Abstract
Lipopolysaccharide (LPS) activates innate immune responses through TLR4·MD-2. LPS binds to the MD-2 hydrophobic pocket and bridges the dimerization of two TLR4·MD-2 complexes to activate intracellular signaling. However, exactly how lipid A, the endotoxic moiety of LPS, activates myeloid lineage cells remains unknown. Lipid IVA, a tetra-acylated lipid A precursor, has been used widely as a model for lipid A activation. For unknown reasons, lipid IVA activates proinflammatory responses in rodent cells but inhibits the activity of LPS in human cells. Using stable TLR4-expressing cell lines and purified monomeric MD-2, as well as MD-2-deficient bone marrow-derived macrophages, we found that both mouse TLR4 and mouse MD-2 are required for lipid IVA activation. Computational studies suggested that unique ionic interactions exist between lipid IVA and TLR4 at the dimerization interface in the mouse complex only. The negatively charged 4′-phosphate on lipid IVA interacts with two positively charged residues on the opposing mouse, but not human, TLR4 (Lys367 and Arg434) at the dimerization interface. When replaced with their negatively charged human counterparts Glu369 and Gln436, mouse TLR4 was no longer responsive to lipid IVA. In contrast, human TLR4 gained lipid IVA responsiveness when ionic interactions were enabled by charge reversal at the dimerization interface, defining the basis of lipid IVA species specificity. Thus, using lipid IVA as a selective lipid A agonist, we successfully decoupled and coupled two sequential events required for intracellular signaling: receptor engagement and dimerization, underscoring the functional role of ionic interactions in receptor activation.
Keywords: Immunology/Innate Immunity, Immunology/LPS, Immunology/Toll Receptors, Membrane/Proteins, Methods/Computer Modeling, Methods/Fluorescence, Methods/Site-directed Mutagenesis, Protein/Structure
Introduction
Innate immunity is the first line of defense against exogenous pathogens. Activation of the innate immune system produces inflammatory cytokines such as tumor necrosis factor α and interleukin-1β, both of which help prevent infection and enhance the adaptive immune response (1). The effect of these cytokines can become detrimental when they are produced in abundance, resulting in sepsis syndrome. Indeed, Gram-negative septic shock still has high mortality (∼20%) and remains a leading cause of death in noncoronary intensive care units in the United States (2, 3).
During Gram-negative bacterial infection, lipopolysaccharide (LPS),4 the major component of the bacterial outer membrane, activates the innate immune response through the Toll-like receptor 4 (TLR4) and MD-2 complex. The active component of LPS is lipid A, a partially conserved glycolipid that anchors LPS into the outer membrane of Gram-negative bacteria. Stimulatory lipid As, such as Escherichia coli lipid A, usually have 6–8 acyl chains covalently linked to the diglucosamine bis-phosphorylated backbone. The phosphate groups at the 1,4′-position are essential for the agonist activity of lipid A, because monophosphorylated lipid A is greatly reduced in its proinflammatory activity (4). In addition, both the number and the length of the acyl chains are essential for the full agonist activity of lipid A (5–7). In fact, the production of a hypoacylated lipid A and the resulting evasion of innate immunity may be associated with virulence in pathogens such as Yersinia pestis (8). E. coli hexa-acylated lipid A acts as a pan-agonist for all mammalian cells that express a complete LPS receptor complex. The precursor of E. coli lipid A, tetra-acylated lipid IVA (9), is only an agonist for some species of mammals (10).
Although commonly referred to as the LPS receptor, TLR4 does not directly bind LPS or any other LPS analog with high avidity. Instead, MD-2, a 25-kDa co-receptor that physically associates with TLR4 directly binds the lipid A moiety of LPS (or its analogs) through the central hydrophobic pocket (11–13). This hydrophobic pocket can accommodate up to five acyl chains. In contrast to commonly held notions that the MD-2 pocket would expand to accommodate additional acyl chains from stimulatory lipid A, the resolution of a co-crystal structure of human TLR4 (hTLR4), human MD-2 (hMD-2), and LPS (12) revealed that the sixth acyl chain of LPS is excluded from the hydrophobic pocket and present on MD-2 surface. Both hMD-2 and hTLR4 undergo “induced fit” conformational changes to allow dimerization to occur (12).
Although an LPS antagonist in human cells, lipid IVA is an LPS mimetic when tested with mouse cells (14, 15). Several studies have been dedicated to understanding the molecular determinants of this species specificity. The results of these studies, however, are contradictory. Our group (16), as well as that of Beutler and co-workers (17), proposed that TLR4 is responsible for the species specificity of lipid IVA. Yet, based on similar approaches, Miyake and co-workers (14) and Miller and co-workers (18) reported that MD-2 is responsible for the species-specific responses to lipid IVA. A full interpretation of these studies was not possible because neither group could ever characterize the activity of human TLR4 with mouse MD-2 (mMD-2), probably because the latter protein is so poorly expressed in transfected cell lines. Using a slightly different system, i.e. comparing human versus equine genes, Bryant and co-workers (19) demonstrated that under defined conditions, MD-2 and TLR4 were both required for the species-specific activation of lipid IVA, partially reconciling the contradiction between the two theories.
The present study addresses both the molecular determinants and the underlying mechanism of the species-specific activation of lipid IVA in an attempt to truly understand the mystery of lipid IVA activity and to extend our knowledge on the mechanism of lipid A activation. We found that both mouse TLR4 (mTLR4) and mMD-2 are required to confer LPS agonist activity to lipid IVA, both in HEK293 cell lines that stably express hTLR4 or mTLR4 and in MD-2-deficient bone marrow-derived macrophages (BMDMs). We used computational docking and modeling to generate a dimeric mTLR4·mMD-2·lipid IVA model to understand the underlying mechanism. We found that unique ionic interactions exist between lipid IVA and TLR4 in the mouse complex only. When these ionic interactions were disrupted by mutagenesis, lipid IVA responsiveness was severely impaired. In contrast, hTLR4 gained lipid IVA responsiveness when ionic interactions were enabled by charge reversal at the dimerization interface, defining the basis for lipid IVA species specificity. Because lipid A also lacks the core polysaccharide, ionic interactions between the phosphates on the lipid A diglucosamine backbone and the positively charged residues on TLR4 at the dimerization interface are likely to play a key role in receptor dimerization and activation. Thus, we provide direct evidence that ionic interactions between the lipids and TLR4, especially at the dimerization interface, play an essential role in triggering the dimerization and activation of the LPS receptor.
