Abstract
FKBP38 is a member of the family of FK506-binding proteins that acts as an inhibitor of the mammalian target of rapamycin (mTOR). The inhibitory action of FKBP38 is antagonized by Rheb, an oncogenic small GTPase, which interacts with FKBP38 and prevents its association with mTOR. In addition to the role in mTOR regulation, FKBP38 is also involved in binding and recruiting Bcl-2 and Bcl-XL, two anti-apoptotic proteins, to mitochondria. In this study, we investigated the possibility that Rheb controls apoptosis by regulating the interaction of FKBP38 with Bcl-2 and Bcl-XL. We demonstrate in vitro that the interaction of FKBP38 with Bcl-2 is regulated by Rheb in a GTP-dependent manner. In cultured cells, the interaction is controlled by Rheb in response to changes in amino acid and growth factor conditions. Importantly, we found that the Rheb-dependent release of Bcl-XL from FKBP38 facilitates the association of this anti-apoptotic protein with the pro-apoptotic protein Bak. Consequently, when Rheb activity increases, cells become more resistant to apoptotic inducers. Our findings reveal a novel mechanism through which growth factors and amino acids control apoptosis.
Keywords: Amino Acid, Apoptosis, G Proteins, Oncogene, Tuberous Sclerosis (TSC), Bcl-2, Bcl-XL, FKBP38, Rheb
Introduction
Tuberous sclerosis complex (TSC)3 is an autosomal dominant disorder manifested by the occurrence of benign tumors in multiple organs (1). The disease condition is caused by inactivating mutations in either the TSC1 or TSC2 tumor suppressor gene (2). The TSC1 and TSC2 gene products form a heterodimeric complex that functions as a GTPase-activating protein against a Ras-like small GTPase, Rheb (3, 4). When the GTPase-activating protein activity of the complex is compromised by mutations in either TSC1 or TSC2, Rheb becomes hyperactive, which is believed to contribute to tumorigenesis in TSC (5, 6).
Rheb is an activator of the mammalian target of rapamycin (mTOR), a central regulator of cell growth that controls a wide spectrum of cellular processes by integrating nutrient and growth factor signals (7, 8). mTOR elicits its rapamycin-sensitive functions in a multiprotein complex named mTORC1 (mTOR complex 1) (9–11). The action of Rheb on mTORC1 is mediated, at least in part, by FKBP38, a member of the FK506-binding protein family that functions as an endogenous inhibitor that binds and down-regulates mTORC1 activity under amino acid starvation or growth factor deprivation conditions (12, 13). In its GTP-bound form, Rheb interacts with FKBP38 and prevents its association with mTORC1, which frees mTORC1 for activation (12). Although this FKBP38-dependent mechanism has been suggested to be responsible for Rheb-mediated mTORC1 activation, several studies have shown that Rheb may control mTORC1 through direct interaction (14–16).
In addition to mTORC1, FKBP38 also interacts with Bcl-2 and Bcl-XL, two anti-apoptotic proteins of the Bcl-2 family. Bcl-2 and Bcl-XL are membrane proteins that reside mainly on mitochondria (17–19). The two proteins exert their anti-apoptotic function by antagonizing pro-apoptotic proteins, such as Bak and Bax, and preventing mitochondrial membrane permeabilization (20). FKBP38 has been shown to bind directly with Bcl-2 and Bcl-XL and is involved in targeting these two proteins to mitochondria, where they perform their anti-apoptotic function (21–23). In light of a recent finding that FKBP38 is an effector of Rheb (24), a possibility arises that the function and mitochondrial targeting of Bcl-2 and Bcl-XL are regulated by Rheb through FKBP38. In this study, we investigate the potential role of Rheb in the regulation of apoptosis.
MATERIALS AND METHODS
Reagents and Plasmids
Anti-hemagglutinin antibody (12CA5), anti-Myc antibody (9E10), and anti-Myc antibody-conjugated Protein A beads were purchased from Santa Cruz Biotechnology; anti-FLAG M2 antibody from Sigma; anti-Bcl-2 antibody from Dako; anti-Bcl-XL (54H6), anti-Bak, anti-Rheb, anti-4E-BP1, anti-phospho-4E-BP1 (Thr-37/46), anti-S6 kinase, and anti-phospho-S6 kinase (Thr-389) antibodies from Cell Signaling; anti-His antibody from BD Biosciences; and anti-glutathione S-transferase (anti-GST) antibody from GenScript. Anti-FKBP38 antibody was generated in rabbits using a polypeptide corresponding to the first 207 amino acids of human FKBP38 as antigen. Protease inhibitor mixtures were purchased from Roche Applied Science. The Promega CellTiter-Glo® luminescent cell viability assay kit was used to assay cell viability. Expression vectors for Myc-FKBP38 and FLAG-Rheb were described (24). The hemagglutinin-Bcl-XL plasmid was constructed by cloning a PCR-amplified Bcl-XL into pcDNA3.1-HA (Invitrogen). The Bcl-2 expression vector was kindly provided by Dr. Lin Zhang of the University of Pittsburgh. FKBP38- and Rheb-specific small interfering RNAs (siRNAs), as well as the scrambled control siRNAs, were purchased from Santa Cruz Biotechnology and used as described (12). The Rag GTPase expression vectors (FLAG-pLJM1) and the empty control vector (pLKO.1) were obtained from Addgene. The Rag mutants used in this study include RagB(Q99L), designated as RagBGTP; RagB(T54L), as RagBGDP; RagD(Q121), as RagDGTP; and RagD(S77L), as RagDGDP, which were described previously (25).
