Abstract
Bacterial acyl carrier protein (ACP) is essential for the synthesis of fatty acids and serves as the major acyl donor for the formation of phospholipids and other lipid products. Acyl-ACP encloses attached fatty acyl groups in a hydrophobic pocket within a four-helix bundle, but must at least partially unfold to present the acyl chain to the active sites of its multiple enzyme partners. To further examine the constraints of ACP structure and function, we have constructed a cyclic version of Vibrio harveyi ACP, using split-intein technology to covalently join its closely apposed N and C termini. Cyclization stabilized ACP in a folded helical conformation as indicated by gel electrophoresis, circular dichroism, fluorescence, and mass spectrometry. Molecular dynamics simulations also indicated overall decreased polypeptide chain mobility in cyclic ACP, although no major conformational rearrangements over a 10-ns period were noted. In vivo complementation assays revealed that cyclic ACP can functionally replace the linear wild-type protein and support growth of an Escherichia coli ACP-null mutant strain. Cyclization of a folding-deficient ACP mutant (F50A) both restored its ability to adopt a folded conformation and enhanced complementation of growth. Our results thus suggest that ACP must be able to adopt a folded conformation for biological activity, and that its function does not require complete unfolding of the protein.
Keywords: Methods/Circular Dichroism CD, Methods/Fluorescence, Methods/Mass Spectrometry, Protein/Conformation, Protein/Stability, Molecular Dynamics, Acyl Carrier Protein, Complementation, Inteins
Introduction
Bacterial acyl carrier protein (ACP)2 is a small (typically 70–80 residue) protein required for the synthesis and transfer of fatty acyl chains in the production of phospholipids and other specialized products, including lipid A, lipoic acid, acylhomoserine lactones, and hemolysin (reviewed in Ref. 1). Over two dozen nuclear magnetic resonance (NMR) and x-ray crystal structures have revealed a conserved “ACP fold” consisting of a four-helix bundle (1). Fatty acids covalently attached to the phosphopantetheine prosthetic group at the N-terminal end of helix II are enclosed within the hydrophobic interior of this bundle, interacting predominantly with residues on helices II-IV (2–5). Further computational (6, 7), crystallographic (8, 9), and mutagenic (10–13) analyses have implicated the acidic central helix II as a “recognition helix” for interaction with most of the ACP enzyme partners. Subtle conformational alterations in this region likely also contribute to discrimination by ACP-dependent enzymes among the various acyl-ACP derivatives (4, 14).
The acidic nature of ACP (pI ∼ 4) contributes to a highly dynamic and flexible structure in solution, and some ACPs exhibit features characteristic of natively unfolded proteins (1). For example, Vibrio harveyi ACP is largely unfolded at neutral pH, but its helical conformation can be stabilized by charge neutralization (i.e. at low pH or by residue replacements) (13), by binding of divalent cations to helix II (10), or by interaction with partner enzymes (15). Molecular dynamic simulations (16) and NMR experiments (17, 18) have indicated greatest mobility in the long loop I connecting helices I and II, in the helix II-helix III region, and at the N and C termini, which are in close proximity. Clearly, for acyl transfer to occur from ACP to a partner enzyme a significant conformational change must occur to allow release of the sequestered acyl chain to the enzyme active site. This unfolding event thus represents a hallmark of ACP function and suggests that the dynamic nature of ACP is essential for cell survival.
An important question is the mechanism and extent to which ACP must unfold to transfer its acyl chain to or from enzyme partners. In this study, we have created cyclic analogues of V. harveyi ACP to examine the effects on conformational stability and constraints on biological activity. Cyclization was achieved by split-intein mediated in vivo ligation of the ACP N- and C termini (19). Several biophysical approaches indicated that the cyclic ACP is stabilized in a folded conformation. Strikingly, expression of the cyclic ACP in Escherichia coli lacking endogenous (i.e. linear) ACP resulted in complementation of growth close to wild-type levels, indicating that this major structural constraint does not abolish interactions with any of the ACP multiple essential partner enzymes. Our data provide insight into ACP structure-function relationships and the dynamic properties relevant to its biological function.
EXPERIMENTAL PROCEDURES
Plasmid Construction
PCR reactions were carried out using Phusion DNA Polymerase (Finnzymes) and commercially synthesized primers (Integrated DNA Technologies). All PCR products were subcloned into cloning vector pJET1.2 (Fermentas), except for products from inverse PCR reactions, which were directly circularized by blunt-end ligation. Complete primer sequences are given in supplemental Table S1; bacterial strains and plasmids used in this study are listed in supplemental Table S2.
The coding regions of the C- and N-terminal splicing domains of the Ssp GyrB intein were amplified from pDSG3 with primer pairs IC-for/IC-rev and IN-for/INHIS-rev, respectively; the products were fused by anneal-extension PCR and subsequently amplified using primers IC-for/INHIS-rev. The complete product was cloned into pT (a derivative of the pTWIN1 vector from New England Biolabs)4 using SacI and PstI, creating pTIC(NS)INH. The coding regions of the V. harveyi ACP L46W and F50A mutants were amplified from pGEX-L46W (15) and pGEX-F50A (20), respectively, using primers ACP-for/ACP-rev and cloned into pTIC(NS)INH using NheI and SapI, resulting in pTCYC-L46W and pTCYC-F50A. The complete open-reading frames of the cyclization constructs from the latter plasmids were transferred into the pMAL vector (New England Biolabs) using NdeI and PstI, thereby replacing the maltose-binding protein (MBP) gene. All splicing-deficient L46W constructs (with C1A/N435A mutations in the intein sequence) were made similarly, but using primer pairs IC-for/ICMUT-rev and INMUT-for/INHIS-rev in the initial PCR.
For in vitro purification of the linL46W-encoded DNA, the C-terminal portion of GST (starting at an internal MscI-site), and the L46W sequence were amplified using primer pairs GSTC-for/GSTC-rev and LINL46W-for/LINL46W-rev, respectively; products were fused by anneal-extension PCR and subsequently amplified using primers GSTC-for/LINL46W-rev. This GSTC-L46W fragment was then cloned into pGEX-L46W using MscI and AfeI, creating pGEX-linL46W. For in vivo expression of linL46W, the coding sequence was amplified from pGEX-linL46W with primers LINL46W-for2/LINL46W-rev2, and cloned into pMAL using NdeI and PstI, resulting in plasmid pMLIN-L46W. For expression of linF50A in vivo, inverse PCR was performed on the linL46W sequence (present in pJET1.2) using primers LINF50A-for/LINF50A-rev, which were 5′-phosphorylated using T4 phosphonucleotide kinase (Fermentas). The linF50A sequence was then cloned into pMAL using NdeI and PstI, to give pMLIN-F50A.
