Abstract
BanLec is a jacalin-related lectin isolated from the fruit of bananas, Musa acuminata. This lectin binds to high mannose carbohydrate structures, including those found on viruses containing glycosylated envelope proteins such as human immunodeficiency virus type-1 (HIV-1). Therefore, we hypothesized that BanLec might inhibit HIV-1 through binding of the glycosylated HIV-1 envelope protein, gp120. We determined that BanLec inhibits primary and laboratory-adapted HIV-1 isolates of different tropisms and subtypes. BanLec possesses potent anti-HIV activity, with IC50 values in the low nanomolar to picomolar range. The mechanism for BanLec-mediated antiviral activity was investigated by determining if this lectin can directly bind the HIV-1 envelope protein and block entry of the virus into the cell. An enzyme-linked immunosorbent assay confirmed direct binding of BanLec to gp120 and indicated that BanLec can recognize the high mannose structures that are recognized by the monoclonal antibody 2G12. Furthermore, BanLec is able to block HIV-1 cellular entry as indicated by temperature-sensitive viral entry studies and by the decreased levels of the strong-stop product of early reverse transcription seen in the presence of BanLec. Thus, our data indicate that BanLec inhibits HIV-1 infection by binding to the glycosylated viral envelope and blocking cellular entry. The relative anti-HIV activity of BanLec compared favorably to other anti-HIV lectins, such as snowdrop lectin and Griffithsin, and to T-20 and maraviroc, two anti-HIV drugs currently in clinical use. Based on these results, BanLec is a potential component for an anti-viral microbicide that could be used to prevent the sexual transmission of HIV-1.
Keywords: Carbohydrate/Lectin, Viruses/Antiviral agents, Viruses/Entry, Viruses/HIV, Viruses/Immunodeficiency, BanLec, ELISA, gp120, Strong-stop DNA
Introduction
Despite the development of more than 25 approved anti-HIV2 drugs and improvements in the availability of antiretroviral drugs in low and middle income countries, the rate of new HIV-1 infections is outpacing the rate of new individuals receiving antiretroviral therapy by 2.5:1 (1). At present, it appears that an efficacious HIV vaccine is still many years away. Therefore, other methods for halting the spread of HIV are vitally needed. This has raised the possibility of developing either intravaginally or intrarectally applied microbicides to halt the spread of HIV during sexual intercourse. This type of intervention is particularly needed in the developing world, such as sub-Saharan Africa, where more than 20 million people are living with HIV/AIDS (1). Although abstinence has been suggested by some groups, campaigns to encourage this method of halting transmission have not been effective (2). Although condoms are quite effective against the spread of HIV and some other sexually transmitted diseases, they are only effective if they are used consistently and correctly, which is often not the case (3, 4). This is particularly true in the developing world, where women have relatively little control over sexual encounters and, thus, have not been able to enforce condom usage (5), so the development of a long-lasting, self-applied, microbicide is very attractive. In fact, it is estimated that 20% coverage with a microbicide that is only 60% effective against HIV may prevent up to 2.5 million HIV infections over three years (6). Therefore, even modest success with microbicides could save millions of lives.
Some of the most promising compounds for inhibiting vaginal or rectal HIV transmission are agents that block HIV before integration of the viral genome into the target cell. Thus, the viral entry step is one potential target for a microbicide. Entry inhibitors that have been proposed for use in a vaginal microbicide include long chain and ionic polymers (such as Pro 2000) as well as dendrimers, lipid membrane modifiers, and anti-CD4 antibodies. HIV-binding peptides and small molecule inhibitors have also been considered, including the fusion inhibitor T-20 (enfuvirtide) and the CCR5 blocker maraviroc, which are already in clinical use for the treatment of HIV infection. In addition, lectins are a growing class of HIV-1 inhibitors under consideration as microbicide candidates (7, 8). Lectins inhibit HIV-1 entry by binding to carbohydrate structures found on the viral envelope. Examples of anti-HIV lectins include Cyanovirin-N (CV-N) (9), Griffithsin (GRFT) (10), and snowdrop lectin (GNA) (11–13).
The HIV-1 envelope protein gp120 contains 20–30 possible N-linked glycosylation sites. These carbohydrate structures make up ∼50% of the molecular weight of the protein (14–16). Glycosylation affects aspects of the viral life cycle including protein folding (17), cellular transport, binding to cellular receptors (14, 18, 19), trans-infection by dendritic cells (19), and shielding from the immune response (20). Because glycosylation is essential to the virus, it presents an attractive therapeutic target.
The lectin termed BanLec, isolated from the ripened fruit of the banana (Musa acuminata cultivars), exists as a dimer with a molecular mass of ∼30 kDa (21). It is a member of the jacalin-related lectin family and can recognize high mannose structures (22–24). Lectins in this family are characterized by the presence of a β-prism 1 structure composed of three Greek Key turn motifs. Greek Keys 1 and 2 are both involved in binding carbohydrates and contain a GXXXD binding motif, whereas Key 3 does not contain the binding motif (25, 26). However, this loop can assist ligand binding and determine lectin specificity (27). Because of its affinity for high mannose structures (15), we sought to investigate whether BanLec might bind the mannose-rich envelope of HIV-1 and thereby block HIV infection. The results presented below demonstrate that BanLec is a potent inhibitor of HIV infection that markedly reduces the replication of a range of HIV-1 isolates and has potential to be further developed for use as a vaginal microbicide.
MATERIALS AND METHODS
Proteins and Anti-HIV Compounds
BanLec was isolated from bananas by modification of previously described methods (21, 22).3 Snowdrop lectin (GNA) was isolated from crude extracts of snowdrop bulbs and purified over a mannose-agarose column as described previously (28). Recombinant HIV-1 gp120, human monoclonal antibody 2G12, recombinant His-tagged GRFT, T-20, maraviroc, and CD4-IgG2 were obtained from the NIH AIDS Research and Reference Reagent Program (10). Recombinant, glycosylated gp120 was produced in HEK-293 cells (human embryonic kidney cells), the recombinant human antibody 2G12 was produced in Chinese hamster ovary cells, and recombinant GRFT was produced in Escherichia coli. The purity of all of the proteins was found to be >95% as determined by SDS-PAGE.
