Abstract
Human embryonic stem cells (hESCs) are unique pluripotent cells capable of self-renewal and differentiation into all three germ layers. To date, more cell surface markers capable of reliably identifying hESCs are needed. The epithelial cell adhesion molecule (EpCAM) is a type I transmembrane glycoprotein expressed in several progenitor cell populations and cancers. It has been used to enrich cells with tumor-initiating activity in xenograft transplantation studies. Here, we comprehensively profile the expression of EpCAM by immunofluorescence microscopy, Western blotting, and flow cytometry using an anti-EpCAM monoclonal antibody (mAb) OC98-1. We found EpCAM to be highly and selectively expressed by undifferentiated rather than differentiated hESCs. The protein and transcript level of EpCAM rapidly diminished as soon as hESC had differentiated. This silencing was closely and exclusively associated with the radical transformation of histone modification at the EpCAM promoter. Moreover, we demonstrated that the dynamic pattern of lysine 27 trimethylation of histone 3 was conferred by the interplay of SUZ12 and JMJD3, both of which were involved in maintaining hESC pluripotency. In addition, we used chromatin immunoprecipitation analysis to elucidate the direct regulation by EpCAM of several reprogramming genes, including c-MYC, OCT-4, NANOG, SOX2, and KLF4, to help maintain the undifferentiation of hESCs. Collectively, our results suggest that EpCAM might be used as a surface marker for hESC. The expression of EpCAM may be regulated by epigenetic mechanisms, and it is strongly associated with the maintenance of the undifferentiated state of hESCs.
Keywords: Chromatin/Regulation, DNA/Methylation, Development, Embryonic Stem Cell, Gene Silencing, EpCAM, Cell Surface Marker, Epigenetic Regulation, Gene Regulation, Human Embryonic Stem Cell
Introduction
Human embryonic stem cells (hESCs)4 are derived from the inner cell mass of blastocyst-stage embryos. They retain the unlimited proliferation and developmental pluripotency from their progenitors and are able to self-renew and give rise to differentiated progeny of all three germ layers (1). Therefore, hESCs have potential clinical applications and can be used to explore our knowledge of basic developmental biology. The identification of selectively expressed cell surface molecules is essential for the purification and characterization of pluripotent hESCs, playing an important role in helping us understand the mechanisms involved in stem cell differentiation and self-renewal.
We recently established a very specific monoclonal antibody (mAb), OC98-1, against the cell surface protein EpCAM and found that EpCAM was highly expressed in undifferentiated hESCs. EpCAM (also known as 17-1A, GA733-2, KSA, ESA, and EGP-40) is a homophilic, calcium-independent cell adhesion molecule of 39–42 kDa (2, 3) expressed by most epithelial tissues and is abundantly and homogeneously expressed in human carcinomas. EpCAM is a type I transmembrane glycoprotein encoded by the TACSTD1 gene. The EpCAM protein consists of a total of 314 amino acids, containing an extracellular domain (EpEX) with a nidogen-like domain as well as thyroglobulin- and epidermal growth factor-like repeats (265 amino acids), a single transmembrane part, and a short intracellular domain (EpICD) of 26 amino acids. It is not structurally related to any of the major families of the adhesion molecules (cadherins, selectins, integrins, or cell adhesion molecules of the Ig superfamily) (4). The level of EpCAM expression has been correlated with dedifferentiation and malignant proliferation of epithelial cells (5, 6). It is frequently detected in cancer-initiating cells (7, 8) and tissue-specific normal stem or progenitor cells (9–13). For example, EpCAM is expressed in the mammalian germ line (12) and is frequently present at the surface of human hepatic multipotent progenitors (9), hepatic stem cells (11), and cancer stem cells (8). Very recently, EpCAM expression on ESCs has been reported by some studies (14–17), suggesting that EpCAM might serve as a potential surface marker for these pluripotent cells.
Little is known about molecular mechanisms underlying the regulation of EpCAM expression in hESC. For the past few years, more has been learned about the influence of DNA methylation and histone modifications on regulating gene expression and genome function. Several studies have discussed the DNA methylation status of EpCAM promoter in lung, colon, prostate, liver, bladder, ovary, and breast cancer cells and tissues (18–21). Post-translational modifications of histone tails, including phosphorylation, acetylation, ubiquitination, and methylation, have been validated as dynamic regulators of gene expression. In order to gain insight into the epigenetic transitions responsible for EpCAM expression in hESC, we studied the 5′-flanking region of EpCAM promoter by evaluating CpG status using methylation-specific PCR (MSP), bisulfite sequencing, and histone modification by chromatin immunoprecipitation (ChIP).
The polycomb group (PcG) proteins are important chromatin modifiers that play a pivotal role in the epigenetic regulation of the development, differentiation, and maintenance of cell fates (22). Dynamic repression of developmental pathways by PcG may be required for maintaining ES cell pluripotency and plasticity during embryonic development (23). The polycomb repressive complex 2 (PRC2) mediates transcriptional repression by catalyzing the trimethylation of Lys27 on histone H3 (H3K27me3) (24). Suppressor of Zeste 12 homolog (SUZ12), one of the PRC2 components, is essential for histone methyltransferase PRC2 activity on H3K27me3 methylation (25–27). The recent identification of JmjC domain-containing histone lysine demethylase JMJD3 suggests that there may be positive and negative regulators simultaneously controlling chromatin structure dynamics through histone methylation mark alterations. JMJD3 specifically removes methyl marks of H3K27me3 in mammalian cells to antagonize PcG gene silencing and permit gene transcription. JMJD3 is highly expressed in ES cells and is responsible for the rapid decrease of the H3K27me3 mark during specific stages of embryogenesis and stem cell differentiation (23, 28). These findings suggest that EpCAM may be regulated by both SUZ12 and JMJD3 during hESC differentiation.
Understanding the downstream targets of EpCAM would help define the molecular function of this gene. However, such studies have been hindered by the obscure signaling mode of EpCAM until the very recent discovery of regulated intramembrane proteolysis and nuclear translocation of its intracellular domain EpICD. Released EpICD associates with FHL2, β-catenin, and Lef-1 and participates in gene regulation in the nucleus (6). One of the EpCAM downstream targets, c-MYC, has been found to be regulated by EpCAM in both normal and cancer cells (29). c-MYC is a member of the four reprogramming factors involved in the induced pluripotent stem cell formation (30–34). Understanding the regulation of EpCAM on c-MYC and even other reprogramming genes like OCT-4, NANOG, SOX2, and KLF4 in hESC may add to our understanding of how EpCAM contributes the long term maintenance of the ES cell phenotype.
This study is a comprehensive analysis of EpCAM expression in undifferentiated hESCs using immunofluorescence microscopy, Western blotting, and flow cytometry. Loss of EpCAM expression in differentiated hESCs through epigenetic silencing is elucidated by ChIP. Because several reprogramming genes are under the regulation of EpCAM, we propose that EpCAM maintains hESC “stemness” through sustaining these reprogramming genes.
EXPERIMENTAL PROCEDURES
Cell Culture
Human embryonic stem cell line H9, hES5, HUES3, and HUES6 cells were maintained in an undifferentiated state by co-culture with mitomycin-treated MEF feeder layers in hESC medium: Dulbecco's modified Eagle's medium/F-12 (Invitrogen) with 20% knock-out serum replacer (Invitrogen), 4 ng/ml basic fibroblast growth factor-2 (Invitrogen), 1 mm β-mercaptoethanol (Sigma), 1% nonessential amino acids (Invitrogen), and 1 mm l-glutamine (Invitrogen). Cells were routinely passaged at a 1:3 dilution by treatment with 1 mg/ml collagenase IV (Invitrogen).
To induce hESC differentiation, we first allowed the hESCs to form embryoid bodies. Confluent ES cells in a 6-well plate were dissociated with 1 mg/ml collagenase IV, and then small clumps of the cells were cultured suspended in hESC medium without basic fibroblast growth factor-2 for 2 days to form embryoid bodies. The embryoid bodies were seeded on gelatin-coated plates in Dulbecco's modified Eagle's medium/F-12 with 20% fetal calf serum, 1 mm β-mercaptoethanol, 1% nonessential amino acids, and 1 mm l-glutamine for further spontaneous differentiation.