Cell Culture and DNA Transfection
HeLa cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (Atlanta Biological). To introduce plasmid DNA or siRNAs, cells were transfected using LipofectamineTM 2000 following the manufacturer's instructions (Invitrogen). For ectopic gene expression, transfected cells were incubated for 24 h before being subjected to any treatments. For the effect of siRNA, transfected cells were incubated for 72 h before any treatments. For amino acid starvation and repletion, cells were grown to ∼90% confluence on culture dishes before being switched to Dulbecco's modified Eagle's medium free of amino acids (Sigma). Upon incubation for 1 h, 50× amino acid stock mixture (Sigma) was added to the medium to a final concentration of 1×. Cells were harvested and lysed after a 30-min incubation. For serum deprivation and readdition, cells were grown to ∼70% confluence under normal conditions and shifted to Dulbecco's modified Eagle's medium containing 0.1% serum for 20 h. Serum was then added back to the medium to a final concentration of 20%. Cells were harvested and lysed 30 min after the addition of serum. For analysis of the interaction of Bcl-XL and Bak on mitochondria, cells were shifted to Dulbecco's modified Eagle's medium containing 0.1% serum for 20 h and then to the same medium but free of amino acids for 3 h. Upon refeeding with amino acids for 30 min, cells were harvested and lysed.
Co-immunoprecipitation of FKBP38 with Bcl-2 and Bcl-XL
HeLa Cells were transfected with plasmids expressing different genes. Cells were washed with cold phosphate-buffered saline on ice and lysed in buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture. Lysates (1 mg) were incubated with 15 μl of anti-Myc antibody (9E10)-linked Protein A-agarose beads for 3 h at 4 °C with agitation. Beads were washed four times with lysis buffer, resuspended in 60 μl of 2× SDS sample buffer, and boiled for 5 min. Samples were subjected to SDS-PAGE. Western blotting was performed by standard protocols using ECL reagents (Amersham Biosciences).
Mitochondrial Isolation and Co-immunoprecipitation of Bcl-XL and Bak
HeLa cells on 10-cm culture dishes (∼90% confluence) were treated briefly with trypsin, washed once with cold phosphate-buffered saline, and resuspended in 1 ml of mitochondrial isolation buffer containing 10 mm Hepes-KOH (pH 7.4), 200 mm mannitol, 50 mm sucrose, 10 mm KCl, 1 mm EDTA, and 1× protease inhibitor mixture. Cells were lysed by being passed through a 28-gauge needle 20 times. Nuclei and unbroken cells were removed by centrifugation at 1000 × g at 4 °C for 10 min. Clarified lysates were subjected to centrifugation at 12,000 × g at 4 °C for 15 min. The mitochondrion-containing pellets were resuspended in buffer containing 50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, and 1× protease inhibitor mixture and incubated on ice for 30 min. After removal of insoluble debris by centrifugation at 12,000 × g for 5 min at 4 °C, supernatants were incubated with anti-Bcl-XL antibody for 3 h at 4 °C, followed by addition of Protein A beads for another 1 h. Samples were then treated as described above.
GST Pulldown
Purified GST-tagged recombinant FKBP38 at a final concentration of 200 nm was incubated on ice with an equal amount of purified His6-tagged recombinant Bcl-2 in 500 μl of phosphate-buffered saline containing 0.5% Triton X-100. Purified His6-tagged Rheb loaded with either GDP or GTPγS was added to the reaction at various concentrations. The reaction mixture was incubated on ice for 30 min. Glutathione-conjugated Sepharose beads (20 μl) were then added, followed by incubation at 4 °C for 15 min. Beads were washed four times with phosphate-buffered saline containing 1% Triton X-100 and once with 20 mm Tris-HCl (pH 7.4) prior to SDS-PAGE analysis. Freshly purified His-Rheb was loaded with GDP or GTPγS as described (12).
RESULTS
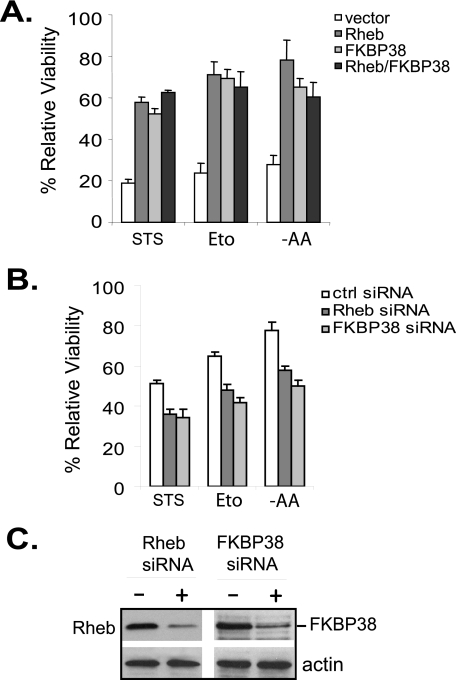
To determine the role of Rheb in apoptosis, we first examined whether an increase in Rheb activity protects cells against stress-induced cell death. Accordingly, we overexpressed Rheb in HeLa cells and monitored the viability of the cells after exposing them to various stress conditions that are known to induce apoptosis. As shown in Fig. 1A, under conditions that induced death in ∼80% of cells expressing control vector, a majority of the cells expressing Rheb remained viable. Overproduction of FKBP38 also protected the cells from stress-induced death, which is consistent with previous observations (21, 22). Similarly, we found that overexpression of Rheb or FKBP38 enhanced the resistance of HCT116 cells to stress conditions (supplemental Fig. S1). In contrast, reducing the expression levels of Rheb or FKBP38 in HeLa cells by siRNA knockdown sensitized them to stress conditions (Fig. 1, B and C). Collectively, these results demonstrate an anti-apoptotic role for Rheb and FKBP38.
