Abstract
Our previous study demonstrated that transforming growth factor (TGF)-β activates β-catenin signaling through Smad3 interaction with β-catenin in chondrocytes. In the present studies, we further investigated the detailed molecular mechanism of the cross-talk between TGF-β/Smad3 and Wnt/β-catenin signaling pathways. We found that C-terminal Smad3 interacted with both the N-terminal region and the middle region of β-catenin protein in a TGF-β-dependent manner. Both Smad3 and Smad4 were required for the interaction with β-catenin and protected β-catenin from an ubiquitin-proteasome-dependent degradation. In addition, the formation of the Smad3-Smad4-β-catenin protein complex also mediated β-catenin nuclear translocation. This Smad3-mediated regulatory mechanism of β-catenin protein stability enhanced the activity of β-catenin to activate downstream target genes during chondrogenesis. Our findings demonstrate a novel mechanism between TGF-β and Wnt/β-catenin signaling pathways during chondrocyte development.
Keywords: β-Catenin, Bone, Protein Degradation, SMAD Transcription Factor, Ubiquitination, Chondrocyte, Smad3, TGF-β
Introduction
Endochondral bone formation is a tightly regulated process. The process starts from mesenchymal cell condensation with subsequent formation of chondrocytes and goes to chondrocyte proliferation and differentiation into hypertrophic chondrocytes. A number of growth factors and signaling molecules are involved in the regulation of chondrogenesis, including Sox9, Ihh, parathyroid hormone-related protein, bonemorphogenetic protein (BMP),2 TGF-β, and Wnt signaling proteins (1).
β-Catenin is a central molecule in canonical Wnt pathway. In the absence of Wnt ligands, cytoplasmic β-catenin is constitutively phosphorylated by a multi-protein complex containing kinases such as GSK-3β, CK1, and scaffolding proteins including Axins, APC, and Dsh. The phosphorylated β-catenin is recognized and modified by ubiquitin-protein isopeptide ligase complex, such as Skp1-β-TrCP (2, 3), and finally degraded by the 26 S proteasome (4). During skeletal development, Wnt/β-catenin signaling controls mesenchymal progenitor cells selectively to differentiate into osteoblasts. In the absence of β-catenin, the progenitor cells differentiate into chondrocytes instead of osteoblasts (5), indicating that β-catenin is required to suppress early mesenchymal cell differentiation into chondrocytes. However, the canonical Wnt pathway has also been reported to play a critical role in chondrocyte proliferation and hypertrophy. Chondrocyte-specific β-catenin deletion (targeted by Col2a1-Cre) leads to decreased chondrocyte proliferation and delayed hypertrophic chondrocyte differentiation (6, 7). In contrast, constitutive activation of β-catenin in chondrocytes through deletion of exon 3 of the β-catenin gene leads to severely compromised cartilage formation (6). Other signaling molecules could interact with β-catenin and indirectly regulate chondrocyte function (8, 9).
The TGF-β signaling is involved in multiple cellular processes including cell proliferation, differentiation, and apoptosis. TGF-β ligand transduces its signal through binding with type II and type I receptors leading to Smad2/3 phosphorylation. Phosphorylated Smad2/3 interact with Smad4 and then translocate into nucleus where they act as transcription factor and participate in transcriptional regulation of target genes. Cumulative evidence demonstrates that TGF-β stimulates cell proliferation during early chondrocyte formation. Specific expression of the dominant-negative form of type II TGF-β receptor in articular cartilage and synovium promotes chondrocyte differentiation leading to osteoarthritis (10). More studies are required to further understand the detailed molecular mechanism of TGF-β during chondrogenesis.
Recent studies suggest that TGF-β interacts with Wnt signaling during chondrogenesis (11), but the detailed molecular mechanism remains poorly understood. Our previous studies demonstrated that TGF-β activates β-catenin signaling through Smad3 (12). In the present studies, we further investigated the role of Smad3 and Smad4 in the protection of β-catenin degradation and β-catenin nuclear translocation in chondrocytes.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
COS cells and 293T cells were cultured in Dulbecco's modified Eagle's medium, and RCJ3.1C5.18 cells were cultured in α-minimum essential medium supplemented with 10% fetal calf serum at 37 °C under 5% CO2. DNA plasmids were transiently transfected into COS, 293T, or RCJ3.1C5.18 in 6-cm culture dishes using Lipofectamine 2000 (Invitrogen). Empty vector was used to keep the total amount of transfected DNA plasmid constant in each group in all experiments. FLAG-EGFP plasmid was co-transfected as an internal control to evaluate transfection efficiency. Western blotting and immunoprecipitation (IP) assays were performed 24 h after transfection.
Western Blotting, Immunoprecipitation, and Immunostaining
Western blotting and IP were performed as described previously (12). The interaction between endogenous β-catenin and Smad3/4 and the ubiquitination of endogenous β-catenin protein was determined in chondrogenic RCJ3.1C5.18 cells. Proteasome inhibitor MG132 (10 μm) was added to the cell culture 4 h before the cells were harvested in β-catenin ubiquitination assay. The rat anti-β-catenin monoclonal antibody (clone 14) was purchased from BD Biosciences (San Jose, CA). Rabbit anti-Smad3 polyclonal antibody (Zymed Laboratories Inc.) was used to detect Smad3 expression. β-Catenin and Smad3 immunostaining was performed as previously described (12).
In Vivo Protein Decay Assay
RCJ3.1C5.18 cells were seeded in 15-cm culture dishes, and 100 mm Smad3 siRNA or 10 μg of pCMV5B-FLAG-Smad3 were used for transfection. 24 h after transfection, the cells were trypsinized and split into five 10-cm dishes. 12 h after recovery, the cells were cultured in regular medium with 10 μg/ml cycloheximide (Calbiochem) for 0, 30, 60, 120, and 300 min before harvesting. Western blotting was performed to detect the decay of endogenous β-catenin proteins.
