Abstract
The ubiquitin-proteasome pathway plays a crucial role in many cellular processes by degrading substrates tagged by polyubiquitin chains, linked mostly through lysine 48 of ubiquitin. Although polymerization of ubiquitin via its six other lysine residues exists in vivo as part of various physiological pathways, the molecular mechanisms that determine the type of polyubiquitin chains remained largely unknown. We undertook a systematic, in vitro, approach to evaluate the role of E2 enzymes in determining the topology of polyubiquitin. Because this study was performed in the absence of an E3 enzyme, our data indicate that the E2 enzymes are capable of directing the ubiquitination process to distinct subsets of ubiquitin lysines, depending on the specific E2 utilized. Moreover, our findings are in complete agreement with prior analyses of lysine preference assigned to certain E2s in the context of E3 (in vitro and in vivo). Finally, our findings support the rising notion that the functional unit of E2 is a dimer. To our knowledge, this is the first systematic indication for the involvement of E2 enzymes in specifying polyubiquitin chain assembly.
Keywords: Proteasome, Protein Degradation, Ubiquitin, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination, Lysine-specific Ubiquitin Conjugation
Introduction
In eukaryotic cells, most proteins are degraded by the 26 S proteasome, which hydrolyzes in an ATP-dependant manner, both ubiquitin-conjugated and certain non-ubiquitinated proteins. In addition to its role in the turnover of damaged or misfolded proteins, the proteasome controls the cell cycle and other processes through the degradation of critical regulatory components and transcription factors (1–3). Upon association of ubiquitinated targets with the proteasome, ubiquitin molecules are proteolytically removed for reuse, whereas the unfolded substrates are fed into the 20 S catalytic core, where they are digested into small peptides (4, 5).
Protein ubiquitination is a multistep process orchestrated by the concerted action of three enzymes. The chain reaction begins with a ubiquitin-activating enzyme (E1), which initially adenylates the C-terminal glycine of ubiquitin. Next, a thioester bond is formed between the activated C terminus of ubiquitin and a cysteine residue of the E1. A ubiquitin-conjugating enzyme (E2) acquires the activated ubiquitin through a trans-thioesterification reaction. Finally, a RING ubiquitin-protein ligase (E3) recruits the substrate and guides the transfer of the ubiquitin from the E2 active site cysteine to the substrate. An ϵ-amine of a lysine residue on the substrate (or of additional ubiquitin) attacks the thioester bond between the ubiquitin and the E2 enzyme, forming an isopeptide bond with the C-terminal glycine of the ubiquitin (6–8). Alternatively, when a HECT E3 catalyzes the transfer of the ubiquitin from the E2 to the target, an intermediate complex, of the activated ubiquitin and the active site cysteine of the HECT domain E3, is formed (9).
Several forms of ubiquitination have been identified (10). Single or multiple monoubiquitinations have been described where a single or multiple ubiquitin moieties are conjugated to distinct lysine residues on the substrates, but they do not polymerize. These forms of ubiquitination were implicated in diverse cellular pathways, which include endocytosis and sorting of proteins to different cellular compartments (11, 12), yet, recently it was also shown to participate in several cases of proteasomal activity, such as the processing of the p105 precursor of the NF-κB transcriptional regulator (13). However, polyubiquitination, which is the covalent assembly of a chain of ubiquitin molecules on one or multiple lysine residues of the substrate, is the most common form of post-translational modification of proteins destined for degradation (14).
Traditionally, polyubiquitin elongation was thought to proceed as a sequential reaction by the addition of ubiquitin molecules, one at a time, in a cyclic manner, first to a lysine residue on the target protein and from then on to a lysine residue on the distal end of the growing ubiquitin chain. However, in view of recent findings, several alternative mechanisms have been proposed (see Ref. 15 for a review). Li et al. (16) demonstrated, in vitro, a mode of action in which polyubiquitin is initially assembled on the active site cysteine of an E2 (E2G2), presumably by the action of additional E2 molecules. Once polyubiquitin is constructed, a RING E3 enzyme (gp78) catalyzes the transfer of the polyubiquitin, as a module, to a lysine residue of the target substrate (the C terminus of HERP, a known substrate of these E2/E3 enzymes). However, it is unclear whether en block transfer of polyubiquitin also occurs in vivo. In a related study, Ravid and Hochstrasser (17) proposed that the active site polyubiquitination of Ubc7 (the yeast ortholog of E2G2) is recognized by the proteasome and may serve as a degradation signal in an autoregulation feedback loop.
Because ubiquitin harbors seven lysine residues (Lys6, Lys11, Lys27, Lys29, Lys33, Lys48, and Lys63), in principle, chains of polyubiquitin can be formed via a bond between the C-terminal glycine of one ubiquitin molecule and an ϵ-amine of any of the seven lysine residues of another ubiquitin. Accordingly, seven different topologies of polyubiquitin can be generated (excluding mixed topologies) (18). Although most ubiquitin-dependent proteasomal degradations were found to be mediated by Lys48 polyubiquitin chains, certain evidence suggests that Lys6 (19), Lys11 (19, 20), and Lys29 (21) may also target substrates for proteasomal degradation. Polyubiquitinations through non-Lys48 lysines exist in vivo and play a role in a range of cellular processes (22, 23). For example, chains polymerized via Lys63 were shown to participate in the DNA damage response (24, 25). Lys63 linkages were also shown to be required for endolysosomal degradation of class I major histocompatibility complex molecules, for NF-κB activation, and for targeting epidermal growth factor receptor to the lysosome (26–28). Lys11 conjugates were shown to be catalyzed by the APC E3 complex and E2C (29), whereas Lys6 and Lys11 chain topologies were found on the aggregated PHF-Tau in Alzheimer disease (30). Lys6 was also implicated in DNA repair, whereas Lys29 may function in proteasome degradation (21, 22).
To systematically study the mode of action of the E2 family of enzymes, we cloned, expressed, and purified all of the annotated human E2 enzymes. As a control, we expressed the well documented yeast Ubc13-UEV1a and the Ubc5 family members (Ubc5A, Ubc5B, and Ubc5C). Initially, we monitored the ability of the various purified E2 enzymes to function in vitro in autoubiquitination reactions and then corroborated our findings by characterizing substrate ubiquitination by these E2s. We determined by mass spectrometry the lysine specificity in ubiquitin-conjugates generated by the different E2 and the site of ubiquitination. In conjunction, we preformed polyubiquitination reactions in the presence of a gallery of single lysine or arginine ubiquitin derivatives (20). This rigorous analysis gave rise to the realization that the E2 enzymes may have a role in selecting the target's lysine.
EXPERIMENTAL PROCEDURES
Purification of the Human E2 Ubiquitin-conjugating Enzymes
The complete coding sequence of the annotated human E2 ubiquitin-conjugating enzymes were subcloned from a human cDNA library by PCR. The amplified genes were inserted into the bacterial expression vector pET22 in frame to a C-terminal His6 tag. The desired plasmid was transformed into Escherichia coli (BL-21 DE3). The bacteria were grown overnight in 50 ml of LB medium complemented with 100 μg/ml ampicillin. This starter served to inoculate 2 liters of culture that were grown until an optical density of 0.6 at 600 nm was reached. Protein expression was induced by the addition of isopropyl 1-thio-β-d-galactopyranoside (0.5 mm final concentration) and further growth for 3 h. The bacteria were harvested and resuspended in buffer A (20 mm Hepes, pH 7.5, 20 mm immidazole) and lysed by sonication. The lysates were cleared by centrifugation (14,000 rpm, 30 min, 4 °C), and the supernatant was loaded on a homemade Ni2+-NTA3 column. Following absorption, the column was washed with buffer B (20 mm Hepes, pH 7.5, 1 m NaCl, 1% Tween 20) and buffer A, the bound proteins were eluted with 0.5 m immidazole in phosphate-buffered saline. The pure protein was dialyzed overnight against phosphate-buffered saline and stored at −80 °C.