EXPERIMENTAL PROCEDURES
MD-2 Expression and Purification
The MD-2 constructs expressing hMD-2 and mMD-2 with a C-terminal protein A tag were gifts from Dr. Jie-Oh Lee (11). MD-2 proteins were expressed and purified as described earlier (11) (supplemental Fig. S1). The MD-2 concentrations were determined by UV absorbance at 280 nm using the extinction coefficients 18,490 and 14,650 liters·m−1·cm−1 for hMD-2 and mMD-2, respectively (20).
Luciferase Assay
The HEK293/mTLR4YFP cell line was constructed as described earlier (21). The E. coli precursor, lipid IVA, was synthesized as described (22). The synthetic compound, Eritoran, was a gift from the Eisai Research Institute (Andover, MA). LPS from E. coli strain O111:B4 (Sigma) was repurified by a repeat phenol chloroform extraction (23). HEK293/hTLR4YFP and HEK293/mTLR4YFP cells were plated in 96-well dishes at a density of 10,000 cells/well. The next day, the cells were transfected with a plasmid encoding for an NF-κB-luciferase gene and a control plasmid expressing Renilla luciferase. After overnight transfection, the supernatants were removed, and the cells were washed twice with phosphate-buffered saline and replenished with serum-free medium consisting of DMEM only. The cells were stimulated with 1) the indicated concentrations of LPS or lipid IVA and 2) increasing concentrations of purified monomeric MD-2. After overnight stimulation, the supernatants were removed, and the luciferase activity was measured in cell lysates. Renilla luciferase was used for normalization.
The single, double, and triple mutants that swap surface charges between hTLR4 and mTLR4 at the dimerization interface were created by site-directed mutagenesis per the manufacturer's instructions (QuikChange). All of the mutations were verified by DNA sequencing (Genewiz, Inc., South Plainfield, NJ). The effects of these mutations on LPS and lipid IVA signaling were tested in HEK293 cells by transient transfection. HEK293 cells were plated in 96-well plates at a density of 20,000 cells/well. The next day, the cells were transfected with 1) one of the hTLR4YFP or mTLR4YFP mutants (1 ng/well), 2) an NF-κB-luciferase plasmid (24), and 3) the Renilla luciferase plasmid. After overnight transfection, the supernatant was removed, and the cells were washed twice with phosphate-buffered saline and replenished with complete DMEM without serum. The cells were stimulated with the indicated concentrations of mMD-2 and LPS·lipid IVA overnight prior to the determination of luciferase activity.
Tumor Necrosis Factor α Production
Mice deficient in MD-2 were a gift from Dr. Kensuke Miyake (25). BMDMs were differentiated in the presence of 20% L929 conditioned medium for 10 days. The cells were then plated in 96-well plates at a concentration of 50,000/well in complete DMEM supplemented with 10% fetal bovine serum. After attachment, the supernatants were removed, and the cells were washed four times with phosphate-buffered saline and replenished with serum-free DMEM. The cells were then stimulated with 1) a fixed concentration of lipid IVA or LPS, 2) decreasing concentrations of purified, monomeric MD-2, and 3) 1 μg of CD14/ml (26). After overnight stimulation, the supernatants were saved, and the tumor necrosis factor α levels were measured by enzyme-linked immunosorbent assay.
Docking Procedure between Lipid IVA and Human or Mouse MD-2
Docking studies were performed using AutoDock 4.0.1 (27, 28) with an AutoDockTool (29). The coordinates of lipid IVA were subtracted from the co-crystal structure of hMD-2 and lipid IVA (Protein Data Bank code 2E59) (13). To reduce expectation bias, the crystal structure of hMD-2 alone (Protein Data Bank code 2E56) (13) instead of from the hMD-2·lipid IVA co-crystal structure (Protein Data Bank code 2E59) (13) was used for docking with lipid IVA. The coordinates of mMD-2 were subtracted from the co-crystal structure of mMD-2 and mTLR4 (Protein Data Bank code 2Z64) (11). Both hMD-2 and mMD-2 were treated as rigid, and lipid IVA was treated as flexible with 30 torsions during docking. The AutoGrid parameters are illustrated in detail in supplemental Table S1. Twenty structures were generated using genetic algorithm searches. A default protocol was applied, with an initial population of 150 randomly placed individuals, a maximum number of 2.5 × 106 energy evaluations, and a maximum number of 2.7 × 104 generations. A mutation rate of 0.02 and a cross-over rate of 0.8 were used.
Generation of a Dimeric mTLR4·mMD-2·Lipid IVA Complex
The coordinates of the docked mMD-2·lipid IVA complex were combined with the coordinates of mTLR4 extracted from the mTLR4/mMD-2 co-crystal structure (Protein Data Bank code 2Z64) (11) to generate a monomeric mTLR4·mMD-2·lipid IVA complex. Two copies of mTLR4·mMD-2·lipid IVA complexes were then aligned to the hTLR4·hMD-2·LPS co-crystal structure to generate a dimeric mTLR4·mMD-2·lipid IVA complex. The detailed alignment procedure is as follows: the mTLR4 sequences from the first mTLR4·mMD-2·lipid IVA complex were aligned to one hTLR4 sequences in the dimeric hTLR4·hMD-2·LPS co-crystal structure, and the mTLR4 sequences from the second mTLR4·mMD-2·lipid IVA complex were aligned to the other hTLR4 sequences in the co-crystal structure. The sequences of MD-2 and lipids were not used for the alignment. A dimeric mTLR4·mMD-2·lipid IVA complex was readily observed after the alignment.
RESULTS
The Full Mouse Receptor Complex, Consisting of Mouse TLR4 and Mouse MD-2, Is Required for the Full Agonist Activity of Lipid IVA
To clarify the contradictions among different studies concerning lipid IVA species specificity, we developed stable cell line systems in which hTLR4 (21) or mTLR4 was stably expressed on the cell surface, allowing us to study each component separately and consistently. Monomeric soluble MD-2 of both human and mouse origin were purified (supplemental Fig. S1) and used as the source for MD-2.