Luciferase and Real Time PCR Assays
The plasmids of reporter constructs were co-transfected with wild type (WT) Smad3 or mutant forms of Smad3 expression plasmid into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with Wnt3a (100 ng/ml) for 36 h or with TGF-β (2 ng/ml) for 4 h. The cell lysates were then collected, and luciferase activity was measured using a Promega Dual Luciferase reporter assay kit (Promega, Madison, WI). Real time PCR was performed as described previously (12). The primers used in this study were as follow: β-actin, 5′-TGT TAC CAA CTG GGA CGA CA (upper primer) and 3′-CTG GGT CAT CTT TTC ACG GT (lower primer); Smad4, 5′-TCG ATT CAA ACC ATC CAA CA (upper primer) and 3′-GCC CTG AAG CTA TCT GCA AC (lower primer); Dkk1, 5′-AAT CGA GGA AGG CAT CAT TG (upper primer) and 3′-GCT TGG TGC ATA CCT GAC CT (lower primer); Axin2, 5′-CTC TAA CGC TAG GCG GAA TG (upper primer) and 3′-CCA GAA GTC CAG GGT ATC CA (lower primer); and cyclin D1, 5′-GCG TAC CCT GAC ACC AAT CT (upper primer) and 3′-GGC TCC AGA GAC AAG AAA CG (lower primer). The PCR conditions included a denaturation step at 95 °C for 10 min, followed by 40 cycles of denaturation at 95 °C for 10 s, annealing at 58 °C for 15 s, and extension at 72 °C for 10 s. Detection of the fluorescent product was carried out at the end of the 72 °C extension period. The PCR products were subjected to a melting curve analysis, and the data were analyzed and quantified with the Rotor-Gene analysis software. Dynamic tube normalization and noise slope correction were used to remove background fluorescence. Each sample was tested at least in triplicate and repeated using three independent cell preparations.
Statistics
Statistical comparison between two groups was performed using unpaired Student's t test. p < 0.05 was considered significant and is denoted in the figures.
RESULTS
Interaction Domains of Smad3 and β-Catenin
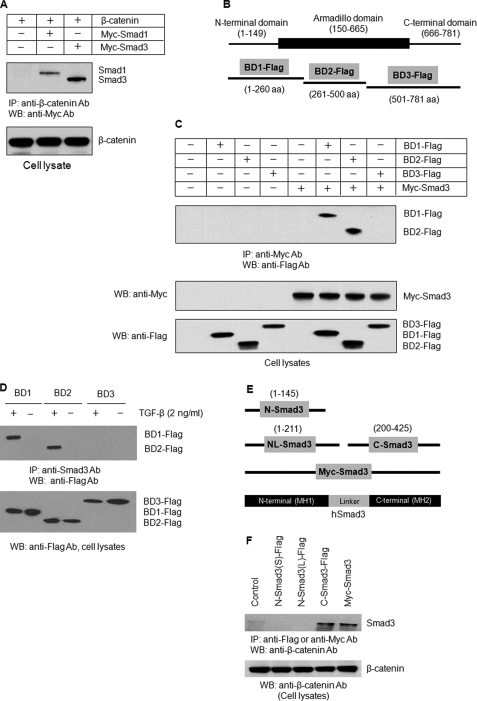
Our previous study demonstrated that Smad3 interacts with β-catenin in chondrocytes (12). In the present studies, we first mapped the interaction domains of β-catenin and Smad3. COS cells were overexpressed with Smad1 or Smad3 together with β-catenin, cell lysates were immunoprecipitated with an anti-β-catenin antibody, and expression of Smad1/3 proteins was detected by Western blotting. Both Smad1 and Smad3 interacted with β-catenin, and Smad3 exhibited much stronger interaction with β-catenin than Smad1 (Fig. 1A). β-Catenin protein contains an N-terminal domain (amino acids 1–149), a central armadillo repeat domain (amino acids 150–665), and a C-terminal p300 binding domain (amino acids 666–781) (13). In this study, we generated three mutant β-catenin constructs that encode different segments of β-catenin protein (BD1, amino acids 1–260; BD2, amino acids 261–500; and BD3, amino acids 501–781) (Fig. 1B). These three truncated forms of β-catenin expression constructs were co-transfected with Smad3 expression plasmid into COS cells. The BD1 and BD2 fragments of β-catenin interacted with Smad3 (Fig. 1C), demonstrating that Smad3 interacts with both the N-terminal and the mid-region of β-catenin protein. We then investigated whether β-catenin fragments interact with endogenous Smad3 in chondrocytes and whether this interaction is TGF-β-dependent. The rat chondrogenic progenitor cell line RCJ3.1C5.18 cells were transfected with three truncated forms of β-catenin and treated with or without TGF-β. Endogenous Smad3 protein was immunoprecipitated, and expression of truncated forms of β-catenin was detected by Western blotting. The results demonstrated that Smad3 interacted with β-catenin BD1 and BD2 fragments in a TGF-β-dependent manner in RCJ3.1C5.18 cells (Fig. 1D). To clarify which domain of Smad3 interacts with β-catenin, we generated three truncated forms of Smad3 expression constructs: N-Smad3 (amino acids 1–145), NL-Smad3 (amino acids 1–211), and C-Smad3 (amino acids 200–425) (Fig. 1E). These three truncated forms of Smad3 were co-transfected with β-catenin in RCJ3.1C5.18 cells, and IP assay was performed. C-terminal Smad3 but not N-Smad3 or NL-Smad3 interacted with β-catenin (Fig. 1F). Taken together, the results demonstrate that the C-terminal domain of Smad3 interacts with both the N-terminal and the mid-region of β-catenin, and this interaction is TGF-β-dependent.
