Abstract
Factor VIII (FVIII) plays a critical role in blood coagulation by forming the tenase complex with factor IXa and calcium ions on a membrane surface containing negatively charged phospholipids. The tenase complex activates factor X during blood coagulation. The carboxyl-terminal C2 domain of FVIII is the main membrane-binding and von Willebrand factor-binding region of the protein. Mutations of FVIII cause hemophilia A, whereas elevation of FVIII activity is a risk factor for thromboembolic diseases. The C2 domain-membrane interaction has been proposed as a target of intervention for regulation of blood coagulation. A number of molecules that interrupt FVIII or factor V (FV) binding to cell membranes have been identified through high throughput screening or structure-based design. We report crystal structures of the FVIII C2 domain under three new crystallization conditions, and a high resolution (1.15 Å) crystal structure of the FVIII C2 domain bound to a small molecular inhibitor. The latter structure shows that the inhibitor binds to the surface of an exposed β-strand of the C2 domain, Trp2313-His2315. This result indicates that the Trp2313-His2315 segment is an important constituent of the membrane-binding motif and provides a model to understand the molecular mechanism of the C2 domain membrane interaction.
Keywords: Blood Coagulation/Coagulation Factors, Protein/Cell Surface, Protein/Domains, Protein/Drug Interactions, Protein/Ligand Binding, Protein/Structure
Introduction
Blood coagulation factor VIII (FVIII)2 is synthesized as a single polypeptide chain, including a 19-residue signal peptide. The mature FVIII contains 2,332 amino acid residues arranged within five domains organized as A1-A2-B-A3-C1-C2 (1, 2). FVIII circulates in the blood as a heterodimer: the A1, A2, and variable portions of the B domain forming the heavy chain with a molecular weight varying between 90,000 and 200,000; and the A3, C1 and C2 domains forming the light chain with a molecular weight of 80,000. The noncovalent assembly of the heterodimer is facilitated by divalent metal ions, as revealed by recent crystallographic studies (3, 4). In plasma, FVIII circulates as a complex with von Willebrand factor which stabilizes FVIII by preventing its rapid clearance from the blood circulation (5). During blood coagulation, FVIII is cleaved by thrombin at Arg372, Arg740, and Arg1689 (6), which converts it to a fully active cofactor, FVIIIa. FVIIIa dissociates from von Willebrand factor and binds to membrane surfaces where it assembles with the serine protease factor IXa. The presence of FVIIIa enhances the Vmax for factor X activation by FIXa by 200,000-fold (7). This large amplification requires that the FVIII level in blood be controlled. Mutations of FVIII cause hemophilia A, an X-linked bleeding disorder (8). The molecular mechanisms of FVIII binding to membranes are not fully understood.
Several lines of evidence suggest that the light chain of FVIII, in particular the C2 domain, is responsible for the specific binding to membrane surfaces (9, 10). A 1.5-Å x-ray crystal structure of the FVIII C2 domain and a mutagenesis study suggested that two hydrophobic loops formed by Met2199-Phe2200 and Leu2251-Leu2255 play an important role (11, 12). Two additional loops formed by Trp2313-His2315 and Gln2222-Lys2227) were also proposed to be involved in membrane binding based on an electron microscopy study (13).
C2 domain membrane-binding sites were proposed as a drug target to regulate the functional concentrations of coagulation factors, FVIII and FV, that contain a C2 domain (14). A number of small organic molecules were identified that disrupt C2 domain membrane anchoring of FVIII (14) and FV (15). One of these inhibitors, 005B10, identified from in silico screening, was found to inhibit FVIII interaction with negatively charged phospholipids (15). In this study, we report the high resolution crystal structure (1.15 Å) of the human FVIII C2 domain in complex with this small molecule. The inhibitor was found to bind to an unexpected surface-exposed segment of the C2 domain, Trp2313-His2315. This study not only suggests that Trp2313-His2315 is a membrane-binding site of the C2 domain but can also become useful for further refinement of in silico screening strategies.
MATERIALS AND METHODS
Materials
SP-Sepharose fast flow cation-exchange resins were purchased from Amersham Biosciences. Vectors and cell strains of the Pichia pastoris expression system were from Invitrogen. The small molecule compound (Hit2Lead, 005B10-7688319) was purchased from Chembridge Corp. Purified human FVIII was kindly provided by Dr. P. Tureck (Baxter AG, Vienna).
Expression and Purification
The C2 domain cDNA for human factor VIII (residues 2174–2326) was amplified by PCR from human factor VIII cDNA (ATCC 10085779) using a forward primer with a SnaB1 site, 5′- CTGTATACGTAATGGGCGTTGATTTAAATAGT-3′ and a reverse primer with a Not1 site 5′-AAGATGCGGCCGCTCAGTAGAGGTCCT-3′. Restriction sites are underlined. The PCR fragment was subcloned into the yeast expression vector pPIC9K under the control of the alcohol oxidase promoter. An α-factor signal peptide was placed 5′ of the C2 domain coding region to facilitate the secretion of the recombinant protein into the medium. The vector was transformed into the P. pastoris strain GS115 by electroporation (1500 V, 25 μF, and two pulses for 4.5 ms). The expression strain was inoculated into BGMY medium for amplification and was transferred to BMMY medium for methanol-induced expression (1% methanol once a day for 4 days). Secreted protein was isolated from the medium by ammonium sulfate precipitation (40% saturated ammonium sulfate) and collected by centrifugation at 12,000 × g for 30 min. The precipitate was resuspended in 10 mm sodium phosphate buffer, pH 7.0, containing 10 mm NaCl. The product was then loaded onto a SP-Sepharose fast flow column, and eluted with a gradient from 50 mm to 500 mm NaCl in 10 mm sodium phosphate buffer, pH 7.0, at a flow rate of 4 ml·min−1. The expression yield was ∼0.05 g per liter of medium.