Flow Cytometry Analysis
An anti-EpCAM mAb (OC98-1) was generated in our laboratory as described previously (35). The target of OC98-1 was confirmed by liquid chromatography-tandem mass spectrometry, co-immunoprecipitation, and successful recognition of OC98-1 against recombinant EpCAM protein. hESCs and MEFs were dissociated with 0.25% trypsin-EDTA (1 mm) (Invitrogen) for 3 min. Cells were washed with fluorescence-activated cell sorting buffer (PBS containing 1% fetal calf serum) and then incubated for 1 h at 4 °C in fluorescence-activated cell sorting buffer with the corresponding mAb: anti-EpCAM mAb (OC98-1) at dilutions that ranged from 0.0001 to 1 μg/ml, anti-SSEA4 mAb (2 μg/ml; 90231; Chemicon, Temecula, CA), and anti-CD29 mAb conjugated to Alexa647 (dilution 1:10; MCA2298A647; Serotec (Oxford, UK)). Phycoerythrin-conjugated goat anti-mouse IgG was used as a secondary antibody (dilution 1:250; Jackson ImmunoResearch Laboratories (West Grove, PA)). For cell sorting, primary antibodies were used as follows: anti-EpCAM mAb (OC98-1) at 0.1 μg/ml, anti-SSEA4 mAb conjugated to phycoerythrin (dilution 1:30; FAB/435P; R&D Systems (Minneapolis, MN)), and anti-CD29 mAb conjugated to Alexa647 (dilution 1:10; MCA2298A647; Serotech). Fluorescein isothiocyanate-conjugated goat anti-mouse IgG was used as a secondary antibody (dilution 1:250; Jackson ImmunoResearch Laboratories). Flow cytometry analysis was performed with a BD FACSCanto II flow cytometer (BD Biosciences).
Protein Extraction and Western Blot Analysis
Cells were lysed in the lysis buffer (150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 1% Nonidet P-40) plus proteinase inhibitor mixture (Roche Applied Science). Proteins were electrophoresed on 10% SDS-PAGE and then transferred to a nitrocellulose membrane (Hybond-C Super, Amersham Biosciences). The membranes were incubated with anti-EpCAM (2 μg/ml; OC98-1) or anti-α-tubulin (1:10,000; Sigma) mAbs. Horseradish peroxidase-conjugated goat anti-mouse IgG (H + L) (Jackson ImmunoResearch Laboratories) was used as the secondary antibody. Bound antibodies were detected using ECL reagents (Amersham Biosciences).
Immunofluorescence Assay
hESCs cultured on coverslips were fixed in 2% paraformaldehyde in PBS for 10 min, washed, and then blocked with 1% bovine serum albumin in PBS for 10 min. Cells were incubated at room temperature with primary antibody anti-EpCAM mAb (1 μg/ml; OC98-1), anti-EpICD mAb (100× dilution; 1144-1; Epitomics (Burlingame, CA)) or anti-Oct-4 mAb (2 μg/ml; sc5279; Santa Cruz Biotechnology, Inc. (Santa Cruz, CA)) in 1% bovine serum albumin. After a 1-h incubation, cells were washed and incubated with fluorescein isothiocyanate goat anti-mouse secondary antibody (Jackson ImmunoResearch Laboratories) for another 1 h at room temperature. Unbound antibodies were removed by three washes in PBS. 4′,6-Diamidino-2-phenylindole was added for cell counterstaining. To further confirm that the two commercial antibodies (1144-1 (Epitomics) and A-20 (Santa Cruz Biotechnology, Inc.)) recognized EpICD, human synthetic EpICD peptide (1 μg/ml) was coated onto the plate, and anti-EpICD Abs (5 μg/ml) were used to detect EpICD antigen using an enzyme-linked immunosorbent assay (ELISA).
RNA Extraction and Quantitative Real-time RT-PCR
Total RNAs were prepared from the cell lines using ULTRASPEC RNA isolation reagent (Biotecx Laboratories, Houston, TX). cDNAs were reverse-transcribed with oligo(dT) primer (Fermentas, Glen Burnie, MD) from 4 μg of total RNA using SuperScript III RNase H-reverse transcriptase (Invitrogen) according to the manufacturer's instructions. The forward and reverse primers for PCR are listed in supplemental Table S1. Quantitative RT-PCR was performed by using the LightCycler480 System (Roche Applied Science). The gene expression level of each sample was normalized to the expression level of GAPDH in the same sample. The ratios of EpCAM/GAPDH, OCT-4/GAPDH, c-MYC/GAPDH, NANOG/GAPDH, SOX2/GAPDH, and KLF4/GAPDH of the H9 cell line and that of collagen, type III, α1 (COL3A1)/GAPDH of 15-day differentiated H9 were set as 1.0. The other ratio values of each gene were recalculated accordingly. The reactions were performed in triplicate, and S.D. values were calculated.
ELISA
96-well plates (Corning Costar, St. Louis, MO) were seeded with H9, hES5, HUES3, HUES6, and MEF cells. The plates were washed with PBS and blocked with 1% bovine serum albumin. Anti-EpCAM mAb (1 μg/ml; OC98–1) was added to the plates of cells and incubated for 1 h. The plates were then washed with PBS containing 0.1% (w/v) Tween 20 (PBST0.1) and incubated with horseradish peroxidase-conjugated anti-mouse IgG (Jackson ImmunoResearch Laboratories) for another 1 h. After washing, the plates were incubated with substrate solution o-phenylenediamine dihydrochloride (Sigma). The reaction was stopped by adding 3 n HCl, and the plates were read using a microplate reader at 490 nm.
Genomic DNA Isolation, Bisulfite Modification, and MSP
The CpG methylation status of OPG promoter was evaluated by MSP. The genomic DNA was purified using the Wizard genomic DNA purification kit (Promega, Madison, WI). The bisulfite reaction was performed on 500 ng of DNA and further subjected to bisulfite modification by the EZ DNA methylation kit according to the manufacturer's directions (Zymo Research, Orange, CA). The primers used for the MSP amplifications are listed in supplemental Table S1. PCRs were performed in a thermocycler (Bio-Rad) for 40 cycles at 95 °C for 1 s, 51 °C for 5 s, and 72 °C for 25 s, followed by a final extension at 72 °C for 10 s to amplify bisulfite-modified DNA.
Bisulfite Sequencing
Genomic DNA (500 ng) was treated with the EZ DNA modification kit (Zymo Research) according to the manufacturer's recommendations. Completely methylated and unmethylated control genomic DNA was purchased from Qiagen (Qiagen Inc., Valencia, CA). The promoter regions of EpCAM and Oct-4 genes were amplified by PCR. Primer sequences used for PCR amplification are listed in supplemental Table S1. The PCR products were subjected to purification using the QIAquick PCR purification kit (Qiagen) following the manufacturer's instructions. The purified PCR products were then subcloned into a TA cloning vector (pGEM-T Easy vector; Promega). Twenty clones (for control DNA) or 10 clones of each sample were verified by sequencing with the T7 universal primer.
Chromatin Immunoprecipitation and Quantitative Real-time PCR
ChIP was performed as previously described (36) with some modifications. Briefly, ChIP assays were carried out on 1 × 105 of H9 and differentiated H9 (H9-Diff.) cells for anti-H3K4me3, anti-H3K9K14Ac, anti-H3K27me3, and anti-H3K9me3 Abs or on 1 × 106 H9 and H9-Diff. cells for anti-SUZ12, anti-JMJD3, and anti-EpICD Abs. The protein-DNA complexes were fixed using 1% formaldehyde, and the cross-linking fixation was quenched by adding glycine to a final concentration of 200 mm. The chromatin complexes were then sonicated to an average size of 250 bp by a MISONIX Sonicator 3000. We used 2.4 μg of anti-H3K4me3 (ab8580; Abcam, Cambridge, MA), 2.4 μg of anti-H3K9K14Ac (06-599; Upstate-Millipore, Charlottesville, VA), 5 μg of anti-H3K27me3 (ab6002; Abcam), 5 μg of anti-H3K9me3 (07-442; Upstate), 2 μg of anti-SUZ12 (ab12073; Abcam), 4 μg of anti-JMJD3 (AP1022a; Abgent, San Diego, CA), and 4 μg of anti-EpICD (A-20; Santa Cruz Biotechnology, Inc.) for immunoprecipitations, which were performed at 4 °C with the indicated antibodies by incubation with Protein A beads (Invitrogen) for 2 h. The immunocomplexes were further incubated with chromatin for another 2 h. The bound fraction was isolated by Protein A beads according to the manufacturer's instructions, and the immunocomplexes were subsequently subjected to reverse cross-linking. The immunoprecipitated DNA was recovered by a PCR purification kit (Qiagen) according to the manufacturer's instructions, and the target DNA amount was detected by real-time PCR using the LightCycler480 system (Roche Applied Science). The amplification primers are listed in supplemental Table S1. For each sample, PCR analysis was performed in triplicate, and the bound fraction was compared with input DNA of 1 × 104 cells. The results are reported as the ratio of immunoprecipitated (IP) DNA to input DNA (IP/input). To obtain relative occupancy values, the IP/input was further normalized to the level observed at a control promoter region of HBB (H3K4me3, H3K9K14Ac, and SUZ12) or of GAPDH (H3K27me3, H3K9me3, JMJD3, and EpCAM), which was defined as 1.0.
