly, the full sequence of the uORF appears to be important in determining instability (not stability). The distinct features of the IFRD1 uORF are consistent with prior findings of multiple forms of UPF1-mediated mRNA decay. For example some targets of NMD are distinguished by differential dependence on UPF2 and UPF3 (61, 62). In addition, UPF1 has been found to participate in non-NMD decay mechanisms (35, 63).
There are several examples of yeast mRNAs where uORFs confer instability based upon mechanisms that depend at least in part upon ribosome stalling at or near the termination codon (26, 27). In the case of the CPA1 mRNA, the uORF encodes a specific peptide, which, in the presence of arginine, promotes the degradation of the mRNA. Because the IFRD1 uORF need not encode a specific peptide, this mechanism seems unlikely. Although the YAP2 mRNA sequence at and downstream of the termination codon for the uORF slows ribosome scanning, this region of the IFRD1 mRNA is not requisite to the instability mechanism. Collectively, the features that characterize the decay of IFRD1 TR1 appear to define a relatively novel mechanism for repressing expression of this gene in unstressed cells.
There is only a limited literature covering the function of the IFRD1 gene product. A number of studies support the role for the mouse TIS7 gene product as a co-regulator of target gene transcription contributing to the control of cell type-specific differentiation or development (2, 4, 6–8). Furthermore TIS7 expression has been shown to be elevated in tissues following ischemia/reperfusion and traumatic injury (4, 11–13). IFRD1 has been recently identified as a modifier gene in cystic fibrosis (10), based upon its capacity to regulate inflammatory activities in polymorphonuclear leukocytes following bacterial exposure; this appears to reflect control of NFκB-induced transcription via a histone deacetylase-dependent mechanism. It is tempting to speculate that the IFRD1 gene product may play an important role in regulating response to tissue injury in association with accompanying cell stress. In this regard, the repair of muscle injury in TIS7-deficient mice is compromised (4). Further experimental analysis will be necessary to test such hypotheses.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant CA62220 from USPHS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum
- uORF
- upstream open reading frame
- mORF
- major open reading frame
- IFRD1
- interferon-related developmental factor 1
- TR1
- IFRD1 transcript 1
- TR2
- IFRD1 transcript 2
- Tm
- tunicamycin
- tet
- tetracycline
- Dox
- doxycycline
- PERK
- PKR-like ER-localized kinase
- NMD
- nonsense-mediated decay
- MOPS
- 4-morpholinepropanesulfonic acid
- ELISA
- enzyme-linked immunosorbent assay
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- siRNA
- small interference RNA
- NKE
- normal kidney epithelial cell
- RT
- reverse transcription
- UTR
- untranslated repeat
- RBG
- rabbit β-globin.
REFERENCES
- 1.Arenander A. T., Lim R. W., Varnum B. C., Cole R., de Vellis J., Herschman H. R. (1989) J Neurosci. Res. 23, 247–256 [DOI] [PubMed] [Google Scholar]
- 2.Guardavaccaro D., Ciotti M. T., Schafer B. W., Montagnoli A., Tirone F. (1995) Cell Growth & Differ. 6, 159–169 [PubMed] [Google Scholar]
- 3.Iacopetti P., Barsacchi G., Tirone F., Cremisi F. (1996) Brain Res. 707, 293–297 [DOI] [PubMed] [Google Scholar]
- 4.Vadivelu S. K., Kurzbauer R., Dieplinger B., Zweyer M., Schafer R., Wernig A., Vietor I., Huber L. A. (2004) Mol. Cell. Biol. 24, 3514–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y., Iordanov H., Swietlicki E. A., Wang L., Fritsch C., Coleman T., Semenkovich C. F., Levin M. S., Rubin D. C. (2005) J. Biol. Chem. 280, 34764–34775 [DOI] [PubMed] [Google Scholar]