FIGURE 1.
Interaction domains of Smad3 and β-catenin. A, β-catenin expression plasmid was co-transfected with Myc-Smad1 or Myc-Smad3 into COS cells. 24 h after transfection, the cell lysates were collected. IP was performed using the anti-β-catenin antibody followed by Western blotting (WB) using an anti-Myc antibody (top panel). Although both Smad1 and Smad3 interact with β-catenin, Smad3 had stronger interaction with β-catenin. B, β-catenin protein structure and truncated β-catenin constructs. C, COS cells were transfected with truncated β-catenin constructs: FLAG-BD1 (amino acids 1–260), FLAG-BD2 (amino acids 261–500), and FLAG-BD3 (amino acids 501–781) with Myc-Smad3. 24 h after transfection, the cell lysates were collected. IP was performed using the anti-Myc antibody followed by Western blotting using the anti-FLAG antibody (top panel). The N-terminal domain and mid-region of β-catenin interacted with Smad3. D, RCJ3.1C5.18 cells were transfected with three truncated β-catenin constructs FLAG-BD1, -BD2, and -BD3. 24 h after transfection, the cells were treated with TGF-β (2 ng/ml) for 4 h. IP was performed using the anti-Smad3 antibody followed by Western blotting using the anti-FLAG antibody (top panel). The N-terminal domain and mid-region of β-catenin interacted with Smad3 in a TGF-β-dependent manner in RCJ3.1C5.18 cells. E, a diagram shows Smad3 structure and truncated Smad3 constructs. F, WT and truncated Smad3 constructs: FLAG-N-Smad3 (amino acids 1–145), FLAG-NL-Smad3 (amino acids 1–211), and FLAG-C-Smad3 (amino acids 200–425) were transfected into 293T cells. 24 h after transfection, the cell lysates were collected. IP was performed using the anti-FLAG or anti-Myc antibody followed by Western blotting using the anti-β-catenin antibody (top panel). The C-terminal domain of Smad3 interacted with β-catenin. IP, immunoprecipitation.
Smad3 Protects β-Catenin Degradation in Chondrocytes
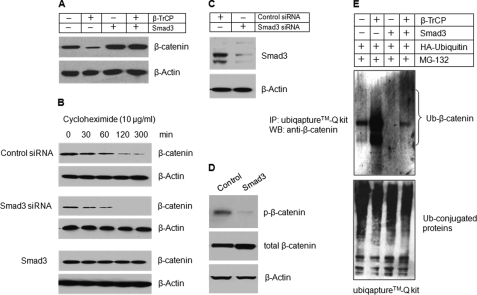
β-Transducin repeat-containing protein (β-TrCP) is an E3 ubiquitin ligase in the SCFβ-TrCP protein complex, which induces β-catenin degradation in many cell types (4, 15–17). To determine whether β-TrCP induces β-catenin degradation in chondrocytes and whether Smad3 plays a role in β-catenin degradation, we transfected β-TrCP expression plasmid with or without Smad3 into RCJ3.1C5.18 cells. Overexpression of β-TrCP mediated β-catenin degradation; however, Smad3 blocked the effect of β-TrCP (Fig. 2A). To determine the effect of Smad3 on β-catenin degradation, the RCJ3.1C5.18 cells were overexpressed with Smad3 or transfected with Smad3 siRNA. 24 h after the transfection, protein synthesis was stopped by the addition of protein synthesis inhibitor cycloheximide. β-Catenin protein levels in the control siRNA transfected group began to decline 60 min after treatment with cycloheximide, whereas β-catenin protein levels declined much faster at the very beginning of cycloheximide treatment, and the protein levels decreased dramatically in the Smad3 siRNA transfected group (Fig. 2B). In contrast, overexpression of Smad3 protected β-catenin degradation. β-Catenin protein levels remained stable during the entire period of cycloheximide treatment (Fig. 2B). The silencing efficiency of Smad3 siRNA was over 70% (Fig. 2C). To determine whether Smad3 inhibits β-catenin phosphorylation, RCJ3.1C5.18 cells were transiently transfected with Smad3 expression plasmid, and expression of phospho-β-catenin was detected. Overexpression of Smad3 significantly inhibited β-catenin phosphorylation (Fig. 2D). We also performed the ubiquitination assay using UbiQaptureTM-Q kit. After 4 h of treatment with MG-132, the cell lysates were immunoprecipitated with the anti-ubiquitin-conjugated beads, and ubiquitinated β-catenin protein levels were detected through Western blotting. Overexpression with β-TrCP induced the ubiquitination of β-catenin (Fig. 2E, second lane), whereas transfection of Smad3 reversed the effect of β-TrCP on β-catenin degradation (Fig. 2E, fourth lane).
FIGURE 2.