Cloning and Purification of Single Lysine and Single Arginine Derivatives of Ubiquitin
Plasmid Construction
The human wild type ubiquitin was cloned into pET22 and used as a template for site-directed mutagenesis. Initially, we substituted all seven lysine residues of ubiquitin by arginines (0K ubiquitin). We then added back each of the seven lysines to its original position, one at a time (denoted as Kx where the x represents for the position of the single lysine added) (20). Single arginine ubiquitin mutants were generated by substituting each lysine of the wild type ubiquitin (one at a time) to arginine using site-directed mutagenesis (denoted as Rx where the x represents the position of the single arginine substituted).
Purification of Wild Type and Single Arginine Ubiquitin Derivatives
All single arginine ubiquitin mutants and wild type ubiquitin were transformed into E. coli BL-21 DE3, grown and induced as described under “Plasmid Constriction.” The bacteria were harvested by centrifugation, resuspended in 20 mm Hepes (pH 7.5), and lysed by sonication. Following centrifugation (16,000 × g), the supernatant was dialyzed overnight against 100 mm acetic acid calibrated by NaOH to pH 5. The lysate was then cleared by centrifugation (14,000 rpm, 30 min, 4 °C), and the supernatant was fractionated on Mono-S anion exchange column using HPLC. The ubiquitin was eluted by a salt gradient at about 350 mm NaCl. The relevant fractions were collected and concentrated on a nitrogen-based ultrafiltration device using a 1-kDa cut-off membrane. The sample was then fractionated on an S-30 gel filtration column using 50 mm Hepes, pH 7.5, and 100 mm NaCl. The relevant fractions were collected, concentrated again by a 1 kDa cut-off ultrafiltration device, and stored at −80 °C.
Purification of Single Lysine Mutants
Following induction, the bacteria were harvested by centrifugation and resuspended in 6 m guanidine HCl in 20 mm Hepes, pH 7.5, and lysed by sonication. The lysate was then cleared by centrifugation (14,000 rpm, 30 min, 4 °C), and the supernatant was passed through a 30 kDa cut-off nitrogen-based ultrafiltration device. The flow-through was further concentrated using a 1 kDa cut-off and fractionated on an S-30 gel filtration column using HPLC as described above. The relevant fractions were collected, concentrated by a 1 kDa cut-off nitrogen-based ultrafiltration device, and stored at −80 °C.
In Vitro Polyubiquitination Reactions
In vitro polyubiquitination reactions contained a 30 nm concentration of the human recombinant E1 (Boston Biochem), 0.5 μm indicated E2 enzyme, and 10 μm wild type ubiquitin or one of the desired purified single lysine/arginine mutants. The polymerization reactions were carried out in 50 μl of buffer C (30 mm Hepes, pH 7.5, 5 mm MgCl2, 2 mm ATP, 0.2 mm DTT, 10 mm sodium citrate, 10 mm creatine phosphate, and 0.2 μg/ml creatine kinase; final concentrations in the reaction mixtures). The reaction was prepared on ice and incubated for 4 h at 37 °C. For analysis, the samples were boiled with sample buffer, separated on an SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and Western blotted with the indicated antibody.
Distinguishing between Lysine and Cysteine Self-ubiquitination of the E2 Enzymes
In vitro polyubiquitination reactions were performed as described above. Ni2+-NTA beads were prewashed in buffer D (20 mm Hepes, pH 7.5) and then added to the reaction mixture. Absorption to the beads was performed at 4 °C on a rotating platform (950 rpm) for 2 h. The beads were sedimented by centrifugation and washed twice with buffer D, twice with buffer B, and two additional times with buffer D. One-half of the beads were boiled in sample buffer containing 100 mm DTT, and the other half were boiled in sample buffer without DTT. Corresponding pairs of reduced and non-reduced samples were separated on SDS-PAGE and analyzed by anti-His and anti-ubiquitin antibodies.
Polyubiquitination of His-Red Fluorescent Protein (RFP)-bound Derivatives
His-RFP derivatives (RFP-Ub, RFP, and RFP-Ub-GW) were conjugated to Ni2+-NTA beads to saturation. The RFP-conjugated beads were washed twice with buffer D, twice with buffer B, and two more times with buffer D. 15 μl of saturated beads were used for each polyubiquitination reaction in the presence of wild type ubiquitin as described above. The reactions were incubated for 4 h at 37 °C on a rotating platform (950 rpm). RFP derivatives were separated from the bulk solution by centrifugation and washed twice with buffer D, twice with buffer B, and two times with buffer D. The beads were resuspended and boiled in sample buffer, separated on SDS-PAGE, and analyzed by Western blot with anti-RFP (MBL).
TEV Protease Digestion of Polyubiquitinated RFP-Ub
Polyubiquitinated RFP-Ub bound to Ni2+-NTA beads (prepared as described above) was washed twice with TEV reaction buffer (50 mm Tris, pH 8, 1 mm DTT). Each reaction was supplemented with 15 μg of TEV protease and incubated at 4 °C for 24 h. The beads were then separated from the reaction mixture by centrifugation, washed twice with 50 mm Tris, pH 8, boiled in sample buffer, separated on SDS-PAGE, and analyzed by Western blot with anti-RFP (MBL).
Mass Spectrometry Detection of Ubiquitinated Lysine Residues
Large scale polyubiquitination reactions were separated on SDS-polyacrylamide gel and stained with Imperial Blue (Pierce). Certain regions and discrete bands were excised from the gel, reduced (10 mm DTT), modified with 40 mm iodoacetamide, and treated with trypsin or chymotrypsin (modified trypsin; Promega) at a 1:100 enzyme/substrate ratio. The resulting tryptic peptides were resolved by reversed phase chromatography on 0.075 × 200-mm fused silica capillaries (J&W) packed with Reprosil reversed phase material (Dr. Maisch GmbH). The peptides were eluted with linear (over 50 min) gradients of 5–45% acetonitrile, 0.1% formic acid, and the tightly bound peptides were collected by a 15-min wash by 95% acetonitrile, 0.1% formic acid in water. All through the run, the flow rate was 0.25 μl/min. Mass spectrometry was performed by an ion trap mass spectrometer (Orbitrap, Thermo) in a positive mode using repetitively full MS scan followed by collision-induced dissociation of the seven most dominant ions selected from the first MS scan. The mass spectrometry data were analyzed using the Sequest 3.31 software (J. Eng and J. Yates, University of Washington and Finnigan (San Jose, CA)) searching against the human part of the NR-NCBI data base. Identification thresholds of an Xcorr value above 2.0 for doubly charged peptides and 2.5 for triply charged peptides were employed. In addition, the identification of the ubiquitinated peptides was assessed visually by a trained operator. Ubiquitination was identified as a mass addition of 114 Da to the relevant peptide (the GG from the C terminus of ubiquitin attached to a lysine residue).
E2 Dimerization Assay
About 0.05 μg (∼50 nm) of each of the purified E2 enzymes, incubated in 50 μl of 20 mm Hepes, pH 7.5, complemented with a 0.05 μm final concentration of cross-linker 3,3′-dithiobis(sulfosuccinimidyl propionate) (Pierce) was added to each sample. The samples were left shaking for 1 h at room temperature. The reactions were quenched with 20 mm Tris, pH 7.5. The samples were then divided into two portions. Half of the reaction mixture was boiled with sample buffer containing 100 mm DTT, whereas the second portion was added boiled in sample buffer without reducing agent. The sample pairs were separated on an SDS-polyacrylamide gel and analyzed by Western blot using α-His antibody.