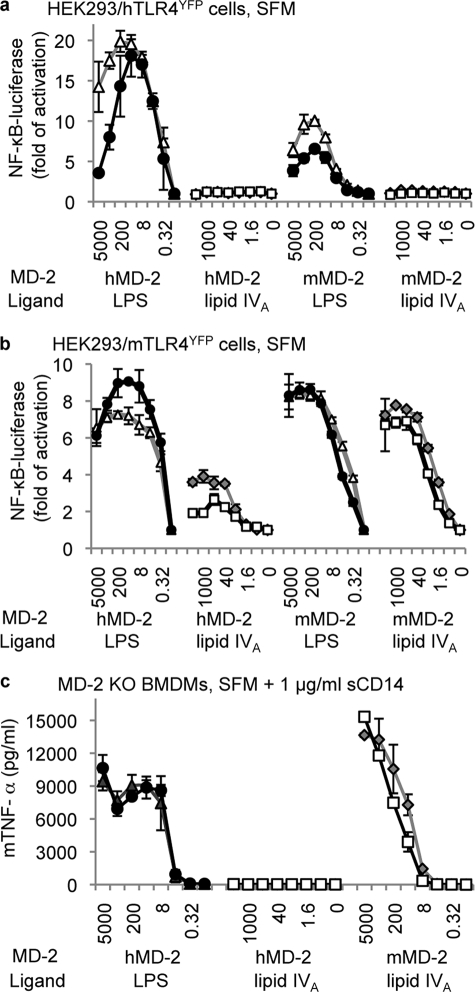
We examined the inducible luciferase activity using an NF-κB reporter construct transiently transfected in HEK293 cells. Both HEK293/hTLR4YFP (Fig. 1a) and HEK293/mTLR4YFP cells (Fig. 1b) responded to LPS stimulation in an MD-2 dose-dependent manner, regardless of the species of MD-2 included. A peak response was observed at the MD-2 concentration of 40 ng/ml (1.3 nm), which likely reflects the optimal concentration of MD-2 to interact with TLR4 in HEK293 cells to activate intracellular signaling. Note that we have consistently observed that HEK293/hTLR4YFP cells responded to LPS more vigorously with hMD-2 than with mMD-2, whereas HEK293/mTLR4YFP cells responded to LPS similarly with hMD-2 and mMD-2.
FIGURE 1.
Both mTLR4 and mMD-2 are required for the full agonist activity of lipid IVA. HEK293/hTLR4YFP cells (a), HEK293/mTLR4YFP cells (b), and MD-2-deficient BMDMs (c) were stimulated with LPS·lipid IVA and MD-2 under serum-free conditions. 1 μg/ml soluble CD14 (sCD14) was included for stimulations in MD-2-deficient BMDMs. After overnight stimulation, luciferase activity was measured in HEK293 cell lysates, and mouse tumor necrosis factor α levels were measured in the supernatant of MD-2-deficient BMDMs. The data are reported as the means ± S.D. of three independent wells for each data point. The luciferase activities in a and b were normalized using Renilla luciferase. One representative data set from four replicates is shown in the figure. ▵, 1 μg/ml LPS; ●, 0.1 μg/ml LPS; ◇, 1 μg/ml lipid IVA; □, 0.1 μg/ml lipid IVA. The MD-2 concentrations used in a and b, from left to right, were 5000, 1000, 200, 40, 8, 1.6, 0.32, and 0 ng/ml.
When lipid IVA was used as the stimulant, HEK293/hTLR4YFP cells did not respond to lipid IVA in the presence of either hMD-2 or mMD-2 at all of the tested concentrations (Fig. 1a); under these conditions, lipid IVA functioned as an LPS antagonist (supplemental Fig. S2). This is consistent with our previously reported data that lipid IVA acts as an antagonist in the presence of hTLR4 (16). In comparison, HEK293/mTLR4YFP cells (Fig. 1b) responded to lipid IVA stimulation (gray diamond and open square) in a (mMD-2) dose-dependent manner that peaked at the mMD-2 concentration of 200 ng/ml (6.7 nm). This suggests that when both mMD-2 and mTLR4 are present, lipid IVA functions as an LPS agonist.
The titration experiments with hMD-2 in HEK293/mTLR4YFP cells (Fig. 1b) show that at low concentrations of hMD-2 (<1.6 ng/ml or 53 pm), lipid IVA did not activate HEK293/mTLR4YFP cells. At higher concentrations of hMD-2 (>40 ng/ml, or 1.3 nm), lipid IVA activated HEK293/mTLR4YFP cells ∼3–4-fold, about half of the response in comparison with LPS stimulation. This indicates that lipid IVA can function as a partial agonist in HEK293/mTLR4YFP cells and that under the right conditions, both MD-2 and TLR4 seem to be qualitatively dictating the biological responses.
BMDMs from MD-2 Knock-out Mice Required the Presence of Mouse MD-2 to Respond to Lipid IVA
HEK293/hTLRYFP and HEK293/mTLRYFP cell lines provide a stable environment to quantitatively assess the activity of MD-2. However, these lines are immortalized and genetically engineered; furthermore, they express levels of TLR4 that are higher than true immune cells (supplemental Fig. S3). In addition, HEK293 cells are not professional phagocytes and have a limited profile of inducible proinflammatory cytokines. BMDMs from MD-2 knock-out mice were therefore tested for their responsiveness to lipid IVA in the presence of purified mMD-2 or hMD-2.
MD-2-null BMDMs responded to LPS stimulation in an hMD-2 dose-dependent manner, which reached a plateau at the hMD-2 concentration of 40 ng/ml (Fig. 1c). This concentration likely reflects the amount of MD-2 necessary to saturate TLR4 on the cell surface and to activate intracellular signaling under physiological conditions. Similarly, MD-2-null BMDMs responded to lipid IVA stimulation in the presence of mMD-2 in a dose-dependent manner until the maximally assessed mMD-2 concentration (Fig. 1c) but did not respond to lipid IVA stimulation when hMD-2 was included regardless of the hMD-2 concentration added (Fig. 1c). This indicates that with hMD-2, lipid IVA does not function as an LPS agonist toward the mouse cells. The partial activation seen in HEK293/mTLR4YFP cells by lipid IVA at high concentrations of hMD-2 was not observed, which is perhaps a reflection of the physiological levels of mouse TLR4 expression. Thus, both mMD-2 and mTLR4 are required for the species-specific activation of lipid IVA under physiological conditions.
