Abstract
Huntingtin proteolysis has been implicated in the molecular pathogenesis of Huntington disease (HD). Despite an intense effort, the identity of the pathogenic smallest N-terminal fragment has not been determined. Using a panel of anti-huntingtin antibodies, we employed an unbiased approach to generate proteolytic cleavage maps of mutant and wild-type huntingtin in the HdhQ150 knock-in mouse model of HD. We identified 14 prominent N-terminal fragments, which, in addition to the full-length protein, can be readily detected in cytoplasmic but not nuclear fractions. These fragments were detected at all ages and are not a consequence of the pathogenic process. We demonstrated that the smallest fragment is an exon 1 huntingtin protein, known to contain a potent nuclear export signal. Prior to the onset of behavioral phenotypes, the exon 1 protein, and possibly other small fragments, accumulate in neuronal nuclei in the form of a detergent insoluble complex, visualized as diffuse granular nuclear staining in tissue sections. This methodology can be used to validate the inhibition of specific proteases as therapeutic targets for HD by pharmacological or genetic approaches.
Keywords: Huntington Disease, Mouse, Neurodegeneration, Polyglutamine Disease, Proteolytic Enzymes, HdhQ150 Knock-in Mouse, R6/2 Mouse, Huntingtin
Introduction
Huntington disease (HD)2 is an inherited neurodegenerative disorder with onset in midlife for which symptoms include motor disturbances, personality changes, and cognitive decline (1). The mutation is a CAG/polyglutamine (polyQ) repeat expansion located at the N terminus of huntingtin (Htt), a protein of many diverse functions (2, 3). Individuals with (CAG)35 or less remain unaffected, those with (CAG)40 and above will develop HD within a normal lifespan, whereas repeats above (CAG)70 will invariably cause childhood onset.
Mouse models of HD include transgenic mice that express either N-terminal fragments of, or full-length human Htt, and the more genetically precise knock-in models, in which mutant CAG repeats have been inserted into the mouse Hd gene (Hdh) (1). R6/2 mice expressing mutant exon 1 Htt (4) exhibit an early onset and rapid progression of HD-related phenotypes, which have allowed extensive complementary analyses and enabled this model to be used as a screening tool. In our colony, nuclear inclusions can be readily detected by immunohistochemistry in the cerebral cortex, striatum, and hippocampus by 3 weeks of age (5, 6), Rotarod impairment is apparent by 6 weeks, and end-stage disease occurs at 15 weeks. The HdhQ150 knock-in mouse carries ∼150Q (7). In our homozygous HdhQ150 (HdhQ150/Q150) colony, nuclear inclusions were detected by immunohistochemistry in the striatum and hippocampus by 6 months and the cortex by 8 months, an impaired Rotarod performance was apparent by 18 months of age, and end stage disease occurs at around 22 months (8). Our recent comparison of R6/2 and HdhQ150/Q150 mice at late stage disease (standardized for strain background and CAG repeat size) found that both models exhibit widespread and comparable phenotypes (8–10). Nuclear inclusions and neuropil aggregates were distributed throughout all regions of the brain (8), and the peripheral distribution of nuclear inclusions was highly comparable (10). Microarray expression profiles from both striatum (9) and cerebellum3 of the R6/2 and HdhQ150/Q150 mice were highly correlated, and striatal profiles were also strikingly similar to those obtained from human post-mortem brain (9). The main difference between these two models is the age of phenotype onset and rate of phenotype progression. Therefore, the accumulation of a critical concentration of N-terminal Htt fragments could be the rate-limiting step required for phenotype onset in HdhQ150 mice.
The hypothesis that the toxic protein in HD is an N-terminal Htt fragment originated through the observations that only N-terminal Htt antibodies detect nuclear inclusions and dystrophic neurites in HD patient brains (11), that N-terminal fragments of Htt aggregate much more readily in vitro (12) and in cell models (13), and that Htt contained a caspase cleavage site (14). The ensuing search for sites of proteolytic cleavage of human Htt has revealed active caspase-3 (Htt-513 and Htt-552) and caspase-6 (Htt-586) sites (15) and calpain sites at Htt-469 and Htt-536 (16, 17). Prevention of cleavage at the caspase-6 but not caspase-3 sites has been found to be protective in YAC transgenic mice (18). Small N-terminal fragments that are potentially highly pathogenic have been isolated from HD post-mortem brains (19) and from a number of HD models systems for which the precise cleavage sites and proteases involved remain elusive (20–25).
To identify N-terminal Htt fragments that are relevant to HD-related phenotypes in the HdhQ150 knock-in mouse, we have conducted an unbiased systematic analysis of the N-terminal fragments that are generated from both the mutant and wild-type (WT) Htt proteins throughout the life span of this mouse model. We identified 14 prominent mutant N-terminal fragments that were present in all nine brain regions studied. We demonstrated that the smallest fragment is an exon 1 protein, known to contain a potent nuclear export (NES) signal (26, 27). Levels of exon 1 Htt and possibly other small fragments decrease in cytoplasmic soluble brain fractions from ∼8 months of age, because they accumulate in the nucleus as a detergent-insoluble aggregated complex. It will be possible to use this methodology as an in vivo pharmacological or genetic screen to validate the inhibition of specific proteases as therapeutic targets for HD.
EXPERIMENTAL PROCEDURES
Mouse Breeding, Maintenance, Genotyping, and CAG Repeat Sizing
All mouse experiments were performed under the Animals Scientific Procedures Act (1986) under project and personal licenses approved and issued by the Home Office. The R6/2 and HdhQ150 knock-in mice were maintained, genotyped, and CAG repeat-sized as previously described (28). The mean repeat size was 154 ± 6.6 for HdhQ150 mice and 190 ± 2.1 (±S.D.) for R6/2. Mouse brains or brain regions were snap-frozen in liquid nitrogen and stored at −80 °C.
Immunoprecipitation
Whole brains were homogenized in freshly prepared 2-ml ice-cold HEPES buffer (50 mm HEPES/NaOH (pH 7.0), 150 mm NaCl, 10 mm EDTA, 1.0% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.1% bovine serum albumin) with 10 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride, and complete protease inhibitor mixture. Lysates were used immediately and never frozen. Samples were sonicated on ice for 10 s (Vibracell Sonicator, amplitude 40), spun at 13,000 × g at 4 °C for 30 min, and the supernatant was transferred to a fresh tube. Protein concentrations were determined using a BCA-protein assay kit (Perbio). Prior to immunoprecipitation, lysates were pre-cleared for 1 h at 4 °C on a rotating wheel by incubation with 50 μl (1:1-slurry) protein G-Sepharose-agarose beads (Invitrogen, prewashed 3× in 1× phosphate-buffered saline (PBS), 0.1% bovine serum albumin). Following brief centrifugation (to pellet beads), 1 mg of the pre-cleared supernatant was immunoprecipitated overnight on a rotating wheel. (When developing the methodology for this work, we performed immunoprecipitations for 2 h, 6 h, and overnight and never observed a difference in the pattern of soluble Htt fragments). Each reaction contained 20 μl (1:1-slurry) of protein G-Sepharose-agarose, 1 μg of antibody, and HEPES buffer to a final volume of 1 ml. Following immunoprecipitation, the protein G-Sepharose-agarose was pelleted by brief centrifugation at 13,000 × g for 30 s, washed with 1 ml HEPES buffer (4×), and finally resuspended in 8 μl of 2× Laemmli buffer. Immunoprecipitated complexes were eluted by denaturation at 75 °C for 5 min. For immunoprecipitation of brain regions, each region was homogenized in 250–500 μl of HEPES buffer, and the whole region was immunoprecipitated as above. For immunoprecipitation of cytoplasmic and nuclear fractions, samples were prepared as described, sonicated on ice for 10 s (amplitude 40), and centrifuged at 13,000 × g for 5 min at 4 °C, and finally the supernatant (500 μg of nuclei or 1 mg of cytoplasm) was immunoprecipitated as above.
Western Immunoblotting
Brains were homogenized in ice-cold KCl buffer and processed for Western blotting as described (8). For immunoblotting, 20–40 μg of total protein or 5–8 μl of immunoprecipitate in Laemmli loading buffer was denatured at 75 °C for 5 min and separated by 8-, 10-, 12-, or 15% SDS-PAGE and blotted onto nitrocellulose membrane (Schleicher and Schuell) and immunoprobed as described (8). Primary antibodies, secondary antibodies, and dilutions are summarized in the supplemental tables.