Protein Cyclization Analysis in Vivo
E. coli BL21(DE3)pLysS cells harboring either pTCYC-L46W or pTPRE-L46W-mut were grown in LB medium (containing 50 μg/ml ampicillin and 34 μg/ml chloramphenicol) to an A595 of 0.6. Isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.8 mm, and cells were grown for an additional 18 h at room temperature. Cells were harvested, lysed in reducing SDS sample buffer, and boiled for 5 min. To monitor expression and cyclization total cell lysates were separated by SDS-PAGE (12.5% NEXT gel (Mandel Scientific) in combination with a conventional 4% Laemmli stacking gel), followed by staining with Coomassie Brilliant Blue G-250 or Western transfer to polyvinylidene difluoride membrane. Western blots were probed with a primary mouse anti-His antibody (Sigma) and a secondary rabbit anti-mouse IgG horseradish peroxidase-linked antibody (GE Healthcare). Chemiluminescence was performed with the ECL Plus Western Detection Kit (GE Healthcare), and signals were visualized on x-ray film.
Protein Expression and Purification
To produce linL46W or linF50A protein, BL21(DE3)pLysS cells harboring the pGEX-linL46W (15) or pGEX-F50A (20) plasmid were grown to mid-log phase (A595 = 0.5–0.6) in LB medium (containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol) at 37 °C and induced with 1 mm IPTG at 30 °C to an A660 of 1.5–2.0. Cells were harvested and protein purified as previously (20) with the additional cell lysis step using progressively smaller bore needles prior to sonication. The eluate of the glutathione-Sepharose 4B (GE LifeSciences) was brought to 100 mm NaCl, 1 mm CaCl2, and incubated with Factor Xa (Hematological Technologies Inc.; 16 μg/mg of protein) at 4 °C for 48 h. Following incubation, the solution was heated (5 min, 95 °C) and subjected to centrifugation (23 °C, 13,000 × g, 10 min) to pellet precipitated GST and Factor Xa. The resultant supernatant was dialyzed into buffer A (10 mm MES (pH 6.0), 2 mm dithiothreitol) and then filtered (0.22 μm) prior to Source15Q (GE Healthcare, 6 ml packed bed volume per 100 ml of culture) anion exchange chromatography using a Waters 650 protein chromatography system. Bound protein was eluted using a linear gradient (1 ml/min flow rate) from 0–1 m NaCl in buffer A. SDS-15% PAGE (160 V, 60 min) and native-20% PAGE (160 V, 90 min) (20) were used to identify fractions containing apo- and/or holo-ACPs.
To obtain cycL46W or cycF50A protein, BL21(DE3)pLysS cells harboring pTCYC-L46W or pTCYC-F50A were induced, harvested, resuspended in buffer A, and lysed as above. After centrifugation (4 °C, 27,000 × g, 20 min), the soluble protein fraction was heated for 10 min at 95 °C, cooled on ice for 10 min, and subjected to centrifugation (4 °C, 14,000 × g, 10 min). The supernatant containing heat-stabile ACP was filtered (0.22 μm) and injected on a Superose 6 10/300 GL size exclusion column (GE Healthcare) equilibrated with buffer A. Fractions found to contain cycL46W were pooled and further purified using SOURCE 15Q anion exchange chromatography as above. Cyclic ACP thus prepared was predominantly in its apo form, and was used for biophysical comparison with the corresponding apo forms of linear ACP. Protein concentrations were determined using the microBCA protein assay (Thermo Scientific).
Steady-state Tryptophan Fluorescence Spectroscopy
All fluorescence measurements were carried out on a Photon Technology International (PTI) QuantaMaster-4CW in a 3 mm semi-micro cuvette (Hellma) at 25 °C. Slit widths were set at 5 nm for both excitation and emission monochromators in all fluorescence experiments. ACPs diluted to 1 μm in 10 mm sodium phosphate (pH 7.0), 0.1 mm EDTA were excited at 296 nm with emission spectra recorded from 300–450 nm in photon-counting mode with an integration time of 1 s both before and after the addition of MgSO4 (10 mm final concentration). All fluorescence intensities were corrected for the Raman band from the solvent.
Circular Dichroism Spectroscopy
Spectra were recorded on a J-810 spectropolarimeter (Jasco) at 25 °C using a 0.1 cm water-jacketed cell. ACPs were diluted to 1 μm in 10 mm sodium phosphate (pH 7.0), 0.1 mm EDTA, and spectra were recorded from 190–260 nm in continuous mode with a speed of 20 nm/min before or immediately after the addition of MgSO4 (10 mm final concentration).
Protein Analysis by Mass Spectrometry
LC-MS/MS was performed using a nanoflow Ultimate system (LC Packings) interfaced to the nanoflow ESI source of a hybrid triple quadrupole linear ion trap (Qtrap) mass spectrometer (Applied Biosystems). Samples were sprayed through a distal coated fused silica needle, 75-μm ID with 15-μm ID tip (New Objectives). Solvent A consisted of 0.1% (v/v) formic acid in water/acetonitrile (98:2), while solvent B consisted of 0.1% (v/v) formic acid in acetonitrile/water (98:2).
For peptide analysis, native PAGE-resolved protein was digested with sequencing-grade trypsin (Promega) for 7.5 h at 37 °C, and the tryptic peptides were resuspended in 30 μl of 5% acetonitrile, 0.5% formic acid. Proteolysis products were injected onto an Onyx monolithic C18 capillary column (0.1 × 150 mm, Phenomenex) and eluted with a 3–30% solvent B gradient over 35 min at a flow rate of 1 μl/min. Spectra were acquired using Information Dependent Acquisition mode.