HIV-1 Production
The HIV-1 isolate Bru was produced in peripheral blood lymphocytes (PBL), whereas the HIV-1 BaL (29) isolate was produced in macrophages. Primary, dual-tropic isolates ASM44 and ASM54 were expanded in peripheral blood mononuclear cells (PBMCs) containing both lymphocytes and macrophages. Production of pseudotyped HIV-1 was performed by transfecting HEK-293FT cells (a fast growing cell line derived from HEK-293 that contains the SV40 large T antigen) with plasmids coding for an HIV-1 envelope from either subtype B (30) or C along with an envelope-deleted proviral clone, pSG3Δenv (31). Proviral plasmid DNA clones pNL4-3 (32), pNL(AD8) (33), p81A-4 (34–36), and p89.6 (37) were transfected into HEK-293FT cells with Lipofectamine 2000 (Invitrogen). The media were changed 24 h post-transfection, and at 48 h post-transfection the supernatants were harvested and frozen at −80 °C. The concentration of virus in the stocks was determined by the HIV-1 p24 Antigen Capture Assay ELISA (AIDS and Cancer Virus Program) or by determining the infectious titer. HIV-1 Bru was treated with 10 units/μl of RNase-free DNase I (Roche Applied Science) before use in the experiments in which the products of early reverse transcription were assayed.
HIV-1 Indicator Assays
HIV-1 infection was quantified using TZM-bl cells, which express a luciferase and β-galactosidase gene under the control of the HIV-1 LTR promoter (31, 38–40). The day before infection of 5000 TZM-bl cells/ml in 100 μl of Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 25 mm HEPES, and 50 μg/ml Geneticin were added to the wells of white, opaque, 96-well tissue culture plates (Falcon). Cells were pretreated with BanLec for 30 min before infection with 100 TCID50 units of virus (∼15,000 relative luminescence units) to a final volume of 200 μl/well. Cells were exposed to virus and lectin for either 2 days with replication competent viruses or for 3 days with pseudotyped, replication defective virus. A Steady-Glo® luciferase assay system (Promega) and a plate reader containing a luminometer (Tecan) were used to measure luminescence, which was indicative of viral infection.
MAGI-CCR5 cells (41, 42) were plated in 24-well tissue culture plates with 40,000 cells per well in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, penicillin, and streptomycin. The cells were pretreated with lectin for 30 min and then infected with different viral isolates at concentrations that yielded ∼100 positively infected cells per well. Forty hours post-infection, cells were stained for β-galactosidase activity as described within the reagent data sheet, and positive cells were counted visually.
Isolation and Culture of Primary Cells
PBMCs were isolated from healthy donors by venipuncture. Briefly, blood was drawn into a 60-ml syringe containing 7 ml of 250 mm sodium citrate and 10 ml of 6% dextran solution and mixed by inversion. After 30 min, to allow for the sedimentation of red blood cells, the supernatant was separated using Hypaque-Ficoll, and the buffy coat layer was removed, washed twice with cold PBS containing 0.2% bovine serum albumin, and centrifuged at 350 × g for 10 min. The cell pellet was resuspended in RPMI 1640 media at a concentration of 5 × 106 cells per ml and seeded into non-tissue culture-treated plates. PBL were removed from the adherent monocytes and washed three times with PBS. For the differentiation of monocytes to macrophages for HIV-1 infection, the monocytes were cultured with Iscove's modified Dulbecco's media containing 10% heat-inactivated human AB sera for 7 days.
Infection of Monocyte-derived Macrophages (MDM)
MDM were washed with PBS three times followed by the addition of fresh media containing BanLec or PBS 30 min before infection. Cells were infected with ∼100 TCID50 of NL(AD8) for 24 h, and the residual virus was removed by three PBS washes followed by the addition of fresh media. Every 3 days, a sample was removed and replaced with fresh media containing the appropriate amount of BanLec for 15 days. The samples were stored at −80 °C until viral replication was determined by the HIV-1 p24 Antigen Capture Assay ELISA (AIDS and Cancer Virus Program). A similar experiment was done in which samples were not removed until the end of the experiment on day 7. For both experiments, an MTT ((3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) reduction assay was performed on the final day to assess cellular viability.
Detection of Early Products of HIV-1 Reverse Transcription (Strong-Stop DNA) in Peripheral Blood Lymphocytes
PBL were stimulated with phytohemagglutinin for 3 days in RPMI media containing 10% heat-inactivated fetal bovine serum and interleukin-2. The cells were washed with PBS and resuspended in RPMI media containing 10% heat-inactivated fetal bovine serum. Lectins were added 30 min before centrifuge-mediated infection (spin-infection) with DNase I-treated HIV-1 Bru (43). Three hours post-infection the cells were harvested, washed with PBS, and then stored at −80 °C. Cellular DNA, including genomic and viral DNA products, was isolated using the QIAamp DNA Blood Mini kit (Qiagen). Strong-stop DNA, the first product of HIV-1 reverse transcription, is used for the assessment of viral entry (13, 44). This reverse transcription product was quantified by performing real-time PCR with primers specific for strong-stop DNA, the DNA concentration of the each sample was normalized, and equal DNA loading was confirmed with primers for α-tubulin (44).
Determination of BanLec and Glycosylated HIV-1 gp120 Interaction by ELISA
96-Well ELISA plates were coated by adding 50 μl of 5 μg/ml BanLec per well and incubated overnight at room temperature. The next day plates were blocked for 1.5 h at room temperature with PBS containing 1% bovine serum albumin, 5% sucrose, and 0.05% sodium azide and then rinsed with wash buffer (PBS containing 0.05% Tween 20, pH 7.4) 3 times before the addition of recombinant, glycosylated gp120 protein diluted in blocking buffer. After a 1-h incubation at room temperature, the plates were washed 3 times before the addition of the detection antibodies. A sheep anti-gp120 antibody (AIDS Research and Reference Reagent Program) was diluted 1:2000 in dilution buffer (wash buffer containing 0.1% bovine serum albumin) and added to the wells and incubated for 1 h. The plate was washed again before a 1-h incubation with an anti-sheep antibody conjugated to alkaline phosphatase (Sigma) diluted 1:40,000 in dilution buffer. After the plate was washed, p-nitrophenyl phosphate (Sigma) was added for colorimetric analysis, and the absorbance was measured at 405 nm.