In Vitro Calpain and Caspase Cleavage
Mouse brains were dissected into two hemispheres, snap-frozen, and stored as described. One hemisphere was lysed in HEPES buffer and immunoprecipitated as above. The other hemisphere was lysed in cleavage buffer (15), sonicated, and spun at 13,000 × g at 4 °C for 30 min, and the protein concentrations of the supernatants were determined as above. For in vitro calpain cleavage, 200 μg was incubated with 0, 0.5, 2.5, 3.5, and 4.5 units of purified calpain I or II (Calbiochem), ± 5 mm CaCl2, in 50 μl of cleavage buffer (minus protease inhibitors) at 30 °C for 15 min. For in vitro caspase cleavage, 100 μg was incubated with 100 units of purified caspase-3 or -6 (Biomol) in 20 μl of cleavage buffer (minus protease inhibitors) at 37 °C for 1 h. Following incubation, 5× Laemmli loading buffer was added to quench the reactions, and samples were denatured at 75 °C for 5 min and analyzed by SDS-PAGE.
Site-directed mutagenesis of 23Q and 148Q pTet-splice-full-length Htt (25) was performed using primers F:5′-CTTGAGCCACAGCTCCTGACAGGTCAGCGCCGTCCC-3′ and R:5′-GGGACGGCGCTGACCTGTCAGGAGCTGTGGCTCAAG-3′. PCRs were performed using 50 ng of DNA, 5.0 μl of 10× Pfu buffer (Stratagene), 0.2 mm dNTPs (Promega), 125 ng each of forward and reverse primers (Integrated DNA Technologies), 5.0% Me2SO, and 1.0 μl of Pfu polymerase (Stratagene) for 18 cycles at 96 °C for 1 min, 55 °C for 1 min, and 68 °C for 45 min, then 68 °C for 10 min. Plasmids were DpnI (Promega)-treated, transformed into XL1-Blue supercompetent cells (Stratagene), and purified using the QIAprep Spin Miniprep Kit (Qiagen). Mutations were confirmed by DNA sequencing. The construct was expressed in 293T cells using SuperFect according to the manufacturer's instructions and lysed in m-PER reagent (Pierce).
Cytoplasmic and Nuclear Fractionation
Cytoplasmic and nuclear fractions were prepared as previously described (29).
TR-FRET
Time-resolved Förster resonance energy transfer (TR-FRET) was performed on cytoplasmic and nuclear fractions from HdhQ150 and R6/2 brains as described previously (30, 31). Statistical significance was determined by Student's t test.
Aggregate Dissociation in SDS and Formic Acid
Aggregates in cytoplasmic and nuclear fractions were dissociated sequentially in SDS and formic acid (19). Cytoplasmic samples: 50 μg was spun at 13,000 × g at room temperature for 40 min, and the supernatant (cytosol) was discarded. The pellet was resuspended in 50 μl of 1× SDS-buffer (2% SDS, 5% β-mercaptoethanol, 15% glycerol) and boiled for 10 min. Nuclear samples: 50 μg was resuspended in 2× SDS-buffer to a final volume of 50 μl and boiled for 10 min. Cytoplasmic and nuclear samples were sonicated on ice for 20 s (Vibracell Sonicator, amplitude 10), spun at 13,000 × g at 4 °C for 15 min, and the supernatants (SDS-soluble fractions) were transferred to a fresh tube. The pellets were briefly washed twice in 0.1 ml of 1× SDS-buffer, centrifuged at 13,000 × g for 5 min, briefly vortexed in 0.1 ml of formic acid, incubated on a shaking platform (250 rpm) at 37 °C for 1 h, and dried in a SpeedVac for 4 h. For SDS-PAGE analysis, 8 μl of the SDS-soluble fractions and all of the formic acid-treated fractions were resuspended in 5 μl of 2× Laemmli buffer, and 4 μl of 1 m Trizma base (Tris base, Sigma). (Note: nuclear samples should be blue in color. If they appeared yellow, 1 m Trizma base was added to neutralize the acidic pH.) Samples were denatured by boiling for 10 min prior to SDS-PAGE analysis.
Immunohistochemical Detection of Polyglutamine and Confocal Microscopy
For immunohistochemical detection of aggregates with formic acid, 6-month paraformaldehyde perfusion-fixed HdhQ150/Q150 brains were sectioned at 35 μm and processed as previously described (32). Sections were treated with formic acid, followed by sodium borohydride, and reacted overnight with either biotinylated 4H7H7 (100 ng/ml), 3B5H10 (100 ng/ml), or 1H6 (1 μg/ml). Following biotin tyramide amplification, biotin was detected with Vectastain ABC Elite reagent using a nickel/3,3′-diaminobenzidine substrate (Vector Laboratories). For confocal analysis, 15-month HdhQ150/Q150 brains were snap-frozen in isopentane on dry ice, and the striatum was sectioned at 15 μm onto poly-l-lysine-coated slides using a cryostat (Bright Instruments Ltd., UK). Sections were fixed in 4% paraformaldehyde for 30 min and washed in PBS (pH 7.4) twice for 15 min and then incubated in blocking buffer (PBS, 2% bovine serum albumin, mouse Ig blocking agent (MOM, Vector Laboratories)) with 0.1% Triton X-100 for 15 min. To test for co-localization with S830, sections were incubated with S830 in combination with each additional anti-Htt primary antibody in blocking buffer overnight at 4 °C. Sections were washed twice in PBS for 15 min, incubated with secondary fluorescent antibodies and TO-PRO-3 nuclear stain (Molecular Probes 1:1000) in blocking buffer for 1 h. Primary and secondary antibody dilutions are detailed in the supplemental tables. Finally, sections were washed twice in PBS for 15 min and mounted in Mowiol mounting media. Sections were examined and imaged on a Zeiss Meta-510 confocal microscope.
COS-1 Cell Transfection
Exon-1 Htt constructs (12) were sequentially mutated using the QuikChange site-directed mutagenesis kit (Stratagene) and subcloned into pSG5 (Stratagene). Mutations were confirmed by DNA sequencing. COS-1 cells were transiently transfected with 1 μg of DNA using FuGENE 6 (Roche Applied Science), cells were harvested after 48 h, and lysates were prepared in m-PER reagent (Pierce). 20 μg of lysate was analyzed by Western blot.
RESULTS
Map of N-terminal Mutant Htt Fragments Present in HdhQ150 Knock-in Mice
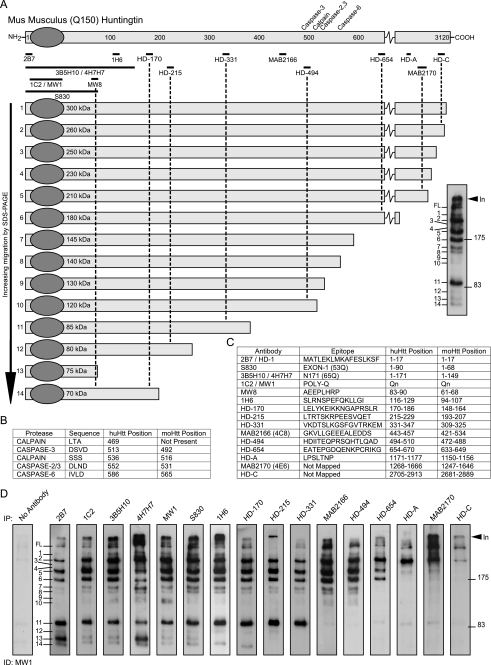
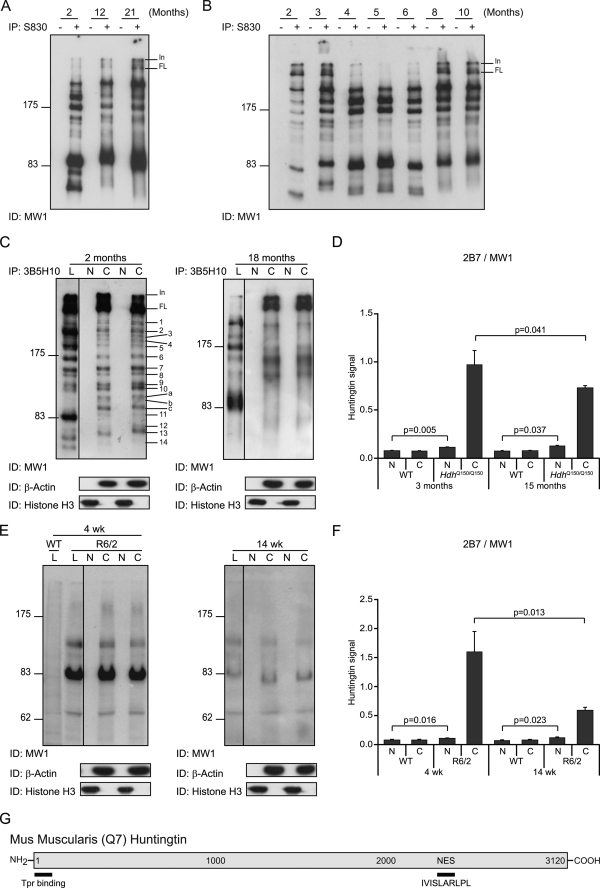
To identify N-terminal Htt fragments that are present in the brains of the HdhQ150 knock-in mice, we employed a combination of immunoprecipitation and Western blotting of whole brain lysates prepared from heterozygous HdhQ150 (Hdh+/Q150) animals aged 2 months (Fig. 1). We chose this approach, because, although Htt fragments can be detected directly on Western blots of brain lysates, construction of an N-terminal map is complicated by the identification of internal fragments recognized by many antibodies and by cross-reaction to other proteins (supplemental Fig. S1). We established a collection of antibodies across the Htt protein (Fig. 1, A and C, and supplemental data) and optimized our brain lysate preparation protocols (supplemental Fig. S2). To prepare an N-terminal fragment map of the mutant protein, Htt was immunoprecipitated with antibodies that recognize epitopes that span the protein and immunodetected with MW1, an antibody that recognizes the expanded polyQ tract and, therefore, does not detect WT Htt (Fig. 1D and supplemental Fig. S3). It was not possible to use HdhQ150/Q150 brains for this purpose, because the CAG repeats in each allele are generally of different lengths resulting in a doublet for each N-terminal fragment, which complicates the analysis. We were able to readily identify 14 fragments (Fig. 1A) that migrated by SDS-PAGE with sizes corresponding to 70–350 kDa. This pattern is robust, because it was obtained by immunoprecipitation with seven different N-terminal antibodies (2B7 through MW8) (Fig. 1 and supplemental Fig. S4). As has been demonstrated in multiple experiments, it is not possible to estimate the molecular weight of a mutant Htt fragment from its migration by SDS-PAGE, because this is retarded by the presence of the polyQ tract (12). For example, Fragments 11–14 all migrated between 70 and 85 kDa, whereas their estimated size range (assuming 150Q) would be 27–66 kDa. This analysis also uncovered the complex manner in which fragment length and polyQ tract interact, because the smallest fragment (Fragment 13) was not the fastest to migrate through the gel. Comparison with the location of the previously described caspase and calpain cleavage sites (Fig. 1B) indicated that Fragments 7–10 could terminate at the caspase Htt-513, Htt-552, Htt-586, and calpain Htt-536 sites, respectively. None of the fragments could terminate at the mouse equivalent of the human calpain Htt-469 site (Fig. 1B). Fragment 13 was the only fragment that could correspond to the previously described CpA (19, 25) or Cp1 (23), because all others are immunoprecipitated with 1H6 and HD-170.