In summary, it appears that when mouse TLR4 was expressed, the activity of lipid IVA depended upon the species of MD-2. That is, when mouse TLR4 was matched with mouse MD-2, lipid IVA was an agonist; when mouse TLR4 was matched with human MD-2, lipid IVA had no activity (except of at higher levels of mTLR4 in HEK293 cells) and hence functioned as an antagonist. Thus, in the presence of mouse TLR4, the species from which MD-2 was derived determined the character of the response. In contrast, when human TLR4 was expressed, lipid IVA was not an LPS agonist regardless of the species of MD-2. Hence, both TLR4 and MD-2 appear to be responsible for the species-specific response to lipid IVA, with the dominance of one over the other depending on the specific combination and expression levels of TLR4·MD-2 examined.
Lipid IVA Packs Superficially into the Hydrophobic Pocket of Mouse MD-2, with the Diglucosamine Backbone Tilted toward the Cys95–Cys105 Loop
Computational docking was performed between lipid IVA and mMD-2 to understand the essential role of mMD-2 in the species-specific activation of lipid IVA. Mouse TLR4 was omitted from initial docking because only MD-2, but not TLR4, has been demonstrated to bind LPS or its analogs directly (13). We used the AutoDock docking software, because this approach produces ligand·receptor complexes that closely resemble co-crystal structures (30). In a quality control experiment (supplemental Fig. S4), AutoDock reproduced an hMD-2·lipid IVA complex highly similar to the co-crystal structure (13), with a root mean square deviation of 0.74 Å and an inhibitory constant (Ki) of 144.85 pm. This Ki is very close to the experimental Kd value between MD-2 and LPS (31).
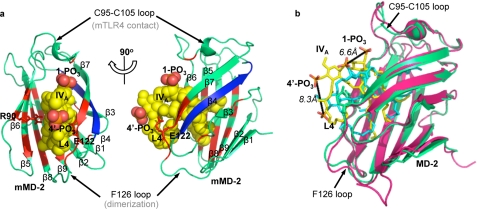
In the docked lipid IVA·mMD-2 complex, lipid IVA packs well into the hydrophobic pocket of mMD-2 (Fig. 2a). The acyl chains of lipid IVA interact extensively with residues lining the hydrophobic pocket, especially residues adjacent to the Phe126 loop that has been implicated essential for receptor dimerization (12). The 1,4′-phosphates of lipid IVA interact weakly with charged residues (e.g. Arg90 and Glu122) at the pocket entrance (Fig. 2a), thus restraining the diglucosamine backbone of lipid IVA to the pocket entrance. In comparison, none of the β4 strand residues, which have been suggested to be essential for the species-specific recognition of lipid IVA (19), are directly involved in lipid IVA interaction (Fig. 2a).
FIGURE 2.
Lipid IVA packs tilted and shallowly in the mouse MD-2 hydrophobic pocket in the docked mMD-2·lipid IVA complex. The docked mMD-2·lipid IVA complex is shown in two perpendicular views in a and overlaid to the hMD-2·lipid IVA co-crystal structure in b. MD-2 is shown in ribbon views in a and b, and lipid IVA is shown in sphere view in a and stick view in b. The residues interacting with lipid IVA are colored red, and the β4 strand is colored blue in a. The fourth acyl chain of lipid IVA is labeled L4 in both a and b. Green, mMD-2; yellow, lipid IVA in mMD-2; pink, hMD-2; cyan, lipid IVA in hMD-2. The graphics were created using PyMol (DeLano Scientific).
Compared with the hMD-2·lipid IVA co-crystal structure (13), lipid IVA packs more superficially in the hydrophobic pocket of mMD-2, with the diglucosamine backbone of lipid IVA tilted toward the Cys95–Cys105 loop (Fig. 2b). Consequently, in the docked mMD-2·lipid IVA complex, the 1-PO3 and 4′-PO3 of lipid IVA are shifted 6.6 and 8.3 Å closer, respectively, to the Cys95–Cys105 loop that directly binds mTLR4 (11). In addition, the fourth acyl chain (L4) of lipid IVA moves toward the surface (Fig. 2b) and hence is more exposed to the solvent. The binding kinetics generated by AutoDock between mMD-2 and lipid IVA was −2.12 kcal mol−1 for the expected binding energy (ΔGbin) and 28.05 mm for the expected inhibitory constant (Ki, 298 K).
The 4′-PO3 of Lipid IVA Bound to TLR4·MD-2 Is Adjacent to a Positively Charged Patch on the Opposite mTLR4 at the Dimerization Interface
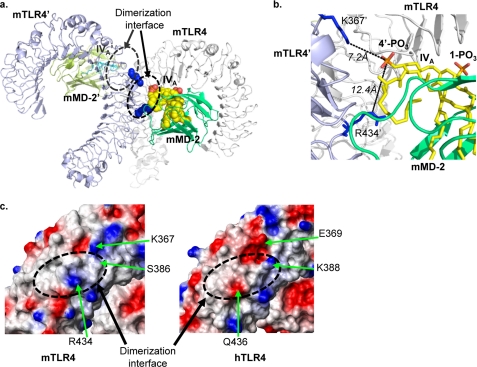
The coordinates of the mMD-2·lipid IVA complex were then combined with the crystal structure of mTLR4 to generate an mTLR4·mMD-2·lipid IVA complex. This complex is very similar to the monomeric hTLR4·hMD-2·LPS complex isolated from the co-crystal structure (see supplemental Fig. S5a for the comparison) (12). Except for the opposite orientation of the lipids, the diglucosamine backbone and the two phosphates on the lipids are in very similar positions in the two complexes (supplemental Fig. S5b). Thus, the docked mTLR4·mMD-2·lipid IVA complex is in a favorable conformation to interact with another copy of mTLR4·mMD-2·lipid IVA complex (denoted as mTLR4′/mMD-2′/lipid IVA′ from this point on, to differentiate from the original mTLR4·mMD-2·lipid IVA complex) to form a dimer. Indeed, when the sequences of mTLR4 from the predicted mTLR4·mMD-2·lipid IVA dimer were superimposed onto the crystal structure of hTLR4 that was defined in the published hTLR4·hMD-2·LPS (see details under “Experimental Procedures”), a dimeric mTLR4·mMD-2·lipid IVA complex was readily observed (Fig. 3a).