To determine whether BanLec could block the recognition of gp120 by the anti-HIV monoclonal antibody 2G12, ELISA plates were coated overnight with 100 μl of a 1 μg/ml solution of recombinant gp120 diluted in PBS. The plates were blocked as described above, washed, and then treated with serial dilutions of BanLec in dilution buffer. After a 1-h incubation the plates were washed to remove unbound BanLec. The 2G12 antibody was added at a concentration of 100 ng/ml to allow for binding to the gp120 protein. One hour later the plates were washed and incubated with biotinylated anti-human antibody (Jackson ImmunoResearch) followed by another series of washes and the addition of streptavidin-conjugated alkaline phosphatase (Jackson ImmunoResearch). The alkaline phosphatase dilution buffer contained 500 mm methyl-α-d-mannopyranoside, which was added to reduce any potential nonspecific binding of the 2G12 antibody to BanLec. The plates were washed again, and the substrate p-nitrophenyl phosphate (Sigma) was added for colorimetric analysis of 2G12 binding. The amount of antibody that remained bound was quantified by comparison to a standard curve consisting of serial 2-fold dilutions of 2G12.
Determination of Anti-HIV Activity Post-cellular Attachment
For measuring the inhibition of HIV-1 infection post-cellular attachment, TZM-bl cells were plated in 96-well plates as described above and spin-infected with a pseudotyped virus with pConCgp160-opt, a consensus subtype C envelope sequence, at 1250 × g for 2 h at 16 °C 1 day after plating (43, 45). The cells were placed on ice, the supernatant was removed, and unbound virus was removed by washing 2 times with PBS containing 0.2% bovine serum albumin. Cell culture media containing inhibitors (CD4-IgG2, T-20, maraviroc, or BanLec) was added to the cells and incubated for 30 min on ice. The cells were then moved to a 37 °C incubator, and luciferase activity was measured 3 days later. These results were compared with cells that were infected with virus that had been pretreated with the same inhibitors described above on ice for 30 min and then incubated at 37 °C (i.e. standard infection conditions).
RESULTS
BanLec Is a Potent Inhibitor of Multiple HIV-1 Isolates
To determine the anti-HIV activity of BanLec, different concentrations of the lectin were incubated with TZM-bl indicator cells before infection with various HIV-1 isolates. Because a microbicide would need to inhibit HIV-1 of different tropisms and because glycosylation can play a role in determining viral tropism (18), we tested the ability of BanLec to inhibit several different HIV-1 isolates. The viral clones 81A-4 and NL(AD8) are both derivatives of NL4-3 in which a portion of the envelope is swapped with the envelope region from either the R5 HIV-1 isolates BaL or ADA, respectively. 81A-4 and NL(AD8) use CCR5 as a cellular co-receptor (R5 tropic), whereas NL4-3 uses CXCR4 (X4 tropic). These isolates allow for the assessment of different HIV-1 envelope sensitivity to BanLec while keeping the remainder of the NL4-3 viral components unchanged. The dual-tropic isolate 89.6 was also assessed for susceptibility to BanLec. We observed dose-dependent inhibition of viral infection with IC50 values calculated in the low nanomolar range against viral isolates with different tropisms (Fig. 1A). These results suggest that sensitivity of HIV-1 isolates to BanLec is independent of viral tropism.
FIGURE 1.
BanLec has antiviral activity against multiple HIV-1 isolates with different tropisms. A, TZM-bl cells were pretreated with different concentrations of BanLec before infection with the R5 tropic isolates NL(AD8) and 81A-4, dual tropic 89.6, and X4 tropic NL4-3. Forty-eight hours after exposure to virus, luciferase activity was determined by measuring relative luminescent units (RLU). The averages from three separate experiments were used for the calculation of IC50 values, which were determined by nonlinear regression. The IC50 for viral inhibition were as follows: NL(AD8) = 2.06 nm, 81A-4 = 0.69 nm, 89.6 = 0.48 nm, NL4-3 = 0.49 nm. B, Magi-CCR5 indicator cells were used to determine anti-viral activity of BanLec against multiple strains of HIV-1. Forty hours after exposure to virus, infected cells were quantified by staining for β-galactosidase activity. Infectivity of BanLec-treated virus is presented as a percent of positively infected cells as compared with the PBS control. Error bars represent S.D. from three separate experiments.
We further confirmed the anti-HIV activity of BanLec with the HIV-1 indicator cell line, MAGI-CCR5. With this cell line, we tested the ability of BanLec to inhibit infection by the laboratory-adapted isolates BaL (R5) and Bru (X4) and the primary isolates ASM 44 (R5X4) and ASM 54 (R5X4) and determined that all were inhibited by BanLec (Fig. 1B). The virus used in this experiment was generated by infection of PBMC, whereas the experiment shown in Fig. 1A used virus produced by transfection of HEK-293FT cells with a proviral plasmid clone. These results further support our initial studies by showing that BanLec can inhibit HIV isolates both independent of viral tropism and of the cell type used to produce virus.
R5 tropic viruses are the dominant form found in sexually transmitted HIV-1 and, therefore, would have to be neutralized by microbicides. In addition, anti-HIV microbicides will need to inhibit infection by viral isolates from different subtypes. Although one would assume that all clades could be neutralized due to the conservation of gp120 glycosylation, a difference in the susceptibility of the viral subtypes B and C to the anti-HIV, high mannose-recognizing antibody 2G12 has been observed (46). To determine whether BanLec could inhibit additional primary isolates from different clades, we tested BanLec for inhibition of HIV-1 pseudotyped with envelopes derived from primary isolates of subtypes B and C. These subtypes are commonly found in North and Central America (subtype B) and parts of Africa and India (subtype C). We observed potent, subnanomolar inhibition of viral replication by BanLec (Fig. 2 and Table 1), and no significant difference was observed when the average IC50 values from the two different subtypes were compared by Student's t test (p < 0.56). This suggests that BanLec can effectively inhibit infection by viral isolates prominent in regions where a microbicide would be most valuable.
FIGURE 2.
BanLec inhibits infection of HIV-1 pseudotyped with envelopes from multiple primary isolates. TZM-bl cells were infected with HIV-1 pseudotyped with primary HIV-1 envelope proteins from subtype B (A) and subtype C (B) in the presence of different concentrations of BanLec. Forty-eight hours later, luciferase activity was assessed. The IC50 values were determined as in Fig. 1 and are shown in Table 1. Results shown are the average of three independent experiments, and error bars represent the S.D. RLU, relative luminescent units.
TABLE 1.