FIGURE 1.
Map of proteolytic cleavage sites that generate the N-terminal mutant Htt fragments present in HdhQ150 knock-in mouse brains. A, schematic representation of mutant mouse Htt protein showing the position of the previously mapped caspase and calpain sites (above) and the antibodies used for immunoprecipitation (below). Htt was immunoprecipitated with each antibody from 2-month Hdh+/Q150 whole brain extracts and immunoprobed with the N-terminal antibody MW1 after fractionation by 8% SDS-PAGE. Fourteen prominent N-terminal fragments can be detected (right panel). The schematic shows the order in which the fragments migrate on the SDS gel, their size (in kilodaltons based on the co-migration of size markers), and their predicted length (based on the location of the antibodies with which they have been immunoprecipitated). B, details of the previously mapped caspase and calpain sites. The position in both human (with 23Q) and mouse Htt (with 7Q) is given. C, details of the antibodies used to generate the protease cleavage map: the epitope that they recognize and its location in both human (23Q) and mouse (7Q) Htt. D, series of SDS-PAGE immunoblots from which the map is derived. Mutant Htt was immunoprecipitated from a single 2-month Hdh+/Q150 brain with the panel of antibodies as indicated and immunodetected with MW1. Results were confirmed using multiple additional 2-month brains. The control immunoprecipitation from a WT brain was routinely performed and showed no signal (supplemental Fig. S3). In = interface between stacking and resolving gel; FL = full-length protein.
Map of N-terminal Wild-type Htt Fragments
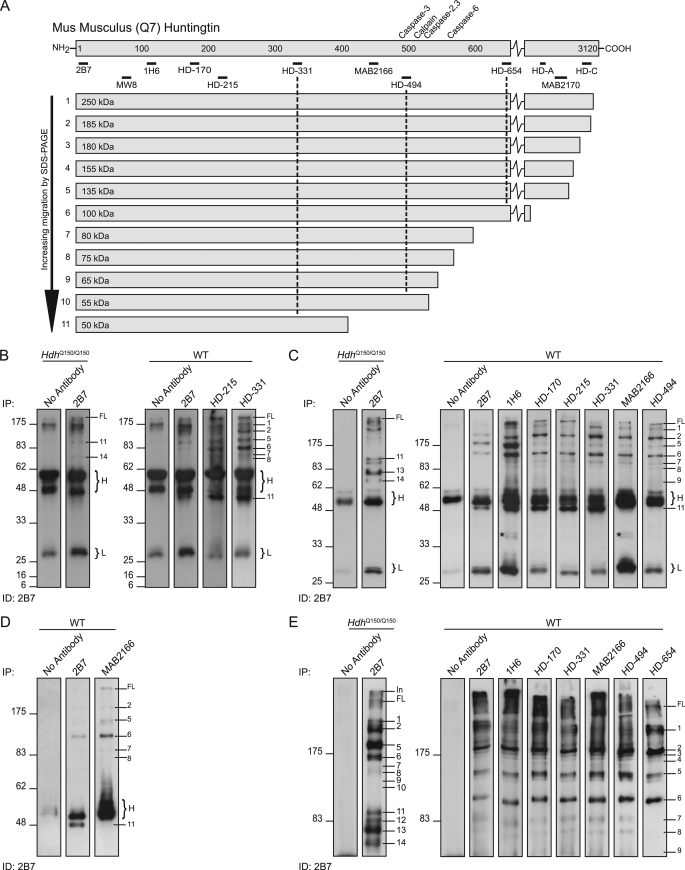
To generate the corresponding map of proteolytic sites in WT Htt, fragments were immunoprecipitated with antibodies recognizing epitopes across the WT protein and immunodetected with 2B7 or 1H6 (MW1 could not be used for these experiments, as it only detects mutant Htt). Identical maps were generated with both antibodies using brains from 2-month-old (Fig. 2) and 18-month-old (data not shown) WT mice. We were able to identify Fragments 1–11 from the mutant Htt map (Fig. 2A). Fragments 12–14, if generated from WT Htt, would be expected to migrate between 8 and 36 kDa, and we were unable to detect any fragments in this size range (Fig. 2B). As had been the case for mutant Htt, Fragment 11 was recognized by antibodies 2B7 to HD-331 (Fig. 2, C and D), and Fragments 7–10 by antibodies 2B7 to HD-494 (Fig. 2E). Fractionation of the WT fragments by 8% SDS-PAGE revealed several additional protease cleavage sites within the C-terminal region of the protein that generate N-terminal fragments larger than Fragment 2 (Fig. 2E). We did not have access to antibodies that would allow us to refine the map location of these more C-terminal cleavage sites.
FIGURE 2.
Map of proteolytic cleavage sites that generate N-terminal WT Htt fragments. A, schematic representation of WT mouse Htt showing the position of the previously mapped caspase and calpain sites (above) and the antibodies used for immunoprecipitation (below). Htt was immunoprecipitated with each antibody from 2-month WT whole brain extracts and immunoprobed with 2B7. The schematic shows the order in which the fragments migrate on the SDS gel, their size (in kilodaltons based on the co-migration of size markers), and their predicted length (based on the location of the antibodies with which they have been immunoprecipitated). B–E, series of SDS-PAGE immunoblots from which the map is derived. B, right-hand panel: fragments were resolved by 12% SDS-PAGE to identify the smallest N-terminal fragments, however, Fragments 12–14, predicted to be 8–36 kDa in size, could not be detected. Left-hand panel: no antibody and HdhQ150/Q150 controls for comparison with the position of mutant Htt Fragments 11 and 14 indicated for reference. C, right-hand panel: fragments were resolved by 10% SDS-PAGE, and Fragment 11 can be detected with antibodies 2B7 to HD-331, but not MAB2166 and HD-494. Left-hand panel: no antibody and HdhQ150/Q150 controls for comparison with the position of mutant Htt Fragments 11, 13, and 14 indicated for reference. D, fragments were separated by 8% SDS-PAGE to better resolve Fragment 11 from the IgG heavy chain. E, right-hand panel: fragments were resolved by 8% SDS-PAGE. Fragments 7–9 can be detected with antibodies 2B7 to HD-494 and not HD-654. Many more N-terminal fragments of a size between that of Fragment 2 and the full-length protein can be detected on this gel. Left-hand panel: no antibody and HdhQ150/Q150 controls for comparison with the position of mutant Htt fragments indicated for reference. H = IgG heavy chain; L = IgG light chain; *, bands detected by the secondary anti-mouse antibody.