Far Western
Following cross-linking (see above) the dimerization reactions were boiled in sample buffer without DTT, separated on SDS-PAGE, and transferred to a nitrocellulose membrane. The membrane was then washed with ubiquitination buffer (buffer C) three times. E1 (250 nm) and wild type ubiquitin (100 μm) were added, and the membranes were incubated for an additional 1 h at room temperature. The reactions were stopped by extensive washing with TBST, blocked with 5% milk, and analyzed by Western blot with anti-ubiquitin.
RESULTS
E2 Enzymes Autoubiquitinate Both Their Active Site Cysteine and Lysine Residues
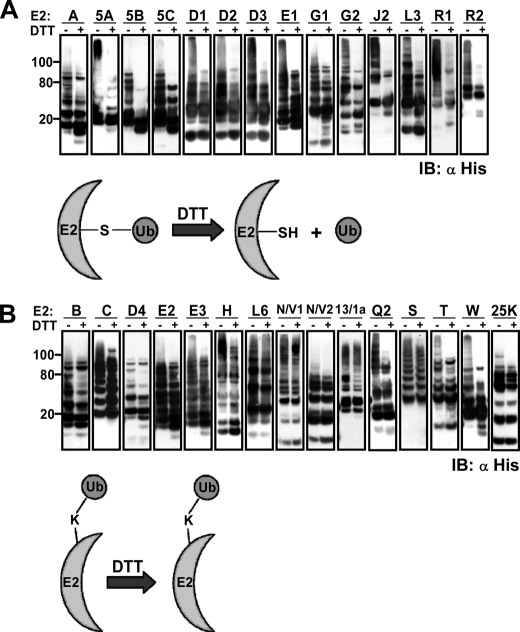
The human E2 family enzymes were cloned and expressed in bacteria genetically fused to a His6 tag to aid in purification and detection (supplemental Fig. S1A and Table S1). Initially, we optimized an in vitro self-ubiquitination reaction for E2 enzymes that possess an active site cysteine within the Ubc domain (the conserved catalytic motif of the E2 family). All E2 enzymes tested catalyzed polyubiquitin chain formation in the absence of an E3 and a substrate (supplemental Fig. S1B). As expected, E2M, E2F, and E2I, which are E2 enzymes of ubiquitin-like proteins, did not form ubiquitin conjugates in vitro. We excluded the possibility that polyubiquitin was assembled on the E2 enzymes by an E2-independent action of the E1 enzyme in control reactions using the naturally occurring active site cysteine mutant E2s (e.g. FTS and UEV1/2).
Next, we determined if free polyubiquitin chains accumulated in the reaction mixture or whether the detected ubiquitin conjugates were linked to the E2 enzymes themselves, either to the active site cysteine or to lysine residues. For this purpose, following the polyubiquitination reaction, the E2 enzymes were captured onto Ni2+-NTA beads. Western blot analysis of the unbound material by an anti-ubiquitin antibody indicated that almost all of the ubiquitin polymers were confined to the beads (data not shown). In parallel, half of the washed beads were boiled in sample buffer containing DTT (100 mm), whereas the second part was boiled in sample buffer without DTT. The boiled pairs of mixtures corresponding to all of the examined E2 enzymes were then separated on SDS-PAGE, and the E2 species were detected by anti-His tag antibody. Polyubiquitin moiety, linked to the active site cysteine, should detach from the E2 upon boiling in the DTT-containing sample buffer. In contrast, polyubiquitin linked to a lysine side chain should be unaffected by the DTT. In the case of E2A, Ubc5A, Ubc5B, Ubc5C, E2D1, E2D2, E2D3, E2E1, E2G1, E2G2, E2J2, E2L3, E2R1, and E2R2 (group 1), the DTT in the sample buffer liberated most of the polyubiquitin from these E2 enzymes, and the high molecular weight species of E2s were lost (Fig. 1A). The size distribution profiles of E2B, E2C, E2D4, E2E2, E2E3, E2H, E2L6, E2N-E2V1, E2N-E2V2, E2Q2, E2S, E2T, E2W, E2-25K, and the yeast Ubc13-UEV1a (group 2) were unaffected or partially affected by the presence of DTT, suggesting that polyubiquitin moieties in these reactions were mostly bound to lysine residues of the E2 enzymes or both to the active site cysteine and lysine residues (Fig. 1B). These findings were supported by mass spectrometry analyses, which detected ubiquitin-modified lysine residues on most DTT-insensitive ubiquitinated E2s and did not identify such residues when the DTT-sensitive ubiquitinated E2s were analyzed (see below; see Table 1). An example of a complete gallery of the peptides detected by mass spectrometry following a ubiquitination reaction is presented in supplemental Table S2.
FIGURE 1.
Recombinant human E2 enzymes catalyze self-ubiquitination in vitro. Shown is autoubiquitination of E2s absorbed to Ni2+-NTA beads. Following ubiquitination, the washed beads were boiled in sample buffer with and without DTT. Pairs of boiled beads corresponding to all E2 enzymes were separated by SDS-PAGE and analyzed for co-migration of the ubiquitin and the E2 enzymes by anti-His antibody. A, E2 enzymes in which the self-ubiquitination was mainly limited to the active site cysteine. B, E2 enzymes that autoubiquitinated mainly on a lysine residue. The categorization of E2s was also supported by mass spectrometry detection of ubiquitin-modified lysine residues only of the E2s belonging to group 2. IB, immunoblot.
TABLE 1.
Summary of human E2 enzyme activity
The lysine preference of the different E2s in autoubiquitination toward the RFP domain in RFP-Ub or in ubiquitin-ubiquitin conjugation was determined by mass spectrometry and single lysine/arginine in vitro polyubiquitination reactions. The regions and the specific bands in the stained gel used for the mass spectrometry analyses are enumerated (supplemental Fig. S3). ND, not determined.
| E2 | MS (supplemental Fig. S3 and Table S2) |
DTT sensitivity (Fig. 1) |
Lysine specificity |
Origin of analyzed MS samples (supplemental Fig. S3) | |||
|---|---|---|---|---|---|---|---|
| Ubiquitination on RFP (substrate) | Self-ubiquitination on lysine | Ubiquitination on active site cysteine | Self-ubiquitination on lysine | Single lysine/arginine (Fig. 2 and supplemental Figs. S4 and S5) | MS (supplemental Fig. S3 and Table S2) | ||
| UBE2B | + | + | − | + | 11 | 11, 48, 63 | 2, 20, 21, 42 |
| UBE2C | + | + | − | + | 11, 48 | 11, 48 | 3, 22, 43, 44 |
| UBE2T | + | + | + | + | All | 11, 48, 27, 63 | 14, 15 |
| UBE2D4 | + | − | + | + | All | 11, 48 | 30, 50 |
| Ubc5C | + | − | + | − | 11, 27, 33, 48 | 11, 48, 63 | 6, 26, 27, 28, 47 |
| Ubc5B | + | − | + | − | 6, 11 | 11, 48, 63 | 5, 24, 25, 46 |
| Ubc5A | − | − | + | − | 11 | 11, 48 | 4, 23, 45 |
| UBE2E2 | − | + | − | + | 11, 48 | 11, 48, 63 | 8, 32, 51, 52 |
| UBE2N/UBE2V1 | − | + (UBE2V1) | − | + | 63 | 63 | 58 |
| UBE2S | − | + | − | + | All but 48 | 11 | 13, 39, 40, 60 |
| UBE2L6 | ND | ND | − | + | ND | ND | ND |
| UBE2H | − | + | − | + | ND | 11, 48 | 35, 56, 57 |
| UBE2W | − | + | + | + | Mono +11 | 11 | 16 |
| UBE2N/UBE2V2 | − | − | − | + | 63 | 63 | Data not shown |
| 25K | − | − | + | + | 48 | 48 | 17 |
| UBE2A | − | − | + | + | 11 | 11, 48 | 1, 18, 19, 41 |
| UBE2D1 | − | − | + | + | 48 | 48 | 29, 48 |
| UBE2D2 | − | − | + | + | 48 | 48 | 49 |
| UBE2D3 | − | − | + | + | 11, 48 | 11 | Data not shown |
| UBE2L3 | ND | ND | + | − | 11 | ND | ND |
| UBE2E1 | − | − | + | + | 48 | 48 | 7, 31 |
| UBE2E3 | − | − | + | + | 11, 48 | 11, 48, 63 | 9, 33, 53 |
| UBE2Q2 | − | − | + | + | ND | 48 | Data not shown |
| UBE2G1 | − | − | + | + | 48 | 48, 63 | 10, 54 |
| UBE2G2 | − | − | + | − | 48 | 48 | 11, 55 |
| UBE2R1 | − | − | + | − | 48 | 48 | Data not shown |
| UBE2R2 | − | − | + | − | Mono +48 | 48 | 12, 37, 38, 59 |
| UBE2J2 | ND | ND | + | − | ND | ND | 36 |
A thiol ester-linked ubiquitin to the E2 active site is an intermediate in any polyubiquitination reactions. Accordingly, one possible explanation for non-active site ubiquitination seen in group 2 (Fig. 1B) is that it might result from a subsequent slower, nonspecific transfer of the polyubiquitin from the active site cysteine to a neighboring lysine on the E2. If true, such a nonspecific transfer is anticipated to be significantly enhanced by the long incubation time used here (4 h). To exclude this possibility, we performed representative ubiquitination reactions (group 2) with a shorter time scale (20 min). Although following shorter incubation periods, lower amounts of ubiquitination species were detected, the type and character of the ubiquitination did not change (supplemental Fig. S2). Thus, group 2 capacity to promote autoubiquitination on lysine residues appears to be a genuine function of these E2s, at least in vitro.