Summary of the calculated IC50 values for BanLec inhibition of HIV-1 pseudotyped with HIV-1 envelopes from subtypes B and C
| Subtype B |
Subtype C |
||||
|---|---|---|---|---|---|
| Envelope | IC50 | 95% Confidence intervals | Envelope | IC50 | 95% Confidence intervals |
| nm | nm | ||||
| SVPB5 | 0.30 | 0.27–0.34 | SVPC3 | 0.57 | 0.51–0.63 |
| SVPB6 | 0.85 | 0.74–0.98 | SVPC5 | 0.30 | 0.21–0.42 |
| SVPB11 | 0.71 | 0.65–0.77 | SVPC6 | 0.28 | 0.25–0.31 |
| SVPB17 | 0.33 | 0.26–0.41 | SVPC7 | 2.3 | 1.8–2.9 |
BanLec Blocks Infection of MDM
Macrophages are susceptible to HIV-1 infection and can become viral reservoirs that cannot be eliminated by highly active antiretroviral therapy. The role of vaginal macrophages in HIV-1 pathogenesis has not been fully characterized, but recent evidence indicates that these cells are permissive for HIV-1 infection (47). We tested the ability of BanLec to inhibit HIV-1 infection of MDM. As shown in Fig. 3A, nanomolar concentrations of BanLec inhibited HIV replication in MDM over a period of 15 days. Furthermore, BanLec had no effect on cellular viability as determined by MTT assay performed on day 15 (data not shown); therefore, this effect was not due to cellular toxicity. When BanLec remained in the culture supernatant for 7 days without changing the media or adding additional lectin, the IC50 value for BanLec inhibition of HIV-1 replication was 9.72 nm (Fig. 3B), demonstrating that BanLec remains a potent and stable inhibitor in a long term culture system at 37 °C.
FIGURE 3.
BanLec inhibits HIV-1 infection of MDM. A, MDM were pretreated with BanLec for 30 min before the addition of 100 TCID50 of HIV-1 NL(AD8). Twenty-four hours later, the media was removed, and the cells were washed with PBS to eliminate remaining virus. Fresh media containing BanLec or PBS was added to the cells. A sample of culture supernatant was taken every 3 days for p24 quantification by ELISA and replaced with new media containing lectin in PBS or PBS alone as a control. On day 15, viability was assessed by an MTT assay, which indicated no cellular toxicity (data not shown). B, MDM were pretreated and infected with HIV-1 as described above. 24 h post-infection the cells were washed with PBS to remove residual virus and cultured in media containing BanLec or PBS. Seven days post-infection, supernatants were removed for determination of p24 antigen as detected by ELISA. The concentration for a 50% reduction in p24 production was calculated to be 9.72 nm. Cellular viability was assessed by an MTT assay, and no toxicity was observed (data not shown). Results shown in panels A and B are representative of three and two separate experiments, respectively.
BanLec Appears to Block HIV-1 Infection at the Viral Entry Step
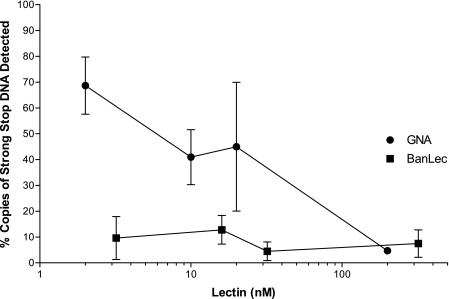
Having shown that BanLec can inhibit HIV replication in MDM, we tested the ability of BanLec to block cellular entry of HIV-1 in PBL. We hypothesized that BanLec binds to high mannose structures found on the HIV-1 envelope, preventing entry and, thus, infection. If so, little or none of the strong-stop DNA product of early HIV-1 reverse transcription (see “Materials and Methods”) should be detected when cells are exposed to HIV-1 in the presence of BanLec (44, 48). To test this hypothesis, we incubated PBL with the HIV-1 Bru isolate in the presence of different concentrations of BanLec. As a positive control and for comparison, a similar experiment with the lectin GNA was performed in parallel. Real-time PCR was used to detect strong-stop DNA, which is a reverse transcription product that can be detected early after viral entry before viral uncoating takes place (49–51). Strong-stop DNA that may have been present in the virus stock was removed by treatment with DNase I to eliminate false detection of reverse transcription products. Treatment with BanLec resulted in a marked decrease in strong-stop DNA at low lectin concentrations (Fig. 4) indicating that, in addition to inhibiting viral replication in MDM, BanLec blocks HIV-1 infection in PBL. Furthermore, this inhibition occurs at a step before early replication events, apparently at the level of viral entry. Although it appears unlikely that BanLec inhibits reverse transcription, these PCR results do not exclude this possibility, as similar results could be produced by an inhibitor of HIV-1 reverse transcriptase. This issue is addressed further below.
FIGURE 4.
BanLec inhibits production of early HIV-1 reverse transcription products in peripheral blood lymphocytes. Peripheral blood lymphocytes were treated with different lectin concentrations 30 min before infection with HIV-1 Bru. Three hours post-infection, cellular DNA of the infected cells was harvested, and strong-stop DNA was quantified by real-time PCR. The number of copies was normalized to a PBS-treated control (100%). The known anti-HIV lectin GNA (circles) was used as a positive control and to assess the relative molar potency of BanLec (squares).
BanLec Binds to Glycosylated gp120
BanLec is known to bind to mannose, and thus, we hypothesized that BanLec binds the high mannose structures found on the glycosylated gp120 envelope protein and blocks entry of HIV-1 into cells. We prepared a BanLec-based ELISA to measure binding of glycosylated HIV-1 gp120 to BanLec. We observed that BanLec does indeed bind to gp120 in a concentration-dependent manner (Fig. 5A). Furthermore, a known BanLec ligand, methyl-α-d-mannopyranoside, inhibited such binding in a concentration-dependent manner (Fig. 5B). Not surprisingly, a high concentration of methyl-α-d-mannopyranoside ligand was needed to compete for binding to gp120 because of the high density of carbohydrate residues on the HIV-1 envelope protein. These results corroborate our hypothesis that BanLec inhibits HIV-1 cellular entry by binding to high mannose structures found on the virus.
FIGURE 5.
BanLec binds to glycosylated gp120. A, shown is dose-dependent binding of BanLec to glycosylated gp120. BanLec was used to coat a 96-well ELISA plate. Serial dilutions of gp120 were added in duplicate to the wells. gp120 was detected with an anti-gp120 antibody. Results are representative of four independent experiments. B, methyl α-d-mannopyranoside inhibits interaction of BanLec with gp120. ELISA plates were coated with BanLec as in panel A. Serial dilutions of methyl α-d-mannopyranoside were added to wells along with a constant amount of gp120. The amount of gp120 bound was determined using the standard curve produced in panel A. Abs, absorbance.