For intact protein LC-MS (21), purified protein was desalted over POROS R2 media (Applied Biosystems) using the manufacturer's guidelines, and resuspended in 10% acetonitrile, 0.5% formic acid. Protein was subjected to LC-MS on an Onyx monolithic C18 capillary column (0.1 × 50 mm, Phenomenex) at a flow rate of 3 μl/min. The gradient was 10–70% solvent B over 20 min, and spectra were acquired in EMS mode (400–1700 m/z).
Molecular Dynamics Simulations
To create initial in silico structures for linL46W and cycL46W, one of the 20 NMR solution structures of E. coli apo-ACP (PDB ID 2K92 (4)) was selected. The mutation equivalent to V. harveyi L46W was made in 2K92 using VMD 1.8.7 (22), and the structure was loaded into PyMol 1.2r1 (23) to complete the construction. For cycL46W, Gly, Ser, and Ala were added sequentially to the C terminus of L46W adjusting the bond dihedrals to bring the termini as close as possible. MD simulations were performed with NAMD V 2.7b1 (24) using CHARMM22 (25) force fields. Each simulation was carried out in a sphere of explicit water molecules at 310 K using Langevin temperature control and was set up with two periods: steepest descent minimization (1 ns; 500,000 steps) and equilibration (10 ns; 5,000,000 steps). To ensure constructs had adopted an equilibration structure, the average root mean square deviation (for backbone atoms) was calculated over the course of the simulation. Sausage representation figures for linL46W and cycL46W were created using MOLMOL 2.6.0 (26).
Construction of the acpP Deletion Strain CY1861 and in Vivo Complementation
Strain CY1861 carries plasmid pCY765 that was constructed from plasmids pNRD25 (27), pBAD30 (28), and p34S-Sm2 (29). The acpP coding sequence plus flanking sequences was excised from pNRD25 with BamHI and FspI and ligated to pBAD30 cut with the same enzymes. The resulting intermediate plasmid was digested with FspI and ligated to a 2067-bp HincII fragment encoding spectinomycin resistance from p34S-Sm2 to give pCY765 (insertion into the FspI site inactivated the pBAD30 β-lactamase gene). CY1878, the host strain for the plasmid was derived from strain DY329 (30) by two steps of transduction with P1vir lysates. First, the temperature-sensitive λ phage and its closely linked nadA::Tn10 element were removed from the chromosome by transduction with a P1 stock grown on a wild-type strain. Transductants that grew in the absence of biotin and nicotinic acid were selected and screened for growth at 42 °C. One of these transductants was then transformed to spectinomycin resistance with pCY765. A transformant was then transduced to chloramphenicol resistance in the presence of arabinose with a P1 phage lysate grown on the ΔacpP fabF::cml strain, NRD62 (27). Transductants resistant to chloramphenicol and spectinomycin were screened for arabinose-dependent growth to give strain CY1861.
For in vivo complementation assays, single colonies of CY1861 transformed with indicated plasmids were resuspended in 50 μl of LB medium supplemented with 50 μg/ml spectinomycin and 100 μg/ml ampicillin (LB-SA). Five microliters of this resuspension were used to inoculate 245 μl of LB-SA additionally supplemented with either 0.2% arabinose or 0.2% glucose/0.8 mm IPTG in a 96-well plate. The optical density (A595) was recorded (t = 0), and the plate incubated at 37 °C and 400 rpm with measurements of A595 taken every 20–30 min. Absorbance values were subtracted from the t = 0 readings of individual wells, and multiplied by 4 to normalize values. IPTG had no effect on growth in 0.2% arabinose, indicating that none of the constructs are toxic.
RESULTS
Construction of Cyclic ACP
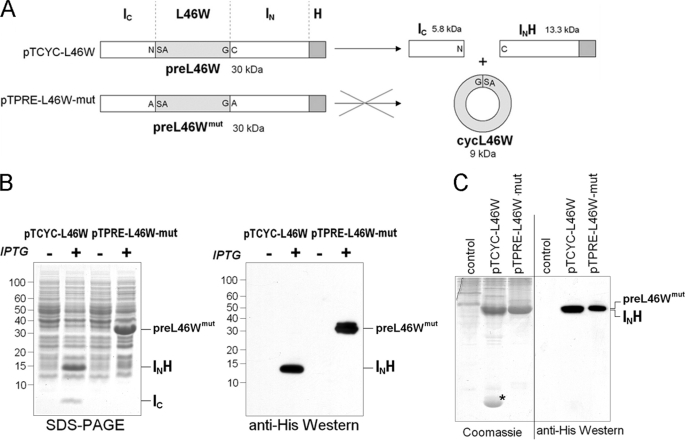
The flexibility and dynamic properties of ACP are thought to be essential for its function (6). As the N and C termini of ACP are both mobile (16) and in relatively close proximity, we used an intein-mediated approach to constrain this flexibility by constructing a cyclic version of V. harveyi ACP. The N-terminal splicing domain of the Ssp GyrB intein (residues 1–111, followed by a hepta-histidine tag; INH) (31) was fused to the C terminus of ACP, while the C-terminal splicing domain (residues 393–435; IC) was fused to the ACP N terminus (plasmid pTCYC-L46W). The L46W mutant of ACP was chosen because the tryptophan at position 46 (replacing leucine in the wild-type V. harveyi ACP) has been shown to be sensitive to the folding state of the protein, without affecting its secondary structure or substrate properties with a variety of ACP-dependent enzymes (15). After trans-splicing, the cyclic ACP L46W mutant (cycL46W) would therefore have its N and C termini connected by a three amino acid linker (Gly-Ser-Ala) (Fig. 1A). Furthermore, a control construct was made (pTPRE-L46W-mut), in which the first and last intein residues were mutated to Ala to inhibit intein-mediated trans-splicing and possible cleavage reactions.
FIGURE 1.
A, schematic representation of cyclization constructs. Plasmid pTCYC-L46W encodes a 30-kDa protein (preL46W), which after in vivo cyclization yields the split-intein products IC and IN, as well as the cyclic V. harveyi ACP L46W protein (cycL46W). Amino acid residues at both the C- and N-terminal splice junction are indicated in single letter code. The ligation site of the ACP C and N termini in cycL46W is the peptide bond between Gly and Ser. Plasmid pTPRE-L46W-mut serves as an expression control, as it encodes for a 30-kDa protein (preL46Wmut) carrying mutations in IC (N → A) and IN (C → A), which prevent intein-mediated cyclization. B, E. coli BL21(DE3)pLysS cells harboring plasmids pTCYC-L46W or pTPRE-L46W-mut were induced with IPTG (+) or grown in the absence of IPTG (−). Total cell lysates were analyzed by SDS-PAGE and Coomassie Blue staining (left) and Western blot analysis using anti-His antibodies (right). Relevant protein species are indicated, and molecular masses are given in kDa. C, native PAGE analysis of soluble protein extracts derived from induced BL21(DE3)pLysS cells harboring indicated plasmids. Gels were either stained with Coomassie Blue or subjected to Western blotting with anti-His antibodies. The control lanes show lysate from BL21(DE3)pLysS cells not overexpressing ACP-related proteins.