E2 Enzymes Catalyze Polyubiquitin Formation through Preferred Lysine Residues
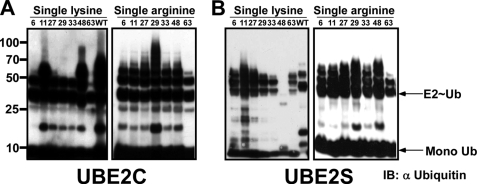
Global in vivo analysis of the specific lysine residues engaged in polyubiquitination indicated that all seven lysine residues of ubiquitin participate in ubiquitin polymerization (23, 31). However, it is unclear how the selection of a specific lysine residue transpires in a given polyubiquitination reaction. To explore whether the E2 directs the type of polyubiquitination, we analyzed the conjugates that were generated by each E2 enzyme by mass spectrometry. Briefly, following in vitro scaled-up self-polyubiquitination reactions in the presence of wild type ubiquitin, the reaction mixtures were separated on SDS-PAGE, and the gel was stained (Imperial, Pierce). Regions or specific bands were subjected to mass spectrometry analysis (supplemental Fig. S3 and Table 1). To complement our mass spectrometry analysis, we also performed similar polyubiquitination reactions in the presence of a panel of single lysine and single arginine variants of ubiquitin (20). Western blot analysis of these reactions with an anti-ubiquitin antibody indicated that all seven lysine residues of ubiquitin could serve in ubiquitin-ubiquitin conjugation (Table 1). As expected, different E2 enzymes could only utilize a subset of the single lysine/arginine derivatives complementary with the specific lysine preference of the E2 enzyme. For example, E2–25K efficiently catalyzes polyubiquitination in the presence of wild type ubiquitin, and the Lys48 single lysine derivative (supplemental Fig. S4A, left), whereas in the single arginine experiments, this E2 enzyme utilized all single arginine derivatives, excluding the Arg48 ubiquitin (supplemental Fig. S4A, right), as reported previously (32). The mammalian ortholog of the yeast Ubc13-UEV1a, the E2N-E2V1 complex, promoted polyubiquitination using only the Lys63 single lysine derivative (supplemental Fig. S4B, left) as described before (33). Consequently, it catalyzed polyubiquitination in the presence of all the single arginine derivatives, except Arg63 (supplemental Fig. S4B, right). E2C (UbcH10) generated polyubiquitin chains via Lys11 and Lys48 single lysine ubiquitin (Fig. 2A, left) and all single arginine derivatives (Fig. 2A, right). This result is in line with previous studies showing that E2C catalyzes Lys11 and Lys48 chain conjugates when acting with the APC E3 complex (34). In agreement with Baboshina et al. (20), our mass spectrometry analysis indicated that E2S promoted formation of long conjugates through lysine 11 (Table 1). However, our single lysine/arginine assays also suggest that E2S can utilize the other single lysine derivatives of ubiquitin to some extent, except Lys48 ubiquitin (Fig. 2B, left). Accordingly, in the complementary single arginine experiments, all single arginine derivatives of ubiquitin supported polyubiquitination (Fig. 2B, right). In agreement with the mass spectrometry analysis, most of the E2 enzymes showed preference to a specific lysine (or a small subset of lysine residues) when catalyzing polyubiquitination. Interestingly, both methods detected a clear preference toward Lys48, Lys11, and Lys63.
FIGURE 2.
Different E2 enzymes could only utilize distinct subsets of the single lysine/arginine derivatives of ubiquitin to produce ubiquitin conjugates. Each E2 enzyme was incubated in a reaction mixture containing the indicated single lysine/arginine ubiquitin derivative. Formation of ubiquitin conjugates was estimated by Western blot analysis with anti-ubiquitin antibody. Representative analyses of such lysine specificity determinations for E2C (A) and E2S (B) are presented. IB, immunoblot.
A notable observation is the frequent involvement of Lys11 of ubiquitin in polyubiquitination. This phenomenon was observed both in the mass spectrometry analyses and in the single lysine/arginine experiments (Table 1 and supplemental Fig. S5). Although the specific role of Lys11 polyubiquitin topology is unclear, it was found to be conjugated to the accumulated PHF-Tau in Alzheimer disease (30) and was shown to be recognized by the 26 S proteasome (20). Moreover, recently, Lys11 linkages were also proposed to have a function in the endoplasmic reticulum-associated degradation pathway (31). Indeed, when whole cell lysate was analyzed by mass spectrometry for the abundance of the different lysines involved in polyubiquitin, Lys11 conjugates appeared as abundant as Lys48 conjugates, both more prevalent than the other topologies (23).
Tethering of a Substrate to an E2 Promotes Its Polyubiquitination
As demonstrated, all purified human E2 enzymes catalyzed in vitro polyubiquitin assembly in the absence of an E3 ubiquitin ligase. However, most of these ubiquitin conjugates were in the form of autoubiquitination of the E2 enzymes themselves (Fig. 1). We were unable to catalyze the transfer of ubiquitin to a model substrate (e.g. mutated form of TCRα, β-casein, bovine serum albumin, and RFP) by our purified E2 enzymes in the absence of an E3 ligase (data not shown). We therefore hypothesized that in the absence of a substrate-specific E3 enzyme, the ubiquitin-bound E2 does not form a complex with the substrate, and thus, the activated ubiquitin cannot be acquired efficiently by the substrate; rather, it is transferred from one E2 monomer to the other, promoting self-ubiquitination. Based on this proposed mode of action (16) and to explore this possibility further and characterize the nature of the functional link between an E2 enzyme and a substrate, we established a model system in which a given substrate could be directly linked to any E2. We genetically engineered a derivative of the RFP with a His6 tag at its N terminus and the human wild type ubiquitin at its C terminus (His-RFP-ubiquitin, abbreviated RFP-Ub). A TEV recognition site was inserted between the RFP and the ubiquitin domain (supplemental Fig. S6). We hypothesized that the ubiquitin would recruit one E2 molecule by binding to its active site cysteine through the action of an E1 enzyme (in analogy to native ubiquitin). An additional E2 monomer would then catalyze the polyubiquitination of the RFP-ubiquitin fusion on either the ubiquitin or the RFP portion of the RFP-Ub in a manner similar to the assembly of polyubiquitin on E2G2 (16).