FIGURE 1.
Overexpression of Rheb enhances cell survival under stress conditions. A, HeLa cells expressing an empty vector, FLAG-Rheb, Myc-FKBP38, or FLAG-Rheb and Myc-FKBP38 together were treated with 1 μm staurosporine (STS) for 6 h or 100 μm etoposide (Eto) for 20 h or starved for amino acids (−AA) for 16 h. Viability of the cells at the end of the treatment was assayed. B, HeLa cells were transfected with control (ctrl) siRNA or siRNAs specific for Rheb or FKBP38. Transfected cells were incubated for 72 before being treated with 500 nm staurosporine for 6 h or 100 μm etoposide for 16 h or starved for amino acids for 6 h. Data represent mean ± S.D. from three independent experiments. C, the levels of Rheb and FKBP38 in cells transfected with control siRNA (−) or Rheb-specific or FKBP38-specific siRNA (+) shown in B were determined by Western blotting using anti-Rheb or anti-FKBP38 antibody.
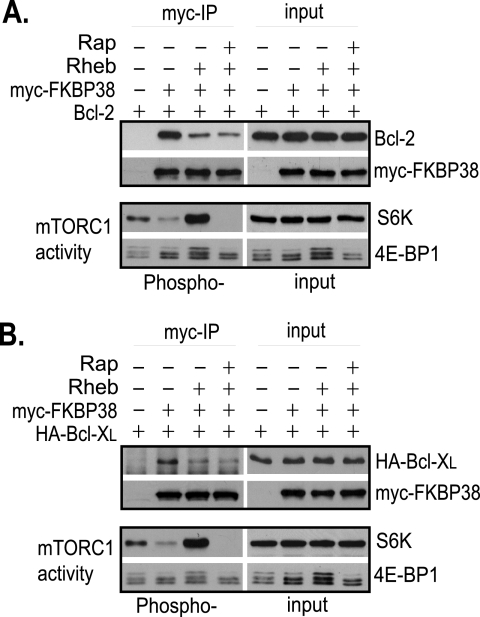
Previous studies have shown that FKBP38 interacts with Bcl-2 and Bcl-XL, and the interaction is critical for the function of the two anti-apoptotic proteins on mitochondria (21). To determine whether the interaction is regulated by Rheb, we examined the effect of Rheb overproduction on the interaction by co-immunoprecipitation. Because the available antibodies for FKBP38 were ineffective in precipitating endogenously expressed FKBP38, we expressed Myc-tagged FKBP38 and untagged Bcl-2 in HeLa cells and immunoprecipitated Myc-FKBP38 with anti-Myc antibody. In cells without Rheb overproduction, we found that Bcl-2 co-precipitated with Myc-FKBP38 (Fig. 2A), which is consistent with the previous observation (21). However, in cells overexpressing Rheb, less Bcl-2 was co-precipitated with FKBP38 (Fig. 2A). The reduction in the association of Bcl-2 with FKBP38 was not affected by rapamycin treatment, suggesting that the reduction was not caused by Rheb-stimulated mTORC1 activation. Similarly, overexpression of Rheb also reduced the amount of Bcl-XL co-purified with FKBP38 independent of mTORC1 (Fig. 2B). These findings suggest that the interaction of FKBP38 with the two anti-apoptotic proteins is negatively regulated by Rheb.
FIGURE 2.
Rheb controls the interaction of FKBP38 with Bcl-2 and Bcl-XL. HeLa cells were transfected with the indicated genes. Transfected cells were treated with rapamycin (Rap+) or drug vehicle control (Rap−) for 30 min. Cells were lysed, and lysates were precipitated with anti-Myc antibody cross-linked to Protein A-agarose beads. The levels of Myc-FKBP38 and Bcl-2 (A) and hemagglutinin (HA)-Bcl-XL (B) in the precipitates (myc-IP) and lysates (input); the phosphorylation levels of S6 kinase (S6K) at Thr-389 and 4E-BP1 at Thr-37/46 (Phospho-); and their protein levels (input) in the lysates were determined by Western blotting.
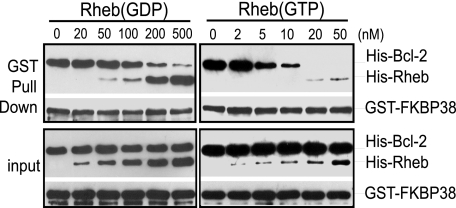
Because the activity of Rheb is controlled by guanine nucleotides, we further determined whether the ability of Rheb to regulate the interaction of FKBP38 with Bcl-2 is nucleotide-dependent. To this end, we used an in vitro GST pulldown assay to examine the effect of both GDP- and GTP-loaded Rheb on the interaction of FKBP38 and Bcl-2. As shown in Fig. 3, we found that recombinant His-tagged Bcl-2 was effectively co-precipitated with GST-tagged FKBP38 in the absence of Rheb. The presence of His-tagged GDP-loaded Rheb had no obvious effect on co-precipitation when its concentration was lower than 100 nm. At higher concentrations, GDP-loaded Rheb reduced the amount of His-Bcl-2 co-purified with GST-FKBP38, which correlated inversely with the amount of His-Rheb found in the precipitates. On the other hand, the amount of His-Bcl-2 co-purified with GST-FKBP38 was undetectable in the presence of GTP-loaded Rheb at a concentration as low as 20 nm. These observations demonstrate that the interaction of FKBP38 with Bcl-2 is negatively regulated by Rheb in a GTP-dependent manner.