When these constructs were expressed in E. coli BL21(DE3)pLysS, the control construct showed an induced protein species at ∼31 kDa on SDS-PAGE that reacted with anti-His tag antibodies (Fig. 1B) and thus corresponds to the splicing- and cleavage-deficient split-intein L46W construct (preL46Wmut) protein. In contrast, the construct competent for cyclization revealed two induced protein species, one migrating at ∼14 kDa and the other below 10 kDa (Fig. 1B). The 14-kDa protein reacted with anti-His antibodies and thus likely corresponds to the INH split-intein part (13.3 kDa). The smaller protein was identified as the IC split-intein part (5.1 kDa) based on in-gel trypsin digestion and MS/MS analysis. The detection of both split-intein parts as separate protein species indicated that the cyclization construct underwent a complete reaction in vivo as judged from the absence of any unspliced protein precursor (preL46W). However, an induced band corresponding to ACP was not apparent.
ACP is a highly acidic protein that resolves well in native PAGE (32) and hence we used this method to further examine soluble protein extracts from E. coli expressing the cyclization and control constructs (Fig. 1C). The splicing control construct (pTPRE-L46W-mut) lysate revealed a heavily stained protein band that reacted with anti-His antibodies in Western blotting, and thus represents the splicing-deficient preL46Wmut protein. The cyclization construct (pTCYC-L46W) lysate showed a protein band of similar intensity and mobility, which likely corresponds to the His-tagged IN split-intein fragment (INH) based on Western blotting and co-migration with INH purified by Ni-NTA affinity chromatography. Interestingly, and in contrast to the pTPRE-L46W-mut lysate, the pTCYC-L46W lysate showed the presence of an additional protein species near the bottom of the native gel, which could represent the cyclized L46W (cycL46W) protein.
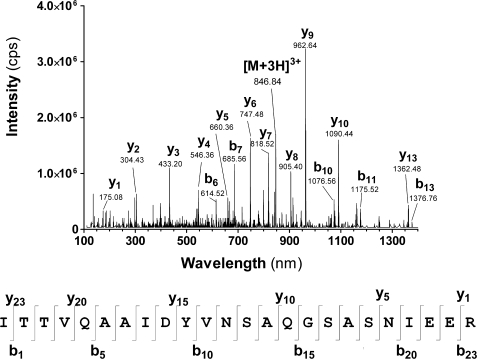
To verify this, the protein suspected to be cycL46W was digested with trypsin (in-gel) and analyzed by LC-MS/MS. An [M+3H]3+ peptide ion with an m/z value of 846.84 was identified, for which the MS/MS spectrum was in excellent agreement with the amino acid sequence ITTVQAAIDYVNSAQGSASNIEER (Fig. 2). In this peptide, residues 1–15 (ITTVQAAIDYVNSAQ) correspond to the C-terminal residues of V. harveyi ACP, whereas residues 19–24 (SNIEER) represent the N terminus. The ACP C and N termini are linked by the residues GSA. This peptide thus has the expected amino acid sequence of the cycL46W protein trypsin fragment extending across the region of cyclization, with the peptide bond between Gly and Ser being the exact ligation point between the N and C termini. These results show that the cycL46W protein was successfully created in vivo by split-intein-mediated trans-splicing.
FIGURE 2.
Tandem mass spectrometry (MS/MS) analysis of a peptide obtained after trypsin digestion of the protein marked with an asterisk in Fig. 1C. The MS/MS spectrum (top) shows the [M+3H]3+ ion, as well as selected y and b ions derived through its fragmentation. The peptide corresponds to the sequence shown below the MS/MS spectrum, with selected y and b ions indicated (bottom).
Biophysical Characterization of cycL46W
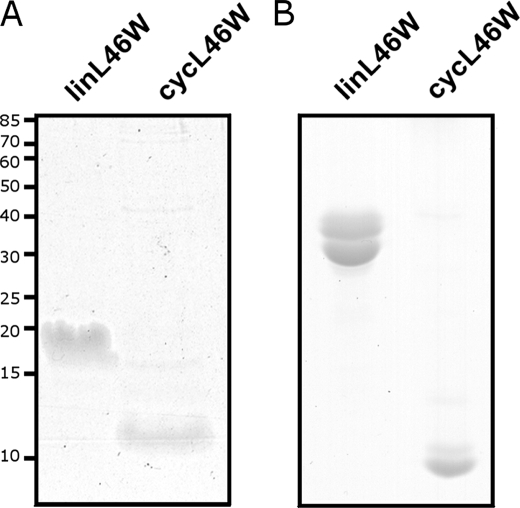
As the cycL46W protein was not clearly evident when lysates were fractionated by SDS-PAGE, we examined the migration behavior of purified cycL46W in comparison to the corresponding linear ACP bearing the exact same amino acid sequence (linL46W). CycL46W migrated as a diffuse band with an apparent molecular mass of ∼12 kDa (Fig. 3A). The fact that cycL46W stains poorly with Coomassie Blue and its migration overlaps with that of the INH split-intein part (13.3 kDa) may explain why the protein was not clearly visible upon separation of the whole cell lysates by SDS-PAGE (Fig. 1B). Notably, the cycL46W protein migrated faster than the linL46W protein (∼20 kDa, Fig. 3A), which like many ACPs exhibits anomalous SDS-PAGE mobility (1), indicating that cycL46W has a more compact structure. This conclusion is supported by the observation that cycL46W also migrated faster than linL46W in a conformationally sensitive native PAGE system (Fig. 3B).
FIGURE 3.
Migration behavior of purified linL46W and cycL46W in SDS-PAGE (A) and native PAGE (B). Molecular masses in A are given in kDa.