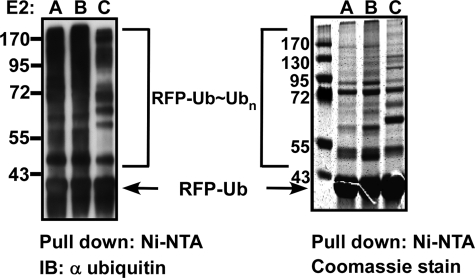
To monitor the level of polyubiquitination on the substrate, agarose-Ni2+-NTA beads were saturated with RFP-Ub and used for in vitro polyubiquitination reactions on beads, in the presence of E1, ubiquitin, and the different E2 enzymes. Once the reactions were completed, the polyubiquitinated RFP-Ub-bound beads were separated from the bulk solution and thoroughly washed. The beads were then boiled with sample buffer and separated on SDS-PAGE. All of the E2 enzymes, with the exception of E2L3, efficiently polyubiquitinated RFP-Ub (see Fig. 3 (left) for representative reactions and supplemental Fig. S7A for the complete data). Next, we scaled up the RFP-Ub polyubiquitination reactions, separated the reaction products by SDS-PAGE, and stained the gel (see Fig. 3 (right) for representative reactions and supplemental Fig. S7B for the complete data). Regions of polyubiquitinated species as well as specific bands were excised from the gel and analyzed by mass spectrometry to determine which lysine residues on the RFP-Ub fusion were conjugated to ubiquitin.
FIGURE 3.
Polyubiquitination of RFP-Ub proceeds in an E3-independent manner. RFP-Ub was absorbed onto Ni2+-NTA-agarose beads, and the beads were then added to the polyubiquitination reactions with each of the E2 enzymes. Left, Western blotting analysis with anti-RFP antibody of representative reactions. Right, similar scaled up reactions stained with Coomassie Blue. RFP-Ub ubiquitination reactions by all other E2s are documented in supplemental Fig. S7. IB, immunoblot.
Only a small subset of the E2 enzymes (E2B, E2C, Ubc5B, Ubc5C, E2D4, and E2T) directly ubiquitinated the RFP portion of the chimera (Table 1 and supplemental Fig. S8). The rest of the E2 enzymes could only link ubiquitin to the ubiquitin portion of the RFP-Ub fusion. Both types of ubiquitinations were highly specific in the selection of lysine residues modified by ubiquitin (Table 1) and in complete agreement with the lysine preference determined in the E2 self-ubiquitination mass spectrometry analyses. To estimate the ratio of the ubiquitination linked to the ubiquitin portion of the RFP-Ub relative to the direct ubiquitination of the RFP domain, we separated the ubiquitin and the RFP portions using the TEV protease. Briefly, following the polyubiquitination reaction, the RFP-Ub beads were spun down and washed extensively. Following incubation of half of the beads with TEV protease, pairs of mixtures (with and without TEV treatment) corresponding to all of the examined E2 enzymes were separated on SDS-PAGE and analyzed by anti-RFP antibodies (Fig. 4). Our results indicate that even in ubiquitination reactions where the E2s ubiquitinate the RFP itself, almost all of the ubiquitination occurred on the ubiquitin domain and not the RFP. Accordingly, after TEV digestion, most of the ubiquitinations were removed from the RFP (Fig. 4 and supplemental Fig. S9).
FIGURE 4.
Ubiquitin is conjugated preferentially to the ubiquitin domain of the RFP-Ub. A, illustration of the three possible scenarios for RFP-Ub ubiquitination. In the left panel, ubiquitin conjugates only to a lysine residue on the ubiquitin portion of the chimera. In the middle panel, ubiquitin is linked to both the RFP and the ubiquitin domains of the RFP-Ub fusion. The right panel represents a mode of action in which ubiquitin is linked to the RFP and the ubiquitin domains of the RFP-Ub. B, polyubiquitination reactions of RFP-Ub beads were conducted as described in the legend to Fig. 3. Each reaction was divided, and one-half was subjected to TEV protease digestion. The TEV-treated and -untreated portions were separated side by side on SDS-PAGE and analyzed by Western blot analysis with anti-RFP antibody. Left, representative reactions of E2s that did not directly ubiquitinate the RFP portion (E2A and E2E2); right, E2s that directly add ubiquitin to the RFP portion (E2C and Ubc5C). Similar analysis of all other E2s is depicted in supplemental Fig. S9. IB, immunoblot.
Although RFP has 23 lysine residues (2 buried and 21 exposed), only five of these were ubiquitinated in our in vitro assay using the various E2s (supplemental Fig. S8). The ubiquitinated lysine residues on the RFP were not necessarily the closest ones to the ubiquitin domain of the molecule but were positioned in all cases on flexible loops at the edges of secondary structures. It is noteworthy that exactly the same characteristics of ubiquitination sites were evaluated in vivo by Catic et al. (35). Again, ubiquitnation sites were found to be located on flexible loops and on the edges of α-helix structures.
Integrity of the Ubiquitin Portion of RFP-Ubiquitin Is Essential for Ubiquitination
To verify the role of the ubiquitin portion of the RFP-Ub fusion in the polyubiquitination reaction, we generated two more derivatives of RFP: a His-RFP that does not have a ubiquitin domain (RFP) and an RFP-Ub in which the C-terminal glycine was substituted by tryptophan (RFP-Ub-GW). These derivatives were conjugated to Ni2+-NTA beads and served in polyubiquitination reactions (as described for the RFP-Ub). Following polyubiquitination, the beads were thoroughly washed, boiled with sample buffer, separated on SDS-PAGE, and analyzed by Western blot with both anti-RFP and anti-ubiquitin antibodies. As seen in supplemental Fig. S11A, when RFP beads were used as a substrate, high molecular weight species of RFP were hardly detected. Thus, the ubiquitin portion of the fusion was essential for polyubiquitination of the RFP. If our model is correct and the RFP-Ub fusion is anchored to the E2 enzymes via their active site cysteine, then the presence of a C-terminal glycine in the ubiquitin should be crucial for efficient polyubiquitination of the RFP. To evaluate this possibility, we used the second derivative (RFP-Ub-GW). As shown in supplemental Fig. S11B, Ni2+-NTA beads preconjugated to RFP-Ub-GW did not serve as substrates for polyubiquitination by the different E2 enzymes, as evident from the lack of high molecular weight species of RFP. Taken together, these results suggest that tethering of the RFP to specific E2 via the active site cysteine is sufficient to promote efficient polyubiquitination of the bound substrate.
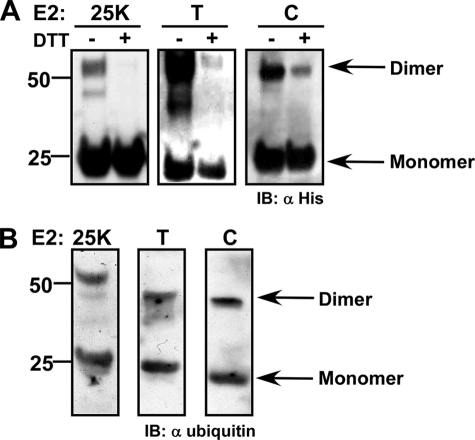
E2 Enzymes Spontaneously Dimerize in Vitro
As mentioned above, one way to juxtapose a pair of ubiquitin molecules for ubiquitin-ubiquitin formation might be through dimerization of E2 enzymes (36–38). Because our experiments with RFP-Ub possibly support the notion that the E2 enzymes functionally act as dimers, we set out to examine whether E2 enzymes form spontaneous dimers by conducting an in vitro cross-linking experiment. Similar amounts of the different purified E2 enzymes (∼5 nm) were incubated in 20 nm Hepes buffer (pH 7.5) in the presence of 10 μm DTSSP (Pierce), a primary amine-reactive homobifunctional cross-linker in which two N-hydroxysuccinimide ester groups are linked through a disulfide bridge. As such, any complex cross-linked by DTSSP should dissociate upon exposure to a reducing agent, such as DTT. Following 1 h of incubation on a rotating platform, the cross-linker was quenched with 200 mm Tris, and the reaction mixture was divided in two. To one portion we added sample buffer with DTT, whereas the other half of the sample was boiled in sample buffer without reducing agent. All of the E2 reaction pairs were separated on SDS-PAGE and analyzed by Western blot with anti-His antibodies. As seen in Fig. 5A and supplemental Fig. S10, almost all E2 enzymes spontaneously formed dimers in solution (in the absence of a charged ubiquitin), and this dimerization was reproducibly captured by cross-linking. Ubc5C, UBE2E1, UBE2E2, and UBE2E3 did not consistently form dimers under these conditions. Possibly, these E2 enzymes form a more labile dimer.