Pattern of N-terminal Mutant Htt Fragments Is Consistent throughout the Brain
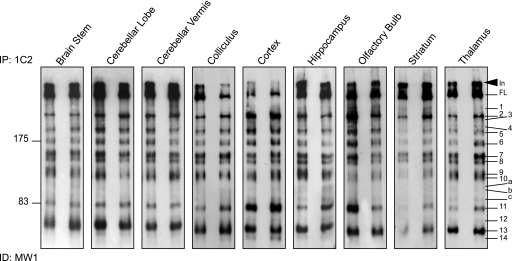
We next compared the pattern of mutant N-terminal Htt fragments upon immunoprecipitation with 1C2 from the brain regions of Hdh+/Q150 aged 2 months (Fig. 3). There was a striking similarity in the pattern of N-terminal fragments that can be detected across brain regions, although the relative intensity of individual fragments within any brain region was found to vary. For example, Fragment 11 appeared to be relatively more abundant in the cortex than in most other brain regions, and Fragment 14 although prominent in the cortex was very faint in the brain stem, striatum, and thalamus. Three comparatively weak bands that had not been identified in total brain lysate (Fragments a–c) were present in a number of brain regions and prominent in hippocampus and thalamus (Fig. 3).
FIGURE 3.
The spatial distribution of N-terminal mutant Htt fragments throughout the mouse brain. N-terminal Htt fragments were immunoprecipitated from the dissected brain regions of two Hdh+/Q150 mice at 2 months of age with antibody 1C2. Immunoprecipitates from both mice were fractionated by 8% SDS-PAGE and immunodetected with MW1. In = interface between stacking and resolving gel; FL = full-length protein.
At Least Four N-terminal Htt Fragments May Terminate at Caspase or Calpain Sites
To identify the C termini of the Htt N-terminal fragments we attempted to use mass spectrometric approaches but were unable to purify sufficient material by immunoprecipitation from mouse brain extracts for this purpose. Similarly, neo-epitope antibodies that recognize Htt caspase (Htt-513, Htt-552, and Htt-586) (15, 33) and calpain (Htt-536) (16) fragments in vitro and in cell culture failed to give a signal on Western blots of Htt fragments that had been immunoprecipitated from Hdh+/Q150 brains. Our interpretation of these negative results was that the neo-epitope antibodies had either relatively weak affinity for the caspase- and calpain-generated Htt fragments or that post-translational modifications blocked reactivity.
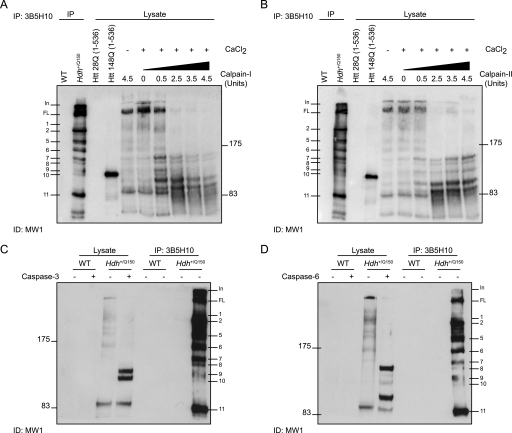
Therefore, in an attempt to determine the source of some of the N-terminal Htt fragments we performed a series of proteolytic digests. Digestion of lysates from a single Hdh+/Q150 brain aged 2 months with calpain-I and calpain-II indicated that Fragment 7 terminates at a previously unidentified calpain cleavage site that would map between amino acid 488 and 633 in the mouse protein (510–654 in human Htt with 23Q) (Fig. 4, A and B). Other fragments enriched by digestion with calpains are difficult to interpret. Two prominent fragments are generated with sizes between those of Fragments 10 and 11 that could correspond to Fragments b and c identified in Fig. 3, terminating between amino acids 325 and 472 in the mouse protein (347 and 494 in human). Digestion with caspase-3 produced two prominent bands most likely terminating at the Htt-513 and Htt-552 sites. Comparison with N-terminal Htt fragments present in the same mouse suggests that Fragment 10 may end at the Htt-513 caspase site and that a fragment terminating at Htt-552 is not present (Fig. 4C). Finally, digestion with caspase-6 indicated that Fragment 8 terminates at the Htt-586 caspase-6 cleavage site (Fig. 4D). An additional band released by caspase-6 cleavage could correspond to Fragment c (Fig. 3).
FIGURE 4.
Identification of Htt fragments generated by calpain or caspase proteolysis. A and B, the lysate from a 2-month-old Hdh+/Q150 mice was digested with increasing concentrations of calpain-I (A) or calpain-II (B) in the presence of CaCl2, fractionated by 8% SDS-PAGE, and immunoprobed with MW1. Control lanes included: (i) WT and Hdh+/Q150 lysates that had been immunoprecipitated with 3B5H10 (the same Hdh+/Q150 brain as digested with calpains), (ii) Htt fragments that terminate at amino acid 536 and contain 23Q or 148Q; and (iii) lysate digested with the maximum concentration (4.5 units) of calpain-I or -II in the absence of CaCl2. C and D, lysates from 2-month-old WT and Hdh+/Q150 mice were digested with or without either caspase-3 (C) or caspase-6 (D) and fractionated by 8% SDS-PAGE and immunoprobed with MW1. Control lanes were lysates that had been immunoprecipitated with 3B5H10 from the same WT and Hdh+/Q150 mice.
Ability to Detect the Smaller N-terminal Mutant Htt Fragments Decreases with Age
We then determined whether the mutant Htt fragments that were present in total brain lysates remained constant over the course of a 21-month period. Immunoprecipitation of N-terminal fragments with S830 and immunodetection with MW1 indicated that Fragments 12–14 could not be readily detected in whole brain lysates from Hdh+/Q150 mice aged 12 and 21 months (Fig. 5A). Also, the resolution of Fragments 7–11 was less distinct at later ages possibly because somatic instability of the CAG repeat results in a mosaic of fragment sizes (34). We conducted a time course to identify the age at which the inability to detect Fragments 12–14 first occurs (Fig. 5B). These three fragments were readily detected at 6 months but had considerably diminished by 8 months and were barely detectable at 10 months. This age transition corresponds to the time at which it is possible to detect statistically significant levels of aggregated Htt using the Seprion ligand enzyme-linked immunosorbent assay (28) and occurs prior to the onset of behavioral and physiological phenotypes (8).
FIGURE 5.
Detection of native N-terminal fragments of mutant Htt in the nucleus. A, N-terminal Htt fragments were immunoprecipitated with or without S830 from whole brain lysates of Hdh+/Q150 mice at 2, 12, and 21 months of age and immunodetected with MW1. The three smaller fragments (Fragments 12–14) cannot be detected in the lysates from mice aged 12 and 21 months. The resolution of the larger fragments is less distinct in the older mice. B, N-terminal Htt fragments were immunoprecipitated with or without S830 from Hdh+/Q150 whole brain lysates aged from 2 to 10 months and immunodetected with MW1. Fragments 12–14 are prominent up to and including 6 months of age but have diminished by 8 months. The variability in the migration of specific fragments between ages is a consequence of the difference in the CAG repeat length in the mice used in panels A and B. C, cytoplasmic and nuclear fractions were prepared from whole brains from two Hdh+/Q150 mice at each of 2 and 18 months of age and N-terminal Htt fragments were immunoprecipitated with 3B5H10 and immunodetected with MW1. These were compared with the fragments that were obtained by immunoprecipitation from whole brain lysates. Purity of the cytoplasmic and nuclear preparations was determined by immunoprobing with antibodies to β-actin and histone H3, respectively. D, TR-FRET (2B7-MW1) was performed on cytoplasmic and nuclear preparations from WT and HdhQ150/Q150 brains at 3 months and 15 months of age (n = 3/genotype/age). E, cytoplasmic and nuclear fractions were prepared from whole brains from two R6/2 mice at each of 4 and 14 weeks of age, the R6/2 transprotein was immunoprobed with MW1 and compared with the signal obtained from whole brain lysates. Purity of the cytoplasmic and nuclear preparations was determined by immunoprobing with antibodies to β-actin and histone H3, respectively. F, TR-FRET (2B7-MW1) was performed on cytoplasmic and nuclear preparations from WT and R6/2 brains at 4 and 14 weeks of age (n = 3/genotype/age). G, schematic of the position of putative nuclear export (NES) sites in the mouse Htt protein. N = nuclear, C = cytoplasm, L = lysates. In = interface between stacking and resolving gel; FL = full-length protein; Tpr = translocated promoter region.
Soluble Native N-terminal Fragments and Full-length Htt Are Not Detected in the Nucleus by Immunoprecipitation and Western Blotting
To probe the subcellular localization of the N-terminal fragments, we prepared whole brain cytoplasmic and nuclear fractions from two Hdh+/Q150 mice aged 2 months (presymptomatic) and two aged 18 months (symptomatic) and immunoprecipitated and immunodetected N-terminal Htt fragments with 3B5H10 and MW1, respectively (Fig. 5C). At both 2 and 18 months, the relative intensity of N-terminal fragments differed between the cytoplasmic fractions and those obtained by using the same two antibodies from whole brain lysates (Fig. 5C), reflecting the influence of the preparation buffer on epitope exposure. Fragments that were previously faint or barely detectable (Fragments 3, 4, 9, 10, a, b, and c) were prominent in the cytoplasmic fraction and, conversely, fragments that had always appeared abundant were comparatively faint (Fragment 11). At 18 months, as was the case for the whole brain lysates (Fig. 5A), the smaller fragments were no longer detected in the cytoplasm and the larger fragments resolved as more diffuse bands. We failed to detect any mutant Htt N-terminal fragments in the nuclear fractions by immunoprecipitation and subsequent Western blotting at 2 or 18 months of age (Fig. 5C).