Smad3 protects β-catenin degradation in chondrocytes. A, Smad3 expression plasmid was co-transfected with β-TrCP into RCJ3.1C5.18 cells. 24 h after transfection, the cell lysates were collected, and Western blotting (WB) was performed using the anti-β-catenin antibody. Transfection of Smad3 prevented β-catenin degradation. B, RCJ3.1C5.18 cells were transfected with control or Smad3 siRNA or Smad3 expression plasmid. The cell lysates were collected 0, 30, 60, 120, or 300 min after cycloheximide treatment (10 μg/ml), and the β-catenin protein levels were detected by Western blotting. Silencing of Smad3 accelerated β-catenin degradation and transfection of Smad3 prevented β-catenin degradation. C, the silencing efficiency of Smad3 siRNA was determined by Western blotting. The efficiency is over 70%. D, RCJ3.1C5.18 cells were transiently transfected with Smad3 expression plasmid. 24 h after transfection, the cell lysates were collected, and Western blotting was performed using the anti-p-β-catenin antibody. Smad3 significantly inhibited β-catenin phosphorylation. E, Smad3 and HA-ubiquitin expression plasmids were co-transfected with β-TrCP expression plasmid into RCJ3.1C5.18 cells in the presence of proteasome inhibitor MG132 (10 μm, 4 h of incubation). 24 h after transfection, the cell lysates were collected, and ubiquitinated proteins were pulled down using an UbiQaptureTM-Q kit, and ubiquitinated β-catenin was detected using the anti-β-catenin antibody. Transfection of β-TrCP induced β-catenin ubiquitination (second lane) and co-transfection of Smad3 prevented β-catenin ubiquitination (fourth lane). Ub, ubiquitin; IP, immunoprecipitation.
Smad4 Is Involved in the Protection of β-Catenin Degradation
In TGF-β signaling pathway, Smad4 acts as co-Smad to bind with Smad2/3 and facilitates the nuclear translocation of the heteromeric complex. To determine whether Smad4 interacts with β-catenin, immunoprecipitation was performed using the anti-β-catenin antibody, Smad4 expression was detected by Western blotting. The interaction between Smad4 and β-catenin was detected in a Smad3-dependent manner. Overexpression of Smad3 facilitated the interaction between Smad4 and β-catenin, whereas silencing of Smad3 blocked this interaction (Fig. 3A). To determine whether this interaction is TGF-β-dependent, RCJ3.1C5.18 cells were treated with TGF-β, and the IP assay was performed. Under TGF-β induction, the interaction between Smad4 and β-catenin was enhanced (Fig. 3B). To determine whether silencing of Smad4 accelerates β-catenin degradation, the β-catenin protein levels were detected after Smad4 siRNA was transfected. Transfection of Smad4 siRNA effectively inhibited Smad4 mRNA expression (Fig. 3C). Silencing of Smad4 attenuated the β-catenin protein level, even in the presence of Smad3. Transfection of Smad3 completely reversed β-TrCP-induced β-catenin degradation. However, Smad3 failed to reverse β-TrCP-induced β-catenin degradation when Smad4 siRNA was transfected into these cells (Fig. 3D). These results suggest that Smad4, together with Smad3, forms a complex with β-catenin and protects β-catenin degradation. Smad2 is another R-Smad protein that is phosphorylated by TGF-β receptors and then interacts with Smad4. The interaction between Smad2 and β-catenin has been demonstrated (18). In this study, we determined whether Smad2 protects β-catenin degradation by Western blotting. Smad2 partially reversed the effect of β-TrCP-induced β-catenin degradation. When the Smad3 expression was knocked down, Smad2 showed minimal effect on protection of β-catenin degradation (Fig. 3E). These results demonstrate that Smad3 plays dominant role in the protection of β-catenin degradation.
FIGURE 3.
Smad4 is involved in the protection of β-catenin degradation. A, Smad3 expression plasmid or Smad3 siRNA were co-transfected with HA-Smad4 expression plasmid into RCJ3.1C5.18 cells. Immunoprecipitation was performed using the anti-β-catenin antibody followed by Western blotting (WB) using the anti-HA antibody (top panel). As a control, IP was performed using the anti-Smad3 antibody followed by Western blotting using the anti-HA antibody (middle panel). Smad4 interacted with β-catenin in a Smad3-dependent manner. B, cell lysates were collected from RCJ3.1C5.18 cells treated with or without TGF-β (2 ng/ml) for 4 h. IP was performed using the anti-β-catenin antibody followed by Western blotting using the anti-HA antibody (top panel). As a control, IP was also performed using the anti-Smad3 antibody followed by Western blotting using the anti-HA antibody (middle panel). Smad4 interacted with β-catenin in a TGF-β-dependent manner in RCJ3.1C5.18 cells. C, Smad4 siRNA significantly inhibited Smad4 mRNA expression (∼70% inhibition). D, RCJ3.1C5.18 cells were transfected with Smad3 and β-TrCP expression plasmids in the presence or absence of Smad4 siRNA. 24 h after transfection, the cell lysates were collected, and Western blotting was performed using the anti-β-catenin antibody. Smad4 siRNA inhibited the protective effect of Smad3 on β-catenin degradation. E, RCJ3.1C5.18 cells were co-transfected with Smad2 and β-TrCP expression plasmids with or without Smad3 siRNA. 24 h after transfection, the cell lysates were collected, and Western blotting was performed using the anti-β-catenin antibody. Smad2 showed a limited effect on protection of β-catenin degradation when Smad3 gene was silenced. IP, immunoprecipitation.