FIGURE 5.
The E2s may act as dimers. A, spontaneous dimerization profile of E2–25K (left), E2T (middle), and E2C (right) captured by cross-linking with DTSSP. Reduced and non-reduced equal portions of the cross-linked mixtures were separated on SDS-PAGE and analyzed by Western blot with anti-His. The entire repertoire of the E2 dimerization profile is presented in supplemental Fig. S10. B, both the monomeric and dimeric forms of the E2 can acquire ubiquitin. Following cross-linking, E2-25K, E2T, and E2C were separated to dimeric and monomeric forms on non-reducing SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was incubated with E1 and wild type ubiquitin in standard ubiquitination conditions. Charging of the monomeric and dimeric forms was determined by Western blot analysis with anti-ubiquitin. IB, immunoblot.
Next, we set out to examine which form of the E2 (monomeric or dimeric) could be charged with ubiquitin by the action of an E1. For this end, we performed a far Western experiment following cross-linking of E2C, E2T, and E2–25K with the DTSSP. Briefly, the dimeric and monomeric forms of the cross-linked enzymes were separated on an SDS-polyacrylamide gel and blotted onto a nitrocellulose membrane. The membrane was washed with polyubiquitination buffer (buffer C) and immersed in the minimal volume possible of the same buffer in the presence of E1 and wild type ubiquitin. The reaction was incubated with slow shaking for 4 h and terminated by extensive washing with TBST. The membrane was then blocked with 5% milk and blotted with anti-ubiquitin. As seen in Fig. 5B, for all three enzymes (E2C, E2T, and E2–25K), both the monomeric and dimeric forms were detected with the anti-ubiquitin, illustrating that both monomeric and dimeric forms could be charged with ubiquitin by the action of the E1.
Uniform Versus Mixed Topologies in Polyubiquitination
The fact that certain E2 enzymes may polymerize ubiquitin through more than one lysine residue in ubiquitin raises the possibility that a given E2 enzyme may promote polyubiquitination in three possible chain topologies: homogenous (i.e. all of the ubiquitin molecules in a given polyubiquitin chain are linked through the same lysine), mixed (where different ubiquitin molecules in the same polyubiquitin are linked through different lysine residues (27, 39)), or branched (heterogeneous) (i.e. some of the ubiquitin molecules in the polyubiquitin are bound to other ubiquitins on multiple lysine residues (40)). In addition, more complex configurations of various mixtures of these topologies are also theoretically possible. In an attempt to address these possibilities, we resolved on SDS-PAGE the reaction products of a large scale polyubiquitination reaction of all active E2 enzymes. The gel was stained, and distinct bands (rather than regions) were analyzed by mass spectrometry. Interestingly, specific polyubiquitin species (bands) generated by E2 enzymes, which showed promiscuous specificity, contained uniform polyubiquitin topologies, conjugated through only one of the lysines. For example, analysis of a specific band (E2T lower band; supplemental Fig. S3A, band 15) detected only Lys11 conjugates from a range of in vitro polyubiquitin conjugates generated by E2T (Lys11, Lys27, Lys48, and Lys63). In addition, although our mass spectrometry analysis detected fragments of ubiquitin with more than one lysine, we did not detect ubiquitin-derived peptides containing two conjugated lysine residues. In contrast, we did detect multiple peptides of E2S, in which two consecutive lysine residues were conjugated to ubiquitin (supplemental Table S2B). This finding suggests that in vitro E2 rarely (if at all) catalyzes the formation of branched polyubiquitin chains.
DISCUSSION
E2 Enzymes Have a Role in Determining the Lysine Preference in Ubiquitination
Our results suggest that most E2 enzymes can only generate polyubiquitin chains through a specific lysine or a limited subset of lysine residues of ubiquitin (Table 1). The lysine preference was assessed using three independent experimental approaches: mass spectrometry analyses of both E2 enzymes following an autoubiquitination reaction and of ubiquitin conjugates on a substrate (RFP-Ub) as well as biochemical analysis of ubiquitination with single lysine/arginine ubiquitin mutants. Combined, these results demonstrate that the E2s themselves possess an inherent preference for specific lysine residues when promoting polyubiquitin chain assembly. Furthermore, comparing the lysine specificity of the E2 enzymes examined here with several studies where the lysine specificity of an E2 was determined in conjunction with an E3 enzyme clearly shows that the ability of a given E2 enzyme to polymerize ubiquitin through a specific lysine is not altered by the E3. This notion is illustrated in the following examples: BRCA/BARD, an E3 ligase was shown to act with E2–25K and generate Lys48-conjugated chains, whereas with E2N-E2V1 (Ubc13-UEV1a), it catalyzes the formation of polyubiquitin chains conjugated through Lys63 (41). Pellino1 E3 also acts with E2N-E2V1 to generate lysine Lys63 chains, whereas when acting with E2R (Ubc3/CDC34), it catalyzes the formation of Lys48 chains. When this E3 functions with the E2D family (Ubc4/5), it promotes the formation of Lys11 and Lys48 chains (42). Polyubiquitinations through Lys11 have also been shown to be preferentially generated when E2C (UbcH10) and E2S are recruited to the APC E3 complex (34). In our in vitro analysis, E2-25K and E2R (CDC34) generate Lys48 chains, whereas Ubc13-UEV1a could conjugate ubiquitin only trough Lys63. The E2D family utilizes both Lys11 and Lys48, whereas E2C (UbcH10) preferentially used Lys11 and Lys48 when catalyzing polyubiquitin chain formation. Thus, our in vitro results are in perfect agreement with the previously published data, where the E2 enzymes were studied in the context of an E3 in vitro and in vivo.
Even when multiple lysine preferences were found for an examined E2 (depending on the specific E3 it functions with), these preferences coincided with the multiple capacities to conjugate ubiquitin that were assigned to this E2 in our in vitro approach. For example, E2D (UbcH5) catalyzes Lys48 chain topologies when acting with the E3 E6-AP, and Lys63 conjugates with Nedd4; both are HECT domain E3 ligases (40). Likewise, the yeast homolog of Nedd4, RSP5, catalyzes Lys63 chains on various substrates when acting with Ubc4 and Ubc5 (E2D) (43, 44). In line with these reports, we show that in vitro E2D can utilize both Lys48 and Lys63 in polyubiquitination. A broader range of lysine specificity was detected when the Rad6 (E2A/B) was used in polyubiquitination with various E3s (Bre1, Ubr1, and Rad18) (20, 34, 45), each selecting different lysine specificity from the potential conjugation abilities of Rad6 (Lys11 and Lys48). In aggregate, the clear overlap in the lysine preferences demonstrated by a given E2, independent of the presence or absence of an E3 enzyme, is a strong indicator of the central role of the E2 enzymes in the determination of the lysine preference in ubiquitination. We conclude that although the role of the E3 is to select the E2 enzyme and perhaps its mode of action (i.e. select specific lysine preference when multiple lysine residues of ubiquitin could be used by the E2), the latter determines the type of ubiquitin chains formed.