Lentivirus-mediated Short Hairpin RNA (shRNA) Knockdown
For virus production, 1 × 106 293T cells were seeded in a 60-mm dish and incubated overnight at 37 °C, 5% CO2. The shRNA vector V2LHS-134158 or V2LHS-134160 targeting EpCAM (Open Biosystems, Huntsville, AL), the envelope plasmid pMD2.G, and the packing plasmid pCMVΔR8.91 were cotransfected at a ratio of 10:1:9 into 293T cells using Arrest-in (Open Biosystems). After an 18-h incubation period, the transfection reagent was replaced with fresh Dulbecco's modified Eagle's medium with 10% fetal calf serum. The supernatant containing virus particles was harvested after 24 h. HCT116 colon cancer cells were seeded at 1 × 106 cells/60-mm dish 1 day before transduction. The medium was replaced with virus-containing supernatant supplemented with 8 μg/ml Polybrene (Sigma) to cells for 24 h. After transduction, cells were replaced with growth medium containing puromycin (2 μg/ml; Sigma) and were incubated for 48 h.
Statistical Analyses
Statistical analyses were made using unpaired Student's t tests as appropriate. p < 0.05 was considered significant.
RESULTS
EpCAM Is Selectively Expressed in hESCs
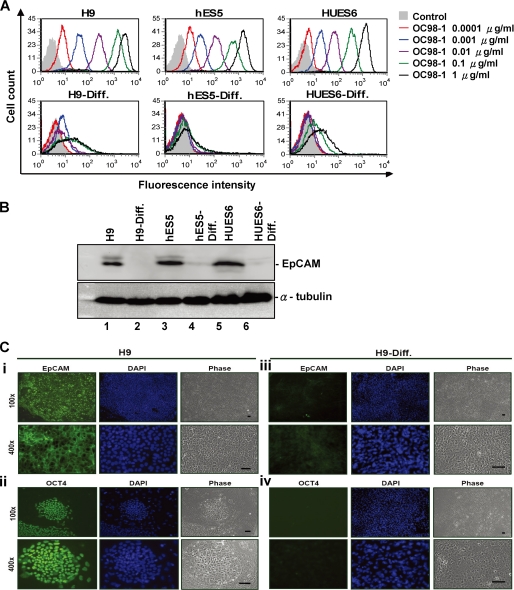
EpCAM has been previously identified as differentially expressed in hepatic stem cells (11). To determine whether hESCs expressed EpCAM, we analyzed the cell surface expression of EpCAM protein using fluorescent flow cytometry in undifferentiated hESCs and 30-day differentiated hESCs using our mAb OC98-1 to recognize the extracellular portion of EpCAM (Fig. 1A). We found that the undifferentiated H9, hES5, and HUES6 ES cells cultured on irradiated MEFs (feeder layer) expressed cell surface EpCAM in a dose-dependent binding intensity proportionate to OC98-1 concentration (concentrations ranging from 0.0001 to 1 μg/ml); this molecule was absent from most of the differentiated counterparts (Fig. 1A). SSEA4 labeling in undifferentiated and differentiated hESCs served as experimental control (Fig. S1). In our Western blot analysis, we found EpCAM to be strongly detected in undifferentiated H9, hES5, and HUES6 ES cells but to be absent in hESCs after differentiation (Fig. 1B), indicating that EpCAM was expressed in undifferentiated hESCs exclusively.
FIGURE 1.
EpCAM is selectively expressed by hESCs. A, dose-dependent increase in relative fluorescence intensity was associated with increasing the concentration of anti-EpCAM mAb (OC98-1) against the cell surface of undifferentiated hESCs (top; H9, hES5, and HUES6). Only basal level of EpCAM binding to 30 day-differentiated hESCs (H9-Diff., hES5-Diff., and HUES6-Diff.) can be seen in the lower panel. The differentiation of hESCs was as described under ”Experimental Procedures.“ B, expression of the EpCAM protein in undifferentiated and differentiated hESCs. Lysates from various hESCs cell lines were analyzed by Western blot analysis with the anti-EpCAM and anti-α tubulin mAb. α-Tubulin was used as an internal control. C, immunofluorescent analysis of EpCAM (i and iii) and OCT-4 protein (ii and iv) expression in undifferentiated H9 (i and ii) and differentiated H9 (iii and iv) cells. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) (blue). H9 cells staining for OCT-4 manifested that these hESCs maintained an undifferentiated state. EpCAM expression was correlated to that of OCT-4 in undifferentiated H9 (i and ii). hESC differentiation was associated with loss of both EpCAM and OCT-4 expression (iii and iv).
Undifferentiated H9, hES5, and HUES6 colonies were subjected to spontaneous differentiation by culture in 20% fetal calf serum-containing media for 30 days. Immunofluorescent microscopy was used to detect the expression of EpCAM and Oct-4 proteins for both undifferentiated and differentiated hESCs (Fig. 1C). Differentiated cells lost Oct-4 protein expression, indicating that spontaneous differentiation had occurred. An absence of EpCAM protein correlated with loss of Oct-4. Examination of EpCAM proteins on H9, hES5, and HUES6 showed selective expression of this molecule in hESCs, suggesting that EpCAM may be used as a surface marker for hESCs.
hESC Differentiation Is Associated with a Loss of EpCAM Expression
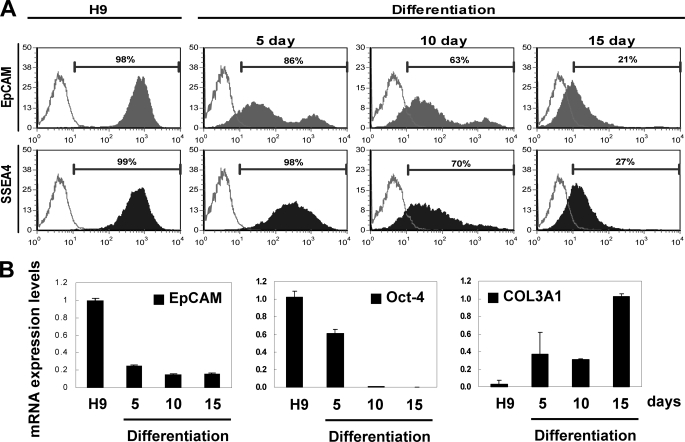
EpCAM protein expression was assessed in undifferentiated and differentiated H9 cells by measuring the amount of EpCAM on the surface of cells by flow cytometry (Fig. 2A). Undifferentiated H9 cells expressed cell surface EpCAM on 98% of the population (Fig. 2A, EpCAM), whereas SSEA4, a known hESC surface antigen (37, 38), was expressed on most (99%) of the cells (Fig. 2A, SSEA4). At day 5 following induction of differentiation of the cells, a fractional proportion of the population (∼14%) lacked cell surface EpCAM (86%; Fig. 2A, EpCAM, day 5), although they retained expression of SSEA4 protein (98%; Fig. 2A, SSEA4, day 5). At day 10 following induction of differentiation, we found a gradual reduction of EpCAM (∼63%; Fig. 2A, EpCAM, day 10) as well as SSEA4 (∼70%; Fig. 2A, SSEA4, day 10) in these cells. At 15 days, a significant proportion of the cells were found to have low levels of cell surface EpCAM protein (∼21%; Fig. 2A, EpCAM, day 15) and SSEA4 protein (∼27%; Fig. 2A, SSEA4, day 15).
FIGURE 2.
Human ES cell differentiation was associated with loss of EpCAM. A, cell surface EpCAM expression histogram was assessed in undifferentiated H9 cells (left) and H9 cells differentiated for 5, 10, and 15 days by fluorescent flow cytometry. EpCAM or SSEA4 was a closed population, and secondary antibody only was an open population. Viable cells were gated using forward and side scatter, and the data represents cells from this population. B, Q-RT-PCR analysis of EpCAM, OCT-4, and COL3A1 transcript expression was assessed in undifferentiated H9 cells and in H9 cells differentiated for 5, 10, and 15 days as described above. GAPDH expression was used to normalize the variability in each template loading.