FIGURE 3.
Rheb regulates the interaction of FKBP38 with Bcl-2 in a GTP-dependent manner. Recombinant GST-FKBP38 was incubated with His-Bcl-2 in reactions containing the indicated concentrations of either GDP-loaded (left panels) or GTP-loaded (right panels) His-Rheb. The reaction mixtures were precipitated with glutathione-conjugated Sepharose beads. The amounts of GST-FKBP38, His-Bcl-2, and His-Rheb in the precipitates (GST Pull Down) and their levels in the reaction mixtures (input) were determined by Western blotting using anti-GST and anti-His antibodies.
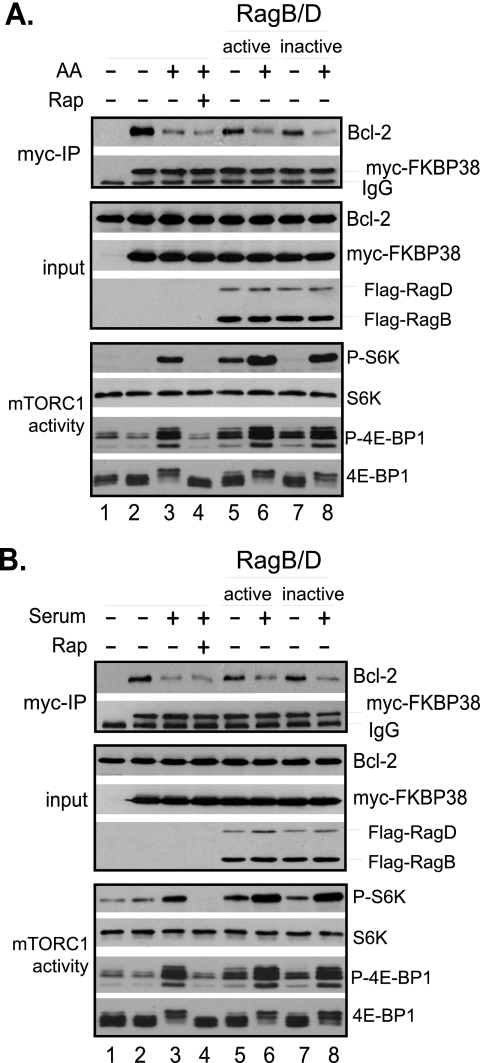
Because Rheb activity in cells is regulated by amino acids and growth factors, we next determined whether changes in amino acid or growth factor conditions affect the interaction between FKBP38 and Bcl-2. Accordingly, we ectopically expressed Bcl-2 and Myc-FKBP38 in HeLa cells and examined the interaction between the expressed proteins upon exposing the cells to changes in amino acid conditions. Under amino acid starvation conditions, Bcl-2 was effectively co-precipitated with Myc-FKBP38. However, the amount of Bcl-2 in the precipitates was drastically reduced when starved cells were refed with amino acids (Fig. 4A, lanes 2 and 3). Preincubating the starved cells with rapamycin before readdition of amino acids did not block the effect of amino acids, indicating that the reduction in the interaction of Myc-FKBP38 with Bcl-2 was not caused by amino acid-stimulated mTORC1 activation (Fig. 4A, bottom panel). Likewise, when serum-deprived cells were restimulated with serum, the association of Myc-FKBP38 with Bcl-2 was reduced. Again, the effect of serum on the interaction was independent of mTORC1 because pretreating the cells with rapamycin failed to block the effect (Fig. 4B). Similar results were obtained when the association of stably expressed Myc-FKBP38 with endogenous Bcl-XL was examined (supplemental Fig. S2). Because amino acid-stimulated mTORC1 activation is mediated through the Rag GTPases (25), we examined whether the GTPases are able to affect the association of FKBP38 with Bcl-2. We found that in cells expressing an active form (RagBGTP-RagDGDP) of the Rag GTPases, as indicated by its ability to stimulate mTORC1, the association of FKBP38 with Bcl-2 was reduced under amino acid starvation conditions (Fig. 4A, compare lanes 2 and 5). However, a similar reduction in the association was also observed in cells expressing an inactive form (RagBGDP-RagDGTP) of the Rag GTPases, as indicated by its inability to stimulate mTORC1 activity (Fig. 4A, top and bottom panels, compare lanes 2, 5, and 7), suggesting that the cause of the reduction was independent of the activity of the Rag GTPases. Furthermore, we found that the association of Bcl-2 with FKBP38 remained sensitive to amino acid stimulation in cells expressing either an active or an inactive form of the Rag GTPases. This observation indicates that amino acids are able to regulate the association of Bcl-2 and FKBP38 independent of the Rag GTPase. Similarly, we found that the Rag GTPases had no obvious effect on serum-induced dissociation of Bcl-2 from FKBP38 (Fig. 4B). Taken, together, these findings demonstrate that the interaction between FKBP38 and Bcl-2 is regulated by amino acid and serum conditions through a mechanism that is independent of mTORC1.
FIGURE 4.
