Abstract
The family of low density lipoprotein (LDL) receptors mediate uptake of a plethora of ligands from the circulation and couple this to signaling, thereby performing a crucial role in physiological processes including embryonic development, cancer development, homeostasis of lipoproteins, viral infection, and neuronal plasticity. Structural integrity of individual ectodomain modules in these receptors depends on calcium, and we showed before that the LDL receptor folds its modules late after synthesis via intermediates with abundant non-native disulfide bonds and structure. Using a radioactive pulse-chase approach, we here show that for proper LDL receptor folding, calcium had to be present from the very early start of folding, which suggests at least some native, essential coordination of calcium ions at the still largely non-native folding phase. As long as the protein was in the endoplasmic reticulum (ER), its folding was reversible, which changed only upon both proper incorporation of calcium and exit from the ER. Coevolution of protein folding with the high calcium concentration in the ER may be the basis for the need for this cation throughout the folding process even though calcium is only stably integrated in native repeats at a later stage.
Keywords: Calcium, Chaperone Chaperonin, Disulfide, Endoplasmic Reticulum (ER), Protein Folding, EGF-like Repeats, LDL Receptor, LDL-A, Disulfide Bond Formation
Introduction
The low density lipoprotein (LDL)5 receptor family is involved in essential physiological processes, including lipoprotein metabolism, embryonic development, cell migration, neuronal plasticity, and homeostasis of various (effector) molecules (1). They were originally found to function as receptors, regulating the concentration of their ligands by binding and endocytosis (2), but they also play an increasingly recognized role in signal transduction, coupling cargo transport to signaling (3, 4). The LDL receptor family has a modular structure, containing an assortment of LDL-A repeats, epidermal growth factor (EGF)-like repeats with β-propeller, a single transmembrane domain, and a small cytosolic tail (5–7).
The LDL receptor mediates uptake of LDL from the circulation via receptor-mediated endocytosis, and mutations cause familial hypercholesterolemia, characterized by elevated levels of plasma LDL that eventually will lead to premature cardiovascular disease (8).
The LDL-A repeat is not limited to the LDL receptor family. Over 6,800 sequences and >500 related protein architectures have been found as of January 2010, and they often function in ligand binding and release. The receptor for Rous sarcoma virus for instance contains an LDL-A repeat (9), as do complement components and various proteases. The LDL receptor binds apoB, apoE, and RAP via its seven LDL-A repeats (LR1 through LR7) (10–12). The structural motif of the LDL-A repeat is a complement-like ligand-binding module stabilized by three disulfide bonds (13) that forms a β-hairpin structure followed by a series of β-turns (6). Structural integrity is imparted by calcium binding to the more C-terminal of two loops (14).
The EGF repeats form a two-stranded β-sheet followed by a loop to a C-terminal short two-stranded sheet, with three conserved disulfide bonds in a different arrangement than the LDL-A repeats (5, 6). They can also bind calcium but may not need it for structural integrity (15). In the LDL receptor two of the three EGF repeats bind calcium, with one of the calcium ions noncanonically bound between the repeats (15, 16). The EGF region in the LDL receptor is important for acid-triggered ligand release, which is coupled to a decrease in Ca2+ affinity (6).
Folding of the LDL receptor and its individual domains, as well as their calcium dependence, has been the subject of many studies (17). Although all LDL-A sequences share >40% identity, calcium-binding sites and disulfide bonds are completely conserved, and although structures (as far as determined) are similar, reports on the role of calcium in folding and function show little consensus. Most studies involve in vitro refolding of individual LDL-A repeats of the LDL receptor family and show different characteristics of calcium binding, ranging from the absolute need of calcium for proper refolding (9) to a need only for ligand binding (18). A study in intact cells with the LDL receptor-related protein (LRP) minireceptor (containing eleven LDL-A repeats and nine EGF repeats) showed that LRP needs calcium to leave the endoplasmic reticulum (ER) (19), indicating that calcium is required for proper folding.
The ER not only is the compartment where secretory proteins and cell surface proteins start their life and where they fold (20), but it also is the major calcium storage compartment of a eukaryotic cell. Determination of the precise concentration of Ca2+ inside the ER has proven to be a difficult task. The reported values for its total Ca2+ concentration vary from as low as 5 μm (21) and 200–500 μm (22, 23) to even 2 mm (24). These differences can be explained by the techniques used for measuring Ca2+ concentrations (25). Many ER-resident proteins involved in protein folding bind calcium through high affinity sites, often with additional low affinity binding sites (26).
The need of the LDL receptor family for calcium for its function is clear from previous studies (14, 27), but detailed insight in calcium incorporation during folding and its requirement for structural integrity in vivo is lacking. We have set up an in vivo folding assay for the LDL receptor (28) in which we addressed these issues.
We showed before that in intact cells the individual modules of the LDL receptor do not fold independently but first collapse into folding intermediates characterized by long distance non-native disulfide bonding and absence of native structure (28). This is indicative of cooperative folding of the repeats. Our findings would predict calcium to be needed only late in folding, after the non-native phase, and long after protein synthesis. We therefore set out to revisit the role of calcium for the folding and structure of the full-length LDL receptor in vivo and to determine the timing of calcium incorporation during folding, its reversibility, and its importance for structural integrity in the intact cell.
We found that the LDL receptor indeed did not adopt its proper conformation without calcium. As long as the protein was in the ER, it misfolded without calcium, but it also lost already native structure when calcium was removed from folded LDL receptors. Resistance to misfolding induced by calcium depletion was only acquired in the Golgi complex and beyond. The LDL receptor did not show reversibility of misfolding; once misfolded because of a lack of calcium, even at very early folding stages, rescue was not possible anymore. We concluded that the LDL receptor required calcium throughout its folding process, even though proper incorporation and formation of native epitopes occur much later.
EXPERIMENTAL PROCEDURES
Cells and Antibodies
The human cervical carcinoma cell line HeLa was cultured in minimal essential medium supplemented with nonessential amino acids, 2 mm Glutamax, 100 units/ml penicillin, 100 μg/ml streptomycin, and 10% fetal calf serum. CHO15B cells were cultured in α-minimal essential medium with 2 mm Glutamax, 100 units/ml penicillin, 100 μg/ml streptomycin, and 8% fetal calf serum. The cells were maintained at 37 °C in humidified air containing 5% CO2. Monoclonal antibodies C7 (29), 6B2, 7H2, 5G2, and 6E2 (30), directed against LDL-A repeats 1 (twice), 3, 5, and 7, respectively (29, 30), and the polyclonal antisera raised against the LDL receptor (121) (28) or influenza virus proteins including the hemagglutinin (31) have been described before.
DNA Constructs
The LDL receptor cDNA sequence expressed from pBluescript (pBS-LDLR) or pcDNA3 (pcDNA3- LDLR) was derived from a pGEM LDL receptor (pGEM-LDLR) construct kindly provided by M. M. Jorgensen. pcDNA3- LDLR-P678L was derived from pBS-LDLR-P678L, which was a kind gift from Drs S. Fouchier and J. Defesche (Academic Medical Center, Amsterdam, The Netherlands).
Transfection and Infection
HeLa cells at 30–40% confluence were transfected with pcDNA3-LDLR using polyethylene-imine (32). Transfections were performed in 6-cm dishes with 4 μg of DNA in a total volume of 2.5 ml of medium containing 5% fetal calf serum. The transfection medium was replaced after 4 h with complete medium for an additional 20 h before pulse-chase experiments. CHO15B cells were infected with X31 influenza virus as described before (33), 5 h before pulse-chase experiments.