To quantify the relative levels of mutant Htt in the cytoplasm and nucleus, we employed the highly sensitive time-resolved FRET (TR-FRET) assay (Fig. 5D) (30). This assay uses a labeled antibody pair: 2B7-europium and MW1-d2 to quantify levels of mutant Htt (the WT mouse protein is not detected by MW1). Cytoplasmic and nuclear preparations from WT and HdhQ150/Q150 brains aged 3 and 15 months were subject to TR-FRET with WT fractions functioning as negative controls. There was a statistically significant reduction in the levels of mutant Htt in cytoplasmic fractions at 15 months as compared with that at 3 months, most likely due to the recruitment of the mutant protein into Htt aggregates. Low but statistically significant levels of mutant Htt could be detected in the nuclear fractions of both 3- and 15-month HdhQ150/Q150 as compared with WT controls. This could represent either soluble Htt, in which case the levels were too low to detect by Western blot, or aggregated forms of Htt, because although MW1 does not detect large fibrillar aggregates, it does recognize smaller oligomeric structures (28).
We previously published that the soluble form of the R6/2 exon 1 transgene protein (transprotein) cannot be detected by Western blot in nuclear fractions prepared from the brains of R6/2 mice at any age (26). We applied TR-FRET to determine whether the transprotein can be detected in R6/2 nuclear brain fractions by this more sensitive approach. Cytoplasmic and nuclear fractions were prepared from the brains of R6/2 mice aged 4 weeks (presymptomatic) and 14 weeks (late-stage disease). Immunodetection with MW1 showed that the soluble transprotein present in whole brain lysates and cytoplasmic fractions decreased with age and could not be detected in the nucleus (Fig. 5E), confirming our previous results. The same samples were subjected to TR-FRET, and, as for the HdhQ150 experiments, the WT fractions served as negative controls (Fig. 5F). The level of transprotein in the cytoplasm significantly decreased with age as a consequence of being recruited into aggregates. There was a small but significant amount of mutant protein in the nucleus, which may represent either the soluble protein or an aggregated form thereof.
Detergent-insoluble Aggregated N-terminal Mutant Htt Fragments Are Present in Cell Nuclei in the Brains of HdhQ150 Knock-in Mice
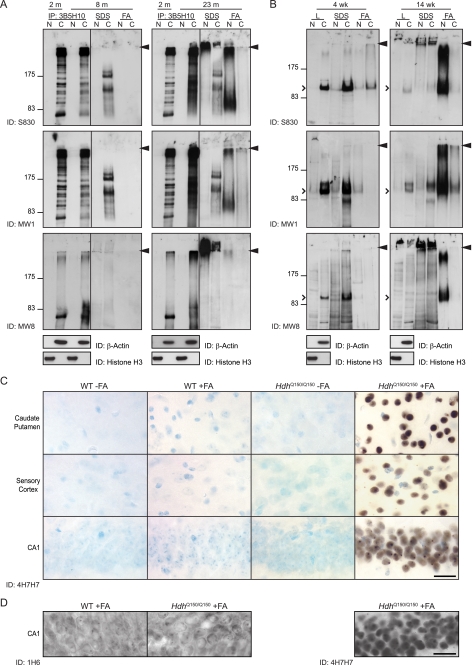
We hypothesized that small mutant N-terminal Htt fragments might have entered the nucleus and been retained there, because they had formed insoluble protein complexes. To investigate this further, Htt aggregates were pelleted from the cytoplasmic and nuclear fractions of Hdh+/Q150 mice aged 8 months (presymptomatic) and 23 months (end stage disease). The supernatants of these lysates were subject to immunoprecipitation with 3B5H10. The pellets were sequentially solubilized in SDS and formic acid. (Note: Htt was not immunoprecipitated from these.) All samples were fractionated by SDS-PAGE alongside immunoprecipitates from the cytoplasmic and nuclear fractions of an Hdh+/Q150 mouse aged 2 months for reference (Fig. 6A). Western blots were then immunoprobed with S830, MW1, or MW8.
FIGURE 6.
Aggregated but not soluble N-terminal mutant Htt fragments are present in the nucleus. A, cytoplasmic and nuclear fractions were prepared from brains from Hdh+/Q150 mice aged 8 and 23 months. After centrifugation, the supernatant was immunoprecipitated with 3B5H10, and the pellet was sequentially solubilized in SDS and formic acid. Samples were fractionated on 8% SDS-PAGE gels alongside N-terminal fragments immunoprecipitated from brain lysates from Hdh+/Q150 mice aged 2 months for comparison. Western blots were probed with S830, MW1, and MW8 as indicated. B, cytoplasmic and nuclear fractions were prepared from the brains of R6/2 mice at 4 and 14 weeks of age. After centrifugation the supernatant was retained, and the pellet was sequentially solubilized in SDS and formic acid. Samples were fractionated by 8% SDS-PAGE, and Western blots were immunoprobed with S830, MW1, and MW8 as indicated. Arrowhead = interface between stacking and resolving gels. N = nuclear, C = cytoplasm; open arrowhead = R6/2 transprotein. C, diffuse nuclear Htt was immunodetected with the polyQ-specific antibody 4H7H7 in sections from HdhQ150/Q150 brains aged 6 months only after prior treatment with formic acid. Staining was absent from formic acid-treated and untreated WT sections. D, diffuse nuclear Htt was immunodetected with the polyQ-specific antibody 4H7H7 in sections from HdhQ150/Q150 brains aged 6 months after prior treatment with formic acid. Staining was absent from formic acid-treated HdhQ150/Q150 and WT sections when immunoprobed with 1H6. FA = formic acid. Scale bars: 20 μm.
Analysis of the immunoprecipitated supernatants indicated that Fragment 13 might represent an exon 1 Htt protein. At 8 months, the pattern of fragments immunoprecipitated from the supernatant with 3B5H10 and immunodetected with S830 and MW1 was comparable to that at 2 months. By 23 months, all fragments resolved as a smear, and the smaller fragments (Fragments 12–14) could no longer be distinguished, possibly reflecting a combination of somatic instability and the accumulation of partially digested, partially soluble complexes. MW8 was raised against an epitope at the C terminus of the exon 1 protein. At 2 months of age MW8 only recognizes Fragment 13, the smallest of the mutant N-terminal fragments, suggesting that it may act as an exon1 C-terminal neo-epitope antibody and that Fragment 13 was an exon 1 protein. At 8 months a less intense fragment and smear were detected and at 23 months, a complex smear indicated that this fragment had formed detergent insoluble structures.
Solubilization of the cytoplasmic and nuclear pellets with SDS and immunodetection with S830 and MW8 revealed a large amount of detergent-insoluble aggregated protein in the stacking gel at 23 months, and trace amounts were present at 8 months. This material was not detected by MW1, which does not recognize fibrillar aggregates (28). The cytoplasmic pellets were partially solubilized with SDS to release a number of fragments that can be detected with both S830 and MW1. Because the solubilized pellets have not been immunoprecipitated prior to immunodetection, some of these bands might represent cross-reacting proteins. However, if that were the case, they should also be present in SDS-solubilized pellets from R6/2 mice (Fig. 6B). The fragments may represent solubilized aggregated material or, alternatively, fragments that had been pelleted with membranes and/or organelles. Solubilization of the nuclear pellet with formic acid released S830 and MW1 immunoreactive material that resolved as a smear and corresponded in size to the smaller Htt fragments. This was not detected by the MW8 C-terminal exon 1 antibody suggesting that the C terminus may have been destroyed by formic acid treatment, or by degradation within the cell.
We performed the same set of experiments with cytoplasmic and nuclear fractions from R6/2 mice aged 4 weeks (presymptomatic) and 14 weeks (late-stage disease) (Fig. 6B), except that the transprotein was not immunoprecipitated prior to fractionation by SDS-PAGE. The R6/2 mice express an exon 1 Htt protein. At 4 weeks, the soluble protein could be readily detected in the cytoplasm with all three antibodies but was absent from the nucleus, and by 14 weeks the cytoplasmic levels had greatly diminished. Analysis with all three antibodies showed that at 4 weeks, the cytoplasmic pellet could be solubilized with SDS, but at 14 weeks, the cytoplasmic SDS-soluble material could only be detected readily with MW1. The MW1 antibody consistently detected bands not seen by S830 and MW8. We have previously shown that MW1 can recognize specific conformers of the Htt exon 1 protein, the epitopes for which are retained upon fractionation through SDS-PAGE gels (28). Formic acid solubilization of the nuclear pellet from 14-week-old mice produced a smear very similar to that obtained from HdhQ150/Q150 mice at 23 months. The size distribution of the smear was mostly larger than that of exon 1 Htt indicating that the aggregates cannot be completely dissociated with formic acid. The smear was detectable with MW8 indicating that the C terminus of the exon 1 Htt protein is formic acid-resistant. Therefore, the absence of this fragment from the formic acid-digested, 23-month HdhQ150/Q150 nuclei might indicate that degradation of the C terminus had occurred over the lifetime of the mouse.