To explore the BanLec binding sites on gp120, we determined the ability of BanLec to block binding by the monoclonal antibody 2G12 using the ELISA-based assay. 2G12 recognizes a cluster of N-linked glycosylation structures at positions Asn-295, -332, and -392 (position numbering is of the HXB2 reference sequence) that are crucial for antibody recognition (52–54). We found that pretreatment of gp120 with BanLec inhibited recognition by 2G12 in a dose-dependent manner, suggesting that BanLec is capable of binding to this antibody's epitope consisting of high mannose structures (Fig. 6).
FIGURE 6.
Binding of gp120 by BanLec blocks access to the anti-HIV monoclonal antibody 2G12. Recombinant gp120-coated ELISA plates were treated with different concentrations of BanLec before incubation with the 2G12 antibody, which recognizes the high mannose structures found at positions Asn-295, -332, and -392 of the gp120 protein. Unbound antibody was removed, and the amount of antibody remaining was determined by comparison to a standard curve. The results shown represent the average from three separate experiments. Error bars represent S.E. The effect of increasing amounts of BanLec was determined to be significant by 1-way analysis of variance testing (p < 0.01).
BanLec Inhibits HIV-1 Infection before Viral Fusion
To further investigate at which point in the viral life cycle BanLec inhibits HIV-1 infection, we tested BanLec for its ability to inhibit HIV-1 infection post-attachment. To do so, we tested if BanLec could inhibit HIV-1 that was already bound to the cell but could not complete fusion due to temperature restriction (55). As controls, we took advantage of the fact that CD4-IgG2 inhibits HIV-1 infection by blocking attachment, whereas T-20 works by blocking fusion. As anticipated, we observed a large decrease in the inhibitory activity of the HIV-1 attachment inhibitor CD4-IgG2 in the post-attachment assay, whereas the bound virus was still essentially completely susceptible to the fusion inhibitor T-20 (Fig. 7 and Table 2). This demonstrates that the assay works as expected, with viral attachment, but not fusion, taking place at 16 °C. Both the CCR5 binding inhibitor maraviroc and BanLec primarily blocked viral replication by inhibiting HIV-1 attachment, but each does appear to also have a modest effect on viral fusion (Fig. 7). Importantly, when we compare the IC50 value of BanLec to those of other anti-HIV compounds, we see that BanLec potency compares quite well to the clinically approved anti-virals T-20 and maraviroc (Table 2). Thus, we conclude that BanLec potently inhibits the attachment of HIV-1 to cells and has a more modest effect on viral fusion.
FIGURE 7.
BanLec primarily inhibits binding of HIV to the cellular membrane. TZM-bl cells were spin-infected at 16 °C, a temperature that allows for attachment of virus but does not allow fusion events to occur. The unbound virus was removed, and the cells were incubated with media containing inhibitors (CD4-IgG2, T-20, maraviroc, or BanLec) on ice for 30 min, and then the plates were shifted to 37 °C to allow for fusion and infection to be completed (○). The results were compared with a standard infection procedure (pre-attachment) in which the virus and inhibitors were incubated together on ice for 30 min and then added to TZM-bl cells and incubated at 37 °C (●). The results shown are the averages from three separate experiments. Nonlinear regression analysis was used for curve fitting and calculation of IC50 values (Table 2). RLU, relative luminescent units.
TABLE 2.
Summary of the calculated IC50 values for inhibition of HIV-1 infection with the addition of drug either pre- or post-attachment to TZM-bl cells
| Compound | Post-attachment |
Pre-attachment |
||
|---|---|---|---|---|
| IC50 | 95% Confidence intervals | IC50 | 95% Confidence intervals | |
| nm | nm | |||
| CD4-IgG2 | –a | 1.39 | 0.667–2.89 | |
| T-20 | 2.78 | 1.96–3.95 | 1.90 | 0.958–3.77 |
| Maraviroc | 10.2 | 7.5–13.8 | 1.23 | 0.584–2.58 |
| BanLec | 13.1 | 9.61–17.7 | 0.224 | 0.168–0.300 |
a Too high to calculate.
The Anti-HIV Activity of BanLec Compares Well to Other Anti-HIV Lectins
Several different lectins have been found to inhibit HIV-1 infection. However, they vary in their degree of anti-viral activity. To assess the relative molar-based potency of BanLec, we compared the anti-HIV activity of BanLec to that of two previously described anti-HIV lectins, GNA and GRFT. Upon comparison, all three lectins showed activity in the nanomolar range (Fig. 8). Taken together, our data suggest that BanLec inhibits infection with a broad range of HIV-1 isolates by blocking viral entry, compares favorably with the potency of previously described lectins and clinically available anti-HIV drugs, and is a potential component for future anti-HIV vaginal microbicides.
FIGURE 8.
Comparison of the anti-HIV activity of BanLec to the anti-HIV lectins GNA and GRFT. TZM-bl cells were pretreated with BanLec, GRFT, or GNA diluted in PBS or PBS alone, as a control, for 30 min before infection by the R5 tropic HIV-1 virus 81-A. Forty-eight hours later, luciferase activity was measured. The results are normalized to infected cells treated with PBS alone. The average of three separate experiments is shown and was used to calculate IC50 values by nonlinear regression. The calculated IC50 values are the following: GNA = 34.3 nm, BanLec 3.18 nm, and GRFT 0.42 nm. RLU, relative luminescent units.
DISCUSSION
The primary mechanism of inhibition by BanLec appears to be blocking cellular attachment of HIV and, thus, viral entry. Our conclusion is based on the findings from our ELISA assays that BanLec can bind to high mannose structures found on HIV-1 gp120, including the high mannose structures that are recognized by the monoclonal antibody 2G12. This was corroborated by the finding that cells treated with BanLec had decreased amounts of an early HIV reverse transcription product, strong-stop DNA, that can be detected shortly after cellular entry of the virus and before viral uncoating. In addition, we performed an assay that took advantage of a temperature-arrested state (16 °C) that prevents HIV-1 fusion and compared the inhibitory activity of BanLec and other anti-HIV drugs pre- and post-cellular attachment of the virus, finding that most of the inhibitory activity of BanLec comes from blocking viral attachment. Interestingly, whereas most of the inhibitory activity of the CCR5 blocker maraviroc, used as a control in these experiments, was due to blocking viral attachment, when maraviroc was added post-attachment we still observed inhibition of HIV, albeit at a reduced level. A similar result was also seen with BanLec, suggesting that these two compounds could have additional inhibitory activity at a post-attachment step, such as fusion of the virus to the cell.