Additional support for the direct role of E2 enzymes in dictating the lysine preference of ubiquitin conjugates comes from analysis of structural motives in E2 enzymes: E2–25K (32), E2D3 (UbcH5C) (46), and Ubc13-UEV1a (E2N-E2V1) (47, 48). They were all shown to contain a region that interacts non-covalently with the attacking ubiquitin in such a manner that the ubiquitin molecule is poised by this interaction to form lysine-specific linkage with the active site-bound ubiquitin (49, 50). In support of this notion, we detected that E2T catalyzed ubiquitin polymerization through Lys27 of ubiquitin (Table 1 and supplemental Fig. S3). Because Lys27 appears buried within the ubiquitin (Protein Data Bank code 1UBQ), the conformation of the attacking ubiquitin is most likely affected by the interaction with E2T, such that its Lys27 would become poised to attack the thioester bond of the E2T active site-bound ubiquitin and acquire the activated ubiquitin.
A major role for the E2 enzymes in determining the lysine preference in polyubiquitination is also compatible with the study of Li et al. (16), who suggested that the polyubiquitin chain is first assembled on the E2 by a “ping-pong” action of a pair of E2 molecules. In this model, only once a polyubiquitin chain is matured, is it transferred as a module to the substrate. Here, the ubiquitin chain topology must be determined by the E2, because the E3 operates only following assembly of the polyubiquitin tree. It is noteworthy that in our self-ubiquitination assay, a significant number of E2 enzymes accumulated polyubiquitin chains on their active site cysteine in the absence of an E3 (Fig. 1A).
E2 Mode of Action
One simple model to account for the initial assembly of polyubiquitin chain on an E2 enzyme prior to its “en block” acquisition by the target (as was demonstrated by Li et al. (16)) is through a ping-pong action of a pair of E2 molecules. In terms of processivity, there is a clear advantage for E2 enzymes to act as dimers because at least one of the E2 monomers may remain associated with the substrate while maintaining continuous additions of ubiquitin monomers as an intramolecular reaction. This can be attributed to the fact that the E1 and the E3 binding sites on the E2 enzyme overlap, and their binding to the E2 is mutually exclusive (51). Thus, the sequential addition of ubiquitin will probably require multiple cycles of E2-E3 binding and release. The presence of a stable E2 dimer in the E2-E3 complex might circumvent the need for complete dissociation of the E3 from the E2, provided that the E1 and E3 bind to different E2 monomers in the dimer. Moreover, this model of ping-pong action of the E2 enzymes in a dimer may conserve certain constrains, which may be important in terms of lysine specificity, length of the polyubiquitin, and other attributes of the polyubiquitin moiety, which might depend on the specific setup (i.e. components and conditions of the reaction). The results of our in vitro E2 dimerization analysis suggest that all E2s can spontaneously form dimers in solution independently of the presence of activated ubiquitin. Additionally, both the dimer and monomer forms can be charged with ubiquitin by the action of an E1. Although not conclusive, these findings support the rising notion that E2s act as dimers when catalyzing polyubiquitination.
It is noteworthy that both in vitro and in vivo analyses performed in the past have recognized an ability of certain E2 enzymes to form dimers, particularly if the E2 is charged with ubiquitin. The homodimerization of the yeast E2, Cdc34 (E2R), is induced by ubiquitin thioester formation and is necessary for its function (52). Heterodimer formation between the yeast E2s Ubc6 (E2J) and Ubc7 (E2G) was suggested by yeast two-hybrid analysis (37) and implicated in endoplasmic reticulum-associated degradation (53). However, the clearest demonstration of a functional dimer in ubiquitin chain assembly was revealed in the structural and functional analysis of the heterodimer formed between Ubc13 (E2N) and the inactive E2 UEV (ubiquitin E2 variant) proteins that catalyzes Lys63 ubiquitin conjugates (33).
It is relatively simple to envision a possible model in which the E2 selects the target lysine in ubiquitin conjugation when a RING E3 is engaged. However, it is probably less intuitive when the E2 is acting with a HECT domain E3. In this scenario, the target lysine on the substrate is determined by the E3 itself (9, 39, 54) unless the polyubiquitin is initially assembled on the active site cysteine of the HECT E3 by the action of the conjugated E2 and only then is acquired by a lysine residue on the target (55). In this respect, the set of E2s known to operate with HECT E3s includes the E2D family, E2L3 (UbcH7) and E2G (Ubc7) (17, 39, 56–59). Interestingly, all of these E2s were found to assemble polyubiquitin on their own active site cysteine in our assay (Fig. 1). This raises the possibility that the lysine preference in a given polyubiquitination reaction is affected by the specific E2 enzyme engaged even when a HECT E3 is coordinating the polyubiquitination reaction, yet a recent study has suggested that the HECT E3s E6AP and RSP5 generate lysine-specific chains regardless of the interacting E2 (60).
Conclusions
Polyubiquitination occurs through all seven lysines of ubiquitin. Here we provide a mechanistic link between specific topologies and the activity of specific E2 enzymes. We suggest that the lysine specificity of the E2 enzymes determined here, in vitro, implies the in vivo capabilities of these E2s. Our data suggest that polyubiquitination through Lys11 of ubiquitin is as common as those of Lys48 and is catalyzed by many of the E2s. Similarly, although not as abundant, polyubiquitination through Lys63 is also frequent.
We propose a mechanism by which the E3 enzyme governs the identity of the recruited E2 enzyme, the substrate specificity, the mode of action (mono- or polyubiquitination), and, perhaps, the general region in the substrate to be ubiquitinated. The E2, in turn, catalyzes the desired type of ubiquitination on a specific lysine residue in that region.
Supplementary Material
Acknowledgments
We thank Dr. Sirano Dhe-Paganon (University of Toronto) for kindly providing UBE2H and UBE2J2 plasmids and Dr. Wei Xiao (University of Saskatchewan) for the UBE2V1 plasmid. We also thank Dr. Boaz Tirosh for fruitful discussion and comments.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S11.
- NTA
- nitrilotriacetic acid
- HPLC
- high pressure liquid chromatography
- DTT
- dithiothreitol
- RFP
- red fluorescent protein
- Ub
- ubiquitin
- TEV
- tobacco etch virus.