Pulse-Chase Analysis
Pulse-chase-based folding assays were performed as described before (28, 34). To deplete calcium from the ER, 5 μm of the calcium ionophore A23187 (ICN) (35) was added to calcium-free starvation, pulse, and chase media, or 100 nm of the Ca2+-ATPase inhibitor thapsigargin (ICN) (36) was added to calcium-containing starvation, pulse, and chase media. For the experiment in Fig. 2a, 200 nm thapsigargin was used. The cells were pulse-labeled for 5 min (for LDL receptor experiments) or 2 min (for influenza virus hemagglutinin (HA) studies) with 125 μCi/ml of Redivue Promix 35S cell labeling mix (Amersham Biosciences). After various chase times, the cells were treated with 20 mm N-ethylmaleimide to block free sulfhydryl groups and prevent any further disulfide bond formation. LDL receptor expressing cells were lysed in 1% Triton X-100, 10 mm HEPES, pH 7.4, 200 mm NaCl, 2 mm CaCl2, 2.5 mm MgCl2, and 2.2% DMSO, whereas HA-expressing cells were lysed in 0.5% Triton X-100 in 100 mm NaCl, 20 mm MES, 30 mm Tris-HCl, pH 7.5. All of the lysis buffers contained 20 mm N-ethylmaleimide, 10 μg/ml each of chymostatin, leupeptin, antipain, and pepstatin, and 1 mm phenylmethylsulfonyl fluoride. For calcium-depleted samples, the lysis buffer did not contain calcium. After pelleting and removing the nuclei, the cell lysates were used for immunoprecipitation as described (28, 34).
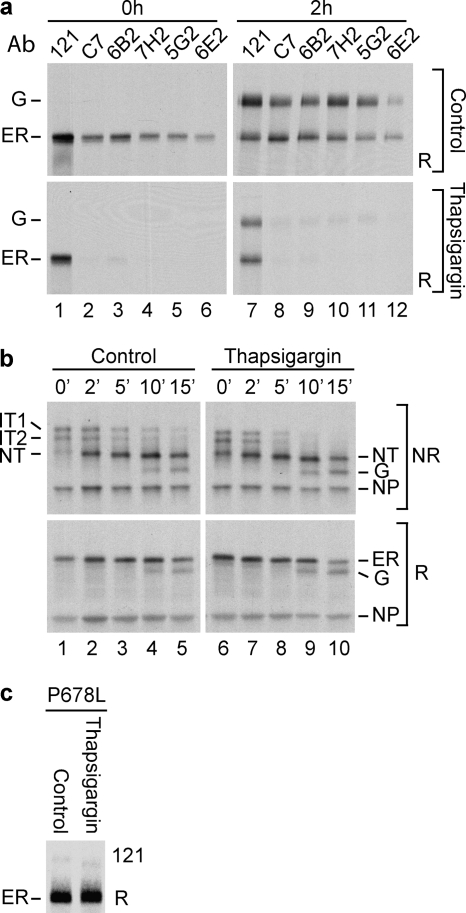
FIGURE 2.
Specificity of ER calcium depletion on LDL receptor folding. a, HeLa cells overexpressing wild-type LDL receptor were pulse-labeled for 4 min and chased for 0 or 2 h in the presence of thapsigargin or DMSO (Control). The LDL receptor was precipitated in parallel with polyclonal antiserum 121 and monoclonal antibodies C7, 6B2, 7H2, 5G2, and 6E2, against properly folded and disulfide-bonded ligand-binding repeats LR1, LR1, LR3, LR5, and LR7, respectively). The samples were analyzed by reducing SDS-PAGE. Ab, antibody. b, influenza virus infected CHO15B cells, expressing as control HA, were pulse-labeled for 2 min and chased for the indicated periods in the presence of DMSO (Control) or thapsigargin. The samples were immunoprecipitated with a polyclonal antibody that recognizes all conformations of HA and were subjected to nonreducing (NR) and reducing (R) SDS-PAGE. IT1 and IT2 are the two HA folding intermediates with incomplete sets of disulfide bonds that fold into NT, which is the native monomer with native epitopes and its six native disulfide bonds. ER represents the ER forms of HA. Upon transport to the Golgi complex, the N-linked glycans of HA are trimmed substantially in these cells, which increases electrophoretic mobility. G represents HA molecules that have reached the Golgi complex and beyond. NP is the viral nucleoprotein, which functions as marker and loading control. c, HeLa cells overexpressing the LDL receptor P678L mutant were pulse-labeled for 5 min and chased for 2 h in the presence of thapsigargin. The samples were immunoprecipitated with polyclonal antibody 121 and subjected to reducing SDS-PAGE.
Immunoprecipitation
Protein A-Sepharose beads (Amersham Biosciences), antisera, and cell lysates were incubated for at least 1 h at 4 °C. The immunoprecipitates were washed twice with wash buffer (for the LDL receptor 1% Triton X-100, 0.5% SDS, 150 mm NaCl, 50 mm Tris-Cl, pH 8.6, for HA 0.05% Triton X-100, 0.1% SDS, 300 mm NaCl, 10 mm Tris-HCl, pH 8.6), resuspended in 10 mm Tris-HCl, pH 6.8, with 1 mm EDTA and heated to 95 °C for 5 min in Laemmli sample buffer (200 mm Tris-Cl, pH 6.8, 3% SDS, 10% glycerol, 1 mm EDTA, and 0.004% bromphenol blue; final concentrations). One-half of each sample was reheated for 5 min at 95 °C in 25 mm DTT. N-Ethylmaleimide was added to a concentration of 100 mm to both reduced and nonreduced samples at room temperature, which then were analyzed by 6% (LDL receptor) or 7.5% (HA) SDS-PAGE.
DTT Resistance Assay
A pulse-chase experiment was performed as above, except that after each chase time, the cells were incubated in chase medium containing 10 mm DTT for an additional 5 min.
RESULTS
Calcium Is Required for Native Disulfide Bond Formation during LDL Receptor Folding
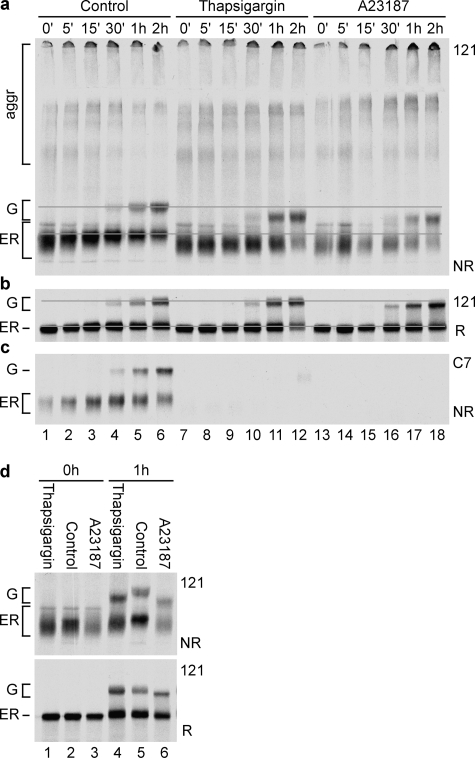
In vitro refolding studies showed that formation of native disulfide bonds in the ligand-binding repeats of the LDL receptor is closely related to calcium incorporation into these repeats (1, 37). Because the LDL receptor acquires its native repeats only late in folding (28), we set out to examine whether and, if so, when and how calcium is important for LDL receptor folding in the intact cell. We used two agents to deplete the ER for calcium: thapsigargin, which causes leakage of Ca2+ from the ER by inhibition of the ER Ca2+-ATPase (sarcoplasmic/endoplasmic reticulum Ca2+-ATPase) pumps (36) and A23187, an ionophore widely used to dissipate Ca2+ gradients in vivo (35). Whereas A23187 equalizes calcium levels of both ER and cytosol with those in the medium, thapsigargin depletes the ER of calcium but as a result may increase calcium levels in cytosol. Similar results obtained with both drugs therefore must be due to the lowered calcium concentration in the ER and not because of pleiotropic effects.