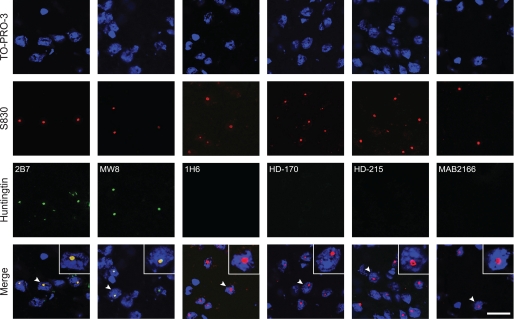
We next performed immunohistochemistry with the polyQ specific antibodies: 4H7H7 and 3B5H10, and with 1H6 on coronal brain sections from HdhQ150/Q150 mice aged 6 months. Mutant Htt was detected as a diffuse “stain” distributed throughout the nucleus in HdhQ150/Q150 but not WT mice at 6 months with both 4H7H7 (Fig. 6C) and 3B5H10 (data not shown) provided the sections had first been treated with formic acid. It was not possible to detect mutant Htt with either antibody in the absence of formic acid treatment. These data indicate that the polyQ tract was present as an H-bonded structure, consistent with the formation of a cross-β-sheet (35). The diffuse nuclear stain was not detected with 1H6 with or without formic acid treatment (Fig. 6D) suggesting that the aggregates may only contain Fragment 13. In addition, we performed immunohistochemistry with 2B7, MW8, 1H6, HD-170, HD-215, and MAB2166 and used confocal microscopy to determine whether these antibodies co-localized with S830. Of these, only 2B7 and MW8 were found to co-localize (Fig. 7), consistent with the above data indicating that aggregates are composed of the smallest N-terminal fragment. However, our immunoprecipitation and Western blotting data showed that our detection of the three smallest Htt fragments in cytoplasmic fractions diminished with age (Fig. 5). Therefore, the epitopes to antibodies, 1H6, HD-170, and HD-215, may be masked from detection by immunohistochemistry.
FIGURE 7.
Nuclear inclusions are immunodetected by antibodies that recognize epitopes N-terminal to 1H6. The exon 1 Htt antibody, S830, readily detects nuclear inclusions in HdhQ150/Q150 mouse brains aged 15 months. To determine which Htt fragments might be present in the nuclear inclusions, frozen sections were fixed in 4% paraformaldehyde, and immunohistochemistry was performed with antibodies that spanned the Htt protein and confocal microscopy used to look for co-localization with S830. The antibodies 2B7 and MW8 that detect exon 1 Htt epitopes, but not 1H6, HD-170, HD-215, and MAB216, were found to co-localize. Therefore, we could only detect epitopes that are present in the smallest N-terminal fragment (Fragment 13) in the nuclear inclusions. Scale bar: 20 μm.
Fragment 13 Is an Exon 1 Htt Protein
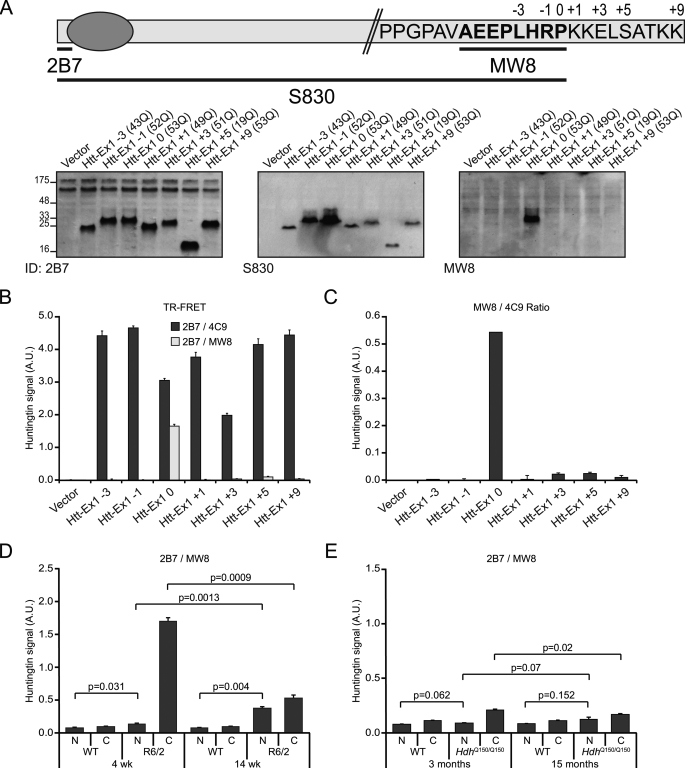
To determine whether MW8 behaves as an exon 1 Htt C-terminal neo-epitope antibody, we mutated a human exon 1 construct by the sequential deletion or addition of amino acids from and to the C-terminal proline (−3 to +9). We cloned a subset of these constructs into the mammalian expression vector pSG5, transiently expressed them in COS-1 cells, performed Western blots, and immunoprobed them with antibodies 2B7, S830, and MW8 (Fig. 8A). 2B7 and S830 both detected all constructs. In contrast, MW8 only recognized the exon 1 protein, failing to detect the −1 and +1 and all other mutant versions (Fig. 8A) indicating that MW8 acts as an exon 1 neo-epitope antibody on Western blots. TR-FRET with 2B7-terbium cryptate and MW8-d2 also specifically detected only the exon 1 protein (Fig. 8B), whereas TR-FRET using 2B7 and 4C9-alexa detected all N-terminal Htt constructs (Fig. 8B). 4C9 is a monoclonal antibody raised against the human specific proline-rich region (amino acid 65–84). Normalization of the 2B7-MW8 signal with the 2B7–4C9 signal on the same lysates confirmed that MW8 specifically recognizes the C terminus of exon 1 (Fig. 8C).
FIGURE 8.
The smallest N-terminal fragment is an exon 1 protein. A, schematic of the exon-1 Htt protein showing the C-terminal amino acid sequence, the positions of the antigens against which 2B7 and S830 were raised, and the epitope recognized by MW8 (39). The numbers above the amino acid sequence indicate the C termini of the various exon 1 mutant constructs. MW8 only recognizes the exon 1 protein in lysates prepared from the constructs when transiently expressed in COS-1 cells, whereas 2B7 and S830 detect exon 1 as well as all of the deletion and addition mutants. B and C, demonstration of the specificity of MW8 against the C terminus of exon 1. Htt specific signal (Htt levels) measured by TR-FRET (B) from cell lysates expressing the mutated exon 1 constructs using 2B7–4C9 (black bars) or 2B7-MW8 (gray bars). Normalization (C) of the MW8 signal with the 4C9 signal is shown. D and E, 2B7-MW8 TR-FRET of cytoplasmic and nuclear fractions from R6/2 mice aged 4 and 14 weeks (D) and HdhQ150/Q150 mice aged 3 and 15 months (E). For comparison, please see Fig. 5 (D and F), where the same tissue samples were analyzed by 2B7-MW1 TR-FRET.
TR-FRET was performed with 2B7-europium and MW8-d2 on nuclear and cytoplasmic fractions from R6/2 brains aged 4 and 14 weeks (Fig. 8D) and HdhQ150/Q150 brains aged 3 and 15 months (Fig. 8E) and compared with 2B7-MW1 FRET that had been performed on lysates from the same tissues (Fig. 5, D and F). The 2B7-MW1 assay is negative in WT mice as MW1 only detects expanded polyQ repeats. The absence of a signal with 2B7-MW8 in WT mice supports our mapping data indicating that Fragment 13 (exon 1 Htt) is not produced from WT huntingtin. As expected, given that the R6/2 mice express a mutant exon 1 protein, the TR-FRET signal between 2B7-MW1 (Fig. 5F) and 2B7-MW8 (Fig. 8D) was similar at both 4 and 14 weeks. In contrast, TR-FRET signals between 2B7-MW1 (Fig. 5D) and 2B7-MW8 (Fig. 8E) in the HdhQ150/Q150 mice were very different. The 2B7-MW1 signals reflect the level of full-length mutant Htt and all N-terminal fragments, whereas the lower signal obtained from 2B7-MW8 is indicative of the level of the exon 1 protein (Fragment 13), in either a soluble or aggregated state, in the HdhQ150/Q150 brains.