FIGURE 3.
Close proximity is observed between the 4′-PO3 on lipid IVA and the positively charged residues on mTLR4 at the dimerization interface. A dimeric mTLR4·mMD-2·lipid IVA model is shown in a. A zoomed in view of the dimerization interface is shown in b. Molecules from the first complex are labeled as mTLR4 (white ribbon), mMD-2 (green ribbon), and lipid IVA (yellow spheres or sticks), and molecules from the second complex are labeled as mTLR4′ (light purple ribbon), mMD-2′ (olive ribbon), and lipid IVA′ (cyan sticks). The phosphate groups on lipid IVA are colored red. Lys367 and Arg434 are shown as blue spheres in a and as blue sticks in b. The electrostatic surface charges (c) of mTLR4 (left panel) and hTLR4 (right panel) were calculated with MolMol (37). Red is for negatively charged surface, blue is for positively charged surface, and white is for noncharged surface. Residues that differ between mTLR4 and hTLR4 are labeled in c. The dashed circles in a and c indicate the dimerization interface.
Close proximity was observed between the 4′-PO3 on lipid IVA and the positively charged residues on mTLR4′ composed of Lys367′ and Arg434′ at the dimerization interface (Fig. 3b). The distances between the phosphorous atom on the 4′-PO3 of lipid IVA and the nitrogen atoms on Lys367′ and Arg434′ (of mTLR4′) were calculated at 7.2 and 12.4 Å, respectively. These distances are likely to be shortened in the actual, active, and dimeric mTLR4·mMD-2·lipid IVA complex, because lipid IVA recognition likely triggers induced fit conformational changes in both mTLR4 and mMD-2 to allow dimerization to occur. These conformational changes likely resemble those observed in the hTLR4·hMD-2·LPS co-crystal structure (12), i.e. the extension of the TLR4 solenoid and the folding back of the Phe126 loop of MD-2. When present in solution, the 4′-PO3 on lipid IVA likely attracts and interacts with the positive patch on mTLR4′ at the dimerization interface to facilitate and enable dimerization and to initiate downstream signaling.
Surface Charge Differences on TLR4 at the Dimerization Interface Account for the Specific Role of TLR4 in the Species-specific Activation of Lipid IVA
The electrostatic surface charges at the dimerization interface were predicted to be different between mTLR4 and hTLR4 (Fig. 3c). Mouse TLR4 has positively charged Lys367 and Arg434 at the dimerization interface, whereas hTLR4 has the negatively charged Glu369 and Gln436 at these positions. In addition, the noncharged Ser386 in mTLR4 corresponds to a positively charged Lys388 in hTLR4. A charge reversal in this area would therefore alter electrostatic forces on the 4′-PO3 on lipid IVA, changing the ionic interactions with lipid IVA at the dimerization interface. We hypothesized that the essential role of TLR4 in the species-specific activation of lipid IVA arises from different electrostatic surface charges at the dimerization interface.
To examine this possibility, we employed site-directed mutagenesis to engineer single, double, and triple mutants that swap surface charges between mTLR4 and hTLR4. The effects of these mutations were examined by measuring the inducible NF-κB luciferase activity after transient transfection and lipid IVA stimulation. Mouse MD-2 (at a concentration of 200 ng/ml) was included in all testing conditions, because only mouse MD-2 functions with both hTLR4 and mTLR4.
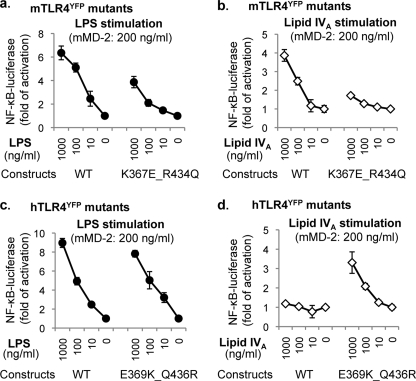
Compared with the wild type mTLR4 construct, the mTLR4 mutants retained most of their LPS responsiveness (supplemental Fig. S6a), indicating that these mutations do not disturb the major structural motif on mTLR4 essential for LPS signaling. Lipid IVA responsiveness, however, was modified by these mutations. As the dimerization interface become less positively charged, the lipid IVA responsiveness decreased (supplemental Fig. S6b), with the most profound effect on the double mutant K367E/R434Q (Fig. 4, a and b). Thus, ionic interactions between the 4′-PO3 of lipid IVA and the positive patch on mTLR4 at the dimerization interface appeared to be required for lipid IVA activation.
FIGURE 4.
Ionic interactions between lipid IVA and TLR4 at the dimerization interface are essential for lipid IVA responsiveness. HEK293 cells were transiently transfected with: 1) one of the four constructs: mTLR4YFP wild type construct (WT), the mTLR4YFP K367E/R434Q mutant (K367E/R434Q), hTLR4YFP wild type construct, and the hTLR4YFP E369K/Q436R mutant (E369K/Q436R); 2) NF-κB luciferase plasmid; and 3) a Renilla luciferase plasmid for luciferase assay. After overnight transfection, the cells were stimulated with indicated concentrations of LPS·lipid IVA and mMD-2 (200 ng/ml) under serum-free conditions. Luciferase activity was measured in cell lysates the next day. The data are reported as the means ± S.D. of three independent wells for each data point. The luciferase activities were normalized from Renilla luciferase activity. One representative data set from four replicates is shown in the figure.
Conversely, all of the hTLR4 mutants retained their LPS responsiveness (supplemental Fig. S7a). However, the engraftment of the positively charged residues from the dimerization interface of mouse TLR4 to its human counterpart had a dramatic effect on lipid IVA signaling (supplemental Fig. S7b). Enhanced responses to lipid IVA were weakly detected in the single mutants E369K and Q436R but were more profoundly observed in the double mutant E369K/Q436R (Fig. 4, c and d) and the triple mutant E369K/K388S/Q436R (supplemental Fig. S7b). In other words, the gain of positive charge at the dimerization interface alone on hTLR4 is sufficient to convert the mMD-2·hTLR4 complex from a lipid IVA nonresponder to a lipid IVA responder. Thus, mMD-2-mediated ionic interactions between the 4′-PO3 on lipid IVA and the positive patch on TLR4 (wild type mTLR4 the hTLR4 double mutant E369K/Q436R) are essential for the LPS mimetic activity of lipid IVA.