HeLa cells overexpressing the LDL receptor were starved for 15 min, pulse-labeled for 5 min, and chased for up to 2 h in the presence of either thapsigargin, the calcium ionophore A23187, or DMSO as control (Figs. 1 and 2). Detergent lysates were precipitated with polyclonal antibody 121 (Figs. 1, a, b, and d, and 2, a and c), which recognizes fully reduced, partly folded, as well as native LDL receptor molecules (28), or with a series of monoclonal antibodies (C7, 6B2, 7H2, 5G2, and 6E2) that recognize epitopes located in four of the seven LDL-A repeats that form the ligand-binding region of the LDL receptor (Figs. 1c and 2a). These monoclonal antibodies only recognize their epitopes when the correct disulfide bonds are formed and calcium is bound (29, 30).
FIGURE 1.
Effect of ER calcium depletion on LDL receptor folding and transport to the Golgi complex. HeLa cells overexpressing wild-type LDL receptor were pulse-labeled for 5 min and chased for the indicated periods in the presence of thapsigargin, A23187, or DMSO (Control). Under the conditions used (see “Experimental Procedures”), thapsigargin and A23187 both deplete ER calcium levels, albeit with different mechanisms and with different effect on cytosolic calcium concentration. a, samples were precipitated with polyclonal antiserum 121, which recognizes all forms of the LDL receptor, whether folded, misfolded, reduced, or denatured, and independent of subcellular location. Immunoprecipitates then were subjected to nonreducing SDS-PAGE. ER represents newly synthesized ER localized LDL receptor molecules, and G represents O-glycosylated “mature” LDL receptor molecules that have reached the Golgi complex or beyond. The gray reference lines help comparison of mobilities. b, the same samples as in a, now reduced. c, as a, except that lysates were immunoprecipitated with monoclonal antibody C7, which recognizes the most N-terminal repeat of the LDL receptor, but only when properly folded and containing its disulfide bonds and bound calcium. d, samples as in a of cells treated with A23187, DMSO (Control), or thapsigargin were analyzed next to each. NR, nonreducing SDS-PAGE; R, reducing SDS-PAGE.
Newly synthesized LDL receptor molecules acquire their native conformation in the ER and were run at 120 kDa on reducing SDS-PAGE (28) (Fig. 1b, lanes 1–6, ER). After exit, they were O-glycosylated in the Golgi complex to a form that runs at 160 kDa before being moved to the plasma membrane (28, 38) (Fig. 1b, lanes 4–6, G). After 2 h of chase in the presence of calcium, almost half of LDL receptor molecules had reached the Golgi complex (Fig. 1b, lane 6).
When these samples were analyzed under nonreducing conditions to uncover changes in disulfide bonding, the LDL receptor first folded into a compact protein that contained non-native disulfide bonds between cysteines far apart in the polypeptide chain (Fig. 1a, lane 1) and lacked native epitopes in the ligand-binding repeats (Fig. 1c, lane 1) (28).6 During isomerization of these transient non-native disulfide bonds into native bonds forming shorter loops, the LDL receptor became more extended, decreased SDS-PAGE mobility (Fig. 1a, lanes 2–6) (28), and acquired its folded repeats; LR1 became and 6B2- and C7-positive (28) (Figs. 1c, lanes 1–7, and 2a), and the other repeats attained their epitopes as well (Fig. 2a, lanes 3–6). The protein thus folded in two phases. The first phase led to a compact non-native form, and in a subsequent reshuffling phase, the individual repeats formed, while the wrong disulfides were replaced by the native ones (as schematically represented in Fig. 3, Control).
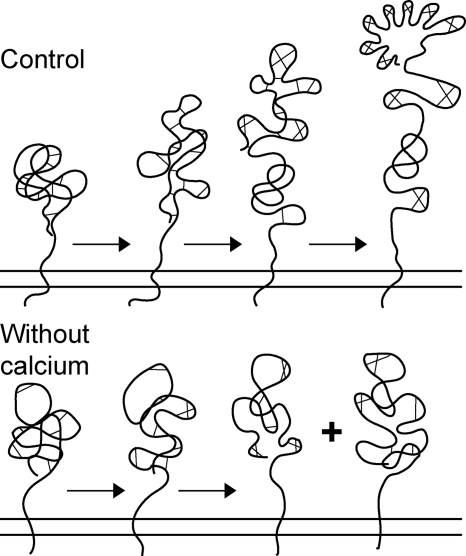
FIGURE 3.
Cartoon representing disulfide bond formation during folding of the LDL receptor in the presence (Control) and absence of calcium. Control cartoon reprinted with permission (28, 34).
In the absence of calcium, a slightly larger fraction of LDL receptor molecules misfolded into disulfide-linked aggregates, but the monomeric ones moved to the Golgi complex with similar or even higher rates (Fig. 1, a and b). They were not properly folded, however, because both the ER and Golgi forms ran with higher mobility than in the presence of calcium, demonstrating the existence of non-native disulfide bonds between cysteines far apart in the polypeptide chain (Fig. 1, a and d). The ER hence released misfolded LDL receptor molecules. The first phase of folding appeared similar with and without calcium (Fig. 1d, lanes 1–3), but after the initial collapse the calcium-depleted molecules barely changed mobility with time (Fig. 1, a, lanes 7–18, and d), and they never acquired a properly folded LR1, LR3, LR5, or LR7 repeat (Figs. 1c, lanes 7–18, and 2a, lanes 2–6 and 8–12, compare control with thapsigargin). This implies that proper folding of the LDL receptor in the ER required calcium, as seen before in vitro (39–41). The cartoon in Fig. 3 depicts the events in vivo.
The A23187 treatment not only changed folding of the LDL receptor, but also its O-glycosylation (Fig. 1d, lanes 4–6), most likely by influencing the activity of glycosylation enzymes in the Golgi complex. Similarly, A23187, but not thapsigargin, slightly inhibited trimming of N-linked glycans on Golgi-localized HA (not shown). We used HA as control for the effect of calcium depletion on the ER, because its folding depends on the calcium-binding lectin chaperone calnexin (42, 43). HA folded via two intermediates with incomplete sets of disulfide bonds that have electrophoretic mobilities between those of reduced HA and native, properly folded HA (NT) when analyzed by nonreducing SDS-PAGE (31) (Fig. 2b, lanes 1–5 and 6–10). Once folded, HA trimerized and left the ER for the Golgi complex, detectable by the extensive trimming of its N-glycans in the CHO15B cells. None of these HA maturation steps were affected by calcium depletion. Transport of misfolded LDL receptor molecules to the Golgi in the absence of calcium was not due to a defective retention system in the ER, because the folding-defective P678L mutant was retained with similar efficiency in the presence or absence of calcium (Fig. 2c). We therefore concluded that transport and quality control in general were not affected by the drugs we used and that the effects on the LDL receptor were caused by its need for calcium during folding in the ER.