Our studies indicate that BanLec is a new and promising member of the group of lectins that are able to inhibit HIV-1 infection through interactions with glycosylation sites found on the viral envelope. The inhibitory activity of BanLec against HIV-1 was broad, independent of tropism, and effective against several subtype B and C envelope sequences. HIV-1 pseudotyped with envelopes derived from primary isolates was inhibited by BanLec in the low nanomolar range. BanLec was also able to inhibit HIV-1 infection of primary cells, and thus, our results are not limited to cell lines.
Based on our findings, it is likely that BanLec will be able to inhibit other HIV-1 subtypes, as they all contain glycosylation sites in their envelope sequences. The isolates used in our experiments differed in the number of predicted N-linked glycosylation sites, supporting the likelihood that BanLec will be effective against most HIV subtypes found in both the developing and developed world. Because glycosylation is not specific to HIV-1, lectins have the potential to inhibit the replication of a broad spectrum of viruses. Indeed, it has been shown that lectins can inhibit other enveloped viruses including Ebola (56, 57), Marburg (57), influenza (58), severe acute respiratory syndrome coronavirus (59), and hepatitis C virus (60, 61).
One potential benefit of the use of lectins as anti-HIV agents is their ability to target multiple different glycosylation sites on the virus, thus making it more difficult for resistance to develop. In support of this prediction, previous studies that determined the resistance profiles of HIV-1 treated with lectin showed that multiple mutations in the envelope sequence were needed for the development of resistance (62). Furthermore, different mutations in N-linked glycosylation sites are required for the development of resistance to different lectins. This suggests that the combinatorial or simultaneous use of multiple lectins can reduce the likelihood of failure of a lectin-based anti-viral therapy due to resistance. If a population of virus develops resistance to BanLec or other anti-HIV lectins, one interesting possible consequence is that the virus will then be more susceptible to neutralization by the human immune response, as the carbohydrate structures found on the HIV-1 envelope are thought to act as a shield against neutralizing antibody responses (63). This glycan shield works by blocking access of epitopes to potentially neutralizing antibodies. Previously published data demonstrate that alterations in glycosylation that result in resistance to lectins can make the virus vulnerable to neutralizing antibody responses (18, 19).
Although several anti-HIV lectins have been described, it is highly unlikely that a majority of them can be developed for therapeutic use. Like all potential drugs, lectins can vary in their degrees of potency and toxicity (64, 65). Also, it has been shown that two anti-HIV lectins can significantly differ in their ability to block attachment of HIV to epithelial cells (66). Concerns have been raised about the potential toxicity of lectins, for example CV-N. This lectin has shown success as a microbicide in in vivo macaque vaginal and rectal transmission models (67, 68), but safety concerns exist. CV-N was found to have mitogenic activity when PBMC cultures were exposed to the lectin for 3 days (65, 69). However, recombinant therapeutic proteins can be attached to polyethylene glycol (PEG) polymer chains to change bioavailability and reduce toxicity. This modification of CV-N has been shown to be effective in reducing mitogenic activity in vitro (70). Although BanLec has also been reported to possess mitogenic activity (71), the relationship between mitogenic activity in vitro and microbicide efficacy has not been elucidated, so it remains possible that recombinant versions of BanLec and other lectins could be developed that retain efficacy but have minimal mitogenic activity. The anti-HIV lectin GRFT has recently been reported not to have a mitogenic effect when added to human PBMCs (72). This observation is of interest, as GRFT is in the same jacalin-related lectin family and has a similar structure to BanLec (73). GRFT has also been shown to be non-inflammatory, non-toxic, and capable of being manufactured on a large scale. Although clinical testing of these newer lectins has yet to be performed, it appears that lectins have potential to be used as anti-HIV agents (72). Because the binding, toxicity, and anti-HIV activity of lectins vary, the identification of novel anti-viral lectins, such as BanLec, will further increase the possibility of successful development of a lectin-based anti-HIV microbicide.
Acknowledgments
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health: TZM-bl cells from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc. (catalog #8129), MAGI-CCR5 cells from Dr. Julie Overbaugh (catalog #3522), Maraviroc (catalog #11580), T-20 fusion inhibitor from Roche Applied Science (catalog #9845), CD4-IgG2 from Progenics Pharmaceuticals (catalog #11780), HIS-Griffithsin from Drs. Barry O'Keefe and James McMahon (catalog #11610), pSG3Δenv from Drs. John C. Kappes and Xiaoyun Wu (catalog #11051), p81A-4 from Dr. Bruce Chesebro (catalog #11440), pAD8(NL4-3) from Dr. Eric O. Freed (catalog #11346), p89.6 from Ronald G. Collman (catalog #3552), pNL4-3 from Dr. Malcolm Martin (catalog #114), standard reference panel of Subtype B HIV-1 Env Clones (catalog #11227), standard reference panel of subtype C HIV-1 Env Clones (catalog #11326), pConC gp160-opt from Dr. Beatrice Hahn (catalog #11407), HIV-1BaL gp120 from Division of AIDS Acquired Immunodeficiency Syndrome, NIAID (catalog #4961), HIV-1 gp120 monoclonal antibody (2G12) from Dr. Hermann Katinger (catalog #1476).
This work was supported, in whole or in part, by National Institutes of Health Grant R01 AI062248 (of D. M. M.). This work was also supported by a Burroughs Wellcome Fund Clinical Scientist Award in Translational Research.
H. C. Winter, K. A. Wearne, and I. J. Goldstein, manuscript in preparation.
- HIV
- human immunodeficiency virus
- ELISA
- enzyme-linked immunosorbent assay
- GRFT
- Griffithsin
- HEK-293 cells
- human embryonic kidney cells
- PBL
- peripheral blood lymphocytes
- PBMC
- peripheral blood mononuclear cell
- MDM
- monocyte-derived macrophages
- MTT
- (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PBS
- phosphate-buffered saline
- TCID50
- tissue culture infective dose 50%.