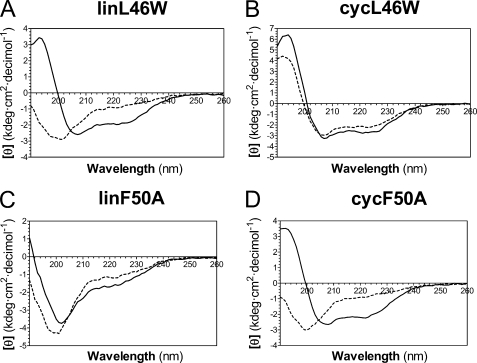
To further examine the effect of cyclization on ACP structure in solution, we used a variety of biophysical methods to analyze the conformation of both cycL46W and linL46W in response to Mg2+ ions, which are known to induce a folded helical conformation upon binding to V. harveyi ACP at physiological pH (10). The apo forms of linear ACPs were used in these studies, as cyclic ACP is predominantly expressed in this form (see below). As expected, circular dichroism (CD) spectroscopy revealed a substantial increase in α-helical content (indicated by increased magnitude of [θ]220) upon addition of Mg2+ to linL46W (Fig. 4A). In contrast, even in the absence of divalent cations, cycL46W exhibited a CD spectrum similar to that of folded ACP, with little further conformational change upon addition of Mg2+ (Fig. 4B).
FIGURE 4.
Circular dichroism analysis of linear and cyclic derivatives of L46W and F50A. CD spectra of the indicated proteins were measured in the absence (dashed line) or presence (solid line) of 10 mm Mg2+ as described in the text.
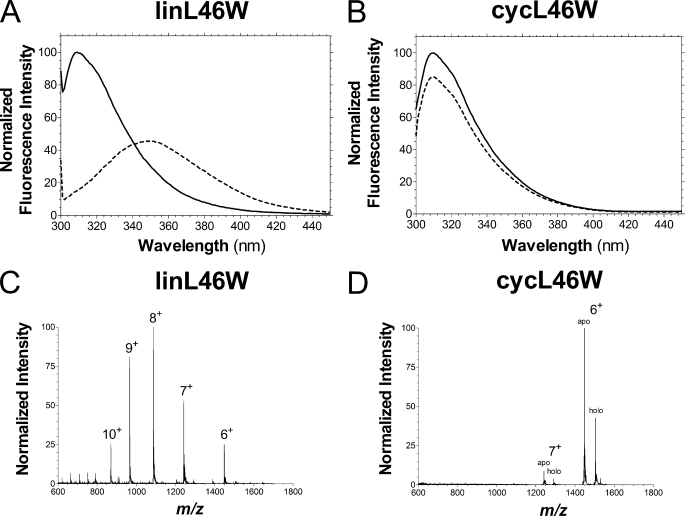
We have recently shown that the L46W mutant of V. harveyi ACP undergoes a pronounced Mg2+-dependent fluorescence blue shift of emission maximum from ∼355 nm to <320 nm, reflecting the movement of Trp-46 in helix II into the hydrophobic interior of the folded protein (15). This was also observed for the linL46W control protein in its apo form (Fig. 5A). In striking contrast, cycL46W had a peak emission at ∼310 nm both in the absence and presence of Mg2+ ions (Fig. 5B), indicating a folded conformation independent of stabilization by divalent cations.
FIGURE 5.
Biophysical characterization of cycL46W and linL46W. Tryptophan fluorescence spectra (λex = 296 nm) of linL46W (A) and cycL46W (B) were obtained in the absence (dashed line) and presence (solid line) of 10 mm Mg2+. Positive mode nano-ionspray MS spectra of intact linL46W (C) and a mixture of apo- and holo-cycL46W (D) proteins were recorded as described in the text. The charge of relevant peaks are indicated.
Finally, electrospray ionization mass spectrometry was utilized to analyze the mass and charge state distribution (CSD) of intact cycL46W and linL46W. Reconstruction of LC-MS data indicated the presence of both apo (8,667 Da) and holo (9,007 Da) forms of cycL46W, i.e. 18 Da less than the corresponding values for linL46W due to loss of H2O during the cyclization reaction. Apo-cycL46W eluted slightly later in the acetonitrile gradient and was typically 5–10 fold more abundant than the holo derivative, although these were not well resolved by either anion-exchange chromatography or native PAGE (data not shown). The CSD of an intact protein has been shown to be sensitive to its solution phase conformation, with unfolded states typically characterized by broad distributions of highly charged ions (21, 33). Apo-linL46W displayed a broad CSD of ions from 6+ to 9+ (maximum at 8+) in positive ESI mode (Fig. 5C), which is similar to that observed for wild-type V. harveyi ACP (34). CycL46W, on the other hand, produced a sharp, narrow CSD of predominantly 6+ and 7+ ions (Fig. 5D), suggesting a more compact structure under these conditions. Both apo and holo cycL46W exhibited similar CSD profiles, indicating that this property is not dependent on the modification state of ACP. Combined, our biophysical analyses indicate that cyclization stabilizes a folded conformation of ACP.
Molecular Dynamics Simulations of Linear and Cyclic ACP
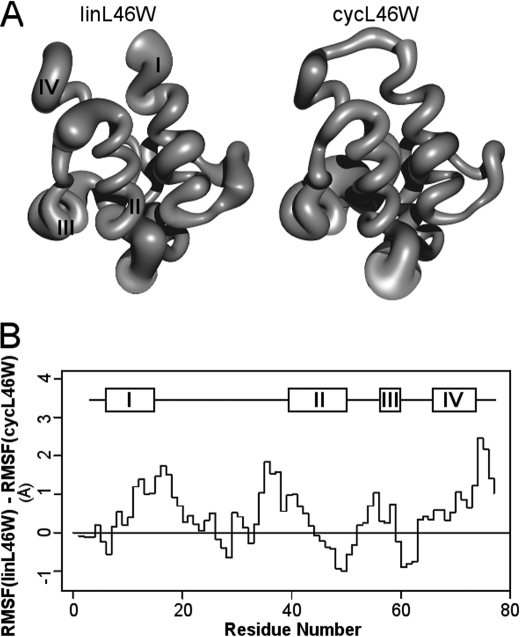
MD simulations of linL46W and cycL46W were performed over a 10-ns interval based on a starting structure obtained from the recent NMR analysis of E. coli apo-ACP (4). After a short initial stabilization period, no substantial polypeptide chain rearrangements were noted in either the linear or cyclic proteins, although the distance between the N and C termini Cα atoms in linL46W increased from 11.3 Å in the starting structure to 15.0 Å at the end of the simulation. Thus, cyclization does not appear to interfere with the four-helix bundle fold of ACP (Fig. 6A), consistent with CD and fluorescence data. As shown previously (16), regions of greatest flexibility in linear ACP included both ends of the long loop between helices I and II, the helix II-III region, and the C terminus. Indeed, these segments were considerably more flexible in linL46W versus cycL46W, whereas the latter exhibited greater fluctuations at the end of helix II and in the helix III-IV region (Fig. 6B).