DTT-resistant Disulfide Bonds Form during LDL Receptor Folding
As a rule, proteins become more packed and rigid during folding. This increase in compactness often coincides with an increase in resistance to in vivo reduction of disulfide bonds (44) and to proteolytic digestion (45). We used a DTT resistance assay to examine conformational differences in early and late LDL receptor folding intermediates in the presence and absence of calcium.
HeLa cells overexpressing the LDL receptor were pulse-labeled for 5 min and chased for 0 or 1 h. The cells then were cooled on ice as control, or in parallel, incubation was continued for an additional 5 min in chase medium containing 10 mm DTT to reduce accessible disulfide bonds in the intact cell before lysis. Detergent cell lysates were immunoprecipitated in parallel with polyclonal antiserum 121 and monoclonal antibody C7 and analyzed using nonreducing and reducing SDS-PAGE (Fig. 4).
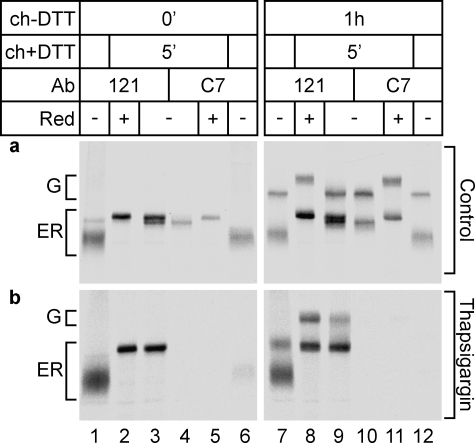
FIGURE 4.
DTT resistance of disulfide bonds during LDL receptor folding. HeLa cells overexpressing the LDL receptor were pulse-labeled for 5 min and chased for 0 or 1 h. The cells then were cooled immediately on ice (ch+DTT 0′) or chased for an additional 5 min in the presence of 10 mm DTT (ch+DTT 5′). The media for starvation, pulse, and chase all contained solvent (DMSO; control) (a) or 100 nm thapsigargin (b). Detergent cell lysates were immunoprecipitated in parallel with polyclonal antiserum 121 and monoclonal antibody C7. The samples were analyzed by reducing (Red, +) and nonreducing (Red, −) SDS-PAGE.
Immediately after the pulse, DTT treatment of the cells separated the LDL receptor folding intermediates (Fig. 4a, lane 1) into two populations (Fig. 4a, lane 3): one completely reduced (upper band) and another population containing DTT-resistant disulfide bonds (lower band), which were completely reduced upon heating in reducing Laemmli sample buffer (lane 2). As expected (28), monoclonal antibody C7 did not recognize the completely reduced LDL receptor (Fig. 4a, compare lane 4 with lane 3), but it did precipitate some partially resistant molecules (lane 4), showing that LR1 was among the correctly folded, DTT-resistant domains. Only a small number of disulfide bonds became resistant because the partially resistant form (in lane 3) ran close to fully reduced LDL receptor (lane 2), and it had a much lower mobility than nonreduced control samples (Fig. 4a, compare lane 3 with lane 1 and lane 4 with lane 6). We concluded that the non-native disulfide bonds that span distant cysteines were DTT-sensitive.
After 1 h of chase, some additional disulfide bonds became DTT-resistant; a smear underneath the lower band appeared upon DTT treatment (Fig. 4a, lane 9). Still, most disulfide bonds remained DTT-sensitive in the ER, because most LDL receptor molecules changed their mobility upon in vivo reduction (from lane 7 to lane 9) and now ran with similar mobility as denatured reduced LDL receptor (compare lanes 8 and 9). LDL receptor molecules that reached the Golgi complex did acquire complete DTT resistance (Fig. 4a, lanes 7, 9, 10, and 12), suggesting that the protein acquired additional compactness in the Golgi, whereas the ER environment kept the LDL receptor in an open, DTT-accessible state.
In the absence of calcium none of the disulfide bonds in the LDL receptor became DTT-resistant, because in vivo reduced LDL receptor (Fig. 4b, lane 3) had similar mobility to denatured reduced protein (Fig. 4b, lane 2), neither after a 1-h chase nor in the Golgi complex (Fig. 4b, lanes 8 and 9). As expected none of the calcium-depleted LDL receptor forms were recognized by the C7 monoclonal antibody (lanes 6 and 12), and this did not change upon reduction in vivo (lanes 4 and 10). Although LDL receptor molecules did escape the ER upon calcium depletion, the conformation of these molecules was drastically different from the native LDL receptor, because they not only lacked many native disulfide bonds and the C7 epitope (also other epitopes Fig. 2a) but also were completely sensitive to reduction by DTT in vivo (Fig. 4b, lanes 7–9).
Calcium Is Required for Maintenance of Native Structure in LDL Receptor Folding Intermediates
We showed that calcium is crucial for correct folding of the LDL receptor in intact cells. It is unclear, however, whether calcium is needed to induce formation of native-like structure and disulfide bonds or whether calcium is required also for their preservation. We therefore determined the resistance of folded LDL receptor and its late folding intermediates to extraction of associated calcium. The protein folded under normal conditions during a 1-h chase, which was followed by an additional 1-h chase in the absence of calcium. The 1-h-old late folding intermediates in the ER (Fig. 5a, lane 2) are characterized by a lower migration rate in nonreducing gel than the early ones (Fig. 5a, lane 1) and by increased recognition of the C7 epitope (compare lane 7 with lane 6). Upon calcium depletion, either by thapsigargin (lane 4) or A23187 (lane 5) treatment, these 1-h-old, late folding intermediates reverted to the phenotype of the early folding intermediates; their migration rates increased again, caused by formation of non-native disulfide bonds between distant cysteines (Fig. 5a, compare lanes 4 and 5 with lane 2), and they lost the already acquired C7 epitope (compare lanes 9 and 10 with lane 7). We concluded that calcium is required for maintenance of native structure in LDL receptor folding intermediates.
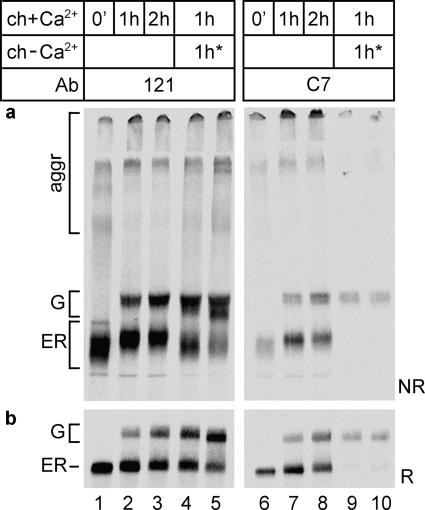
FIGURE 5.
Resistance of the LDL receptor to calcium depletion during folding. HeLa cells overexpressing the LDL receptor were pulse-labeled for 5 min, chased for 1 h in complete chase medium (ch+Ca2+), and then depleted of calcium in an additional 1-h chase (ch-Ca2+) in thapsigargin (lanes 4 and 9) or in calcium-free medium containing A23187 (lanes 5 and 10). Detergent cell lysates were immunoprecipitated in parallel with polyclonal antiserum 121 (lanes 1–5) and monoclonal antibody C7 (lanes 6–10). The control samples were pulse-labeled and chased for the same time intervals in complete medium containing calcium and DMSO (lanes 1–3 and 6–8). The samples were subjected to nonreducing (a) and reducing (b) SDS-PAGE.