Transcript expression in the undifferentiated and differentiated H9 populations was analyzed by Q-RT-PCR (Fig. 2B). EpCAM transcripts were detected from undifferentiated H9 cells and were rapidly down-regulated at days 5–15. OCT-4, which is expressed in undifferentiated hESCs (39), was abundantly detected in undifferentiated H9, with levels decreasing at day 5 and absent at days 10 and 15. In contrast, COL3A1 transcripts, which are expressed in differentiated cells (40), were found to be absent in undifferentiated H9 but present in differentiated cells at all examination time points (days 5–15). Together, these data suggest a close association between hESC differentiation and loss of expression of EpCAM.
Specificity of EpCAM Surface Expression in hESCs
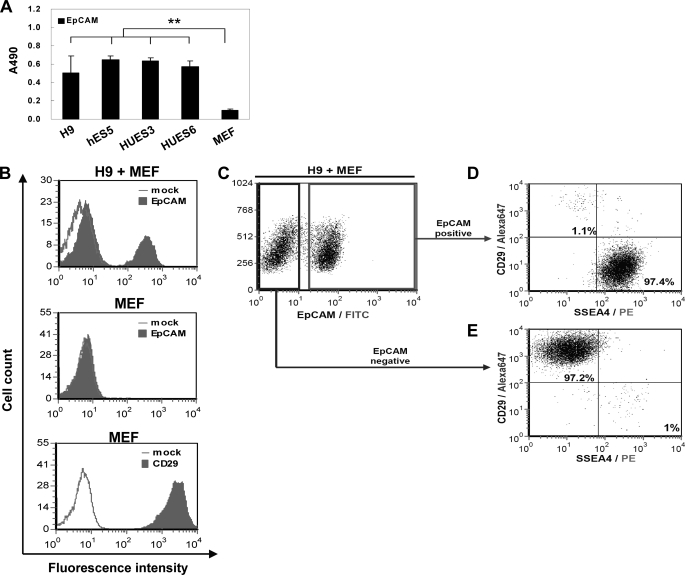
We next investigated whether EpCAM could be used reliably as a marker to identify, isolate, and qualify hESCs in vitro. As shown in Fig. 3A, the results of our ELISA, we found a significant relationship between EpCAM level and hESCs, including undifferentiated H9, hES5, HUES3, and HUES6 cells but not MEFs. Using flow cytometry, we further demonstrated that EpCAM could discriminate hESCs from MEFs in primary H9/MEF co-cultures (Fig. 3B). MEFs exhibited CD29-positive staining (41), which was used as a marker for fibroblast identity (Fig. 3B, bottom).
FIGURE 3.
Cell surface EpCAM expression and isolation of hESCs. A, cell surface EpCAM protein expression by undifferentiated hESCs (H9, hES5, HUES3, and HUES6) and feeder MEF by ELISA using an anti-EpCAM mAb (**, p < 0.01). B, flow cytometry analysis of EpCAM on H9 hESCs co-cultured with MEF (top) and MEF alone (middle) and analysis of CD29 on MEF alone (bottom). C, analysis of cell surface expression of EpCAM on H9 cells co-cultured with MEF by fluorescent flow cytometry. D and E, double labeling of EpCAM positive (D) and EpCAM negative (E) population with anti-SSEA4 and anti-CD29 antibodies on undifferentiated H9 cells co-cultured with MEF.
To find out whether the OC98–1 was able to distinguish between two populations of hESCs cultured on MEFs, we used flow cytometry to analyze EpCAM expression profile on hESCs/MEFs co-cultures. There were two main cell populations, EpCAM+ and EpCAM− (Fig. 3C). When we double-stained these two populations with SSEA4 and CD29, we found SSEA4 to be expressed in the hESC EpCAM+ population (97.4%; Fig. 3D) but not in the fibroblast EpCAM− CD29+ population (97.2%; Fig. 3E). These findings show that EpCAM labeling can be used to separate hESCs from fibroblasts and obtain pure hESC populations.
EpCAM Methylation Status in Undifferentiated and Differentiated hESCs
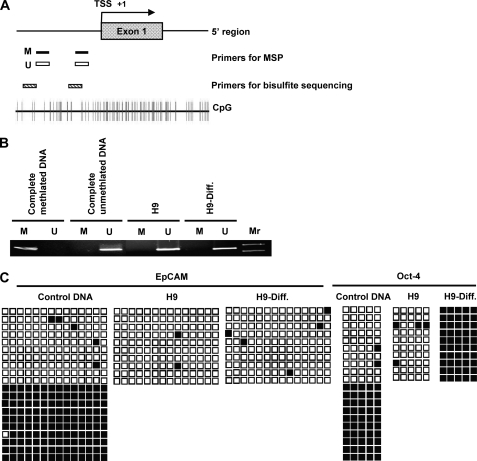
The EpCAM gene expression was completely silenced while hESCs differentiated (Fig. 2). To determine whether EpCAM expression correlated with DNA methylation, we examined the methylation status of EpCAM promoter regions (18, 21) in both undifferentiated and differentiated at day 30 hESCs. Fig. 4A depicts the gene structure and CpG contents of the EpCAM promoter region. Primers for MSP and bisulfite sequencing were designed to target the 5′-flanking region of the EpCAM promoter. MSP assays were performed to determine the methylation status of EpCAM for both undifferentiated and differentiated hESCs. We used commercial available completely methylated and completely unmethylated DNAs as positive controls validating the properties of the primers in this experiment. As shown in Fig. 4B, both undifferentiated and differentiated H9 cells revealed abundant amounts of unmethylated PCR product, whereas in differentiated H9 cells, there were trace amounts of methylated PCR product.
FIGURE 4.
Methylation status of EpCAM promoter regions in undifferentiated and differentiated hESCs. A, schematic representation of the EpCAM gene promoter region. Primers for MSP and bisulfite sequencing used in the study are indicated. B, MSP analysis of the EpCAM gene promoter region in undifferentiated and differentiated H9 cells. The PCR products that were methylated (M) were generated by methylation-specific primers, and those that were unmethylated (U) were generated by primers specific for unmethylated DNA. C, mapping the methylation status of the CpG islands in the promoter region of the EpCAM gene by bisulfite sequencing. Each row of squares represents a single plasmid cloned and sequenced from PCR products generated from amplification of bisulfite-treated DNA. Open squares, unmethylated cytosines; filled squares, methylated cytosines. Most CpGs in the promoter region in both undifferentiated and differentiated H9 cells were unmethylated.
To further ensure the reliability of this finding, we used bisulfite sequencing to evaluate the methylation statuses of CpG in the promoter region of EpCAM. By clone sequencing analysis, we found 98% of the CpG sites of the undifferentiated H9 clones and 96% of those of the differentiated H9 ones to be unmethylated (Fig. 4C). We included a positive control, OCT-4, which is unmethylated in ES cells and methylated in differentiated cells (31) (Fig. 4C). As expected, H9 hESCs were predominantly unmethylated at the OCT-4 promoter, whereas prominent methylation at this region was found in differentiated H9 cells, a finding consistent with previous studies of transcriptional silencing in the differentiated cells (31). These results indicate that EpCAM silencing during hESC differentiation may not be due to changes in the methylation status of the EpCAM promoter in these cells.
Histone Modification of EpCAM Promoter Region in Undifferentiated and Differentiated hESCs
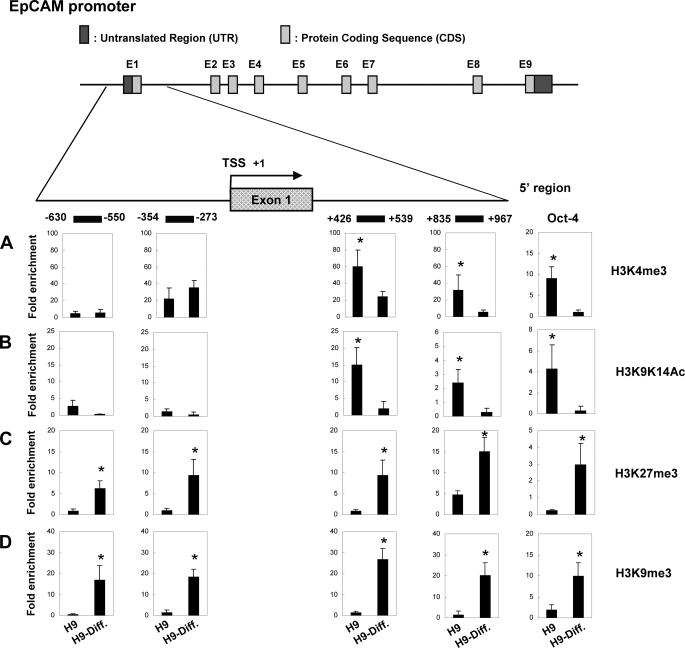
Covalent histone tail modifications of histone 3, including acetylation or methylation, regulate these different states of chromatin configuration and gene transcription (42). To address whether there is an association between the expression of EpCAM expression and chromatin architecture, we determined the profile of histone modification of EpCAM promoter vicinity in undifferentiated and differentiated at day 30 H9 cells. To do this, we performed ChIP assays on four regions of EpCAM promoter: the upstream distal (−630 to −550) and proximal (−354 to −273) and the downstream proximal (+426 to +539) and distal (+835 to +967) relative to the transcription start site (TSS) (43). We also explored the histone marks of H3K4me3, H3K9K14Ac, H3K27me3, and H3K9me3.