DISCUSSION
We have employed an immunoprecipitation and immunodetection strategy to identify N-terminal Htt proteolytic cleavage fragments that are present in the brains of HdhQ150 knock-in mice. The C termini of 14 mutant Htt N-terminal fragments were mapped with respect to antibodies that span the protein (Fragments 1–14). These fragments were present at 2 months, and, in contrast to previous reports (36), their appearance was not related to age or stage of disease. All 14 fragments were present in the nine brain regions studied and three additional fragments (Fragments a–c), not detected in immunoprecipitates from whole brain lysates, were identified. The relative intensity of the fragments varied depending on the antibody and lysis buffer used (Figs. 1D and 5) and, therefore, it is not possible to estimate the relative amounts of these N-terminal fragments in the mouse brain in vivo. We showed that Fragment 7 is generated by proteolysis with calpains, Fragment 8 by caspase-6 cleavage at Htt-586, Fragments 9 and 10 are probably produced by calpain Htt-536 and caspase-3 Htt-513 digestion. Fragments b and c may originate through digestion with a calpain or Fragment c could be the product of caspase-6 digestion. We showed that of the 14 N-terminal fragments, the three smallest (Fragments 12–14) could not be detected in the brains of WT mice. The reasons may be that the WT protein is processed to produce these fragments, that the WT fragments are degraded more rapidly than their mutant counterparts, or that the epitopes required for immunoprecipitation are not accessible in the context of the WT protein.
Htt contains a translocated promoter region binding site within the first 17 amino acids that acts as a very potent NES (27) and has an NES at amino acid 2397–2406 in the human protein (2373–2382 in mouse) (37) (Fig. 5G). We previously generated mice transgenic for a 20Q exon 1 protein with an N-terminal nuclear localization signal (26). The soluble transprotein was only detected in cytoplasmic fractions of brain lysates from these mice, demonstrating that the NES within amino acids 1–17 can override an exogenous nuclear localization signal in vivo (26) as had been shown previously in vitro (27). Here, we confirm that the soluble R6/2 transprotein can only be detected in the cytoplasmic fractions of R6/2 mice and that the nuclear fractions contain detergent-insoluble, formic acid-soluble, aggregated protein that is present in the stacking gels of Western blots. We conclude that the R6/2 transprotein enters the cell nucleus and is only retained there if it has formed detergent-insoluble complexes that interfere with the function of the N-terminal NES. These data suggest that the appearance of the transprotein as diffuse nuclear stain (5), as visualized by immunohistochemistry, represents an oligomeric/aggregated form of the R6/2 transprotein.
This hypothesis is consistent with the results of the present study. We did not detect the soluble form of full-length Htt or of the N-terminal Htt fragments in nuclear lysates from HdhQ150 knock-in brains at any age from 2 to 23 months by Western blots. These results are inconsistent with previous reports of low levels of nuclear full-length Htt detected on long exposures of Western blots from the brains of the HdhQ111 knock-in model (38) and in an inducible Htt transgenic model (25). This discrepancy could reflect differences in lysis conditions or in the subcellular fractionation protocols, because α-tubulin (a cytoplasmic marker) was also present in nuclear preparations (38). To investigate this issue further, we employed the more sensitive 2B7-MW1 TR-FRET assay (Fig. 5). This assay detects a FRET signal between 2B7 (1–17 amino acids) and MW1 (polyQ) (39) and would be expected to recognize soluble and oligomeric forms of Htt but not Htt fibrils (28). Therefore, in addition to formic acid-insoluble aggregates, the nucleus contains low levels of either soluble or oligomeric mutant Htt as recognized by MW1. It follows that, if the Htt-586 caspase-6 fragment, previously reported to be enriched in nuclei in cell culture systems (33), is present in neuronal nuclei in HdhQ150 brains, it is there at very low levels.
Our ability to detect the smallest mutant N-terminal Htt fragments in the cytoplasmic fractions from HdhQ150 brains diminishes with age and occurs in parallel to increased levels of detergent-insoluble, formic acid-soluble aggregated Htt in cell nuclei. The prior treatment of brain sections with formic acid allowed the immunodetection of mutant N-terminal Htt in the form of a “diffuse nuclear stain” by exposing epitopes that had been masked by an H-bonded structure in an aggregated state. That the diffuse nuclear stain might represent an aggregated form of the protein in Hdh knock-in mouse models, rather than an altered conformer of full-length Htt (38), is consistent with the fact that this nuclear immunostain in HdhQ111/Q111 striatal cells was decreased by the administration of anti-aggregation compounds that had been isolated in an N-terminal Htt anti-aggregation screen (40).
We have shown that the smallest N-terminal fragment (Fragment 13) is an exon 1 Htt protein. When used to immunoprobe Western blots, MW8 acts as an exon 1 C-terminal neo-epitope antibody, an interpretation confirmed by our Htt exon 1 mutagenesis experiments and supported by TR-FRET. However, MW8 does not act as a neo-epitope antibody in all situations, because it can immunoprecipitate all 14 N-terminal Htt fragments from tissue lysates (supplemental Fig. S4). The diffuse immunostaining that accumulates in neuronal nuclei and is revealed by formic acid treatment was detected with the polyQ antibodies 4H7H7 and 3B5H10, but not by 1H6, suggesting that it may comprise only the exon 1 Htt fragment. However, we routinely observed that our ability to detect the three smallest Htt fragments in cytoplasmic fractions by immunoprecipitation and Western blotting diminished with age suggesting that the epitopes to antibodies, 1H6, HD-170, and HD-215, might be masked from detection by immunohistochemistry. The demonstration that the smallest N-terminal fragment is an exon 1 Htt protein is extremely important as there are many exon 1 HD models. These may therefore be representative of the nuclear pathogenic process that occurs in the Hdh knock-in mice and that has previously been linked to phenotype severity (41). In addition, cytoplasmic mutant exon 1 Htt contributes to the phenotype of HD mouse models (26) and, therefore, this would also be expected to be pathogenic in the knock-in models.
We have mapped the C termini of the mutant and WT N-terminal Htt fragments that are generated in HdhQ150 knock-in and in WT mice. Both mutant and WT full-length Htt can be cleaved to generate many N-terminal and internal fragments, the physiological role of which is unknown. This methodology can be used to confirm the origin of specific fragments by either genetic or pharmacological approaches e.g. by crossing to caspase-6 knock-out mice. These approaches can also be used to test the cascade hypothesis and determine whether the production of one fragment is essential for the generation of others. The identification of the source of the smaller N-terminal fragments is a high priority. If the three smallest mutant N-terminal fragments are not present in WT mice, they may not have a physiological function and, therefore, blocking their production may not have deleterious consequences.
Supplementary Material
Acknowledgments
We thank Johannes Voshol for advice, Glenn Morris for HD-A and HD-C antibodies, Erich Wanker for GST-exon1 constructs, Michael Hayden and Rona Graham for neo-epitope antibodies and advice on caspase digests, and Jude Nixon and Oliver Pressey for CAG repeat sizing.
This work was supported, in whole or in part, by National Institutes of Health Grant NS40251 (to L. M. E.). This work was also supported by the Huntington's Disease Society of America Coalition for the Cure (to G. P. B.), the CHDI Foundation, the Hereditary Disease Foundation (to A. O.), and the Medical Research Council (Grant G9800001).

The on-line version of this article (available at http://www.jbc.org) contains supplemental tables, Refs. 1–8, and Figs. S1–S4.