Molecules that were in the ER when calcium depletion started but that did reach the Golgi complex during the additional chase without calcium did not acquire the C7 epitope (compare lanes 9 and 10 with lanes 4 and 5) and contained non-native disulfide bonds (Fig. 5a, lanes 4 and 5), similar to the results shown in Figs. 1 and 2. LDL receptor molecules that had escaped the ER to the Golgi already before calcium depletion, however, were not affected. They contained a native-like set of disulfide bonds, a properly folded LR1 repeat, and they acquired apparently normal O-glycans, shown by the native-like mobility (Fig. 5a, lane 3) of these Golgi forms (top part of the G band in Fig. 5a, lanes 4 and 5). That these broad smears were due to heterogeneity in disulfide bonding is clear from the reducing gel in Fig. 5b, where these forms collapsed into a much sharper band (Fig. 5b, lanes 3–5). These results are consistent with the DTT resistance data (Fig. 4), implying that only in the Golgi did the LDL receptor became completely resistant to DTT and to calcium depletion. As long as the protein resided in the ER, its disulfide bonds remained sensitive to DTT, its bound calcium could still be released, and its conformation could be reversed to a state resembling early folding intermediates; the LDL receptor was backtracking its folding pathway.
LDL Receptor Misfolding in the Absence of Ca2+ Is Not Reversible
Native repeats are formed relatively late in the folding pathway of the LDL receptor. Only in the second folding phase did long range disulfide bonds resolve into the native links, which results in formation of the C7 epitope (28) (Figs. 1 and 2). Because calcium was required to maintain native structure in LDL receptor folding intermediates (Fig. 5) and because early folding intermediates appeared similar irrespective of calcium (Fig. 1), we anticipated that calcium ions are incorporated into the protein predominantly during this second folding phase. Hence, LDL receptor molecules that start their folding in the absence of calcium and accumulate in the compact population should be rescued by later addition of calcium. We therefore examined LDL receptor folding in HeLa cells that were treated with A23187 in calcium-free medium during starvation and pulse labeling only, followed by a rescue chase in calcium-containing medium without ionophore. For comparison, HeLa cells were incubated during starvation, pulse, and chase with either calcium-free medium containing A23187 or medium containing DMSO and calcium.
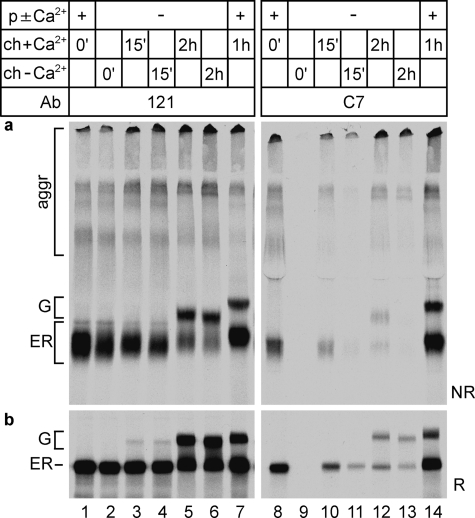
As shown in Fig. 1 as well, the LDL receptor folding intermediates synthesized in the absence of calcium ran as a compact smear (Fig. 6a, lane 2), and the N-terminal LR1 repeat was not folded correctly (lane 9 shows absence of the C7 epitope). During the rescue chase in the presence of calcium, the mobility of LDL receptor folding intermediates gradually decreased (Fig. 6a, from lane 2 to lanes 3 and 5), and the majority lacked a correctly folded LR1 repeat, although a fraction of molecules did obtain the C7 epitope (lanes 10 and 12). The LDL receptor molecules that left the ER did not contain the proper set of disulfide bonds, illustrated by their altered electrophoretic mobility compared with the DMSO control sample (Fig. 6a, lanes 5 and 7) and the virtual absence of the C7 epitope in these molecules (Fig. 6a, lanes 10 and 12). The reversibility of the A23187-induced glycosylation defect (Fig. 6b, lanes 5–7) demonstrated that calcium levels indeed recovered in the secretory pathway. We concluded that LDL receptor molecules that folded initially in the absence of calcium could not be rescued by restoration of ER calcium levels. Even after only a short Ca2+ depletion (when LDL receptor folding was still limited to the first collapsing phase), folding already suffered irreversibly from the absence of this ion. Apparently, calcium is specifically required for both the acquirement and the maintenance of native LDL receptor structure in the ER.
FIGURE 6.
Rescue from early calcium depletion. HeLa cells were pulse-labeled for 5 min (p±Ca2+) and chased for 15 min or 2 h in calcium-free medium containing A23187 (ch - Ca2+). For the rescued samples (ch+Ca2+) the chase medium was replaced by calcium-containing complete chase medium without A23187. Control samples were pulse-labeled for 5 min (p + Ca2+) and chased for 1 h (ch+Ca2+) in complete medium containing DMSO. a, samples precipitated in parallel with polyclonal antiserum 121 (lanes 1–7) or monoclonal antibody C7 (lanes 8–14) were subjected to nonreducing SDS-PAGE. b, the same samples as in a, now reduced.
DISCUSSION
Native LDL receptor folding needs calcium. We established in intact cells that calcium is required not only at each stage of the folding process but also for maintenance of structure at these stages. Although the LDL receptor folds its individual LDL-A repeats with octahedral calcium coordination only late in the folding process, calcium was required from the very beginning, when non-native inter-repeat disulfide bonds predominated. As long as the LDL receptor resided in the ER, DTT treatment reduced most of the formed disulfide bonds. Only a few native disulfide bonds including the ones in the C7 epitope were resistant. Calcium depletion in the second folding phase removed already coordinated calcium, resulting in isomerization of native disulfide bonds into non-native ones between distant cysteines and loss of the conformational C7 epitope. Once in the Golgi complex and beyond, properly folded LDL receptor molecules became completely resistant to unfolding by DTT or by calcium depletion.
Upon calcium depletion many misfolded forms of the LDL receptor left the ER to move on to the Golgi complex. LDL receptor molecules that had folded in the absence of calcium were released from the ER with similar kinetics as in the presence of calcium, but they had a drastically changed conformation. Because the ligand-binding repeats LR1, LR3, LR5, and LR7 had not properly folded in these misfolded forms, they are highly likely to be dysfunctional.
Reversibility of LDL Receptor Folding in the ER
During its stay in the ER, the LDL receptor remained susceptible to unfolding. Only a small number of disulfide bonds (including the ones in LR1) in about half of the LDL receptor molecules in the ER became resistant to reduction by DTT. Calcium depletion even revealed a reversion of LDL receptor folding: disulfide bonds rearranged back into long distance cysteines, C7 recognition was lost, as well as its partial DTT resistance. This demonstrates that protein folding in the ER in principle is a reversible process. Proteins remain malleable and therefore conformationally rescuable while in the ER.
Sensitivity to DTT throughout the folding process is not a general feature of proteins folding in the ER because, for instance, HA becomes completely resistant to in vivo reduction already in the ER (44). The native LDL receptor structure may well explain its DTT-accessible and calcium-extractable conformation during folding. The native ectodomain is extended (6, 28) and has a large surface to volume ratio, and the repeats have a rather small hydrophobic core (46). Disulfide bonds therefore are likely to remain accessible to DTT for a longer time during the folding process. Reducibility of the inter-repeat disulfides is no surprise either, because non-native structures are less packed than native ones and are expected to be susceptible to various denaturing treatments.