H3K4me3 has been positively correlated with gene expression (43). As shown in Fig. 5A, the kinetics of H3K4me3, there was an increase of occupancy in undifferentiated H9 cells expressing EpCAM in both proximal and distal regions downstream of TSS, compared with differentiated H9 cells, but no significant alterations between these two cell populations in proximal and distal regions upstream of TSS. OCT-4 was expressed by undifferentiated hESCs. There was a significant H3K4me3 occupancy in the OCT-4 promoter region in undifferentiated H9 (Fig. 5A) than in differentiated H9 cells. Acetylation at H3K9K14 of the promoter has been found to be enriched at the 5′-end of active genes and has been strongly correlated with methylation of H3K4 (44). In EpCAM-expressing undifferentiated H9 cells, the acetylation level of H3K9K14 was higher in undifferentiated H9 than in differentiated H9 in both proximal and distal downstream regions of TSS, as observed in H3K4me3 (Fig. 5B).
FIGURE 5.
Histone modification at the EpCAM promoter in undifferentiated and differentiated hESCs. Top, schematic representation of the EpCAM gene promoter region, which spanned positions −630 to +967 with respect to the TSS. The ChIP primers used in the study are indicated by horizontal lines. A–D, a combination of ChIP and Q-PCR analyses showing quantitative occupancy of H3K4me3 (A), H3K9K14Ac (B), H3K27me3 (C), and H3K9me3 (D) to EpCAM and OCT-4 promoter in undifferentiated and differentiated H9 cells. OCT-4 was used as a positive control for histone modification binding. Each experiment was done in triplicate (mean ± S.D.). The amount of immunoprecipitated target was quantified by real-time PCR, and the value of immunoprecipitated target was calculated as the ratio of IP DNA to the total amount of input DNA used for the immunoprecipitation (IP/input). To obtain relative -fold enrichment value, the target IP/input was further normalized to the level of a control promoter region of HBB (H3K4me3 or H3K9K14Ac) or of GAPDH (H3K27me3 or H3K9me3). In ChIP analyses, H3K4me3 and H3K9K14Ac enrichment were observed in undifferentiated H9 cells downstream of TSS, whereas H3K27me3 and H3K9me3 occupancy were detected in differentiated H9 cells both upstream and downstream of TSS (*, p < 0.05).
Previous studies have suggested that there is a correlation between H3K27me3 and gene repression (43). Indeed, our study found H3K27me3 signals to be elevated at all upstream and downstream regions of silent promoters of differentiated H9 cells that did not express EpCAM (Fig. 5C). Methylation of lysine 9 of histone 3 is known to facilitate formation of heterochromatin, and elevated levels of H3K9me3 at promoter sequences have been associated with suppression of gene expression (43, 45). Our study found greater increases in H3K9me3 on all four promoter regions in differentiated H9 than in undifferentiated H9 cells (Fig. 5D). This profiling of histone modification by ChIP assay suggests that chromatin remodeling in the 5′-flanking region of the EpCAM promoter may be responsible for EpCAM gene regulation.
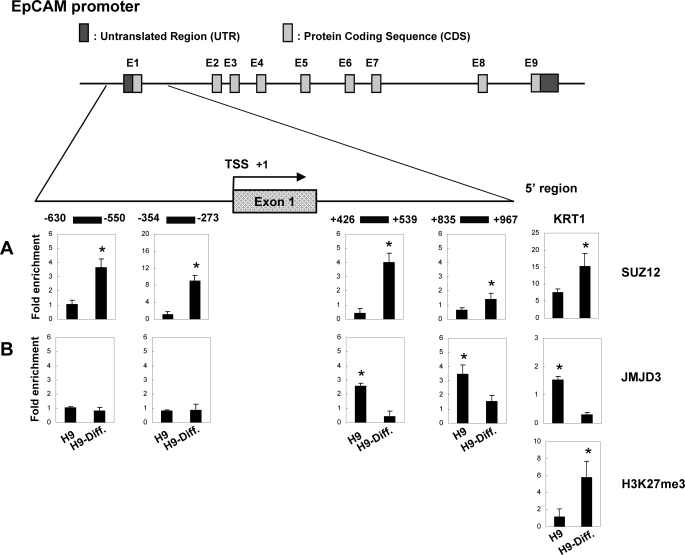
SUZ12 and JMJD3 Are Required for Bidirectional Regulation of H3K27me3 in hESCs
In hESCs, EpCAM was marked by a chromatin signature that was dynamically regulated during differentiation (Fig. 5). Hence, the observation that SUZ12 and JMJD3 are recruited to their promoters prompted us to assess the dynamic profile of H3K27 methylation at the TSS of EpCAM during stem cell differentiation. To test whether SUZ12 conferred the methylation of H3K27 of EpCAM, we performed ChIP of both undifferentiated and differentiated at day 30 H9 cells. Remarkably, we detected higher SUZ12 signals in differentiated H9 than in undifferentiated H9 both upstream and downstream of the EpCAM promoter (Fig. 6A), and these results were strongly mirrored by H3K27me3 occupancy. These results suggest that EpCAM is controlled by PRC2 complex, the major histone methyl transferase responsible for H3K27me3. We provided a parallel control, KRT1 (Fig. 6), an epidermal differentiation gene that is expressed in differentiated cells and whose expression can be regulated by chromatin modification machinery SUZ12/JMJD3 (46). We observed higher SUZ12 and lower JMJD3 occupancy in the differentiated H9 cells than in the undifferentiated H9 cells. This in turn resulted in higher H3K27me3 modification of the KRT1 promoter in the differentiated H9 cells.
FIGURE 6.
Recruitment of chromatin modifier SUZ12 and JMJD3 to EpCAM promoter in undifferentiated and differentiated hESCs. Top, schematic representation of the EpCAM promoter locus, which spanned positions −630 to +967 with respect to the TSS. The ChIP primers used in the study are indicated by horizontal lines. A and B, chromatin samples were immunoprecipitated with anti-SUZ12 antibody (A) or anti-JMJD3 antibody (B), and enrichment of the EpCAM and KRT1 promoter was quantitated by Q-PCR. KRT1 was used as a control for SUZ12/JMJD3/H3K27me3 binding. Each experiment was done in triplicate (mean ± S.D.). The value of immunoprecipitated target was calculated as the ratio of IP DNA to the total input DNA (IP/input). The target IP/input was further normalized to a control promoter region of HBB (SUZ12) or of GAPDH (JMJD3) to obtain -fold enrichment values. By ChIP measurement, the association of SUZ12 with the EpCAM promoter was elevated both upstream and downstream of TSS in differentiated H9 cells. In contrast, quantification of the intensities of JMJD3 binding was increased downstream of TSS in undifferentiated H9 cells (*, p < 0.05).
Downstream of EpCAM TSS of undifferentiated H9 cells, both proximal and distal promoter regions, we found a corresponding rise in JMJD3 binding (Fig. 6B), which coincided with the reduction of H3K27me3 (Fig. 5C), suggesting a direct causal relationship between JMJD3 recruitment and H3K27 demethylation. These findings indicate that EpCAM expression in hESC is maintained by the loss of H3K27me3 and SUZ12 as well as the increase in JMJD3 on the promoters of the EpCAM gene.