DISCUSSION
Several studies have been dedicated to study the underlying mechanism for the species-specific recognition of lipid IVA, because it has long been clear that understanding this pharmacology would offer important insights for how the LPS receptor functions. Surprisingly, these studies have achieved contradictory results. Although we (16) and Beutler and co-workers (17) suggested that TLR4 is responsible for the species-specific activation of lipid IVA, Miller and co-workers (18) and Miyake and co-workers (14) indicated that MD-2 is responsible for this species specificity. In all of these experiments, the animal serum that included variable amounts of bovine MD-2 was included in the assays, which added a confounding variable. In addition, transient transfection was generally used to introduce TLR4 and MD-2 into cells, and therefore, adequate expressions of TLR4 and MD-2 were often assumed. This was especially the case for mouse MD-2, which in our hands was minimally expressed after transfection in any mammalian cell line. Miyake and co-workers (14) appear to have had similar problems. Their report focusing on the species-specific response to lipid IVA revealed that they were unable to observe any response, regardless of the ligand, when mouse MD-2 was transfected with human TLR4 (14). One important advance here is that we were able to provide recombinant protein as the source of MD-2 at defined concentrations. Hence, an important combination of TLR4·MD-2 can now be tested (mouse MD-2 plus human TLR4). We found that mTLR4 is a prerequisite but not the sole determinant for the agonist activity of lipid IVA, which also required the presence of mMD-2. The dual requirement of mTLR4 and mMD-2 for the agonistic activity of lipid IVA was also confirmed in MD-2-deficient BMDMs, a relevant system in which levels of mTLR4 are physiologic.
We then used computational docking and modeling to generate a working model for lipid IVA activation. It appears that both mMD-2 and mTLR4 are required for lipid IVA signaling, because only the mMD-2·mTLR4 complex provides an environment for lipid IVA to efficiently interact with mTLR4′ at the dimerization interface. The essential role of mTLR4 arises mainly from its unique surface charge at the dimerization interface (Lys367 and Arg434) and, hence, its capability to interact with the 4′-PO3 of lipid IVA through ionic interactions.
In comparison, the essential role of mMD-2 arises from its ability to bind lipid IVA so that the ligand sits shallowly in its hydrophobic pocket in a tilted conformation. This provides the proper scaffold for the 4′phosphate of lipid IVA to interact with a positively charged patch on mouse TLR4 at the dimerization interface and trigger receptor activation. This unique scaffolding role of mMD-2 in packaging the tetra-acylated lipid IVA derives from differences in its hydrophobic pocket in comparison with hMD-2. The hydrophobic pockets of hMD-2 and mMD-2 differ in their fine volume, dimension, and adjacent surface charges (supplemental Fig. S8). Although the hydrophobic pocket of hMD-2 looks like a true pocket, i.e. a deep invagination with a sealed bottom, the hydrophobic pocket of mMD-2 is more likely a funnel structure (supplemental Fig. S8a). Its pocket is wider and shallower near the entrance but deeper at the bottom, rendering the appearance of a “hole” at the bottom. The surface charges near the pocket entrance are also slightly different between the two species of MD-2. Human MD-2 has the strongly positively charged Lys122 on the β7 strand that flanks the pocket entrance and Lys125 on the Phe126 loop that mediates dimerization (12). In comparison, mMD-2 has a negatively charged Glu122 at the pocket entrance and the highly hydrophobic Leu125 on the Phe126 loop. The charge reversal from Lys122 to Glu122 likely introduces repulsive forces toward the bis-phosphate diglucosamine backbone of lipid IVA, whereas a gain of hydrophobicity from Lys125 to Leu125 in mMD-2 likely facilitates interaction with the acyl chains of lipid IVA. Consequently, we predict that lipid IVA packs more superficially in the mMD-2 hydrophobic pocket than in the human receptor, with the diglucosamine backbone tilted toward the Cys95–Cys105 loop (which binds MD-2 to TLR4). Supporting data for this prediction have been reported by Muroi and Tanamoto (32), who demonstrated that when Glu122 was mutated to Lys122 on mMD-2, lipid IVA no longer functioned as an agonist. In a sense, it is the hydrophobic pocket of mMD-2 that determines the species-specific pharmacology of lipid IVA with respect to mMD-2.
In the published hTLR4·hMD-2·LPS co-crystal structure (12), extensive hydrophobic interactions were observed at the dimerization interface, involving hydrophobic residues Phe126, Ile124, Leu87, Met85, and Val82 on hMD-2, the sixth acyl chain of LPS, and hydrophobic residues Phe440′, Leu444′, and Phe463′ on the opposing hTLR4′. When Phe126 on hMD-2 was mutated to alanine, receptor dimerization did not occur (11). Hence, the authors proposed that hydrophobic interactions are the primary driving force for receptor dimerization and activation (11, 12). This hypothesis was supported by mutagenesis studies from Jerala and co-workers (33), showing that when the hydrophobic residues at the dimerization interface on MD-2 and TLR4 were mutated to Ala, receptor activation was not observed. Therefore, hydrophobic interactions at the dimerization interface appeared to be the driving force for receptor dimerization. These mutagenesis data on MD-2 and TLR4, however, should not be confused with “mutagenesis on LPS.” These data only demonstrated that hydrophobic interactions arising from hydrophobic residues on MD-2 and TLR4 are essential for receptor dimerization. They did not suggest that hydrophobic interactions derived from the sixth acyl chain of LPS at the dimerization interface are essential for receptor activation. In fact, the Phe126 loop on hMD-2 has to “fold back” to accommodate the extra acyl chain of LPS at the dimerization interface to allow dimerization to occur (11, 12), strongly suggesting a redundancy of hydrophobic interactions at the dimerization interface in regard to receptor dimerization.