FIGURE 6.
Molecular dynamics simulations of linear and cyclic L46W ACP. Panel A shows sausage representations of linL46W (left) and cycL46W (right), where the thickness of the polypeptide chain represents the RMSF of Cα atoms during the final 9 ns of simulation. A scaling factor of ½ was used, and helices I-IV are indicated. Panel B, difference plot of Cα RMSF values of linL46W versus cycL46W as a function of position in the chain. The positions of helical segments of the starting E. coli structure (4) is shown above the plot.
In Vivo Complementation Assays
To explore the potential of cycL46W to serve as a substrate for enzyme partners, and thus as a mediator in cellular processes, we undertook in vivo complementation growth assays using an E. coli acpP-null strain (CY1861). In CY1861, the chromosomal acpP gene is replaced with a chloramphenicol resistance cassette, and growth is rescued by a plasmid borne synthetic acpP gene under control of the araBAD promoter. Plasmids derived from pMAL expressing cycL46W, linL46W, or preL46Wmut from the ptac promoter were transformed into CY1861 (the two plasmids have compatible origins), and the ability to grow in medium containing arabinose was compared with growth in medium containing glucose and IPTG, which leads to shutdown of expression of the acpP gene from the araBAD promoter and induction of protein expression from the ptac promoter, respectively. The plasmid pMAL served as a negative control because it only expresses a protein unrelated to ACP (MBP).
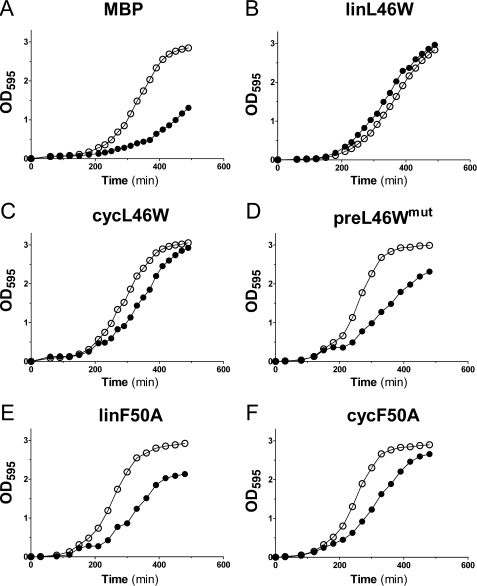
As expected, the negative control (expression of MBP) led to severely delayed growth in the presence of glucose and IPTG, compared with growth in the presence of arabinose (Fig. 7A). Consistent with the finding that V. harveyi wild-type ACP is able to complement growth of an E. coli strain with a temperature-sensitive ACP (NRD224) at the restrictive temperature (35), expression of the linL46W protein led to strong complementation of the CY1861 strain in the presence of glucose and IPTG. Strikingly, expression of the cycL46W protein under similar conditions also led to complementation (Fig. 7C), albeit with a slight delay when compared with linL46W. Production of the splicing-deficient preL46Wmut protein also allowed some growth in the presence of glucose and IPTG (Fig. 7D), but not to the extent observed for the linL46W and cycL46W proteins.
FIGURE 7.
In vivo complementation assay. E. coli CY1861 were transformed with pMAL-derived plasmids encoding the indicated proteins under control of the IPTG-inducible ptac promoter. Cells were grown in medium containing arabinose (open circles) or IPTG and glucose (closed circles), and the absorbance at 595 nm was recorded over 6 h. Growth curves representative of three individual experiments are shown.
Effects of Cyclization of an ACP-folding Mutant
We also investigated complementation by a V. harveyi ACP mutant (F50A) that is incapable of proper Mg2+-induced folding and is also a poor substrate for the ATP-dependent acyl-ACP synthetase (AasS) from this species (20). Production of linear F50A ACP (linF50A) in CY1861 led to weak complementation of growth (Fig. 7E). However, upon induction of the corresponding cyclic F50A protein (cycF50A), growth of CY1861 cells in the presence of glucose, and IPTG was considerably improved (Fig. 7F), to an extent comparable to that observed for cycL46W (Fig. 7C).
To relate this stronger complementation effect of cycF50A to its biophysical properties, we subjected both the linear and cyclic ACP-F50A mutants to CD spectroscopy. Consistent with previous observations (20), addition of Mg2+ was unable to restore the helical content of the linear F50A mutant protein as indicated by CD (Fig. 4C). Moreover, cyclization did not by itself restore the ability of the F50A ACP mutant to adopt a folded conformation in the absence of Mg2+, in contrast to the corresponding L46W protein (Fig. 4D). However, unlike linear F50A, folding of the cyclic F50A derivative was restored in the presence of Mg2+. These results indicate that constraining the N and C termini can work in concert with divalent cation binding to stabilize the folded conformation of this ACP mutant.
DISCUSSION
ACP must exhibit considerable conformational flexibility in interacting with over 30 bacterial enzymes (36) as it carries out its essential role in fatty acid and complex lipid synthesis. It is currently believed that, upon initial interaction with a partner enzyme, ACP must at least partially unfold for the reversible transfer of acyl groups from its hydrophobic core to the enzyme active site (6). However, despite the prediction of highly mobile N and C termini of E. coli ACP (16), the present study clearly shows that ACP function is not severely compromised by constraining its N and C termini close together in a cyclized derivative. Thus, enzyme interaction and acyl transfer do not appear to require complete unfolding or the presence of free amino acids at either end of ACP, at least for enzyme partners that are essential for growth. Indeed, ACP is able to accommodate foreign sequences at either terminus, as demonstrated by the limited growth complementation by the splicing deficient preL46Wmut construct. This is consistent with the fact that bacterial ACPs with C-terminal fusion tags such as TAP/SPA (36) and Flag (37) are also functional in vivo. In this regard, it is worth noting that mammalian ACP can function despite being an internal domain within the large type I fatty acid synthase polypeptide chain found in the cytosol (38).