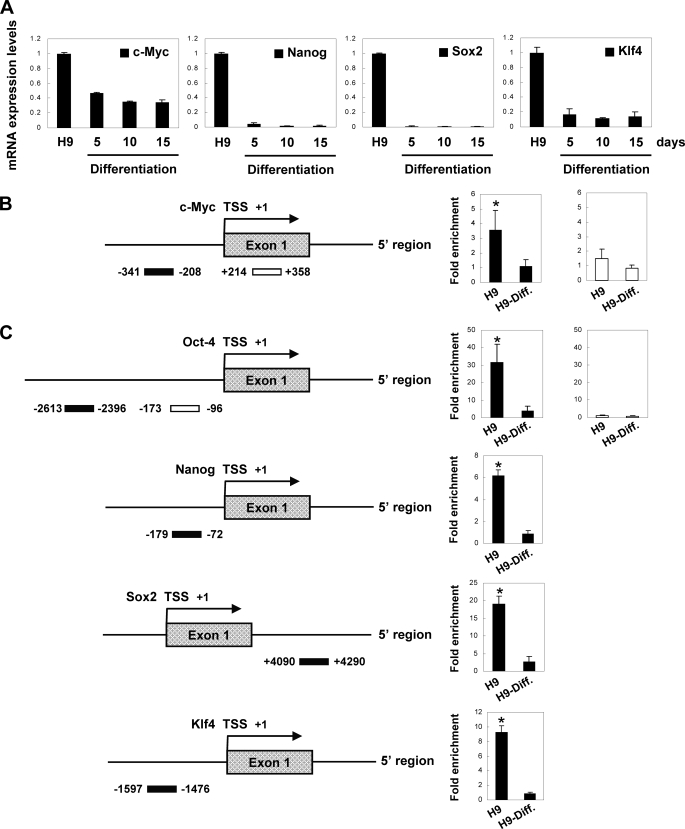
EpCAM Is Involved in ES Cell Maintenance through Its Influence on c-MYC, OCT-4, SOX2, and KLF4
It is of biological significance to study the function of EpCAM at the molecular level in ES cells. In order to correlate EpCAM with its early response molecule c-MYC and other possible downstream targets, such as OCT-4, NANOG, SOX2, and KLF4, we examined expression of these genes using Q-RT-PCR. We also found that all of the four genes were highly expressed in hESCs but collapsed after hESC differentiation was induced, as reported (Figs. 2B and 7A) (47–50). The expression of these genes remained repressed during differentiation at all observation time points (days 5–15; Figs. 2B and 7A). To directly determine whether EpCAM regulated these genes, we performed ChIP assays in both undifferentiated H9 and H9-Diff. cells with the EpICD antibody. Nuclear localization of EpICD was visualized by immunofluorescent staining using domain-specific antibodies. We found positive staining of EpICD in the nucleus of undifferentiated H9 but not in that of H9-Diff. cells (supplemental Fig. S2). The specificity of the two commercial anti-EpCAM Abs (1144-1 and A-20) against EpICD was further confirmed by ELISA (supplemental Fig. S3). There was a 3 times greater increase of EpICD binding in undifferentiated H9 than in H9-Diff. cells at the proximal upstream region of TSS of the c-MYC promoter (51). In contrast, there was no significant EpICD binding to the control downstream exon 1 of the c-MYC in either cells (Fig. 7B).
FIGURE 7.
EpCAM regulates c-MYC, OCT-4, NANOG, SOX2, and KLF4 to help maintain stemness in hESCs. A, Q-RT-PCR analysis of mRNA expression in undifferentiated H9 cells and in H9 cells differentiated for 5, 10, and 15 days. The expression level was normalized to internal control GAPDH. B, quantitative ChIP analysis of EpCAM binding to c-MYC promoter (*, p < 0.05). C, quantitative ChIP analysis of EpCAM binding to OCT-4, NANOG, SOX2, and KLF4 promoters (*, p < 0.05). The experiment was done in triplicate (mean ± S.D.).
It was interesting to observe that there was EpICD binding to OCT-4 (distal upstream region) (52), NANOG (upstream region) (53–55), SOX2 (downstream region) (52), and KLF4 (upstream region) promoters as well, possibly suggesting that that EpCAM modulates ES phenotype through promoting the expression of these downstream targets (Fig. 7C).
Decrease EpCAM Expression Was Associated with Down-regulation of ES Reprogramming Genes
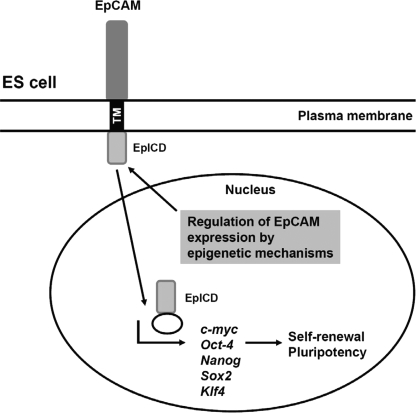
To investigate the relationship between EpCAM and four reprogramming genes, we performed a gene-silencing experiment using a lentivirus encoding shRNA for EpCAM. Q-RT-PCR revealed a significant knockdown (about 66%) in EpCAM expression in the shRNA-treated cells, HCT116/shRNA1 and HCT116/shRNA2 (supplemental Fig. S4). OCT-4, NANOG, SOX2, and KLF4 were found to be concomitantly down-regulated by the EpCAM shRNA treatment (supplemental Fig. S4). Based on these findings, we propose that EpCAM expression in hESCs is regulated by an epigenetic mechanism, and EpCAM activates gene expression of c-MYC, OCT-4, NANOG, SOX2, and KLF4 through transducing signal to the nucleus by EpICD, resulting in self-renewal and the maintenance of pluripotency in ES cells (Fig. 8).
FIGURE 8.
Schematic illustration of signaling pathways of EpCAM. EpCAM expression in hESCs is controlled by epigenetic regulation. The signaling of EpCAM was achieved by EpICD translocation into the nucleus, which contacted promoters of c-MYC, OCT-4, NANOG, SOX2, and KLF4 to exert its impact on maintaining the ES cell stemness condition.
DISCUSSION
The recent successful use of somatic cells that can be directly reprogrammed into pluripotency and self-renewal stem cells (31, 32) has opened a new means of investigating basic biology, developmental biology, and regenerative medicine. Ectopic transduction of terminally differentiated fibroblasts with the original four reprogramming transcription factors OCT-4, SOX2, KLF4, and c-MYC results in induced pluripotent stem cell formation morphologically and genetically (31, 32). These four factors play a pivotal role in ES cell self-renewal and maintenance of pluripotency. However, until this study, it was unknown what was mutually regulating these four genes. This study found that EpICD could directly bind to the promoters of these four genes (Fig. 7) and sustain the expression of these genes in undifferentiated hESCs (Fig. 7). EpCAM expression in undifferentiated hESCs occurred through epigenetic regulation (Figs. 5 and 6). Based on these findings, we propose that the maintenance of pluripotency by these four genes is controlled by the expression of EpCAM, which collaborates with these four factors to maintain self-renewal and pluripotency in ES cells.
The expression of EpCAM has been used to recognize hepatic stem cells in fetal, postnatal, and adult humans (9–11) and is thought to be useful in the selection of cancer-initiating cells (7, 8). Our study found EpCAM to be exclusively expressed in hESCs (Figs. 1 and 3) and its expression to be correlated with the expression of OCT-4 (Figs. 1 and 2), a known stem cell marker. This finding suggests that the cell surface expression property of EpCAM can be used to purify and enrich hESCs effectively, and it can be used as a surrogate for OCT-4 or potentially other hESC markers to simplify and improve the isolation and purity of hESCs. Our data can be compared with a recent publication showing EpCAM expression in undifferentiated hESCs (17). Similar to that report, we show that EpCAM expression was restricted to undifferentiated hESCs, and its silencing was associated with cell differentiation. This property of EpCAM can make it useable as a surface marker for hESCs.
Rigorous hESC isolation is a complicated process requiring various combinations of multiple cell surface markers. Nevertheless, many of the stem cell markers used nowadays cannot be used specifically on hESCs. For example, peanut agglutinin is only applicable as a cell surface marker in murine hematopoietic stem cells and human neural stem cells (56), and CD29 expression only can be used as a stem cell marker in murine skin or liver (10, 57). CD133 (prominin-1) has recently emerged as a major somatic stem cell or progenitor marker (58). Thus, there is a need to expand the current repertoire of hESC markers to enable hESC studies. Recognition by hESC-selective cell surface molecules is required in order to specifically isolate pure hESCs. Particularly, the combination of viable markers has made it possible to separate multiple hESC populations at each distinct differentiation stage and yielded important insights into stem cell differentiation. Because EpCAM is not expressed by differentiated cells, this molecule is probably exerted at the stem or multipotent progenitor cell stage.