Resistance of LDL Receptor to Unfolding in Golgi Complex
Whereas ER forms of the LDL receptor remained prone to unfolding, the properly folded Golgi forms did acquire complete resistance to unfolding by calcium removal or DTT. Thus, when all native disulfides had been formed, the O-glycans attached, and the molecules resided in a different compartment than the ER, the structure could not be unfolded anymore. This cannot be explained by compartmental differences, O-glycosylation of the protein, or a combination of these factors, because the calcium-depleted O-glycosylated LDL receptor remained completely sensitive to DTT. The sharp difference in stability between the ER and Golgi forms of the LDL receptor is more likely due to release from chaperones and oxidoreductases in the ER in combination with a rate-limiting final conformational maturation followed by rapid transport to the Golgi complex and O-glycosylation (28). In the latter scenario the native form is not populated in the ER, consistent with our findings.
Calcium and Native LDL Receptor Folding, Structure, and Function
We anticipated that LDL receptor folding in vivo was dependent on calcium, because crystal and NMR structures of the ectodomain and individual repeats show that calcium is an integral part of LDL receptor structure (6, 14, 40, 46–48). Decreased or altered calcium binding therefore may have an impact on structure and hence function of the LDL receptor. Indeed, familial hypercholesterolemia-related mutations that affect amino acids involved in calcium binding show a lower affinity for calcium, misfolding, which for full-length protein results in (partial) ER retention, lower affinity for ligand on the cell surface, or a combination of these defects (14, 47, 49, 50). The LDL receptor requires repeats LR2-LR7 and in particular LR4 and LR5 (10) for LDL binding, none of which we expected to fold properly without calcium, based on our data and the similarity of all seven LR repeats.
Individual LDL-A repeats that were refolded in vitro without calcium adopted conformations with all possible cysteine pairings (39, 40, 51, 52), indicative of a complete lack of stable native state. Accordingly, formation of the C7 epitope in LR1 requires calcium (29, 53). The structure of the EGF repeats, with their noncanonical calcium-binding site, appears less dependent on this cation (15). Once isolated LDL-A repeats are folded, calcium can be extracted by incubation without calcium or with EGTA or EDTA (39, 41). Calcium affinity measurements demonstrate that bound calcium in a folded repeat indeed is in equilibrium with free calcium (54). It remains unclear whether calcium can be extracted from full-length LDL receptors on the cell surface. Whereas most calcium ions, once bound, are not easily removed from the LDL receptor family member Tva, the Rous sarcoma virus receptor (9), most studies on LDL receptor (domains) itself show that calcium extraction leads to loss of structural integrity and decreased ligand binding (27). Although we found the LDL receptor Golgi form to be resistant to calcium depletion, subtle changes that may lead to reduced ligand binding may well occur. Considering the range of proteins that associate with the LDL receptor (RAP, ApoB, and ApoE), conformational flexibility may well vary with conditions and partners. Moreover, our experiments follow newly synthesized LDL receptor before it starts to function on the cell surface. The intracellular milieu and the plasma membrane of cells in culture is mild for proteins. This changes completely when the protein binds ligand and starts cycling through the endosomal acid bath while undergoing repeated conformational changes. During those cycles, sensitivity to the loss of calcium and the loss of structural integrity is likely to increase.
Calcium and the Non-native Phase of Folding
NMR studies on LR5 refolding have shown a prerequisite formation of the first two disulfide bonds for calcium binding. With calcium bound, the third native disulfide bond closes, and calcium affinity increases (37). Together with the precise coordination of calcium by conserved residues in each repeat, these data imply that native calcium incorporation cannot happen before individual repeats are attaining native structure during folding. Unexpected then was our finding that calcium was required already during the earliest folding phase of the LDL receptor, when little trace of native structure was evident. Calcium was not required for formation of a compact state per se but was required for formation of folding-competent intermediates. Although rescue of LRP folded in the absence of calcium was reported to still be possible upon calcium restoration (19), LRP in this study had been pulse-labeled for 30 min in the presence of calcium first, before depletion started, giving the protein ample chance to pass into an unfolding-resistant stage.
In addition to the negatively charged Asp and Glu side chains that coordinate calcium, additional acidic residues in the LDL receptor form a negatively charged surface in the folded protein that binds RAP and apoB (12, 47). To bring such a large number of negative charges together during folding, small inorganic cations may be needed to prevent repulsion (55). Although we depleted calcium from the ER, other cations were still present. Most of these cations would not have sufficiently high concentrations to bind, because of affinities lower than millimolar. Mg2+, however, which is present in millimolar concentrations, should be sufficient to take care of the charge issue but cannot substitute for calcium in the native fold (39). This implies that calcium performs a specific role during the earliest stages of LDL receptor folding. During folding, calcium affinity increases with each additional disulfide bond in a repeat, and calcium thereby stimulates formation of native-like structure. Within the early compact folding intermediates, the cation may be essential for some rapidly folding isolated native structure elements, or it may bind transiently in native-like coordination to non-native motifs on the proper folding path, perhaps through domain swapping or strand swapping. In that case, the long distance non-native disulfide bonds in early LDL receptor folding intermediates may exist between strand-swapped repeats.
Role of Calcium for ER Function
Other proteins that fold in the ER have been reported to suffer from calcium depletion as well, even when they are not known as calcium-binding proteins. Insulin (56), thyroglobulin, asialoglycoprotein (57), and α1-antitrypsin (58), for instance, are not released efficiently from the ER of calcium-depleted cells. This has been attributed to a secondary effect, namely to less efficient chaperone activities in calcium-depleted ER (56). Because all of the resident ER proteins, the chaperones and folding enzymes, appear to bind calcium, this cation has been assumed to be crucial for proper ER function. Prolonged calcium depletion indeed causes ER stress and activation of PERK, one of the unfolded protein response sensors (59). Calcium was reported to be necessary for activity of many ER-resident calcium-binding chaperones, including Grp94 (60), PDI (61), BiP (62), calnexin (63), and calreticulin (64). This, however, remains a controversial issue, as a detailed study shows that calcium binding does contribute to calreticulin stability but is not required for its function as lectin chaperone (65). Similarly, various laboratories routinely use EDTA in their in vitro assays, showing that a range of oxidoreductases, including QSOX and PDI, catalyze disulfide isomerization reactions in the absence of calcium (66–68).
A wealth of additional data suggests that ER function is not affected by calcium depletion without chelators. (i) Albumin is hardly affected in its biosynthesis (57). (ii) Influenza virus HA folding, trimerization, and transport to the Golgi were not affected by the absence of calcium, although this protein does depend on the calcium-binding chaperone calnexin for folding (42, 43), which implies that calnexin function is intact in a calcium-depleted ER. (iii) We show here that retention of a misfolded LDL receptor molecule (P678L) in the ER was intact. (iv) Calcium waves in the cell cause frequent transient depletion of calcium from the ER, dropping calcium levels from millimolar to 1–50 μm levels (22). The charge change in the ER is compensated by parallel influx of other cations, such as Mg2+ or K+ in muscle (69) and monovalent cations in hepatocytes (70). It is unlikely that the ER would be dysfunctional every time the cell needs its calcium for signaling. (v) ER-resident chaperones have both high (Kd values approximately micromolar) and low affinity (Kd values approximately millimolar) calcium-binding sites, and depletion without chelators would not immediately extract all calcium from the high affinity sites, especially not when calcium levels are in the micromolar range during a calcium wave (71, 72). Calnexin, calreticulin, BiP, Grp94, and PDI are thought to constitute the major calcium store of the ER, in particular because of their abundance. Their affinities for calcium, as far as they have been studied, are similar (60–62, 73), except that of calnexin, which is at least an order of magnitude lower (150 μm) (74). This may permit extrapolation of calnexin functioning in low calcium conditions to the other major ER-resident chaperones. Therefore, unless the calcium concentration is lowered far enough to empty even the high affinity sites in chaperones, a condition that is difficult to reach in the absence of chelators, chaperone activity is not bound to be affected. The calcium binding of resident ER folding factors also may protect newly synthesized calcium-binding proteins like the LDL receptor during calcium oscillations.