Interaction of FKBP38 with Bcl-2 is regulated by amino acid and serum conditions. HeLa cells were transfected with vectors expressing S6 kinase, Bcl-2, and Myc-FKBP38, along with vector expressing FLAG-RagBGTP-RagDGDP (RagB/D active, lanes 5 and 6) or FLAG-RagBGDP-RagDGTP (RagB/D inactive, lanes 7 and 8) or empty vector (lanes 1–4). A, the transfected cells were starved for amino acids for 1 h followed by refeeding with amino acids (AA+) or vehicle control (AA−) for 30 min. B, the transfected cells were deprived of serum for 20 h followed by refeeding with serum (+) or vehicle control (−) for 30 min. Cells were treated with 40 nm rapamycin (Rap+) or drug vehicle (Rap−) for 30 min before refeeding. Cells were lysed at the end of the treatments, and lysates were precipitated with anti-Myc antibody (myc-IP) cross-linked with Protein A-agarose beads. The levels of Myc-FKBP38 and Bcl-2 in the precipitates and those of FKBP38, Bcl-2, and the Rag GTPases in the lysates (input); the phosphorylation levels of S6 kinase at Thr-389 (P-S6K) and 4E-BP1 at Thr-37/46 (P-4E-BP1); and their protein levels in the lysates were determined by Western blotting.
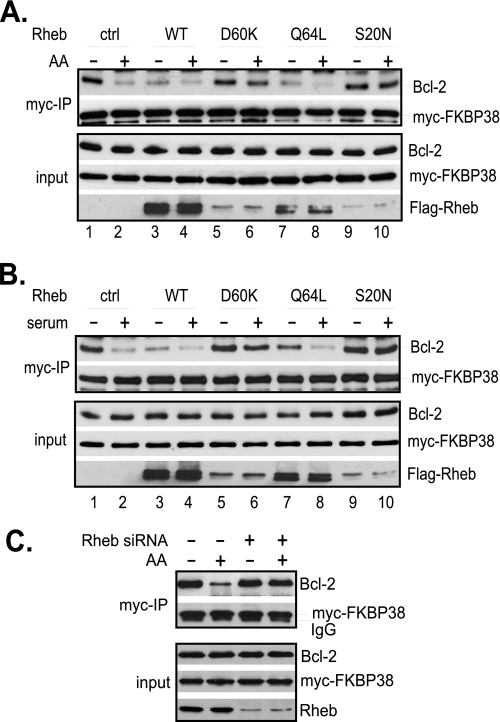
To test whether Rheb mediates the effect of amino acids and serum on the interaction of FKBP38 with Bcl-2, we examined the interaction in HeLa cells overexpressing either dominant active or inactive mutants of Rheb. In cells expressing a control vector, amino acid starvation induced a strong binding between FKBP38 and Bcl-2 (Fig. 5A). This starvation-induced interaction was reduced either by amino acid repletion (Fig. 5A, lane 2) or by expressing wild-type Rheb or an active Rheb mutant (Q64L). On the other hand, amino acid-induced reduction in the interaction was largely blocked by expressing two dominant inactive Rheb mutants, D60K and S20N. Similarly, serum deprivation-induced interaction of FKBP38 with Bcl-2 was reduced by serum refeeding or by expressing active Rheb mutants, whereas serum refeeding-induced dissociation was blocked by dominant inactive Rheb mutants (Fig. 5B). These findings suggest that Rheb activity is able to overwrite the effect of amino acids and serum on the interaction of FKBP38 with Bcl-2, indicating that amino acids and serum signal through Rheb to regulate the interaction of FKBP38 with Bcl-2.
FIGURE 5.
Rheb mediates the effect of amino acids and growth factors on the interaction of FKBP38 with Bcl-2. HeLa cells were transfected with Bcl-2 and Myc-FKBP38 together with wild-type FLAG-Rheb or its mutants. Transfected cells were either starved for amino acids for 1 h followed by amino acid repletion for 30 min (A) or deprived of serum for 20 h followed by readdition of serum for 30 min (B). C, HeLa cells were transfected with Rheb-specific siRNA or scrambled control siRNA. The transfected cells were starved for amino acids for 1 h followed by refeeding with amino acids (AA+) or vehicle control (AA−) for 30 min. Cells were lysed, and lysates were precipitated with anti-Myc antibody cross-linked with Protein A-agarose beads. The amounts of Myc-FKBP38 and Bcl-2 in the precipitates (myc-IP) and lysates (input) were determined by Western blotting. ctrl, control; WT, wild type.
To further ascertain the role of Rheb in the interaction of FKBP38 with Bcl-2, we examined the amino acid-mediated interaction of FKBP38 with Bcl-2 in cells with the level of Rheb reduced by siRNA. As shown in Fig. 5C, we found that, although the interaction was reduced by amino acids in cells transfected with control siRNA, it became largely insensitive to amino acids in cell transfected with Rheb-specific siRNA, which reduced the level of Rheb to ∼20% of that in control cells. Hence, the effect of Rheb knockdown is similar to that of overexpression of dominant negative Rheb mutants, supporting the notion that Rheb mediates the effect of amino acids on the interaction of FKBP38 with Bcl-2.