It should be noted that the present study does not contradict these findings. Except residue 85 which is a Met in hMD-2, but an Ile in mMD-2, all of the hydrophobic residues on MD-2 and TLR4′ at the dimerization interface are completely conserved between the two species. Therefore, hydrophobic interactions between mMD-2 and mTLR4′ are preserved at the dimerization interface, which can still function as the primary driving force for receptor dimerization. These interactions by themselves, however, are not sufficient to provide enough forces for receptor dimerization. Additional forces such as ionic interactions, as described here, are required for the formation of an active receptor complex. Although the sixth acyl chain of E. coli lipid A may enhance receptor activation, it should be noted that penta-acylated lipid A is capable of activating TLR4·MD-2 with the same potency as hexa-acylated lipid A (34). Thus, whereas hydrophobic forces are clearly critical for TLR4·MD-2 activation, they are not sufficient.
A noticeable difference between the hTLR4·hMD-2·LPS crystal structure and our lipid IVA activation model is the opposite orientation of the lipids in the MD-2 hydrophobic pockets. This difference should not result in a significant functional difference, because lipid IVA can be considered as functionally symmetric. Additionally, it is plausible that lipid A inserts into MD-2 in both orientations and that only one orientation was well resolved by crystallographic approaches. This might happen if a particular crystal was analyzed in which one orientation predominated or even if there were a preponderance of one orientation over the other. It is also possible that the current configuration of LPS is favored in the co-crystal structure, because the co-crystallized LPS has an asymmetric core polysaccharide that interacts extensively with TLR4 (12).
Lipid IVA has partial activity in the equine TLR4·MD-2 receptor (19). One explanation for this observation is that the relative orientation between equine MD-2 and equine TLR4 are different from other species. In particular, repulsive forces from the phosphate of lipid IVA combined with residue 122 on MD-2 may not be required to “lift” lipid IVA up in the horse receptor, accounting for the partial activity. Similar to human MD-2, equine MD-2 has a positively charged residue, Arg122, at the hydrophobic pocket entrance. Therefore, repulsive forces cannot be generated at the pocket entrance to lift MD-2 up. However, in a recent separate study, we identified that not only residues near the hydrophobic pocket of MD-2 are essential for lipid IVA activation but also residues at the A patch (11, 12) of MD-2 that physically associate with TLR4 are important for lipid IVA activation. Specifically, we identified that Gly69 and Tyr42 on mMD-2 are required for a full response to lipid IVA.5 Because Gly69 and Tyr42 reside at the opposite surface of the dimerization interface, these residues likely indirectly affect receptor dimerization through affecting the MD-2·TLR4 binding angle. Because equine MD-2 has Ser42 instead of Tyr42, the binding angle between equine MD-2 and equine TLR4 could be slightly different from the human and mouse complexes. Hence, lipid IVA functions as a partial agonist for the equine receptor.
Despite of all its implications, the resolution of the complete TLR4·MD-2/LPS crystal structure did not identify the charge interactions on TLR4 at the dimerization interface that appear to be so important for lipid A activity. In other words, the crystal structure of TLR4·MD-2/LPS did not allow us to make conclusions concerning whether the physical proximity between the charged groups of LPS and hTLR4 play a functional role in LPS activation. As can be seen here, the use of lipid IVA as a selective lipid A agonist allows us to address questions concerning the meaning of the receptor structure that might otherwise be difficult to test. Because both MD-2 and TLR4 are required for lipid IVA signaling, we were able to dissect the roles of TLR4 and MD-2 on receptor activation to identify a key ionic interaction that might otherwise have been missed.
Indeed, a practical application of the results presented here are opportunities for designing better adjuvants. New compounds with alterations in charge and the way in which they bind to MD-2 (via acylation patterns and/or chain length) are likely to affect immunogenicity differentially, and the effects of alterations in lipid A structure should be amenable to prediction using an approach similar to that presented here. The view that minor alterations in adjuvants can have major effects on outcomes has been borne out by recent clinical trials for important new vaccines, such as the circumsporozoite vaccine for malaria (35, 36).
Supplementary Material
Acknowledgments
We thank Kristen Halmen for help on bone marrow-derived macrophages preparation, Brian Monks for help on mutagenesis, and Rosane DeOliveira for help with flow cytometry. We thank Dr. C. James McKnight for critical reading of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM54060 (to D. T. G.) and AI057588 (to E. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S8.
J. Meng, J. R. Drolet, B. Monks, and D. T. Golenbock, unpublished data.
- LPS
- lipopolysaccharide
- TLR
- Toll-like receptor
- h
- human
- BMDM
- bone marrow-derived macrophage
- DMEM
- Dulbecco's modified Eagle's medium
- m
- mouse.