REFERENCES
- 1.Goldberg A. L. (1995) Science 268, 522–523 [DOI] [PubMed] [Google Scholar]
- 2.Hershko A., Ciechanover A. (1998) Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 3.Hochstrasser M. (1996) Annu. Rev. Genet 30, 405–439 [DOI] [PubMed] [Google Scholar]
- 4.Hanna J., Finley D. (2007) FEBS Lett. 581, 2854–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Köhler A., Cascio P., Leggett D. S., Woo K. M., Goldberg A. L., Finley D. (2001) Mol. Cell 7, 1143–1152 [DOI] [PubMed] [Google Scholar]
- 6.Busch H., Goldknopf I. L. (1981) Mol. Cell Biochem. 40, 173–187 [DOI] [PubMed] [Google Scholar]
- 7.Hershko A., Ciechanover A., Rose I. A. (1981) J. Biol. Chem. 256, 1525–1528 [PubMed] [Google Scholar]
- 8.Hershko A., Heller H., Elias S., Ciechanover A. (1983) J. Biol. Chem. 258, 8206–8214 [PubMed] [Google Scholar]
- 9.Ardley H. C., Robinson P. A. (2005) Essays Biochem. 41, 15–30 [DOI] [PubMed] [Google Scholar]
- 10.Hicke L. (2001) Nat. Rev. Mol. Cell Biol. 2, 195–201 [DOI] [PubMed] [Google Scholar]
- 11.Hicke L., Dunn R. (2003) Annu. Rev. Cell Dev. Biol. 19, 141–172 [DOI] [PubMed] [Google Scholar]
- 12.Haglund K., Di Fiore P. P., Dikic I. (2003) Trends Biochem. Sci. 28, 598–603 [DOI] [PubMed] [Google Scholar]
- 13.Kravtsova-Ivantsiv Y., Cohen S., Ciechanover A. (2009) Mol. Cell 33, 496–504 [DOI] [PubMed] [Google Scholar]
- 14.Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 15.Hochstrasser M. (2006) Cell 124, 27–34 [DOI] [PubMed] [Google Scholar]
- 16.Li W., Tu D., Brunger A. T., Ye Y. (2007) Nature 446, 333–337 [DOI] [PubMed] [Google Scholar]
- 17.Ravid T., Hochstrasser M. (2007) Nat. Cell Biol. 9, 422–427 [DOI] [PubMed] [Google Scholar]
- 18.Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. (2006) Mol. Cell 24, 701–711 [DOI] [PubMed] [Google Scholar]
- 19.Shang F., Deng G., Liu Q., Guo W., Haas A. L., Crosas B., Finley D., Taylor A. (2005) J. Biol. Chem. 280, 20365–20374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baboshina O. V., Haas A. L. (1996) J. Biol. Chem. 271, 2823–2831 [DOI] [PubMed] [Google Scholar]
- 21.Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. (1995) J. Biol. Chem. 270, 17442–17456 [DOI] [PubMed] [Google Scholar]
- 22.Pickart C. M., Fushman D. (2004) Curr. Opin. Chem. Biol. 8, 610–616 [DOI] [PubMed] [Google Scholar]
- 23.Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., Marsischky G., Roelofs J., Finley D., Gygi S. P. (2003) Nat. Biotechnol. 21, 921–926 [DOI] [PubMed] [Google Scholar]
- 24.Wen R., Newton L., Li G., Wang H., Xiao W. (2006) Plant. Mol. Biol. 61, 241–253 [DOI] [PubMed] [Google Scholar]
- 25.Fujimuro M., Nishiya T., Nomura Y., Yokosawa H. (2005) Biol. Pharm. Bull. 28, 2315–2318 [DOI] [PubMed] [Google Scholar]
- 26.Duncan L. M., Piper S., Dodd R. B., Saville M. K., Sanderson C. M., Luzio J. P., Lehner P. J. (2006) EMBO J. 25, 1635–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang F., Kirkpatrick D., Jiang X., Gygi S., Sorkin A. (2006) Mol. Cell 21, 737–748 [DOI] [PubMed] [Google Scholar]
- 28.Wu C. J., Conze D. B., Li T., Srinivasula S. M., Ashwell J. D. (2006) Nat. Cell Biol. 8, 398–406 [DOI] [PubMed] [Google Scholar]
- 29.Rape M., Kirschner M. W. (2004) Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 30.Cripps D., Thomas S. N., Jeng Y., Yang F., Davies P., Yang A. J. (2006) J. Biol. Chem. 281, 10825–10838 [DOI] [PubMed] [Google Scholar]
- 31.Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merkley N., Shaw G. S. (2004) J. Biol. Chem. 279, 47139–47147 [DOI] [PubMed] [Google Scholar]
- 33.Pickart C. M. (2002) Nature 419, 120–121 [DOI] [PubMed] [Google Scholar]
- 34.Jin L., Williamson A., Banerjee S., Philipp I., Rape M. (2008) Cell 133, 653–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catic A., Collins C., Church G. M., Ploegh H. L. (2004) Bioinformatics 20, 3302–3307 [DOI] [PubMed] [Google Scholar]
- 36.Silver E. T., Gwozd T. J., Ptak C., Goebl M., Ellison M. J. (1992) EMBO J. 11, 3091–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen P., Johnson P., Sommer T., Jentsch S., Hochstrasser M. (1993) Cell 74, 357–369 [DOI] [PubMed] [Google Scholar]
- 38.Gazdoiu S., Yamoah K., Wu K., Escalante C. R., Tappin I., Bermudez V., Aggarwal A. K., Hurwitz J., Pan Z. Q. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15053–15058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang M., Cheng D., Peng J., Pickart C. M. (2006) EMBO J. 25, 1710–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim H. T., Kim K. P., Lledias F., Kisselev A. F., Scaglione K. M., Skowyra D., Gygi S. P., Goldberg A. L. (2007) J. Biol. Chem. 282, 17375–17386 [DOI] [PubMed] [Google Scholar]
- 41.Christensen D. E., Brzovic P. S., Klevit R. E. (2007) Nat. Struct. Mol. Biol. 14, 941–948 [DOI] [PubMed] [Google Scholar]
- 42.Ordureau A., Smith H., Windheim M., Peggie M., Carrick E., Morrice N., Cohen P. (2008) Biochem. J. 409, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu J. Y., Lin Y. Y., Qian J., Tao S. C., Zhu J., Pickart C., Zhu H. (2008) Mol. Cell Proteomics 7, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fisk H. A., Yaffe M. P. (1999) J. Cell Biol. 145, 1199–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baboshina O. V., Crinelli R., Siepmann T. J., Haas A. L. (2001) J. Biol. Chem. 276, 39428–39437 [DOI] [PubMed] [Google Scholar]
- 46.Brzovic P. S., Lissounov A., Christensen D. E., Hoyt D. W., Klevit R. E. (2006) Mol. Cell 21, 873–880 [DOI] [PubMed] [Google Scholar]
- 47.VanDemark A. P., Hofmann R. M., Tsui C., Pickart C. M., Wolberger C. (2001) Cell 105, 711–720 [DOI] [PubMed] [Google Scholar]
- 48.McKenna S., Moraes T., Pastushok L., Ptak C., Xiao W., Spyracopoulos L., Ellison M. J. (2003) J. Biol. Chem. 278, 13151–13158 [DOI] [PubMed] [Google Scholar]
- 49.Eddins M. J., Carlile C. M., Gomez K. M., Pickart C. M., Wolberger C. (2006) Nat. Struct. Mol. Biol. 13, 915–920 [DOI] [PubMed] [Google Scholar]
- 50.Lewis M. J., Saltibus L. F., Hau D. D., Xiao W., Spyracopoulos L. (2006) J. Biomol. NMR 34, 89–100 [DOI] [PubMed] [Google Scholar]
- 51.Eletr Z. M., Huang D. T., Duda D. M., Schulman B. A., Kuhlman B. (2005) Nat. Struct. Mol. Biol. 12, 933–934 [DOI] [PubMed] [Google Scholar]
- 52.Varelas X., Ptak C., Ellison M. J. (2003) Mol. Cell. Biol. 23, 5388–5400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Biederer T., Volkwein C., Sommer T. (1997) Science 278, 1806–1809 [DOI] [PubMed] [Google Scholar]
- 54.Wang M., Pickart C. M. (2005) EMBO J. 24, 4324–4333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verdecia M. A., Joazeiro C. A., Wells N. J., Ferrer J. L., Bowman M. E., Hunter T., Noel J. P. (2003) Mol. Cell 11, 249–259 [DOI] [PubMed] [Google Scholar]
- 56.Park Y., Yoon S. K., Yoon J. B. (2009) J. Biol. Chem. 284, 1540–1549 [DOI] [PubMed] [Google Scholar]
- 57.Raimondo D., Giorgetti A., Bernassola F., Melino G., Tramontano A. (2008) Biochem. Pharmacol. 76, 1620–1627 [DOI] [PubMed] [Google Scholar]
- 58.Eletr Z. M., Kuhlman B. (2007) J. Mol. Biol. 369, 419–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogunjimi A. A., Briant D. J., Pece-Barbara N., Le Roy C., Di Guglielmo G. M., Kavsak P., Rasmussen R. K., Seet B. T., Sicheri F., Wrana J. L. (2005) Mol. Cell 19, 297–308 [DOI] [PubMed] [Google Scholar]
- 60.Kim H. C., Huibregtse J. M. (2009) Mol. Cell. Biol. 29, 3307–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.