Because FKBP38 is involved in recruiting Bcl-2 and Bcl-XL to mitochondria, the amino acid- and serum-sensitive interaction between FKBP38 and the two anti-apoptotic proteins is expected to affect their anti-apoptotic function. One possibility is that the amino acid and serum-induced release of Bcl-2 and Bcl-XL from FKBP38 facilitates their binding with pro-apoptotic proteins, such as Bax and Bak. To test this possibility, we assessed the ability of Bcl-XL to interact with Bak on mitochondria in response to changes in amino acid conditions. Accordingly, we collected mitochondrion-enriched fractions from cells starved for amino acids and examined the association between endogenous Bcl-XL and Bak by immunoprecipitation. As shown in Fig. 6A, we found that less Bak was associated with Bcl-XL in cells starved for amino acids than in those refed with amino acids. The amino acid-stimulated increase in the association was not abrogated by rapamycin, suggesting that it was not caused by mTORC1 activation. To confirm that the effect of amino acids is mediated through Rheb, we determined whether ectopically expressed Rheb was able to interfere with the effect. As shown in Fig. 6B, we found that in cells starved for amino acids, overexpression of wild-type Rheb or an active Rheb mutant, like amino acid repletion, enhanced the association of Bcl-XL with Bak. On the other hand, expression of dominant inactive Rheb mutants, including D60K and S20N, blocked the amino acid-stimulated increase in the association. These findings demonstrate that the association of Bcl-XL with Bak is regulated by amino acids through Rheb.
FIGURE 6.
Effect of Rheb and amino acids on the association of Bcl-XL with Bak. A, HeLa cells were starved for amino acids (AA) for 3 h and re-fed for 30 min. At 30 min prior to readdition of amino acids, cells were treated with 40 nm rapamycin (Rap) or drug vehicle control (ctrl). Cells were lysed, and mitochondrion-enriched fractions were obtained by fractionation. Upon solubilizing with detergent, the mitochondrial fraction was precipitated with anti-Bcl-XL antibody. The amounts of Bcl-XL and Bak in the precipitates (Bcl-XL-IP) and lysates (input) were determined by Western blotting. B, HeLa cells expressing control vector, FLAGwild-type (WT) FLAG-Rheb, or Rheb mutants were starved for amino acids for 3 h followed by refeeding with amino acids for 30 min. Cells were then collected and analyzed as in A. C, HeLa cells expressing control vector or FLAG-Rheb were exposed to 100 μm etoposide (+) or drug vehicle control (−) for 20 h followed by treatment with 40 nm rapamycin (+) or vehicle control (−) for 30 min. Cells were lysed and analyzed as in A.
To further confirm the role of Rheb in the interaction of Bcl-XL and Bak, we examined the effect of Rheb overexpression on the interaction in cells treated with etoposide, an apoptotic inducer. As shown in Fig. 6C, we found that the drug treatment greatly reduced the interaction of Bcl-XL with Bak in cells expressing a control vector but had no obvious effect on the interaction in those expressing Rheb. Treating the cells with rapamycin did not block the effect of Rheb overexpression, suggesting that the effect was independent of mTORC1. Consistent with the drug-resistant interaction of Bcl-XL with Bak, the cells overexpressing Rheb were resistant to the drug-induced cell death (Fig. 1).
DISCUSSION
In animal models, elevated Rheb activity is able to promote tumorigenesis by enhancing cell proliferation and down-regulating apoptosis (26, 27). The anti-apoptotic function of Rheb has been suggested to be mediated indirectly by mTORC1, which, upon activation, increases the expression of Mcl-1, a Bcl-2-like anti-apoptotic protein (28). In addition, mTORC1 may also control apoptosis through S6 kinase-dependent phosphorylation of Bad, a BH3-containing pro-apoptotic protein (29). However, in this study, we found that Rheb has a direct role in the regulation of apoptosis that is independent of mTORC1. The key factor in the Rheb-mediated apoptosis is FKBP38, an effector of Rheb that interacts with anti-apoptotic proteins Bcl-2 and Bcl-XL.
Bcl-2 and Bcl-XL exert their anti-apoptotic function by binding to pro-apoptotic proteins, Bax and Bak, and preventing their oligomerization on mitochondria (30). Previous studies have shown that Bcl-2 and Bcl-XL are recruited to mitochondria through their interaction with FKBP38, which resides mainly on mitochondria (21). In this study, we found that the interaction of FKBP38 with Bcl-2 and Bcl-XL is regulated by Rheb in an amino acid- and serum-dependent manner (Figs. 2 and 4). This novel role of Rheb in the regulation of two major anti-apoptotic proteins is consistent with our previous observation that Rheb is partially localized on mitochondria (24). In addition, we have shown that the association of Bcl-XL and Bak is enhanced when Rheb activity increases (Fig. 6). These findings hence promote a model for the action of FKBP38 and Rheb in apoptosis. When cells are grown in the presence of amino acids and serum, Bcl-2 and Bcl-XL are recruited to mitochondria by binding to FKBP38 and are subsequently released from FKBP38 by Rheb. The Rheb-dependent release may enhance the anti-apoptotic function of Bcl-2 and Bcl-XL by facilitating their association with pro-apoptotic proteins. Our observation that amino acid availability or Rheb overexpression enhances the association of Bcl-XL with Bak is consistent with this notion. Alternatively, the release may free FKBP38 for binding and recruiting additional Bcl-2 and Bcl-XL to mitochondria. The findings that overexpression of FKBP38 increases the presentation of Bcl-2 and Bcl-XL on mitochondria (21) and enhances the association of Bcl-XL with Bak (supplemental Fig. S3) appear to support the latter possibility. In either case, the action of Rheb and FKBP38 results in an increase in the association of the apoptotic proteins with pro-apoptotic proteins. On the other hand, when cells are starved for amino acids or deprived of growth factors, Bcl-2 and Bcl-XL may still be recruited to mitochondria by binding to FKBP38 but cannot be released because Rheb is inactive, which may restrict their binding with pro-apoptotic proteins or limit their presentation on mitochondria. Because the antagonistic action of the anti- and pro-apoptotic proteins is a key regulation that sets a rheostat control for apoptosis (20), the enhanced association between the two types of proteins of opposing functions is expected to raise the threshold for apoptosis initiation, thus making cells more resistant to apoptotic inducers. It is thus conceivable that under optimal growth conditions or when Rheb is active, cells are less likely to initiate a cell death program, whereas under nutrient starvation or growth factor deprivation, they are prone to stress-induced death.