REFERENCES
- 1.Executive Summary United Nations Programme on HIV/AIDS (UNAIDS) and World Health Organization (WHO) (2008) Report on the Global AIDS Epidemic [Google Scholar]
- 2.Underhill K., Montgomery P., Operario D. (2007) BMJ 335, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Randolph M. E., Pinkerton S. D., Bogart L. M., Cecil H., Abramson P. R. (2007) Arch. Sex. Behav. 36, 844–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crosby R., Yarber W. L., Sanders S. A., Graham C. A. (2005) J. Am. Coll. Health 54, 143–147 [DOI] [PubMed] [Google Scholar]
- 5.Gupta G. R. (2002) BMJ 324, 183–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watts C. (2002) Microbicides, Antwerp, Belgium, May 12–15, 2002 [Google Scholar]
- 7.Balzarini J., Van Damme L. (2007) Lancet 369, 787–797 [DOI] [PubMed] [Google Scholar]
- 8.Balzarini J. (2005) Lancet Infect. Dis. 5, 726–731 [DOI] [PubMed] [Google Scholar]
- 9.Boyd M. R., Gustafson K. R., McMahon J. B., Shoemaker R. H., O'Keefe B. R., Mori T., Gulakowski R. J., Wu L., Rivera M. I., Laurencot C. M., Currens M. J., Cardellina J. H., 2nd, Buckheit R. W., Jr., Nara P. L., Pannell L. K., Sowder R. C., 2nd, Henderson L. E. (1997) Antimicrob. Agents Chemother. 41, 1521–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mori T., O'Keefe B. R., Sowder R. C., 2nd, Bringans S., Gardella R., Berg S., Cochran P., Turpin J. A., Buckheit R. W., Jr., McMahon J. B., Boyd M. R. (2005) J. Biol. Chem. 280, 9345–9353 [DOI] [PubMed] [Google Scholar]
- 11.Balzarini J. (2007) Nat. Rev. Microbiol. 5, 583–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balzarini J., Schols D., Neyts J., Van Damme E., Peumans W., De Clercq E. (1991) Antimicrob. Agents Chemother. 35, 410–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balzarini J., Hatse S., Vermeire K., Princen K., Aquaro S., Perno C. F., De Clercq E., Egberink H., Vanden Mooter G., Peumans W., Van Damme E., Schols D. (2004) Antimicrob. Agents Chemother. 48, 3858–3870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matthews T. J., Weinhold K. J., Lyerly H. K., Langlois A. J., Wigzell H., Bolognesi D. P. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5424–5428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geyer H., Holschbach C., Hunsmann G., Schneider J. (1988) J. Biol. Chem. 263, 11760–11767 [PubMed] [Google Scholar]
- 16.Allan J. S., Coligan J. E., Barin F., McLane M. F., Sodroski J. G., Rosen C. A., Haseltine W. A., Lee T. H., Essex M. (1985) Science 228, 1091–1094 [DOI] [PubMed] [Google Scholar]
- 17.Li Y., Luo L., Rasool N., Kang C. Y. (1993) J. Virol. 67, 584–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clevestig P., Pramanik L., Leitner T., Ehrnst A. (2006) J. Gen. Virol 87, 607–612 [DOI] [PubMed] [Google Scholar]
- 19.Geijtenbeek T. B., Kwon D. S., Torensma R., van Vliet S. J., van Duijnhoven G. C., Middel J., Cornelissen I. L., Nottet H. S., KewalRamani V. N., Littman D. R., Figdor C. G., van Kooyk Y. (2000) Cell 100, 587–597 [DOI] [PubMed] [Google Scholar]
- 20.Back N. K., Smit L., De Jong J. J., Keulen W., Schutten M., Goudsmit J., Tersmette M. (1994) Virology 199, 431–438 [DOI] [PubMed] [Google Scholar]
- 21.Peumans W. J., Zhang W., Barre A., Houlès, Astoul C., Balint-Kurti P. J., Rovira P., Rougé P., May G. D., Van Leuven F., Truffa-Bachi P., Van Damme E. J. (2000) Planta 211, 546–554 [DOI] [PubMed] [Google Scholar]
- 22.Koshte V. L., van Dijk W., van der Stelt M. E., Aalberse R. C. (1990) Biochem. J. 272, 721–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura-Tsuruta S., Uchiyama N., Peumans W. J., Van Damme E. J., Totani K., Ito Y., Hirabayashi J. (2008) FEBS J. 275, 1227–1239 [DOI] [PubMed] [Google Scholar]
- 24.Mo H., Winter H. C., Van Damme E. J., Peumans W. J., Misaki A., Goldstein I. J. (2001) Eur. J. Biochem. 268, 2609–2615 [DOI] [PubMed] [Google Scholar]
- 25.Meagher J. L., Winter H. C., Ezell P., Goldstein I. J., Stuckey J. A. (2005) Glycobiology 15, 1033–1042 [DOI] [PubMed] [Google Scholar]
- 26.Singh D. D., Saikrishnan K., Kumar P., Surolia A., Sekar K., Vijayan M. (2005) Glycobiology 15, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 27.Jeyaprakash A. A., Srivastav A., Surolia A., Vijayan M. (2004) J. Mol. Biol. 338, 757–770 [DOI] [PubMed] [Google Scholar]
- 28.Van Damme E. J. M., Allen A. K., Peumans W. J. (1987) FEBS Lett. 215, 140–144 [Google Scholar]
- 29.Gartner S., Markovits P., Markovitz D. M., Kaplan M. H., Gallo R. C., Popovic M. (1986) Science 233, 215–219 [DOI] [PubMed] [Google Scholar]
- 30.Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C. (2005) J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. (2002) Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adachi A., Gendelman H. E., Koenig S., Folks T., Willey R., Rabson A., Martin M. A. (1986) J. Virol. 59, 284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Freed E. O., Englund G., Martin M. A. (1995) J. Virol. 69, 3949–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toohey K., Wehrly K., Nishio J., Perryman S., Chesebro B. (1995) Virology 213, 70–79 [DOI] [PubMed] [Google Scholar]
- 35.Chesebro B., Nishio J., Perryman S., Cann A., O'Brien W., Chen I. S., Wehrly K. (1991) J. Virol. 65, 5782–5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chesebro B., Wehrly K., Nishio J., Perryman S. (1992) J. Virol. 66, 6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Collman R., Balliet J. W., Gregory S. A., Friedman H., Kolson D. L., Nathanson N., Srinivasan A. (1992) J. Virol. 66, 7517–7521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derdeyn C. A., Decker J. M., Sfakianos J. N., Wu X., O'Brien W. A., Ratner L., Kappes J. C., Shaw G. M., Hunter E. (2000) J. Virol. 74, 8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. (1998) J. Virol. 72, 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeuchi Y., McClure M. O., Pizzato M. (2008) J. Virol. 82, 12585–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chesebro B., Wehrly K., Metcalf J., Griffin D. E. (1991) J. Infect. Dis. 163, 64–70 [DOI] [PubMed] [Google Scholar]