Smad3 Promotes β-Catenin Nuclear Translocation
In previous studies, we found that TGF-β promotes nuclear translocation of Smad3 as well as β-catenin in chondrocytes (12). To determine whether Wnt3a-induced β-catenin nuclear translocation is impaired when Smad3 is inhibited in chondrocytes, Smad3 siRNA were transfected into RCJ3.1C5.18 cells. 24 h after transfection of Smad3 siRNA, the cells were treated with Wnt3a, and immunostaining was performed 4 h later. Compared with the control group, in which Wnt3a facilitated β-catenin nuclear translocation, silencing of Smad3 inhibited β-catenin nuclear translocation (Fig. 4A). To further determine whether Smad3 promotes β-catenin nuclear translocation, we have generated two Smad3 NLS mutant constructs: ΔK40K41 and K43N/K44Q mutants based on previous report (19). Expression of the deletion mutant ΔK40K41 of Smad3 is evenly distributed throughout the cells. The K43N/K44Q mutant of Smad3 is significantly less enriched in the nucleus compared with the WT Smad3 and acts as a dominant-negative inhibitor of TGF-β-mediated transcriptional activation (20). To confirm that mutant Smad3 does serve as a dominant-negative mutant, RCJ3.1C5.18 cells were co-transfected with WT Smad3 or two mutant forms of Smad3 and p3TP-Lux reporter with or without TGF-β treatment. The results from the luciferase assay demonstrated that TGF-β stimulated p3TP-Lux reporter activity, and overexpression of Smad3 further increased the reporter activity. In contrast, two mutant forms of Smad3 (ΔK40K41 and K43N/K44Q) partially inhibited TGF-β-mediated reporter activity (Fig. 4B). To determine whether Smad3 promotes β-catenin nuclear translocation, WT and two mutant Smad3 constructs were transfected into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with or without TGF-β (2 ng/ml) for 4 h, and the nuclear translocation of Smad3 and β-catenin were examined by the immunostaining assay. TGF-β induced the nuclear translocation of Smad3 as well as β-catenin in WT Smad3-transfected cells but failed to induce Smad3 and β-catenin localization in mutant Smad3 (K43N/K44Q and Smad3 ΔK40K41) transfected cells (Fig. 4C). These findings are consistent with the previous report (19). To further confirm the nuclear translocation patterns of Smad3 and β-catenin, RCJ3.1C5.18 cells were transfected with WT Smad3 or two mutant forms of Smad3 with or without TGF-β treatment. Nuclear and cytoplasmic proteins were collected separately, and Western blotting was performed. TGF-β induced nuclear Smad3 and β-catenin protein levels in WT Smad3 transfected cells (Fig. 4D). Overexpression of the mutant forms of Smad3 inhibited the stimulatory effect of TGF-β on β-catenin nuclear translocation (Fig. 4D). RCJ3.1C5.18 cells were also transfected with Smad3 siRNA. 24 h after transfection, the cells were treated with TGF-β for 4 h, and β-catenin localization was determined by immunostaining. Silencing of Smad3 inhibited the stimulatory effect of TGF-β on β-catenin nuclear translocation (Fig. 4E). These results suggest that in addition to the protection of β-catenin degradation, Smad3 also assists β-catenin nuclear translocation.
FIGURE 4.
Smad3 promotes β-catenin nuclear translocation. A, RCJ3.1C5.18 cells were treated with Wnt3a (100 ng/ml) for 24 h. The cells were then fixed and incubated with the anti-β-catenin antibody (Ab) followed by the incubation with a fluorescein isothiocyanate-conjugated secondary antibody. Treatment with Wnt3a induced β-catenin nuclear translocation and silencing of Smad3 inhibited β-catenin nuclear translocation. B, p3TP-Lux reporter construct was co-transfected with WT or mutant forms of Smad3 into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with TGF-β (2 ng/ml) for 4 h. The cell lysates were collected, and luciferase assay was performed. Transfection of mutant Smad3 constructs significantly inhibited TGF-β reporter activity. C, WT or mutant forms of Smad3 were transfected into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with TGF-β (2 ng/ml) for 4 h. The cells were then fixed and incubated with the anti-β-catenin or anti-Smad3 antibody followed by the incubation with fluorescein isothiocyanate-conjugated secondary antibody. Transfection of mutant Smad3 constructs inhibited TGF-β-induced Smad3 and β-catenin nuclear translocation. D, RCJ3.1C5.18 cells were transfected with WT or mutant Smad3 (K43N/K44Q and ΔK40K41) expression plasmids. 24 h after transfection, the cells were treated with TGF-β (2 ng/ml) for 4 h. Nuclear and cytoplasmic proteins were collected using NE-PER nuclear and cytoplasmic extraction reagents (Pierce). Western blotting was performed using the anti-β-catenin antibody and anti-FLAG antibody to detect changes in β-catenin and Smad3 protein levels. Transfection of Smad3 increased nuclear Smad3 as well as β-catenin protein levels. In contrast, transfection of mutant Smad3 constructs inhibited Smad3 as well as β-catenin protein levels even in the presence of TGF-β. E, RCJ3.1C5.18 cells were treated with TGF-β (2 ng/ml) for 4 h. The cells were then fixed and incubated with the anti-β-catenin antibody followed by the incubation with a fluorescein isothiocyanate-conjugated secondary antibody. Treatment with TGF-β induced β-catenin nuclear translocation and silencing of Smad3 inhibited β-catenin nuclear translocation.