REFERENCES
- 1.Janeway C. A., Jr., Medzhitov R. (1998) Semin. Immunol. 10, 349–350 [DOI] [PubMed] [Google Scholar]
- 2.Rangel-Frausto M. S. (2005) Arch. Med. Res. 36, 672–681 [DOI] [PubMed] [Google Scholar]
- 3.Angus D. C., Linde-Zwirble W. T., Lidicker J., Clermont G., Carcillo J., Pinsky M. R. (2001) Crit. Care Med. 29, 1303–1310 [DOI] [PubMed] [Google Scholar]
- 4.Qureshi N., Takayama K., Ribi E. (1982) J. Biol. Chem. 257, 11808–11815 [PubMed] [Google Scholar]
- 5.Homma J. Y., Matsuura M., Kumazawa Y. (1990) Adv. Exp. Med. Biol. 256, 101–119 [DOI] [PubMed] [Google Scholar]
- 6.Takada H., Kotani S. (1989) Crit. Rev. Microbiol. 16, 477–523 [DOI] [PubMed] [Google Scholar]
- 7.Fujimoto Y., Adachi Y., Akamatsu M., Fukase Y., Kataoka M., Suda Y., Fukase K., Kusumoto S. (2005) J. Endotoxin Res. 11, 341–347 [DOI] [PubMed] [Google Scholar]
- 8.Montminy S. W., Khan N., McGrath S., Walkowicz M. J., Sharp F., Conlon J. E., Fukase K., Kusumoto S., Sweet C., Miyake K., Akira S., Cotter R. J., Goguen J. D., Lien E. (2006) Nat. Immunol. 7, 1066–1073 [DOI] [PubMed] [Google Scholar]
- 9.Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golenbock D. T., Hampton R. Y., Qureshi N., Takayama K., Raetz C. R. (1991) J. Biol. Chem. 266, 19490–19498 [PubMed] [Google Scholar]
- 11.Kim H. M., Park B. S., Kim J. I., Kim S. E., Lee J., Oh S. C., Enkhbayar P., Matsushima N., Lee H., Yoo O. J., Lee J. O. (2007) Cell 130, 906–917 [DOI] [PubMed] [Google Scholar]
- 12.Park B. S., Song D. H., Kim H. M., Choi B. S., Lee H., Lee J. O. (2009) Nature [DOI] [PubMed] [Google Scholar]
- 13.Ohto U., Fukase K., Miyake K., Satow Y. (2007) Science 316, 1632–1634 [DOI] [PubMed] [Google Scholar]
- 14.Akashi S., Nagai Y., Ogata H., Oikawa M., Fukase K., Kusumoto S., Kawasaki K., Nishijima M., Hayashi S., Kimoto M., Miyake K. (2001) Int. Immunol. 13, 1595–1599 [DOI] [PubMed] [Google Scholar]
- 15.Schromm A. B., Lien E., Henneke P., Chow J. C., Yoshimura A., Heine H., Latz E., Monks B. G., Schwartz D. A., Miyake K., Golenbock D. T. (2001) J. Exp. Med. 194, 79–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lien E., Means T. K., Heine H., Yoshimura A., Kusumoto S., Fukase K., Fenton M. J., Oikawa M., Qureshi N., Monks B., Finberg R. W., Ingalls R. R., Golenbock D. T. (2000) J. Clin. Invest. 105, 497–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poltorak A., Ricciardi-Castagnoli P., Citterio S., Beutler B. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2163–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajjar A. M., Ernst R. K., Tsai J. H., Wilson C. B., Miller S. I. (2002) Nat. Immunol. 3, 354–359 [DOI] [PubMed] [Google Scholar]
- 19.Walsh C., Gangloff M., Monie T., Smyth T., Wei B., McKinley T. J., Maskell D., Gay N., Bryant C. (2008) J. Immunol. 181, 1245–1254 [DOI] [PubMed] [Google Scholar]
- 20.Edelhoch H. (1967) Biochemistry 6, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 21.Latz E., Visintin A., Lien E., Fitzgerald K. A., Monks B. G., Kurt-Jones E. A., Golenbock D. T., Espevik T. (2002) J. Biol. Chem. 277, 47834–47843 [DOI] [PubMed] [Google Scholar]
- 22.Liu W. C., Oikawa M., Fukase K., Suda Y., Kusumoto S. (1999) Bull. Chem. Soc. Jpn. 72, 1377–1385 [Google Scholar]
- 23.Hirschfeld M., Ma Y., Weis J. H., Vogel S. N., Weis J. J. (2000) J. Immunol. 165, 618–622 [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald K. A., Palsson-McDermott E. M., Bowie A. G., Jefferies C. A., Mansell A. S., Brady G., Brint E., Dunne A., Gray P., Harte M. T., McMurray D., Smith D. E., Sims J. E., Bird T. A., O'Neill L. A. (2001) Nature 413, 78–83 [DOI] [PubMed] [Google Scholar]
- 25.Nagai Y., Akashi S., Nagafuku M., Ogata M., Iwakura Y., Akira S., Kitamura T., Kosugi A., Kimoto M., Miyake K. (2002) Nat. Immunol. 3, 667–672 [DOI] [PubMed] [Google Scholar]
- 26.Meng J., Parroche P., Golenbock D. T., McKnight C. J. (2008) J. Biol. Chem. 283, 3376–3384 [DOI] [PubMed] [Google Scholar]
- 27.Huey R., Morris G. M., Olson A. J., Goodsell D. S. (2007) J. Comput. Chem. 28, 1145–1152 [DOI] [PubMed] [Google Scholar]
- 28.Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 29.Sanner M. F. (1999) J. Mol. Graph Model 17, 57–61 [PubMed] [Google Scholar]
- 30.Moitessier N., Henry C., Maigret B., Chapleur Y. (2004) J. Med. Chem. 47, 4178–4187 [DOI] [PubMed] [Google Scholar]
- 31.Prohinar P., Re F., Widstrom R., Zhang D., Teghanemt A., Weiss J. P., Gioannini T. L. (2007) J. Biol. Chem. 282, 1010–1017 [DOI] [PubMed] [Google Scholar]
- 32.Muroi M., Tanamoto K. (2006) J. Biol. Chem. 281, 5484–5491 [DOI] [PubMed] [Google Scholar]
- 33.Resman N., Vasl J., Oblak A., Pristovsek P., Gioannini T. L., Weiss J. P., Jerala R. (2009) J. Biol. Chem. 284, 15052–15060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rietschel E. T., Brade L., Schade U., Galanos C., Freudenberg M., Lüderitz O., Kusumoto S., Shiba T. (1987) Eur. J. Biochem. 169, 27–31 [DOI] [PubMed] [Google Scholar]
- 35.Bejon P., Lusingu J., Olotu A., Leach A., Lievens M., Vekemans J., Mshamu S., Lang T., Gould J., Dubois M. C., Demoitié M. A., Stallaert J. F., Vansadia P., Carter T., Njuguna P., Awuondo K. O., Malabeja A., Abdul O., Gesase S., Mturi N., Drakeley C. J., Savarese B., Villafana T., Ballou W. R., Cohen J., Riley E. M., Lemnge M. M., Marsh K., von Seidlein L. (2008) N. Engl. J. Med. 359, 2521–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdulla S., Oberholzer R., Juma O., Kubhoja S., Machera F., Membi C., Omari S., Urassa A., Mshinda H., Jumanne A., Salim N., Shomari M., Aebi T., Schellenberg D. M., Carter T., Villafana T., Demoitié M. A., Dubois M. C., Leach A., Lievens M., Vekemans J., Cohen J., Ballou W. R., Tanner M. (2008) N. Engl. J. Med. 359, 2533–2544 [DOI] [PubMed] [Google Scholar]
- 37.Koradi R., Billeter M., Wuthrich K. (1996) J. Mol. Graph 14, 51–55, 29–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.