R. Luthi-Carter and G. P. Bates, unpublished data.
- HD
- Huntington disease
- polyQ
- polyglutamine
- Htt
- Huntingtin
- WT
- wild type
- Hdh+/Q150
- heterozygous HdhQ150 mice
- HdhQ150/Q150
- homozygous HdhQ150 mice
- TR-FRET
- time resolved-Förster resonance energy transfer
- FA
- formic acid
- NES
- nuclear export signal
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Bates G. P., Harper P. S., Jones A. L. (eds) (2002) Huntington's Disease, Oxford University Press, Oxford [Google Scholar]
- 2.Landles C., Bates G. P. (2004) EMBO Rep. 5, 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harjes P., Wanker E. E. (2003) Trends Biochem. Sci. 28, 425–433 [DOI] [PubMed] [Google Scholar]
- 4.Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S. W., Bates G. P. (1996) Cell 87, 493–506 [DOI] [PubMed] [Google Scholar]
- 5.Li H., Li S. H., Cheng A. L., Mangiarini L., Bates G. P., Li X. J. (1999) Hum. Mol. Genet. 8, 1227–1236 [DOI] [PubMed] [Google Scholar]
- 6.Smith D. L., Portier R., Woodman B., Hockly E., Mahal A., Klunk W. E., Li X. J., Wanker E., Murray K. D., Bates G. P. (2001) Neurobiol. Dis. 8, 1017–1026 [DOI] [PubMed] [Google Scholar]
- 7.Lin C. H., Tallaksen-Greene S., Chien W. M., Cearley J. A., Jackson W. S., Crouse A. B., Ren S., Li X. J., Albin R. L., Detloff P. J. (2001) Hum. Mol. Genet. 10, 137–144 [DOI] [PubMed] [Google Scholar]
- 8.Woodman B., Butler R., Landles C., Lupton M. K., Tse J., Hockly E., Moffitt H., Sathasivam K., Bates G. P. (2007) Brain Res. Bull. 72, 83–97 [DOI] [PubMed] [Google Scholar]
- 9.Kuhn A., Goldstein D. R., Hodges A., Strand A. D., Sengstag T., Kooperberg C., Becanovic K., Pouladi M. A., Sathasivam K., Cha J. H., Hannan A. J., Hayden M. R., Leavitt B. R., Dunnett S. B., Ferrante R. J., Albin R., Shelbourne P., Delorenzi M., Augood S. J., Faull R. L., Olson J. M., Bates G. P., Jones L., Luthi-Carter R. (2007) Hum. Mol. Genet. 16, 1845–1861 [DOI] [PubMed] [Google Scholar]
- 10.Moffitt H., McPhail G. D., Woodman B., Hobbs C., Bates G. P. (2009) PLoS One 4, e8025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 12.Scherzinger E., Lurz R., Turmaine M., Mangiarini L., Hollenbach B., Hasenbank R., Bates G. P., Davies S. W., Lehrach H., Wanker E. E. (1997) Cell 90, 549–558 [DOI] [PubMed] [Google Scholar]
- 13.Martindale D., Hackam A., Wieczorek A., Ellerby L., Wellington C., McCutcheon K., Singaraja R., Kazemi-Esfarjani P., Devon R., Kim S. U., Bredesen D. E., Tufaro F., Hayden M. R. (1998) Nat. Genet. 18, 150–154 [DOI] [PubMed] [Google Scholar]
- 14.Goldberg Y. P., Nicholson D. W., Rasper D. M., Kalchman M. A., Koide H. B., Graham R. K., Bromm M., Kazemi-Esfarjani P., Thornberry N. A., Vaillancourt J. P., Hayden M. R. (1996) Nat. Genet. 13, 442–449 [DOI] [PubMed] [Google Scholar]
- 15.Wellington C. L., Ellerby L. M., Gutekunst C. A., Rogers D., Warby S., Graham R. K., Loubser O., van Raamsdonk J., Singaraja R., Yang Y. Z., Gafni J., Bredesen D., Hersch S. M., Leavitt B. R., Roy S., Nicholson D. W., Hayden M. R. (2002) J. Neurosci. 22, 7862–7872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gafni J., Hermel E., Young J. E., Wellington C. L., Hayden M. R., Ellerby L. M. (2004) J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]
- 17.Kim Y. J., Yi Y., Sapp E., Wang Y., Cuiffo B., Kegel K. B., Qin Z. H., Aronin N., DiFiglia M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12784–12789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham R. K., Deng Y., Slow E. J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., Warby S. C., Doty C. N., Roy S., Wellington C. L., Leavitt B. R., Raymond L. A., Nicholson D. W., Hayden M. R. (2006) Cell 125, 1179–1191 [DOI] [PubMed] [Google Scholar]
- 19.Lunkes A., Lindenberg K. S., Ben-Haïem L., Weber C., Devys D., Landwehrmeyer G. B., Mandel J. L., Trottier Y. (2002) Mol. Cell 10, 259–269 [DOI] [PubMed] [Google Scholar]
- 20.Kim Y. J., Sapp E., Cuiffo B. G., Sobin L., Yoder J., Kegel K. B., Qin Z. H., Detloff P., Aronin N., DiFiglia M. (2006) Neurobiol. Dis. 22, 346–356 [DOI] [PubMed] [Google Scholar]
- 21.Schilling G., Klevytska A., Tebbenkamp A. T., Juenemann K., Cooper J., Gonzales V., Slunt H., Poirer M., Ross C. A., Borchelt D. R. (2007) J. Neuropathol. Exp. Neurol. 66, 313–320 [DOI] [PubMed] [Google Scholar]
- 22.Ratovitski T., Nakamura M., D'Ambola J., Chighladze E., Liang Y., Wang W., Graham R., Hayden M. R., Borchelt D. R., Hirschhorn R. R., Ross C. A. (2007) Cell Cycle 6, 2970–2981 [DOI] [PubMed] [Google Scholar]
- 23.Ratovitski T., Gucek M., Jiang H., Chighladze E., Waldron E., D'Ambola J., Hou Z., Liang Y., Poirer M. A., Hirschhorn R. R., Graham R., Hayden M. R., Cole R. N., Ross C. A. (2009) J. Biol. Chem. 284, 10855–10867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun B., Fan W., Balciunas A., Cooper J. K., Bitan G., Steavenson S., Denis P. E., Young Y., Adler B., Daugherty L., Manoukian R., Elliott G., Shen W., Talvenheimo J., Teplow D. B., Haniu M., Haldankar R., Wypych J., Ross C. A., Citron M., Richards W. G. (2002) Neurobiol. Dis. 11, 111–122 [DOI] [PubMed] [Google Scholar]
- 25.Tanaka Y., Igarashi S., Nakamura M., Gafni J., Torcassi C., Schilling G., Crippen D., Wood J. D., Sawa A., Jenkins N. A., Copeland N. G., Borchelt D. R., Ross C. A., Ellerby L. M. (2006) Neurobiol. Dis. 21, 381–391 [DOI] [PubMed] [Google Scholar]
- 26.Benn C. L., Landles C., Li H., Strand A. D., Woodman B., Sathasivam K., Li S. H., Ghazi-Noori S., Hockly E., Faruque S. M., Cha J. H., Sharpe P. T., Olson J. M., Li X. J., Bates G. P. (2005) Hum. Mol. Genet. 14, 3065–3078 [DOI] [PubMed] [Google Scholar]
- 27.Cornett J., Cao F., Wang C. E., Ross C. A., Bates G. P., Li S. H., Li X. J. (2005) Nat. Genet. 37, 198–204 [DOI] [PubMed] [Google Scholar]
- 28.Sathasivam K., Lane A., Legleiter J., Warley A., Woodman B., Finkbeiner S., Paganetti P., Muchowski P. J., Wilson S., Bates G. P. (2010) Hum. Mol. Genet. 19, 65–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies S. W., Sathasivam K., Hobbs C., Doherty P., Mangiarini L., Scherzinger E., Wanker E. E., Bates G. P. (1999) Methods Enzymol. 309, 687–701 [DOI] [PubMed] [Google Scholar]
- 30.Paganetti P., Weiss A., Trapp M., Hammerl I., Bleckmann D., Bodner R. A., Coven-Easter S., Housman D. E., Parker C. N. (2009) Chembiochem 10, 1678–1688 [DOI] [PubMed] [Google Scholar]
- 31.Weiss A., Abramowski D., Bibel M., Bodner R., Chopra V., DiFiglia M., Fox J., Kegel K., Klein C., Grueninger S., Hersch S., Housman D., Régulier E., Rosas H. D., Stefani M., Zeitlin S., Bilbe G., Paganetti P. (2009) Anal. Biochem. 395, 8–15 [DOI] [PubMed] [Google Scholar]
- 32.Osmand A. P., Berthelier V., Wetzel R. (2006) Methods Enzymol. 412, 106–122 [DOI] [PubMed] [Google Scholar]
- 33.Warby S. C., Doty C. N., Graham R. K., Carroll J. B., Yang Y. Z., Singaraja R. R., Overall C. M., Hayden M. R. (2008) Hum. Mol. Genet. 17, 2390–2404 [DOI] [PubMed] [Google Scholar]
- 34.Gonitel R., Moffitt H., Sathasivam K., Woodman B., Detloff P. J., Faull R. L., Bates G. P. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 3467–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perutz M. F., Johnson T., Suzuki M., Finch J. T. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 5355–5358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou H., Cao F., Wang Z., Yu Z. X., Nguyen H. P., Evans J., Li S. H., Li X. J. (2003) J. Cell Biol. 163, 109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xia J., Lee D. H., Taylor J., Vandelft M., Truant R. (2003) Hum. Mol. Genet. 12, 1393–1403 [DOI] [PubMed] [Google Scholar]
- 38.Wheeler V. C., White J. K., Gutekunst C. A., Vrbanac V., Weaver M., Li X. J., Li S. H., Yi H., Vonsattel J. P., Gusella J. F., Hersch S., Auerbach W., Joyner A. L., MacDonald M. E. (2000) Hum. Mol. Genet. 9, 503–513 [DOI] [PubMed] [Google Scholar]
- 39.Ko J., Ou S., Patterson P. H. (2001) Brain Res. Bull. 56, 319–329 [DOI] [PubMed] [Google Scholar]
- 40.Wang J., Gines S., MacDonald M. E., Gusella J. F. (2005) BMC Neurosci. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang C. E., Tydlacka S., Orr A. L., Yang S. H., Graham R. K., Hayden M. R., Li S., Chan A. W., Li X. J. (2008) Hum. Mol. Genet. 17, 2738–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.