Smad3 Enhances β-Catenin Activity in Chondrocytes
To investigate the effect of Smad3 on the activity of β-catenin as a transcription factor, RCJ3.1C5.18 cells were transfected with Smad3 siRNA and treated with Wnt3a. mRNA expression and promoter activity of cyclin D1 and Axin2, downstream target genes of β-catenin, were detected. Silencing of Smad3 inhibited the stimulatory effect of Wnt3a on both cyclin D1 and Axin2 expression (Fig. 5, A and B). In addition, knocking down of Smad3 also inhibited Wnt3a-induced cyclin D1 promoter activity (Fig. 5C). To determine whether β-catenin signaling is affected by NLS mutants of Smad3, RCJ3.1C5.18 cells were co-transfected with WT and two mutant constructs of Smad3 together with TOP-flash reporter and treated with or without Wnt3a. The effect of mutant Smad3 on β-catenin signaling was examined by luciferase assay. Smad3 alone significantly increased TOP-flash reporter activity, which is consistent with our previous results (12). Wnt3a-induced β-catenin reporter activity was enhanced by overexpression of Smad3, suggesting that Smad3 protects β-catenin degradation and activates the transcriptional activity of β-catenin. However, the effect of Wnt3a on TOP-flash reporter activity was significantly inhibited by transfection of NLS mutant forms of Smad3 (Fig. 5D). WT and two NLS mutant forms of Smad3 were then co-transfected with cyclin D1 promoter, −962CD1-Luc, into RCJ3.1C5.18 cells with or without β-catenin co-transfection. Compared with the control group, WT Smad3 increased cyclin D1 promoter activity (∼2.5-fold increase). Overexpression of β-catenin significantly enhanced the effect of Smad3 on cyclin D1 promoter activity (6-fold increase). In contrast, two mutant forms of Smad3 inhibited the stimulatory effect of β-catenin on cyclin D1 promoter activity (Fig. 5E). These results suggest that Smad3 nuclear translocation is required for β-catenin to activate downstream target genes. To determine whether TGF-β stimulates cyclin D1 promoter activity in a β-catenin-dependent manner, WT (−962CD1) and mutant (−962CD1) (with the deletion of four TCF binding sites) (21) cyclin D1 promoter constructs were transfected into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with different concentrations of TGF-β (0, 0.2, 1, and 5 ng/ml). Treatment with TGF-β significantly increased WT cyclin D1 promoter activity (6-fold increase, 1 ng/ml of TGF-β), and deletion of TCF binding sites inhibited the stimulatory effect of TGF-β on cyclin D1 promoter activity (Fig. 5E). These results suggest that TGF-β may regulate target genes indirectly through activation of the β-catenin activity.
FIGURE 5.
Smad3 enhances β-catenin activity in chondrocytes. A and B, RCJ3.1C5.18 cells were transfected with control (Cont) or Smad3 siRNA and treated with Wnt3a. Expression of cyclin D1 and Axin2 was examined by real time PCR. Treatment with Wnt3a significantly increased cyclin D1 and Axin2 expression. Silencing of Smad3 inhibited Wnt3a-induced expression of cyclin D1 and Axin2. C, RCJ3.1C5.18 cells were transfected with control or Smad3 siRNA and cyclin D1 promoter construct (−962CD1) and treated with Wnt3a. Wnt3a stimulated cyclin D1 promoter activity, and Smad3 siRNA significantly inhibited Wnt3a-induced cyclin D1 promoter activity. D, WT or NLS mutant forms of Smad3 (K43NK44Q or ΔK40K41) were co-transfected with the TOP-flash reporter construct into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with Wnt3a (100 ng/ml). The cell lysates were collected 24 h later for luciferase assay. Wnt3a-stimulated TOP-flash reporter activity was enhanced by transfection of Smad3 and inhibited by transfection of mutant Smad3. E, WT or NLS mutant forms of Smad3 were co-transfected with β-catenin expression plasmid and the −962CD1 reporter construct into RCJ3.1C5.18 cells. Cell lysates were collected 24 h later for luciferase assay. β-Catenin-induced cyclin D1 promoter activity was enhanced by Smad3 and inhibited by mutant Smad3. F, WT (−962wtCD1) and mutant cyclin D1 (−962mCD1) (TCF binding site deletion) promoter constructs were transfected into RCJ3.1C5.18 cells. 24 h after transfection, the cells were treated with different concentrations of TGF-β (0, 0.2, 1, and 5 ng/ml). The cell lysates were collected 24 h later for luciferase assay. TGF-β stimulated cyclin D1 promoter activity in a dose-dependent manner. The effect of TGF-β was significantly reduced in cells transfected with mutant cyclin D1 promoter in which four TCF binding sites were deleted.
DISCUSSION
Chondrogenesis is a tightly regulated event involving multiple steps from condensation of the precartilagineous mesenchyme to chondrogenic lineage determination toward chondroblasts and eventually differentiation into hypertrophic chondrocytes. This process is regulated by several key growth factors and signaling molecules, such as BMP, Hedgehog, Wnt, and TGF-β, which play important roles at various stages of chondrogenesis (22, 23). At sites of endochondral bone formation, TGF-β1 and TGF-β3 are detected in the chondrocytes of proliferative and hypertrophic zones, and TGF-β2 is detected in cells of all zones of the cartilage (24). Overexpression of Smad2/3 in upper sternal chondrocytes inhibits colX expression and alkaline phosphatase activity, indicating that the TGF-β pathway inhibits chondrocyte maturation (25, 26). The importance of Smad3 in chondrogenesis has been supported by several lines of evidence (12, 26–29). However, it seems that Smad2 does not play an essential role in chondrogenesis (29, 30). Our recent studies demonstrated that chondrocyte proliferation was inhibited, the chondrocyte differentiation process was promoted, and BMP signaling was up-regulated in chondrocytes derived from Smad3 knock-out mice (12, 26). Compared with TGF-β, canonical Wnt pathway inhibits early chondrogenesis in the developing skeleton but is necessary for the chondrocyte maturation and the lineage determination of osteo-chondrogenic progenitor cells (5, 7). Cross-talk between TGF-β and Wnt/β-catenin signaling pathways during chondrogenesis of mesenchymal cells has been previously reported. It has been demonstrated that TGF-β stimulates Wnt2, 4, 5a, 7a, 10a, and Lrp5 expression. TGF-β also dramatically increases the protein level, protein stability, and nuclear accumulation of β-catenin in human mesenchymal progenitor cells and human bone marrow stromal cells (11, 32), and this pathway is required for the stimulation of bone marrow stromal cell proliferation (33). In the present studies, we further investigated the detailed molecular mechanism on Wnt/β-catenin signaling activation by TGF-β. We found that both Smad3 and Smad4 interact with β-catenin and form a protein complex. This complex not only stabilizes β-catenin to prevent it from proteasome degradation but also assists β-catenin in nuclear translocation and eventually facilitates β-catenin transcriptional activity. This regulatory pattern also indicates that Wnt/β-catenin may act synergistically with the TGF-β pathway or work as a downstream target of TGF-β during a specific stage of chondrogenesis.