The results of studies over the past few years have suggested that epigenetic mechanisms play a key role in these fundamental processes of self-renewal, maintenance of pluripotency, and lineage specification (23, 59). For example, pluripotency-associated genes, such as OCT-4 and NANOG, are stably silenced upon cell differentiation through epigenetic mechanisms (60). Non-transcribed genes in ESCs are repressed by the PcG in mice and humans with the promoters of these genes enriched with repressive histone H3K27 trimethylation. Our investigation on the epigenetic regulations of EpCAM expression during differentiation indicated that the expression of EpCAM was not regulated by DNA methylation (Fig. 4), which would lead to permanent silencing of the gene. Instead, we found that during differentiation, there is a drastic reduction in histone active markers, such as H3K4 trimethylation and H3K9 acetylation, and clear enhancement of repressive markers H3K9 and H3K27 trimethylation at the promoter of EpCAM (Fig. 5). In addition, EpCAM was not controlled by bivalent chromatin modifications (61, 62) because H3K4 and H3K27 trimethylation did not coexist at its promoter before or after the differentiation (Fig. 5), indicating that EpCAM does not belong to the category of lineage commitment or cell fate determination genes (62, 63). Our findings reflect the fact that EpCAM is virtually reintroduced in mature tissues (64), especially in epithelia cells. Therefore, our results suggest that, to facilitate the dynamic expression pattern of EpCAM during development, the chromatin state of its promoter is elaborately modulated by histone-modifying enzymes, such as SUZ12 and JMJD3, but not DNA methylation (Fig. 6) in response to hESC differentiation, which sustains the plasticity of EpCAM expression. SUZ12 has been found to be a key component of polycomb complex, PRC2 (26, 27), and essential for the activity of H3K27 methyltransferase. Conversely, JMJD3, a newly identified histone demethylase for H3K27 methylation, is able to remove the methylation mark from H3 lysine 27 and control cell differentiation (65). We found SUZ12 and JMJD3 to be dynamically associated with EpCAM promoter (Fig. 6). The methylation patterns of H3 Lys27 also changed accordingly (Fig. 5). ChIP combination resulted from the enzyme bindings and the changes of the corresponding histone mark, suggesting that EpCAM is part of a polycomb-mediated differentiation program in human ES cells.
Our study found that EpCAM played an intricate role in the regulation of the ES cell state. This may be achieved by modulating the expression of the EpCAM downstream target gene c-MYC, which, as other studies have reported, is involved in governing cell proliferation and dedifferentiation (29, 50). Both c-MYC and EpCAM are controlled by the Wnt signaling cascade (51, 66–68), in which signal transduction by EpICD interacts with β-catenin/Lef-1 in cohort to regulate c-MYC expression (6). The canonical Wnt signaling pathway has emerged as a critical regulator of ES and hematopoietic lineage stem cells in other studies (69–72). The results of such studies provide a compelling explanation for the high levels of EpCAM in stem or progenitor cells. Through molecular circuitry established around Wnt signaling, EpCAM and c-Myc can collaborate to sustain self-renewal and the pluripotent state of ES cells.
Nuclear translocation of EpICD (supplemental Fig. S2) (6) up-regulates c-MYC, OCT-4, NANOG, SOX2, and KLF4 (Figs. 7 and 8), indicating that EpCAM targeting of these ES cell fate genes may not occur exclusively through Wnt signaling, suggesting that there may be several pathways orchestrating the maintenance of the physical state of ES cells. Further investigations are required to clarify the relationship between Wnt signaling and OCT-4, NANOG, SOX2, and KLF4, because the results of such studies may further our understanding the nature of pluripotency and of self-renewal signals in hESCs.
Supplementary Material
Acknowledgments
We thank Bei-Chia Yang for preparing the human embryonic stem cells and the Core Facility of the Institute of Cellular and Organismic Biology, Academia Sinica, Taiwan for assistance.
This work was supported by Academia Sinica and National Science Council (Taiwan) Grants NSC98-2323-B-001-001 and NSC98-3111-B-001-004 (to H.-C. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- hESC
- human embryonic stem cell
- mAb
- monoclonal antibody
- MSP
- methylation-specific PCR
- ChIP
- chromatin immunoprecipitation
- H3K27
- lysine 27 of histone H3
- H3K27me3
- lysine 27 trimethylation of histone 3
- H3K4me3
- lysine 4 trimethylation of histone 3
- H3K9K14Ac
- lysine 9/14 acetylation of histone 3
- H3K9me3
- lysine 9 trimethylation of histone 3
- H9-Diff.
- H9 cells differentiated at day 30
- PcG
- polycomb group
- ES
- embryonic stem
- MEF
- mouse embryo fibroblast
- PBS
- phosphate-buffered saline
- ELISA
- enzyme-linked immunosorbent assay
- RT
- reverse transcription
- Q-RT
- quantitative reverse transcription
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- IP
- immunoprecipitation
- shRNA
- short hairpin RNA.
REFERENCES
- 1.Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 2.Litvinov S. V., Velders M. P., Bakker H. A., Fleuren G. J., Warnaar S. O. (1994) J. Cell Biol. 125, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Litvinov S. V., Balzar M., Winter M. J., Bakker H. A., Briaire-de Bruijn I. H., Prins F., Fleuren G. J., Warnaar S. O. (1997) J. Cell Biol. 139, 1337–1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balzar M., Winter M. J., de Boer C. J., Litvinov S. V. (1999) J. Mol. Med. 77, 699–712 [DOI] [PubMed] [Google Scholar]
- 5.Litvinov S. V., van Driel W., van Rhijn C. M., Bakker H. A., van Krieken H., Fleuren G. J., Warnaar S. O. (1996) Am. J. Pathol. 148, 865–875 [PMC free article] [PubMed] [Google Scholar]
- 6.Maetzel D., Denzel S., Mack B., Canis M., Went P., Benk M., Kieu C., Papior P., Baeuerle P. A., Munz M., Gires O. (2009) Nat. Cell Biol. 11, 162–171 [DOI] [PubMed] [Google Scholar]
- 7.Al-Hajj M., Wicha M. S., Benito-Hernandez A., Morrison S. J., Clarke M. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3983–3988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalerba P., Dylla S. J., Park I. K., Liu R., Wang X., Cho R. W., Hoey T., Gurney A., Huang E. H., Simeone D. M., Shelton A. A., Parmiani G., Castelli C., Clarke M. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 10158–10163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmelzer E., Wauthier E., Reid L. M. (2006) Stem Cells 24, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 10.Dan Y. Y., Riehle K. J., Lazaro C., Teoh N., Haque J., Campbell J. S., Fausto N. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9912–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmelzer E., Zhang L., Bruce A., Wauthier E., Ludlow J., Yao H. L., Moss N., Melhem A., McClelland R., Turner W., Kulik M., Sherwood S., Tallheden T., Cheng N., Furth M. E., Reid L. M. (2007) J. Exp. Med. 204, 1973–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anderson R., Schaible K., Heasman J., Wylie C. (1999) J. Reprod. Fertil. 116, 379–384 [DOI] [PubMed] [Google Scholar]
- 13.Stingl J., Raouf A., Emerman J. T., Eaves C. J. (2005) J. Mammary Gland Biol. Neoplasia 10, 49–59 [DOI] [PubMed] [Google Scholar]
- 14.Sundberg M., Jansson L., Ketolainen J., Pihlajamäki H., Suuronen R., Skottman H., Inzunza J., Hovatta O., Narkilahti S. (2009) Stem Cell Res. 2, 113–124 [DOI] [PubMed] [Google Scholar]
- 15.Kolle G., Ho M., Zhou Q., Chy H. S., Krishnan K., Cloonan N., Bertoncello I., Laslett A. L., Grimmond S. M. (2009) Stem Cells 27, 2446–2456 [DOI] [PubMed] [Google Scholar]