The intramolecular in vivo cyclization of ACP using Ssp GyrB split inteins was very efficient, as evident by the absence of unspliced precursor (preL46W) protein on SDS-PAGE, and was highly specific toward cyclization, because protein species resulting from inter-molecular trans-splicing (i.e. polymeric structures) were absent from total cell lysates. Furthermore, the efficiency of cyclization was likely the result of the high affinity of the split-intein parts to each other, rather than their close proximity due to intracellular folding of ACP, since cyclization was similarly efficient with the V. harveyi F50A mutant, which is unable to adopt a folded conformation (20). Growth restoration by cyclized ACP is unlikely due to trace levels of unspliced or linearized preL46W protein because expression of both preL46Wmut and linF50W resulted in weaker growth restoration when compared with their cyclic ACP counterparts. Moreover, a time course expression experiment revealed that cyclization occurs rapidly after IPTG induction (IC and INH fragments are detectable after <20 min of IPTG addition to the growth medium), and preL46W is never detected at any time. In any case, the ability of L46W derivatives to complement growth in vivo validates this fluorescent ACP analogue as a physiologically relevant probe of ACP structure and function.
Cyclization of V. harveyi-derived ACP clearly stabilizes a folded helical conformation, as suggested by MD simulations and demonstrated by biophysical methods such as native PAGE, intrinsic tryptophan fluorescence, and circular dichroism. Indeed, the latter two techniques revealed that the cycL46W protein is folded under conditions (i.e. the absence of Mg2+ ions) in which the linear control protein is not. This stabilization is not unexpected, as constraint of the free ACP termini should decrease the conformational entropy of the unfolded relative to the folded state. However, of particular interest here is the difference in charge state distribution (CSD) of cycL46W and linL46W. We recently reported that V. harveyi ACP exhibits a similar broad CSD regardless of its conformation in solution, as manipulated by charge neutralization or fatty acylation (34). We further speculated that ACP may unfold rapidly upon entering the gas phase during ionization, unlike many other proteins (e.g. cytochrome c), which are trapped in folded and unfolded conformers that can be sampled by this technique (21). The narrow CSD for cycL46W observed in the current study suggests that, unlike other factors that stabilize ACP, cyclization may prevent the protein from unfolding on the timescale of the electrospray ionization process. CSD analysis has recently been used to show that cyclization also stabilizes E. coli DnaB against unfolding under acidic conditions (39).
Contrasting the effects of cyclization on growth complementation by L46W versus F50A analogues provides insight into the structural requirements for ACP function. While cyclization had a small detrimental effect on the ability of L46W to restore growth of ACP-deficient cells, it markedly improved the functional properties of the F50A mutant. Based on conformational analysis, we might expect that under physiological conditions of millimolar Mg2+ levels, the cyclic (but not linear) F50A protein would be able to fold properly, thus correlating with the pattern of complementation in vivo. This indicates that the ability to adopt a folded structure is essential for ACP function, a feature that is perhaps linked to a requirement for helix II for recognition by ACP-dependent enzymes.
Cyclization of L46W ACP does appear to moderately hinder its ability to complement E. coli growth in vivo. Whether this is due to the effect of decreased conformational flexibility or to altered interactions with one or more rate-limiting enzymes is currently not known. In general, we anticipate that cyclization would have distinct and different effects on the kinetic properties of individual ACP-dependent enzymes, some of which are essential and might be rate-limiting under various conditions. Interestingly, MD simulation suggests differences in flexibility between cyclic and linear ACP in two key functional regions: along helix II (important in enzyme recognition) (6) and in the helix II-III region (implicated in acyl chain binding and release) (4). To better define the role of conformational flexibility in ACP function, it will be worthwhile to examine the effects of cyclization on the kinetic properties of particular classes of ACP-dependent enzymes, such as holo-ACP synthases, acyltransferases, and acyl-ACP synthetases.
Another question that has not been addressed in this study relates to the location of the fatty acyl chain attached to cyclic ACP. Recent NMR (4) and steered molecular dynamics (40) analyses have suggested that acyl groups may enter and exit the hydrophobic core laterally through a separation of helices II and III, perhaps reflecting the dynamic nature of this region. Molecular dynamics simulations of acyl-ACP also indicate that attached acyl groups can enter the protein interior spontaneously (16), without the necessity for major rearrangements at the N and C termini that would be compromised in a cyclic ACP. Alternatively, cyclization of ACP could prevent accommodation of acyl chains within the hydrophobic core entirely, such that they would be continually exposed to solvent and to partner enzymes alike. This would be analogous to the internal ACP domain of rat type I fatty acid synthase, which does not appear to sequester its acyl chain (41). Work is currently underway to resolve this issue.
Supplementary Material
Acknowledgments
We thank David Chan (University of Calgary) for helpful advice and the Dalhousie University Faculty of Medicine Proteomics Core Facility for assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI15650 (to J. E. C.). This work was also supported by the Natural Sciences and Engineering Research Council of Canada (to D. M. B.), Canadian Institutes of Health Research (to X. Q. L.), and by an IWK Health Centre Studentship (to P. W. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2.
X. Q. Liu, unpublished data.
G. Volkmann, unpublished data.
- ACP
- acyl carrier protein
- CD
- circular dichroism
- CSD
- charge state distribution
- ESI
- electrospray ionization
- GST
- glutathione S-transferase
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- MBP
- maltose-binding protein
- MD
- molecular dynamics
- MES
- 2-[N-morpholino]ethanesulfonic acid
- RMSF
- root mean square fluctuations.