Cross-talk among TGF-β, BMP, Wnt, and other pathways is crucial for stem cell maintenance, body patterning, organogenesis, and homeostasis in adult animals. The cooperation among multiple signaling pathways may contribute to the specification of cell fates (34, 35). It has been reported that BMP-2 cross-talk with Wnt pathway through Smad1/4 interacting with β-catenin/TCF to synergistically activate late osteoblastic differentiation marker genes. Overexpression of Wnt3a suppresses the inhibitory effect of BMP-2 on myogenic differentiation (36). Our present results indicate that in addition to Smad3, Smad4 also interacts with β-catenin in chondrocytes, which is consistent with the previous report that Smad4 forms a complex with Lef1/TCF and β-catenin to activate target gene transcription (37). Our data also showed that in chondrocytes, silencing of Smad4 reversed the effect of Smad3 on β-catenin, suggesting that the interaction of Smad4 with β-catenin also protects β-catenin degradation. Similar observations were found in pancreatic carcinoma cells (38), suggesting that regulation of Smad3/4 on β-catenin may exist in multiple cell types.
Many studies have reported that the cooperation between TGF-β and Wnt/β-catenin pathways permits tight control of critical developmental processes. It has been reported that TGF-β and Wnt pathways synergistically regulate downstream target gene transcription through transcription factors TCF/Lef1 and Smad3 (39). In human bone marrow stromal cells, myostain induces Smad3/β-catenin interaction to activate Wnt/β-catenin pathway, which results in inhibition of adipogenesis (40). It has been demonstrated that TGF-β3 stimulates chondrogenic differentiation partially through Wnt5a-mediated signaling. Up-regulation of Wnt5a promotes expression of cell adhesion molecules through the p38 MAPK pathway in early stage chondrogenesis (41). Smad7 directly interacts with β-catenin and mediates β-catenin degradation by recruiting E3 ligase Smurf2, thereby reducing Wnt activity (42). Axin1, as a negative regulator of Wnt signaling pathway, binds with Smad7, leading to Smad7 ubiquitination and degradation, and subsequently facilitates TGF-β signaling (14). In addition to its indirect effect, Axin1 also directly interacts with Smad3 and facilitates Smad3 phosphorylation and enhances TGF-β transcriptional activity (31).
In summary, our results reveal the novel molecular mechanism that TGF-β activates Wnt/β-catenin pathway through interaction of Smad3/4 with β-catenin. This interaction permits tight regulation of chondrocyte development and leads to a stage-specific effect of TGF-β during chondrogenesis.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AR051189, R01 AR054465, K02 AR052411, and R01 AR055915 (to D. C.). This work was also supported by Grant N08G-070 (to D. C.) from the New York State Department of Health and the Empire State Stem Cell Board.
- BMP
- bone morphogenetic protein
- TGF
- transforming growth factor
- IP
- immunoprecipitation
- WT
- wild type
- β-TrCP
- β-transducin repeat-containing protein
- E3
- ubiquitin-protein isopeptide ligase
- siRNA
- small interfering RNA
- MAPK
- mitogen-activated protein kinase
- HA
- hemagglutinin
- TCF
- T cell factor.
REFERENCES
- 1.Kronenberg H. M. (2003) Nature 423, 332–336 [DOI] [PubMed] [Google Scholar]
- 2.Hart M., Concordet J. P., Lassot I., Albert I., del los Santos R., Durand H., Perret C., Rubinfeld B., Margottin F., Benarous R., Polakis P. (1999) Curr. Biol. 9, 207–210 [DOI] [PubMed] [Google Scholar]
- 3.Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 4.Aberle H., Bauer A., Stappert J., Kispert A., Kemler R. (1997) EMBO J. 16, 3797–3804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hill T. P., Später D., Taketo M. M., Birchmeier W., Hartmann C. (2005) Dev. Cell 8, 727–738 [DOI] [PubMed] [Google Scholar]
- 6.Akiyama H., Lyons J. P., Mori-Akiyama Y., Yang X., Zhang R., Zhang Z., Deng J. M., Taketo M. M., Nakamura T., Behringer R. R., McCrea P. D., de Crombrugghe B. (2004) Genes Dev. 18, 1072–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day T. F., Guo X., Garrett-Beal L., Yang Y. (2005) Dev. Cell 8, 739–750 [DOI] [PubMed] [Google Scholar]
- 8.Monga S. P., Mars W. M., Pediaditakis P., Bell A., Mulé K., Bowen W. C., Wang X., Zarnegar R., Michalopoulos G. K. (2002) Cancer Res. 62, 2064–2071 [PubMed] [Google Scholar]
- 9.Haq S., Michael A., Andreucci M., Bhattacharya K., Dotto P., Walters B., Woodgett J., Kilter H., Force T. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4610–4615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Serra R., Johnson M., Filvaroff E. H., LaBorde J., Sheehan D. M., Derynck R., Moses H. L. (1997) J. Cell Biol. 139, 541–552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tuli R., Tuli S., Nandi S., Huang X., Manner P. A., Hozack W. J., Danielson K. G., Hall D. J., Tuan R. S. (2003) J. Biol. Chem. 278, 41227–41236 [DOI] [PubMed] [Google Scholar]