We have recently shown that FKBP38 acts as an inhibitor of mTORC1 and that its interaction with mTORC1 is regulated by Rheb (12). The mechanism by which Rheb controls the interaction of FKBP38 with Bcl-2 and Bcl-XL appears to be the same as the one controlling the interaction of FKBP38 with mTORC1. Our in vitro binding results show that Rheb prevents the interaction of FKBP38 with Bcl-2 by directly binding to FKBP38 and that the binding is largely GTP-dependent (Fig. 3). In cells, Rheb negatively regulates the interaction in response to amino acid or serum stimulation. When Rheb is active, Bcl-2 and Bcl-XL, like mTOR, are released from FKBP38 (Figs. 2 and 4). Therefore, FKBP38 represents a branch point at which the oncogenic signal of Rheb is diverged to cell growth control and apoptosis regulation, two key events that govern cell proliferation.
In addition to Rheb, FKBP38 is also regulated by calcium and calmodulin, which bind to the calcium/calmodulin-binding domain of FKBP38. Previous studies have shown that calcium and calmodulin promote the binding of FKBP38 with Bcl-2 (22). Consistent with this finding, we found that in an in vitro binding assay, the presence of calcium and calmodulin enhanced the interaction of FKBP38 with Bcl-2 and antagonized the effect of Rheb on the interaction (supplemental Fig. S4). It thus appears that calcium and Rheb represent two opposing inputs that converge on FKBP38 and that the interplay between calcium- and Rheb-dependent regulations may dictate the ability of FKBP38 to interact with Bcl-2 and Bcl-XL.
Rheb interacts with FKBP38 by binding to its FKBP-C domain, a region that is highly related to FKBP12 peptidyprolyl cis/trans-isomerase (12). The same region is also involved in binding of FKBP38 with Bcl-2 (21). It thus appears that Rheb may displace Bcl-2 by competing with it for FKBP38 binding. However, we found that expression of a dominant negative allele of Rheb, Rheb(D60K), which exhibits a strong binding with FKBP38 (12, 24), did not prevent the binding of FKBP38 with Bcl-2 (Fig. 5). This observation suggests that the interactions of FKBP38 with Rheb and Bcl-2 are not mutually exclusive, indicating that Rheb may induce the release of Bcl-2 by altering the conformation of FKBP38. Furthermore, interaction with FKBP38 through its FKBP-C domain seems to be a prerequisite for Rheb-dependent regulation. PHD2, a prolyl hydroxylase that modifies hypoxia-inducible factor 1α, has been shown to interact with FKBP38 by binding to a region outside of the FKBP-C domain (31). We find that the interaction is not regulated by Rheb (data not shown). In addition, the binding of FKBP38 with Bcl-2 and Rheb through its FKBP-C domain also raises a possibility that Rheb may alter the conformation of Bcl-2 through the peptidylprolyl cis/trans-isomerase activity of FKBP38, thus influencing the ability of Bcl-2 to interact with pro-apoptotic proteins.
Elevated Rheb activity, caused by inactivation mutations in the TSC1 and TSC2 tumor suppressor genes, has been associated with tumorigenesis in human (2). Although the activation of mTORC1, a downstream target of Rheb, is a major contributor for the oncogenic activity of Rheb, repression of apoptosis also plays a critical role in Rheb-induced tumorigenesis (26, 27). Our data suggest that Rheb controls apoptosis through FKBP38-dependent regulation of Bcl-2 and Bcl-XL. Hyperactivated Rheb is able to prevent apoptosis by enhancing the ability of these apoptotic proteins to interact with pro-apoptotic proteins, which allow cells to survive conditions that normally trigger apoptosis, leading to deregulated proliferation.
Supplementary Material
Acknowledgments
We thank Lin Zhang (University of Pittsburgh) for providing Bcl-2 expression vectors and Xiaoming Yin for comments and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA129821 and GM068832 (to Y. J.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods” and Figs. S1–S4.
- TSC
- tuberous sclerosis complex
- mTOR
- mammalian target of rapamycin
- GST
- glutathione S-transferase
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- siRNA
- small interfering RNA.