- 16.González B., Denzel S., Mack B., Conrad M., Gires O. (2009) Stem Cells 27, 1782–1791 [DOI] [PubMed] [Google Scholar]
- 17.Ng V. Y., Ang S. N., Chan J. X., Choo A. B. (2010) Stem Cells 28, 29–35 [DOI] [PubMed] [Google Scholar]
- 18.Tai K. Y., Shiah S. G., Shieh Y. S., Kao Y. R., Chi C. Y., Huang E., Lee H. S., Chang L. C., Yang P. C., Wu C. W. (2007) Oncogene 26, 3989–3997 [DOI] [PubMed] [Google Scholar]
- 19.Spizzo G., Gastl G., Obrist P., Fong D., Haun M., Grünewald K., Parson W., Eichmann C., Millinger S., Fiegl H., Margreiter R., Amberger A. (2007) Cancer Lett. 246, 253–261 [DOI] [PubMed] [Google Scholar]
- 20.Yu G., Zhang X., Wang H., Rui D., Yin A., Qiu G., He Y. (2008) Oncol. Rep. 20, 1061–1067 [PubMed] [Google Scholar]
- 21.Shiah S. G., Chang L. C., Tai K. Y., Lee G. H., Wu C. W., Shieh Y. S. (2009) Oral Oncol. 45, e1–e8 [DOI] [PubMed] [Google Scholar]
- 22.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. (2007) Cell 128, 735–745 [DOI] [PubMed] [Google Scholar]
- 23.Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 24.Schwartz Y. B., Pirrotta V. (2007) Nat. Rev. Genet. 8, 9–22 [DOI] [PubMed] [Google Scholar]
- 25.Swigut T., Wysocka J. (2007) Cell 131, 29–32 [DOI] [PubMed] [Google Scholar]
- 26.Cao R., Zhang Y. (2004) Mol. Cell 15, 57–67 [DOI] [PubMed] [Google Scholar]
- 27.Pasini D., Bracken A. P., Jensen M. R., Lazzerini Denchi E., Helin K. (2004) EMBO J. 23, 4061–4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agger K., Cloos P. A., Christensen J., Pasini D., Rose S., Rappsilber J., Issaeva I., Canaani E., Salcini A. E., Helin K. (2007) Nature 449, 731–734 [DOI] [PubMed] [Google Scholar]
- 29.Münz M., Kieu C., Mack B., Schmitt B., Zeidler R., Gires O. (2004) Oncogene 23, 5748–5758 [DOI] [PubMed] [Google Scholar]
- 30.Park I. H., Zhao R., West J. A., Yabuuchi A., Huo H., Ince T. A., Lerou P. H., Lensch M. W., Daley G. Q. (2008) Nature 451, 141–146 [DOI] [PubMed] [Google Scholar]
- 31.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 32.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 33.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 34.Yu J., Hu K., Smuga-Otto K., Tian S., Stewart R., Slukvin, Thomson J. A. (2009) Science 324, 797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y. C., Huang H. N., Lin C. T., Chen Y. F., King C. C., Wu H. C. (2007) Clin. Vaccine Immunol. 14, 404–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lu T. Y., Kao C. F., Lin C. T., Huang D. Y., Chiu C. Y., Huang Y. S., Wu H. C. (2009) J. Cell. Biochem. 108, 315–325 [DOI] [PubMed] [Google Scholar]
- 37.Kannagi R., Cochran N. A., Ishigami F., Hakomori S., Andrews P. W., Knowles B. B., Solter D. (1983) EMBO J. 2, 2355–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brimble S. N., Sherrer E. S., Uhl E. W., Wang E., Kelly S., Merrill A. H., Jr., Robins A. J., Schulz T. C. (2007) Stem Cells 25, 54–62 [DOI] [PubMed] [Google Scholar]
- 39.Gerrard L., Zhao D., Clark A. J., Cui W. (2005) Stem Cells 23, 124–133 [DOI] [PubMed] [Google Scholar]
- 40.Assou S., Le Carrour T., Tondeur S., Ström S., Gabelle A., Marty S., Nadal L., Pantesco V., Réme T., Hugnot J. P., Gasca S., Hovatta O., Hamamah S., Klein B., De Vos J. (2007) Stem Cells 25, 961–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jovic M., Naslavsky N., Rapaport D., Horowitz M., Caplan S. (2007) J. Cell Sci. 120, 802–814 [DOI] [PubMed] [Google Scholar]
- 42.Esteller M. (2007) Nat. Rev. Genet. 8, 286–298 [DOI] [PubMed] [Google Scholar]
- 43.Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 44.Bernstein B. E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D. K., Huebert D. J., McMahon S., Karlsson E. K., Kulbokas E. J., 3rd, Gingeras T. R., Schreiber S. L., Lander E. S. (2005) Cell 120, 169–181 [DOI] [PubMed] [Google Scholar]
- 45.Mutskov V., Felsenfeld G. (2004) EMBO J. 23, 138–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen G. L., Webster D. E., Barragan D. I., Chang H. Y., Khavari P. A. (2008) Genes Dev. 22, 1865–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niwa H., Miyazaki J., Smith A. G. (2000) Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 48.Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 50.Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. (2005) Development 132, 885–896 [DOI] [PubMed] [Google Scholar]
- 51.He T. C., Sparks A. B., Rago C., Hermeking H., Zawel L., da Costa L. T., Morin P. J., Vogelstein B., Kinzler K. W. (1998) Science 281, 1509–1512 [DOI] [PubMed] [Google Scholar]
- 52.Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuroda T., Tada M., Kubota H., Kimura H., Hatano S. Y., Suemori H., Nakatsuji N., Tada T. (2005) Mol. Cell. Biol. 25, 2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chan K. K., Zhang J., Chia N. Y., Chan Y. S., Sim H. S., Tan K. S., Oh S. K., Ng H. H., Choo A. B. (2009) Stem Cells 27, 2114–2125 [DOI] [PubMed] [Google Scholar]
- 55.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005) J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- 56.Rietze R. L., Valcanis H., Brooker G. F., Thomas T., Voss A. K., Bartlett P. F. (2001) Nature 412, 736–739 [DOI] [PubMed] [Google Scholar]
- 57.Shackleton M., Vaillant F., Simpson K. J., Stingl J., Smyth G. K., Asselin-Labat M. L., Wu L., Lindeman G. J., Visvader J. E. (2006) Nature 439, 84–88 [DOI] [PubMed] [Google Scholar]
- 58.Huttner H. B., Janich P., Köhrmann M., Jászai J., Siebzehnrubl F., Blümcke I., Suttorp M., Gahr M., Kuhnt D., Nimsky C., Krex D., Schackert G., Löwenbrück K., Reichmann H., Jüttler E., Hacke W., Schellinger P. D., Schwab S., Wilsch-Bräuninger M., Marzesco A. M., Corbeil D. (2008) Stem Cells 26, 698–705 [DOI] [PubMed] [Google Scholar]
- 59.Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. (2006) Nat. Cell Biol. 8, 188–194 [DOI] [PubMed] [Google Scholar]
- 61.Azuara V., Perry P., Sauer S., Spivakov M., Jørgensen H. F., John R. M., Gouti M., Casanova M., Warnes G., Merkenschlager M., Fisher A. G. (2006) Nat. Cell Biol. 8, 532–538 [DOI] [PubMed] [Google Scholar]
- 62.Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 63.Mikkelsen T. S., Ku M., Jaffe D. B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T. K., Koche R. P., Lee W., Mendenhall E., O'Donovan A., Presser A., Russ C., Xie X., Meissner A., Wernig M., Jaenisch R., Nusbaum C., Lander E. S., Bernstein B. E. (2007) Nature 448, 553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trzpis M., McLaughlin P. M., de Leij L. M., Harmsen M. C. (2007) Am. J. Pathol. 171, 386–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Santa F., Totaro M. G., Prosperini E., Notarbartolo S., Testa G., Natoli G. (2007) Cell 130, 1083–1094 [DOI] [PubMed] [Google Scholar]
- 66.Yamashita T., Budhu A., Forgues M., Wang X. W. (2007) Cancer Res. 67, 10831–10839 [DOI] [PubMed] [Google Scholar]
- 67.van de Wetering M., Sancho E., Verweij C., de Lau W., Oving I., Hurlstone A., van der Horn K., Batlle E., Coudreuse D., Haramis A. P., Tjon-Pon-Fong M., Moerer P., van den Born M., Soete G., Pals S., Eilers M., Medema R., Clevers H. (2002) Cell 111, 241–250 [DOI] [PubMed] [Google Scholar]
- 68.Muncan V., Sansom O. J., Tertoolen L., Phesse T. J., Begthel H., Sancho E., Cole A. M., Gregorieff A., de Alboran I. M., Clevers H., Clarke A. R. (2006) Mol. Cell. Biol. 26, 8418–8426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reya T., Duncan A. W., Ailles L., Domen J., Scherer D. C., Willert K., Hintz L., Nusse R., Weissman I. L. (2003) Nature 423, 409–414 [DOI] [PubMed] [Google Scholar]
- 70.Sato N., Meijer L., Skaltsounis L., Greengard P., Brivanlou A. H. (2004) Nat. Med. 10, 55–63 [DOI] [PubMed] [Google Scholar]
- 71.Reya T., Clevers H. (2005) Nature 434, 843–850 [DOI] [PubMed] [Google Scholar]
- 72.Jones D. L., Wagers A. J. (2008) Nat. Rev. Mol. Cell Biol. 9, 11–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.