In summary, we demonstrated here the crucial importance of calcium for every step of native LDL receptor folding in vivo. This calcium dependence of folding, maintenance of conformation, and hence function will be similar for all LDL-A module containing proteins and will thus be important for the many physiological processes this protein family mediates, ranging from viral infection, to lipoprotein homeostasis, to signaling. Already from the first stages of folding, when calcium binding in native structure was not prominent yet, calcium was required. Our and other studies did not find support for calcium depletion disrupting ER functioning under the conditions we used. The essential role of calcium during the non-native stages of LDL receptor folding suggests that native-like calcium coordination already occurs from the very beginning. On the other hand, even proteins that do not incorporate calcium into their native structure show some metal ion dependence. Any protein with negatively charged surfaces in their native structure may need cations to facilitate folding, whether particular ones or any. The calcium-rich environment of the ER not only includes free calcium, but also involves the proximity of associated folding factors with their many calcium-binding sites. The local calcium concentration a folding protein senses may well be an order of magnitude higher than anticipated, especially considering the heterogeneity of the ER (75). Coevolution of protein folding in the ER with the calcium-rich ER milieu would be a plausible basis for a calcium requirement for proper folding of many proteins in the ER.
Acknowledgments
We thank Dr. Malene Jorgensen for the pGEM-LDLR construct, Jan den Boesterd and Ingrid van Rooijen for advice on the figures, and members of the Braakman lab for helpful discussions. Jürgen Gent is acknowledged for discussions and comments on the manuscript; Sabine Gremme is acknowledged for supporting experiments.
This work was supported by grants from The Netherlands Heart Foundation (to I. B. and A. J.), EuroSCOPE (to I. B. and F. P.), The Netherlands Organization for Scientific Research-Chemical Division (to I. B. and F. P.), and Federation of European Biochemical Societies and Marie Curie (to F. P.).
J. Gent, personal communication.
- LDL
- low density lipoprotein
- ER
- endoplasmic reticulum
- EGF
- epidermal growth factor
- LRP
- LDL receptor-related protein
- HA
- hemagglutinin
- DMSO
- dimethyl sulfoxide
- MES
- 4-morpholineethanesulfonic acid
- DTT
- dithiothreitol.
REFERENCES
- 1.Jeon H., Blacklow S. C. (2005) Annu. Rev. Biochem. 74, 535–562 [DOI] [PubMed] [Google Scholar]
- 2.Brown M. S., Goldstein J. L. (1974) Science 185, 61–63 [DOI] [PubMed] [Google Scholar]
- 3.Willnow T. E., Nykjaer A., Herz J. (1999) Nat. Cell Biol. 1, E157–162 [DOI] [PubMed] [Google Scholar]
- 4.May P., Bock H. H., Herz J. (2003) Sci. STKE 2003, PE12. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto T., Davis C. G., Brown M. S., Schneider W. J., Casey M. L., Goldstein J. L., Russell D. W. (1984) Cell 39, 27–38 [DOI] [PubMed] [Google Scholar]
- 6.Rudenko G., Henry L., Henderson K., Ichtchenko K., Brown M. S., Goldstein J. L., Deisenhofer J. (2002) Science 298, 2353–2358 [DOI] [PubMed] [Google Scholar]
- 7.Gent J., Braakman I. (2004) Cell Mol. Life Sci. 61, 2461–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M. S., Goldstein J. L. (1986) Science 232, 34–47 [DOI] [PubMed] [Google Scholar]
- 9.Yu X., Wang Q. Y., Guo Y., Dolmer K., Young J. A., Gettins P. G., Rong L. (2003) J. Virol. 77, 7517–7526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Russell D. W., Brown M. S., Goldstein J. L. (1989) J. Biol. Chem. 264, 21682–21688 [PubMed] [Google Scholar]
- 11.Fisher C., Abdul-Aziz D., Blacklow S. C. (2004) Biochemistry 43, 1037–1044 [DOI] [PubMed] [Google Scholar]
- 12.Fisher C., Beglova N., Blacklow S. C. (2006) Mol. Cell 22, 277–283 [DOI] [PubMed] [Google Scholar]
- 13.Bieri S., Djordjevic J. T., Jamshidi N., Smith R., Kroon P. A. (1995) FEBS Lett. 371, 341–344 [DOI] [PubMed] [Google Scholar]
- 14.Fass D., Blacklow S., Kim P. S., Berger J. M. (1997) Nature 388, 691–693 [DOI] [PubMed] [Google Scholar]
- 15.Kurniawan N. D., Aliabadizadeh K., Brereton I. M., Kroon P. A., Smith R. (2001) J. Mol. Biol. 311, 341–356 [DOI] [PubMed] [Google Scholar]
- 16.Malby S., Pickering R., Saha S., Smallridge R., Linse S., Downing A. K. (2001) Biochemistry 40, 2555–2563 [DOI] [PubMed] [Google Scholar]
- 17.Beglova N., Blacklow S. C. (2005) Trends Biochem. Sci. 30, 309–317 [DOI] [PubMed] [Google Scholar]
- 18.Schneider W. J., Basu S. K., McPhaul M. J., Goldstein J. L., Brown M. S. (1979) Proc. Natl. Acad. Sci. U.S.A. 76, 5577–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Obermoeller L. M., Chen Z., Schwartz A. L., Bu G. (1998) J. Biol. Chem. 273, 22374–22381 [DOI] [PubMed] [Google Scholar]
- 20.Sitia R., Braakman I. (2003) Nature 426, 891–894 [DOI] [PubMed] [Google Scholar]
- 21.Short A. D., Klein M. G., Schneider M. F., Gill D. L. (1993) J. Biol. Chem. 268, 25887–25893 [PubMed] [Google Scholar]
- 22.Miyawaki A., Llopis J., Heim R., McCaffery J. M., Adams J. A., Ikura M., Tsien R. Y. (1997) Nature 388, 882–887 [DOI] [PubMed] [Google Scholar]
- 23.Combettes L., Cheek T. R., Taylor C. W. (1996) EMBO J. 15, 2086–2093 [PMC free article] [PubMed] [Google Scholar]
- 24.Montero M., Brini M., Marsault R., Alvarez J., Sitia R., Pozzan T., Rizzuto R. (1995) EMBO J. 14, 5467–5475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meldolesi J., Pozzan T. (1998) Trends Biochem. Sci. 23, 10–14 [DOI] [PubMed] [Google Scholar]
- 26.Macer D. R., Koch G. L. (1988) J. Cell Sci. 91, 61–70 [DOI] [PubMed] [Google Scholar]
- 27.Kita T., Brown M. S., Watanabe Y., Goldstein J. L. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 2268–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jansens A., van Duijn E., Braakman I. (2002) Science 298, 2401–2403 [DOI] [PubMed] [Google Scholar]