REFERENCES
- 1.Crino P. B., Nathanson K. L., Henske E. P. (2006) N. Engl. J. Med. 355, 1345–1356 [DOI] [PubMed] [Google Scholar]
- 2.Mak B. C., Yeung R. S. (2004) Cancer Invest. 22, 588–603 [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Corradetti M. N., Inoki K., Guan K. L. (2004) Trends Biochem. Sci. 29, 32–38 [DOI] [PubMed] [Google Scholar]
- 4.Manning B. D., Cantley L. C. (2003) Trends Biochem. Sci. 28, 573–576 [DOI] [PubMed] [Google Scholar]
- 5.Inoki K., Corradetti M. N., Guan K. L. (2005) Nat. Genet. 37, 19–24 [DOI] [PubMed] [Google Scholar]
- 6.Tee A. R., Blenis J. (2005) Semin. Cell Dev. Biol. 16, 29–37 [DOI] [PubMed] [Google Scholar]
- 7.Martin D. E., Hall M. N. (2005) Curr. Opin. Cell Biol. 17, 158–166 [DOI] [PubMed] [Google Scholar]
- 8.Wullschleger S., Loewith R., Hall M. N. (2006) Cell 124, 471–484 [DOI] [PubMed] [Google Scholar]
- 9.Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 10.Kim D. H., Sarbassov D. D., Ali S. M., King J. E., Latek R. R., Erdjument-Bromage H., Tempst P., Sabatini D. M. (2002) Cell 110, 163–175 [DOI] [PubMed] [Google Scholar]
- 11.Loewith R., Jacinto E., Wullschleger S., Lorberg A., Crespo J. L., Bonenfant D., Oppliger W., Jenoe P., Hall M. N. (2002) Mol. Cell 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 12.Bai X., Ma D., Liu A., Shen X., Wang Q. J., Liu Y., Jiang Y. (2007) Science 318, 977–980 [DOI] [PubMed] [Google Scholar]
- 13.Dunlop E. A., Dodd K. M., Seymour L. A., Tee A. R. (2009) Cell. Signal. 21, 1073–1084 [DOI] [PubMed] [Google Scholar]
- 14.Sato T., Nakashima A., Guo L., Tamanoi F. (2009) J. Biol. Chem. 284, 12783–12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uhlenbrock K., Weiwad M., Wetzker R., Fischer G., Wittinghofer A., Rubio I. (2009) FEBS Lett. 583, 965–970 [DOI] [PubMed] [Google Scholar]
- 16.Wang X., Fonseca B. D., Tang H., Liu R., Elia A., Clemens M. J., Bommer U. A., Proud C. G. (2008) J. Biol. Chem. 283, 30482–30492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockenbery D., Nuñez G., Milliman C., Schreiber R. D., Korsmeyer S. J. (1990) Nature 348, 334–336 [DOI] [PubMed] [Google Scholar]
- 18.Krajewski S., Tanaka S., Takayama S., Schibler M. J., Fenton W., Reed J. C. (1993) Cancer Res. 53, 4701–4714 [PubMed] [Google Scholar]
- 19.González-Garcia M., Perez-Ballestero R., Ding L., Duan L., Boise L. H., Thompson C. B., Nuñez G. (1994) Development 120, 3033–3042 [DOI] [PubMed] [Google Scholar]
- 20.Youle R. J., Strasser A. (2008) Nat. Rev. Mol. Cell Biol. 9, 47–59 [DOI] [PubMed] [Google Scholar]
- 21.Shirane M., Nakayama K. I. (2003) Nat. Cell Biol. 5, 28–37 [DOI] [PubMed] [Google Scholar]
- 22.Edlich F., Weiwad M., Erdmann F., Fanghänel J., Jarczowski F., Rahfeld J. U., Fischer G. (2005) EMBO J. 24, 2688–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Germain M., Shore G. C. (2003) Sci. STKE 2003, pe10. [DOI] [PubMed] [Google Scholar]
- 24.Ma D., Bai X., Guo S., Jiang Y. (2008) J. Biol. Chem. 283, 25963–25970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008) Science 320, 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrakis K. J., Zhu H., Silva R. L., Mills J. R., Teruya-Feldstein J., Lowe S. W., Tam W., Pelletier J., Wendel H. G. (2008) Genes Dev. 22, 2178–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nardella C., Chen Z., Salmena L., Carracedo A., Alimonti A., Egia A., Carver B., Gerald W., Cordon-Cardo C., Pandolfi P. P. (2008) Genes Dev. 22, 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wendel H. G., Silva R. L., Malina A., Mills J. R., Zhu H., Ueda T., Watanabe-Fukunaga R., Fukunaga R., Teruya-Feldstein J., Pelletier J., Lowe S. W. (2007) Genes Dev. 21, 3232–3237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freilinger A., Rosner M., Krupitza G., Nishino M., Lubec G., Korsmeyer S. J., Hengstschläger M. (2006) Oncogene 25, 6467–6479 [DOI] [PubMed] [Google Scholar]
- 30.Antignani A., Youle R. J., Reed J. C., van Delft M. F., Huang D. C., Deming P. B., Rathmell J. C., Willis S. N., Adams J. M., Kim R., Lindsten T., Zong W. X., Thompson C. B., Petros A. M., Olejniczak E. T., Fesik S. W., Breckenridge D. G., Germain M., Mathai J. P., Nguyen M., Shore G. C., Degli Esposti M., Dive C., Scorrano L., Korsmeyer S. J., Opferman J. T., Korsmeyer S. J., Korsmeyer S. J., Wei M. C., Saito M., Weiler S., Oh K. J., Schlesinger P. H., Lutz R. J., Tsujimoto Y. (2006) Curr. Opin. Cell Biol. 18, 685–689 [DOI] [PubMed] [Google Scholar]
- 31.Barth S., Nesper J., Hasgall P. A., Wirthner R., Nytko K. J., Edlich F., Katschinski D. M., Stiehl D. P., Wenger R. H., Camenisch G. (2007) Mol. Cell. Biol. 27, 3758–3768 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.