- 12.Li T. F., Chen D., Wu Q., Chen M., Sheu T. J., Schwarz E. M., Drissi H., Zuscik M., O'Keefe R. J. (2006) J. Biol. Chem. 281, 21296–21304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song L. N., Gelmann E. P. (2005) J. Biol. Chem. 280, 37853–37867 [DOI] [PubMed] [Google Scholar]
- 14.Liu W., Rui H., Wang J., Lin S., He Y., Chen M., Li Q., Ye Z., Zhang S., Chan S. C., Chen Y. G., Han J., Lin S. C. (2006) EMBO J. 25, 1646–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitagawa M., Hatakeyama S., Shirane M., Matsumoto M., Ishida N., Hattori K., Nakamichi I., Kikuchi A., Nakayama K., Nakayama K. (1999) EMBO J. 18, 2401–2410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Latres E., Chiaur D. S., Pagano M. (1999) Oncogene 18, 849–854 [DOI] [PubMed] [Google Scholar]
- 17.Winston J. T., Strack P., Beer-Romero P., Chu C. Y., Elledge S. J., Harper J. W. (1999) Genes Dev. 13, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirota M., Watanabe K., Hamada S., Sun Y., Strizzi L., Mancino M., Nagaoka T., Gonzales M., Seno M., Bianco C., Salomon D. S. (2008) Cell Signal. 20, 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Z., Liu X., Lodish H. F. (2000) J. Biol. Chem. 275, 23425–23428 [DOI] [PubMed] [Google Scholar]
- 20.Xiao Z., Liu X., Henis Y. I., Lodish H. F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7853–7858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tetsu O., McCormick F. (1999) Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 22.Kawakami Y., Rodriguez-León J., Izpisúa Belmonte J. C. (2006) Curr. Opin. Cell Biol. 18, 723–729 [DOI] [PubMed] [Google Scholar]
- 23.Tang Y., Wu X., Lei W., Pang L., Wan C., Shi Z., Zhao L., Nagy T. R., Peng X., Hu J., Feng X., Van Hul W., Wan M., Cao X. (2009) Nat. Med. 15, 757–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janssens K., ten Dijke P., Janssens S., Van Hul W. (2005) Endocr. Rev. 26, 743–774 [DOI] [PubMed] [Google Scholar]
- 25.Ferguson C. M., Schwarz E. M., Reynolds P. R., Puzas J. E., Rosier R. N., O'Keefe R. J. (2000) Endocrinology 141, 4728–4735 [DOI] [PubMed] [Google Scholar]
- 26.Li T. F., Darowish M., Zuscik M. J., Chen D., Schwarz E. M., Rosier R. N., Drissi H., O'Keefe R. J. (2006) J. Bone Miner. Res. 21, 4–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X., Chen L., Xu X., Li C., Huang C., Deng C. X. (2001) J. Cell Biol. 153, 35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ionescu A. M., Schwarz E. M., Zuscik M. J., Drissi H., Puzas J. E., Rosier R. N., O'Keefe R. J. (2003) Exp. Cell Res. 288, 198–207 [DOI] [PubMed] [Google Scholar]
- 29.Furumatsu T., Tsuda M., Taniguchi N., Tajima Y., Asahara H. (2005) J. Biol. Chem. 280, 8343–8350 [DOI] [PubMed] [Google Scholar]
- 30.Waldrip W. R., Bikoff E. K., Hoodless P. A., Wrana J. L., Robertson E. J. (1998) Cell 92, 797–808 [DOI] [PubMed] [Google Scholar]
- 31.Furuhashi M., Yagi K., Yamamoto H., Furukawa Y., Shimada S., Nakamura Y., Kikuchi A., Miyazono K., Kato M. (2001) Mol. Cell. Biol. 21, 5132–5141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou S., Eid K., Glowacki J. (2004) J. Bone Miner Res. 19, 463–470 [DOI] [PubMed] [Google Scholar]
- 33.Jian H., Shen X., Liu I., Semenov M., He X., Wang X. F. (2006) Genes Dev. 20, 666–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mishra L., Shetty K., Tang Y., Stuart A., Byers S. W. (2005) Oncogene 24, 5775–5789 [DOI] [PubMed] [Google Scholar]
- 35.Guo X., Wang X. F. (2009) Cell Res. 19, 71–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakashima A., Katagiri T., Tamura M. (2005) J. Biol. Chem. 280, 37660–37668 [DOI] [PubMed] [Google Scholar]
- 37.Nishita M., Hashimoto M. K., Ogata S., Laurent M. N., Ueno N., Shibuya H., Cho K. W. (2000) Nature 403, 781–785 [DOI] [PubMed] [Google Scholar]
- 38.Romero D., Iglesias M., Vary C. P., Quintanilla M. (2008) Carcinogenesis 29, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 39.Labbé E., Letamendia A., Attisano L. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 8358–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W., Flanagan J., Jasuja R., Kirkland J., Jiang L., Bhasin S. (2008) J. Biol. Chem. 283, 9136–9145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jin E. J., Park J. H., Lee S. Y., Chun J. S., Bang O. S., Kang S. S. (2006) Int. J. Biochem. Cell Biol. 38, 183–195 [DOI] [PubMed] [Google Scholar]
- 42.Han G., Li A. G., Liang Y. Y., Owens P., He W., Lu S., Yoshimatsu Y., Wang D., Ten Dijke P., Lin X., Wang X. J. (2006) Dev. Cell 11, 301–312 [DOI] [PubMed] [Google Scholar]