- 6.Vietor I., Vadivelu S. K., Wick N., Hoffman R., Cotten M., Seiser C., Fialka I., Wunderlich W., Haase A., Korinkova G., Brosch G., Huber L. A. (2002) EMBO J. 21, 4621–4631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vietor I., Kurzbauer R., Brosch G., Huber L. A. (2005) J. Biol. Chem. 280, 39795–39801 [DOI] [PubMed] [Google Scholar]
- 8.Wick N., Schleiffer A., Huber L. A., Vietor I. (2004) J. Mol. Biol. 336, 589–595 [DOI] [PubMed] [Google Scholar]
- 9.Micheli L., Leonardi L., Conti F., Buanne P., Canu N., Caruso M., Tirone F. (2005) Mol. Cell. Biol. 25, 2242–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu Y., Harley I. T., Henderson L. B., Aronow B. J., Vietor I., Huber L. A., Harley J. B., Kilpatrick J. R., Langefeld C. D., Williams A. H., Jegga A. G., Chen J., Wills-Karp M., Arshad S. H., Ewart S. L., Thio C. L., Flick L. M., Filippi M. D., Grimes H. L., Drumm M. L., Cutting G. R., Knowles M. R., Karp C. L. (2009) Nature 458, 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin D. C., Swietlicki E. A., Wang J. L., Levin M. S. (1998) Am. J. Physiol. 275, G506–G513 [DOI] [PubMed] [Google Scholar]
- 12.Nelson D. P., Wechsler S. B., Miura T., Stagg A., Newburger J. W., Mayer J. E., Jr., Neufeld E. J. (2002) Ann. Thorac. Surg. 73, 156–162 [DOI] [PubMed] [Google Scholar]
- 13.Roth A., Gill R., Certa U. (2003) Mol. Cell Neurosci. 22, 353–364 [DOI] [PubMed] [Google Scholar]
- 14.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 15.Zhang K., Kaufman R. J. (2008) Nature 454, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 17.Marciniak S. J., Ron D. (2006) Physiol. Rev. 86, 1133–1149 [DOI] [PubMed] [Google Scholar]
- 18.Scheuner D., Kaufman R. J. (2008) Endocr. Rev. 29, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kitamura M. (2008) Am. J. Physiol. Renal Physiol. 295, F323–F334 [DOI] [PubMed] [Google Scholar]
- 20.Moenner M., Pluquet O., Bouchecareilh M., Chevet E. (2007) Cancer Res. 67, 10631–10634 [DOI] [PubMed] [Google Scholar]
- 21.Sonenberg N., Hinnebusch A. G. (2009) Cell 136, 731–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wek R. C., Cavener D. R. (2007) Antioxid. Redox. Signal. 9, 2357–2371 [DOI] [PubMed] [Google Scholar]
- 23.Vattem K. M., Wek R. C. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu P. D., Harding H. P., Ron D. (2004) J. Cell Biol. 167, 27–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gardner L. B. (2008) Mol. Cell. Biol. 28, 3729–3741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaba A., Jacobson A., Sachs M. S. (2005) Mol. Cell 20, 449–460 [DOI] [PubMed] [Google Scholar]
- 27.Vilela C., Ramirez C. V., Linz B., Rodrigues-Pousada C., McCarthy J. E. (1999) EMBO J. 18, 3139–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehwinkel J., Raes J., Izaurralde E. (2006) Trends Biochem. Sci. 31, 639–646 [DOI] [PubMed] [Google Scholar]
- 29.Chang Y. F., Imam J. S., Wilkinson M. F. (2007) Annu. Rev. Biochem. 76, 51–74 [DOI] [PubMed] [Google Scholar]
- 30.Zhou D., Palam L. R., Jiang L., Narasimhan J., Staschke K. A., Wek R. C. (2008) J. Biol. Chem. 283, 7064–7073 [DOI] [PubMed] [Google Scholar]
- 31.Yaman I., Fernandez J., Liu H., Caprara M., Komar A. A., Koromilas A. E., Zhou L., Snider M. D., Scheuner D., Kaufman R. J., Hatzoglou M. (2003) Cell 113, 519–531 [DOI] [PubMed] [Google Scholar]
- 32.Lee Y. Y., Cevallos R. C., Jan E. (2009) J. Biol. Chem. 284, 6661–6673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shyu A. B., Wilkinson M. F., van Hoof A. (2008) EMBO J. 27, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebbapragada I., Lykke-Andersen J. (2009) Curr. Opin. Cell Biol. 21, 394–402 [DOI] [PubMed] [Google Scholar]