- 29.Beisiegel U., Schneider W. J., Goldstein J. L., Anderson R. G., Brown M. S. (1981) J. Biol. Chem. 256, 11923–11931 [PubMed] [Google Scholar]
- 30.Nguyen A. T., Hirama T., Chauhan V., Mackenzie R., Milne R. (2006) J. Lipid Res. 47, 1399–1405 [DOI] [PubMed] [Google Scholar]
- 31.Braakman I., Hoover-Litty H., Wagner K. R., Helenius A. (1991) J. Cell Biol. 114, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Durocher Y., Perret S., Kamen A. (2002) Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Braakman I., Helenius J., Helenius A. (1992) EMBO J. 11, 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jansens A., Braakman I. (2003) Methods Mol. Biol. 232, 133–145 [DOI] [PubMed] [Google Scholar]
- 35.Pfeiffer D. R., Lardy H. A. (1976) Biochemistry 15, 935–943 [DOI] [PubMed] [Google Scholar]
- 36.Thastrup O., Cullen P. J., Drobak B. K., Hanley M. R., Dawson A. P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 2466–2470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koduri V., Blacklow S. C. (2001) Biochemistry 40, 12801–12807 [DOI] [PubMed] [Google Scholar]
- 38.Cummings R. D., Kornfeld S., Schneider W. J., Hobgood K. K., Tolleshaug H., Brown M. S., Goldstein J. L. (1983) J. Biol. Chem. 258, 15261–15273 [PubMed] [Google Scholar]
- 39.Atkins A. R., Brereton I. M., Kroon P. A., Lee H. T., Smith R. (1998) Biochemistry 37, 1662–1670 [DOI] [PubMed] [Google Scholar]
- 40.Bieri S., Atkins A. R., Lee H. T., Winzor D. J., Smith R., Kroon P. A. (1998) Biochemistry 37, 10994–11002 [DOI] [PubMed] [Google Scholar]
- 41.Daly N. L., Djordjevic J. T., Kroon P. A., Smith R. (1995) Biochemistry 34, 14474–14481 [DOI] [PubMed] [Google Scholar]
- 42.Molinari M., Eriksson K. K., Calanca V., Galli C., Cresswell P., Michalak M., Helenius A. (2004) Mol. Cell 13, 125–135 [DOI] [PubMed] [Google Scholar]
- 43.Pieren M., Galli C., Denzel A., Molinari M. (2005) J. Biol. Chem. 280, 28265–28271 [DOI] [PubMed] [Google Scholar]
- 44.Tatu U., Braakman I., Helenius A. (1993) EMBO J. 12, 2151–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Copeland C. S., Doms R. W., Bolzau E. M., Webster R. G., Helenius A. (1986) J. Cell Biol. 103, 1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Daly N. L., Scanlon M. J., Djordjevic J. T., Kroon P. A., Smith R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 6334–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.North C. L., Blacklow S. C. (2000) Biochemistry 39, 2564–2571 [DOI] [PubMed] [Google Scholar]
- 48.North C. L., Blacklow S. C. (1999) Biochemistry 38, 3926–3935 [DOI] [PubMed] [Google Scholar]
- 49.Graadt van Roggen J. F., van der Westhuyzen D. R., Coetzee G. A., Marais A. D., Steyn K., Langenhoven E., Kotze M. J. (1995) Arterioscler. Thromb. Vasc. Biol. 15, 765–772 [DOI] [PubMed] [Google Scholar]
- 50.Boswell E. J., Jeon H., Blacklow S. C., Downing A. K. (2004) J. Biol. Chem. 279, 30611–30621 [DOI] [PubMed] [Google Scholar]
- 51.Blacklow S. C., Kim P. S. (1996) Nat. Struct. Biol. 3, 758–762 [DOI] [PubMed] [Google Scholar]
- 52.Arias-Moreno X., Arolas J. L., Aviles F. X., Sancho J., Ventura S. (2008) J. Biol. Chem. 283, 13627–13637 [DOI] [PubMed] [Google Scholar]
- 53.van Driel I. R., Goldstein J. L., Sudhof T. C., Brown M. S. (1987) J. Biol. Chem. 262, 17443–17449 [PubMed] [Google Scholar]
- 54.Abdul-Aziz D., Fisher C., Beglova N., Blacklow S. C. (2005) Biochemistry 44, 5075–5085 [DOI] [PubMed] [Google Scholar]
- 55.Huang W., Dolmer K., Gettins P. G. (1999) J. Biol. Chem. 274, 14130–14136 [DOI] [PubMed] [Google Scholar]
- 56.Guest P. C., Bailyes E. M., Hutton J. C. (1997) Biochem. J. 323, 445–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lodish H. F., Kong N., Wikstrom L. (1992) J. Biol. Chem. 267, 12753–12760 [PubMed] [Google Scholar]
- 58.Cooper G. R., Brostrom C. O., Brostrom M. A. (1997) Biochem. J. 325, 601–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Harding H. P., Zhang Y., Ron D. (1999) Nature 397, 271–274 [DOI] [PubMed] [Google Scholar]
- 60.Biswas C., Ostrovsky O., Makarewich C. A., Wanderling S., Gidalevitz T., Argon Y. (2007) Biochem. J. 405, 233–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lucero H. A., Kaminer B. (1999) J. Biol. Chem. 274, 3243–3251 [DOI] [PubMed] [Google Scholar]
- 62.Lievremont J. P., Rizzuto R., Hendershot L., Meldolesi J. (1997) J. Biol. Chem. 272, 30873–30879 [DOI] [PubMed] [Google Scholar]
- 63.Schrag J. D., Bergeron J. J., Li Y., Borisova S., Hahn M., Thomas D. Y., Cygler M. (2001) Mol. Cell 8, 633–644 [DOI] [PubMed] [Google Scholar]
- 64.Corbett E. F., Michalak K. M., Oikawa K., Johnson S., Campbell I. D., Eggleton P., Kay C., Michalak M. (2000) J. Biol. Chem. 275, 27177–27185 [DOI] [PubMed] [Google Scholar]
- 65.Conte I. L., Keith N., Gutierrez-Gonzalez C., Parodi A. J., Caramelo J. J. (2007) Biochemistry 46, 4671–4680 [DOI] [PubMed] [Google Scholar]
- 66.Nuss J. E., Choksi K. B., DeFord J. H., Papaconstantinou J. (2008) Biochem. Biophys. Res. Commun. 365, 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chandrashekar R., Tsuji N., Morales T., Ozols V., Mehta K. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rancy P. C., Thorpe C. (2008) Biochemistry 47, 12047–12056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baumann O., Walz B., Somlyo A. V., Somlyo A. P. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joseph S. K., Williamson J. R. (1986) J. Biol. Chem. 261, 14658–14664 [PubMed] [Google Scholar]
- 71.Tse F. W., Tse A., Hille B. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 9750–9754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nigam S. K., Goldberg A. L., Ho S., Rohde M. F., Bush K. T., Sherman M. (1994) J. Biol. Chem. 269, 1744–1749 [PubMed] [Google Scholar]
- 73.Baksh S., Michalak M. (1991) J. Biol. Chem. 266, 21458–21465 [PubMed] [Google Scholar]
- 74.Brockmeier A., Williams D. B. (2006) Biochemistry 45, 12906–12916 [DOI] [PubMed] [Google Scholar]
- 75.Papp S., Dziak E., Michalak M., Opas M. (2003) J. Cell Biol. 160, 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]