REFERENCES
- 1.Byers D. M., Gong H. (2007) Biochem. Cell Biol. 85, 649–662 [DOI] [PubMed] [Google Scholar]
- 2.Kim Y., Prestegard J. H. (1990) Proteins 8, 377–385 [DOI] [PubMed] [Google Scholar]
- 3.Zornetzer G. A., Fox B. G., Markley J. L. (2006) Biochemistry 45, 5217–5227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu B. N., Zhang Y. M., Rock C. O., Zheng J. J. (2009) Protein Sci. 18, 240–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roujeinikova A., Baldock C., Simon W. J., Gilroy J., Baker P. J., Stuitje A. R., Rice D. W., Slabas A. R., Rafferty J. B. (2002) Structure 10, 825–835 [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y. M., Rao M. S., Heath R. J., Price A. C., Olson A. J., Rock C. O., White S. W. (2001) J. Biol. Chem. 276, 8231–8238 [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y. M., Marrakchi H., White S. W., Rock C. O. (2003) J. Lipid Res. 44, 1–10 [DOI] [PubMed] [Google Scholar]
- 8.Rafi S., Novichenok P., Kolappan S., Zhang X., Stratton C. F., Rawat R., Kisker C., Simmerling C., Tonge P. J. (2006) J. Biol. Chem. 281, 39285–39293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parris K. D., Lin L., Tam A., Mathew R., Hixon J., Stahl M., Fritz C. C., Seehra J., Somers W. S. (2000) Structure 8, 883–895 [DOI] [PubMed] [Google Scholar]
- 10.Gong H., Murphy A., McMaster C. R., Byers D. M. (2007) J. Biol. Chem. 282, 4494–4503 [DOI] [PubMed] [Google Scholar]
- 11.Gong H., Byers D. M. (2003) Biochem. Biophys. Res. Comm. 302, 35–40 [DOI] [PubMed] [Google Scholar]
- 12.Worsham L. M., Earls L., Jolly C., Langston K. G., Trent M. S., Ernst-Fonberg M. L. (2003) Biochemistry 42, 167–176 [DOI] [PubMed] [Google Scholar]
- 13.Keating M. M., Gong H., Byers D. M. (2002) Biochim. Biophys. Acta 1601, 208–214 [DOI] [PubMed] [Google Scholar]
- 14.Roujeinikova A., Simon W. J., Gilroy J., Rice D. W., Rafferty J. B., Slabas A. R. (2007) J. Mol. Biol. 365, 135–145 [DOI] [PubMed] [Google Scholar]
- 15.Gong H., Murphy P. W., Langille G. M., Minielly S. J., Murphy A., McMaster C. R., Byers D. M. (2008) Biochim. Biophys. Acta 1784, 1835–1843 [DOI] [PubMed] [Google Scholar]
- 16.Chan D. I., Stockner T., Tieleman D. P., Vogel H. J. (2008) J. Biol. Chem. 283, 33620–33629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Andrec M., Hill R. B., Prestegard J. H. (1995) Protein Sci. 4, 983–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y., Kovrigin E. L., Eletr Z. (2006) Biochem. Biophys. Res. Commun. 341, 776–783 [DOI] [PubMed] [Google Scholar]
- 19.Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flaman A. S., Chen J. M., Van Iderstine S. C., Byers D. M. (2001) J. Biol. Chem. 276, 35934–35939 [DOI] [PubMed] [Google Scholar]
- 21.Konermann L., Douglas D. J. (1997) Biochemistry 36, 12296–12302 [DOI] [PubMed] [Google Scholar]
- 22.Humphrey W., Dalke A., Schulten K. (1996) J. Mol. Graphics 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 23.De Lano W. (2002) The PyMol Molecular Graphics System, San Carlos, CA [Google Scholar]
- 24.Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mackerell A. D., Jr., Bashford D., Bellot M., Dunbrack R. L., Jr., Evanseck J. D., Field M. J., Fischer S., Gao J., Gao H., Ha S., Joseph-McCarthy D., Kuchnir L., Kuczera K., Lau F. T. K., Mattos C., Michnick S., Ngo T., Nguyen D. T., Prodhom B., Reiher W. E., III, Roux B., Schlenkrich M., Smith J. C., Stote R., Straub J., Watanabe M., Wiorkeiwicz-Kuczera J., Yin D., Karplus M. (1998) J. Phys. Chem. B. 102, 3586–3616 [DOI] [PubMed] [Google Scholar]
- 26.Koradi R., Billeter M., Wüthrich K. (1996) J. Mol. Graph. 14, 51–55 [DOI] [PubMed] [Google Scholar]
- 27.De Lay N. R., Cronan J. E. (2006) J. Bacteriol. 188, 287–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dennis J. J., Zylstra G. J. (1998) BioTechniques 25, 772–774, 776 [DOI] [PubMed] [Google Scholar]
- 30.Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., Court D. L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perler F. B. (2002) Nucleic Acids Res. 30, 383–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rock C. O., Cronan J. E., Jr., Armitage I. M. (1981) J. Biol. Chem. 256, 2669–2674 [PubMed] [Google Scholar]
- 33.Katta V., Chait B. T. (1991) Rapid Commun. Mass Spectrom. 5, 214–217 [DOI] [PubMed] [Google Scholar]
- 34.Murphy P. W., Rowland E. E., Byers D. M. (2007) J. Am. Soc. Mass Spectrom. 18, 1525–1532 [DOI] [PubMed] [Google Scholar]
- 35.De Lay N. R., Cronan J. E. (2007) J. Biol. Chem. 282, 20319–20328 [DOI] [PubMed] [Google Scholar]
- 36.Butland G., Peregrín-Alvarez J. M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Davey M., Parkinson J., Greenblatt J., Emili A. (2005) Nature 433, 531–537 [DOI] [PubMed] [Google Scholar]
- 37.Battesti A., Bouveret E. (2009) J. Bacteriol. 191, 616–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leibundgut M., Maier T., Jenni S., Ban N. (2008) Curr. Opin. Struct. Biol. 18, 714–725 [DOI] [PubMed] [Google Scholar]
- 39.Watt S. J., Sheil M. M., Beck J. L., Prosselkov P., Otting G., Dixon N. E. (2007) J. Am. Soc. Mass Spectrom. 18, 1605–1611 [DOI] [PubMed] [Google Scholar]
- 40.Colizzi F., Recanatini M., Cavalli A. (2008) J. Chem. Inf. Model. 48, 2289–2293 [DOI] [PubMed] [Google Scholar]
- 41.Ploskoń E., Arthur C. J., Evans S. E., Williams C., Crosby J., Simpson T. J., Crump M. P. (2008) J. Biol. Chem. 283, 518–528 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.