- 35.Kim Y. K., Furic L., Desgroseillers L., Maquat L. E. (2005) Cell 120, 195–208 [DOI] [PubMed] [Google Scholar]
- 36.Novotny M., Datta S., Biswas R., Hamilton T. (2005) J. Biol. Chem. 280, 30166–30174 [DOI] [PubMed] [Google Scholar]
- 37.Thoma C., Bergamini G., Galy B., Hundsdoerfer P., Hentze M. W. (2004) Mol. Cell 15, 925–935 [DOI] [PubMed] [Google Scholar]
- 38.Sun X., Perlick H. A., Dietz H. C., Maquat L. E. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 10009–10014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurova K. V., Hill J. E., Razorenova O. V., Chumakov P. M., Gudkov A. V. (2004) Cancer Res. 64, 1951–1958 [DOI] [PubMed] [Google Scholar]
- 40.Zhao C., Hamilton T. (2007) J. Biol. Chem. 282, 20230–20237 [DOI] [PubMed] [Google Scholar]
- 41.Harding H. P., Calfon M., Urano F., Novoa I., Ron D. (2002) Annu. Rev. Cell Dev. Biol. 18, 575–599 [DOI] [PubMed] [Google Scholar]
- 42.Scheuner D., Song B., McEwen E., Liu C., Laybutt R., Gillespie P., Saunders T., Bonner-Weir S., Kaufman R. J. (2001) Mol. Cell 7, 1165–1176 [DOI] [PubMed] [Google Scholar]
- 43.Perkins D. J., Barber G. N. (2004) Mol. Cell. Biol. 24, 2025–2040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendell J. T., Sharifi N. A., Meyers J. L., Martinez-Murillo F., Dietz H. C. (2004) Nat. Genet. 36, 1073–1078 [DOI] [PubMed] [Google Scholar]
- 45.Meijer H. A., Thomas A. A. (2002) Biochem. J. 367, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Morris D. R., Geballe A. P. (2000) Mol. Cell. Biol. 20, 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wek R. C., Jiang H. Y., Anthony T. G. (2006) Biochem. Soc. Trans. 34, 7–11 [DOI] [PubMed] [Google Scholar]
- 48.Pickering B. M., Willis A. E. (2005) Semin. Cell Dev. Biol. 16, 39–47 [DOI] [PubMed] [Google Scholar]
- 49.Kozak M. (1999) Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 50.Vilela C., McCarthy J. E. (2003) Mol. Microbiol. 49, 859–867 [DOI] [PubMed] [Google Scholar]
- 51.Kozak M. (2002) Gene 299, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinnebusch A. G. (1993) Mol. Microbiol. 10, 215–223 [DOI] [PubMed] [Google Scholar]
- 53.Sachs M. S., Geballe A. P. (2006) Genes Dev. 20, 915–921 [DOI] [PubMed] [Google Scholar]
- 54.Matsui M., Yachie N., Okada Y., Saito R., Tomita M. (2007) FEBS Lett. 581, 4184–4188 [DOI] [PubMed] [Google Scholar]
- 55.Lejeune F., Maquat L. E. (2005) Curr. Opin. Cell Biol. 17, 309–315 [DOI] [PubMed] [Google Scholar]
- 56.Maquat L. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 89–99 [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Echevarría M. J., González C. I., Peltz S. W. (1998) EMBO J. 17, 575–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ruiz-Echevarría M. J., Peltz S. W. (2000) Cell 101, 741–751 [DOI] [PubMed] [Google Scholar]
- 59.Stockklausner C., Breit S., Neu-Yilik G., Echner N., Hentze M. W., Kulozik A. E., Gehring N. H. (2006) Nucleic Acids Res. 34, 2355–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J. E. (1998) Nucleic Acids Res. 26, 1150–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gehring N. H., Kunz J. B., Neu-Yilik G., Breit S., Viegas M. H., Hentze M. W., Kulozik A. E. (2005) Mol. Cell 20, 65–75 [DOI] [PubMed] [Google Scholar]
- 62.Chan W. K., Huang L., Gudikote J. P., Chang Y. F., Imam J. S., MacLean J. A., 2nd, Wilkinson M. F. (2007) EMBO J. 26, 1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaygun H., Marzluff W. F. (2005) Nat. Struct. Mol. Biol. 12, 794–800 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.