- 42.Chackerian B., Long E. M., Luciw P. A., Overbaugh J. (1997) J. Virol. 71, 3932–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Doherty U., Swiggard W. J., Malim M. H. (2000) J. Virol. 74, 10074–10080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidtmayerova H., Alfano M., Nuovo G., Bukrinsky M. (1998) J. Virol. 72, 4633–4642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kothe D. L., Li Y., Decker J. M., Bibollet-Ruche F., Zammit K. P., Salazar M. G., Chen Y., Weng Z., Weaver E. A., Gao F., Haynes B. F., Shaw G. M., Korber B. T., Hahn B. H. (2006) Virology 352, 438–449 [DOI] [PubMed] [Google Scholar]
- 46.Gray E. S., Moore P. L., Pantophlet R. A., Morris L. (2007) J. Virol. 81, 10769–10776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen R., Richter H. E., Clements R. H., Novak L., Huff K., Bimczok D., Sankaran-Walters S., Dandekar S., Clapham P. R., Smythies L. E., Smith P. D. (2009) J. Virol. 83, 3258–3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B. Q., Fu T., Dongyan Y., Mikovits J. A., Ruscetti F. W., Wang J. M. (2000) Biochem. Biophys. Res. Commun. 276, 534–538 [DOI] [PubMed] [Google Scholar]
- 49.Warrilow D., Stenzel D., Harrich D. (2007) Retrovirology 4, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Warrilow D., Meredith L., Davis A., Burrell C., Li P., Harrich D. (2008) J. Virol. 82, 1425–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arhel N. J., Souquere-Besse S., Munier S., Souque P., Guadagnini S., Rutherford S., Prévost M. C., Allen T. D., Charneau P. (2007) EMBO J. 26, 3025–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trkola A., Purtscher M., Muster T., Ballaun C., Buchacher A., Sullivan N., Srinivasan K., Sodroski J., Moore J. P., Katinger H. (1996) J. Virol. 70, 1100–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scanlan C. N., Pantophlet R., Wormald M. R., Ollmann Saphire E., Stanfield R., Wilson I. A., Katinger H., Dwek R. A., Rudd P. M., Burton D. R. (2002) J. Virol. 76, 7306–7321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanders R. W., Venturi M., Schiffner L., Kalyanaraman R., Katinger H., Lloyd K. O., Kwong P. D., Moore J. P. (2002) J. Virol. 76, 7293–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mkrtchyan S. R., Markosyan R. M., Eadon M. T., Moore J. P., Melikyan G. B., Cohen F. S. (2005) J. Virol. 79, 11161–11169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrientos L. G., O'Keefe B. R., Bray M., Sanchez A., Gronenborn A. M., Boyd M. R. (2003) Antiviral. Res. 58, 47–56 [DOI] [PubMed] [Google Scholar]
- 57.Barrientos L. G., Lasala F., Otero J. R., Sanchez A., Delgado R. (2004) J. Infect. Dis. 189, 1440–1443 [DOI] [PubMed] [Google Scholar]
- 58.Smee D. F., Bailey K. W., Wong M. H., O'Keefe B. R., Gustafson K. R., Mishin V. P., Gubareva L. V. (2008) Antiviral Res. 80, 266–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keyaerts E., Vijgen L., Pannecouque C., Van Damme E., Peumans W., Egberink H., Balzarini J., Van Ranst M. (2007) Antiviral Res. 75, 179–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Helle F., Wychowski C., Vu-Dac N., Gustafson K. R., Voisset C., Dubuisson J. (2006) J. Biol. Chem. 281, 25177–25183 [DOI] [PubMed] [Google Scholar]
- 61.Bertaux C., Daelemans D., Meertens L., Cormier E. G., Reinus J. F., Peumans W. J., Van Damme E. J., Igarashi Y., Oki T., Schols D., Dragic T., Balzarini J. (2007) Virology 366, 40–50 [DOI] [PubMed] [Google Scholar]
- 62.Witvrouw M., Fikkert V., Hantson A., Pannecouque C., O'keefe B. R., McMahon J., Stamatatos L., de Clercq E., Bolmstedt A. (2005) J. Virol. 79, 7777–7784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wei X., Decker J. M., Wang S., Hui H., Kappes J. C., Wu X., Salazar-Gonzalez J. F., Salazar M. G., Kilby J. M., Saag M. S., Komarova N. L., Nowak M. A., Hahn B. H., Kwong P. D., Shaw G. M. (2003) Nature 422, 307–312 [DOI] [PubMed] [Google Scholar]
- 64.Lis H., Sharon N. (2003) Lectins, 2nd Ed., Kluwer Academic Publishers Group, Dordrecht, Netherlands [Google Scholar]
- 65.Huskens D., Vermeire K., Vandemeulebroucke E., Balzarini J., Schols D. (2008) Int. J. Biochem. Cell Biol. 40, 2802–2814 [DOI] [PubMed] [Google Scholar]
- 66.Saïdi H., Nasreddine N., Jenabian M. A., Lecerf M., Schols D., Krief C., Balzarini J., Bélec L. (2007) J. Transl. Med. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai C. C., Emau P., Jiang Y., Tian B., Morton W. R., Gustafson K. R., Boyd M. R. (2003) AIDS Res. Hum. Retroviruses 19, 535–541 [DOI] [PubMed] [Google Scholar]
- 68.Tsai C. C., Emau P., Jiang Y., Agy M. B., Shattock R. J., Schmidt A., Morton W. R., Gustafson K. R., Boyd M. R. (2004) AIDS Res. Hum. Retroviruses 20, 11–18 [DOI] [PubMed] [Google Scholar]
- 69.Buffa V., Stieh D., Mamhood N., Hu Q., Fletcher P., Shattock R. J. (2009) J. Gen. Virol. 90, 234–243 [DOI] [PubMed] [Google Scholar]
- 70.Zappe H., Snell M. E., Bossard M. J. (2008) Adv. Drug Deliv. Rev. 60, 79–87 [DOI] [PubMed] [Google Scholar]
- 71.Gavrovic-Jankulovic M., Poulsen K., Brckalo T., Bobic S., Lindner B., Petersen A. (2008) Int. J. Biochem. Cell Biol. 40, 929–941 [DOI] [PubMed] [Google Scholar]
- 72.O'Keefe B. R., Vojdani F., Buffa V., Shattock R. J., Montefiori D. C., Bakke J., Mirsalis J., d'Andrea A. L., Hume S. D., Bratcher B., Saucedo C. J., McMahon J. B., Pogue G. P., Palmer K. E. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6099–6104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ziółkowska N. E., O'Keefe B. R., Mori T., Zhu C., Giomarelli B., Vojdani F., Palmer K. E., McMahon J. B., Wlodawer A. (2006) Structure 14, 1127–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]