Protein Crystallization
For crystallization, the purified FVIII C2 domain was concentrated in a Millipore Ultrafree concentrator to a concentration of 6.0 mg/ml. Crystallization was carried out at room temperature using the hanging drop vapor diffusion method. Three different crystallization conditions were found (see supplemental Fig. S1 for crystal images): (1) 2.8 m NaCl, 3% ethylene glycol, 0.1 m Tris-HCl pH 8.0; (2) 2.4–3.3 m NaCl, 0.1 m MES, pH 6.0; and (3) 30% polyethylene glycol monomethyl ether (PEG-MME) 2000, 0.15 m KBr. The inhibitor 005B10 was dissolved in dimethyl sulfoxide to a final concentration of 5 mg/ml. The complex of protein with the inhibitor was formed by mixing the protein and inhibitor at a molar ratio of 1:5 for 24 h, and crystallized with 2.8 m sodium chloride, 0.1 m Tris-HCl buffer, pH 8.0, 3% ethylene glycol.
X-ray Data Collection and Structure Determination
The crystals were briefly soaked in a cryoprotectant solution consisting of the crystallization mother liquor with 20% glycerol. X-ray diffraction data collection was carried out at 100°K on the Argonne Advanced Photon Source Southeast Regional Collaborative Access Team beam line 22-ID. All structures were determined using the molecular replacement program MOLREP of the CCP4 package (16). The structure of the C2 domain of FVIII (Protein Data Bank (PDB) code 1D7P; Ref. 11) was used as a molecular replacement model for phasing of the x-ray data. Model building was done with the program COOT against σA weighted 2Fo − Fc maps, and the structure was refined by randomly removing 5% of the measurements to monitor the free R-factor (Rfree) to minimize model bias. The electron density for the inhibitor was clearly visible after the rigid body refinement step, and the inhibitor was manually positioned into this electron density. After several cycles of positional and B-factor refinement together with manual adjustments, the results were successfully refined to the final R and Rfreevalues. The structures of two crystal forms of the inhibitor-free FVIII C2 domain were determined in a similar way (Table 1).
TABLE 1.
Data collection and structural refinement statistics of three FVIII C2 domain structures
| Parameters | Value(s) or determination | ||
|---|---|---|---|
| Crystals | FVIII C2 domain | FVIII C2 domain | FVIII C2-005B10 complex |
| Deposited PDB code | 3HNY | 3HOB | 3HNB |
| Crystallization conditions | NaCl, Tris-HCl, pH 8.0 | PEG-MME 2000, KBr | NaCl, Tris-HCl, pH 8.0 |
| Synchrotron wavelength | 1.000 Å | 1.000 Å | 1.000 Å |
| Space group | P212121 | P21 | P212121 |
| Unit cell parameters | 42.2, 55.5, 68.3, α = β = γ = 90° | 43.3, 59.0, 60.1, α = γ = 90°, β = 110.7° | 42.1, 55.8, 68.0, α = β = γ = 90° |
| Independent reflections | 69,391 (6,479)c | 17,899 (1,231) | 50,523 (4,441) |
| Highest resolution (Å) | 1.07 | 2.07 | 1.15 |
| Completeness (%) | 97.7 (92.5) | 82.8 | 93.7 (76.8) |
| Redundancy | 5.7 | 3.1 | 4.7 |
| Solvent content (%) | 52.03 | 46.56 | 51.89 |
| Rmergea | 0.065 (0.349) | 0.07 (0.418) | 0.084 (0.415) |
| I/σ | 29.04 (1.83) | 13.49 (1.21) | 17.53 (2.48) |
| Refinement | |||
| Resolution range | 43.07–1.07 Å | 56.254–2.07 Å | 43.11–1.15 Å |
| Rcryst/Rfreeb % | 18.5, 20 | 21.6, 26.7 | 18.5, 22.0 |
| r.m.s.d. of bond lengths and bond anglesc | 0.009 Å, 1.43° | 0.012 Å, 1.43° | 0.008 Å, 1.45° |
| Ramachandran plot statistics | |||
| % Residues in core, allowed, generous, and disallowed regions | 87.0, 11.6, 0, 1.4 | 86.6, 12.7, 0, 0.7 | 84.8, 13.8, 0, 1.4 |
| Average B factors, Å2 | 13 | 32 | 12 |
a Rmerge = ΣhΣi|Ii(h) − 〈I(h)〉|/ΣhΣiIi(h), where 〈I(h)〉 is the mean intensity of reflection h.
b Numbers in parentheses refer to the highest resolution shell.
c Rcryst = 100 × Σ|Fo(h) − Fc(h)|/ΣFo(h), where Fo(h) and Fc(h) are observed and calculated reflections. Rfree is Rcryst that was calculated using 5% of the data, chosen randomly, and omitted from the subsequent structure refinement.
Surface Plasmon Resonance Measurement of FVIII Binding to Immobilized Phospholipid Vesicles
The preparation of phospholipid vesicles (20/80 phosphatidylserine /phosphatidylcholine and the determination of the binding isotherm of FVIII binding to immobilized phosphatidylserine/phosphatidylcholine vesicles was as described previously (15).
RESULTS
Determination of the Crystal Structures of FVIII C2
Pratt et al. (11) reported the crystal structure of the recombinant human FVIII C2 domain (sequence 2,171–2,329) crystallized using ammonium sulfate as precipitant. We determined that the high concentration of ammonium sulfate in the crystallization buffer interfered with the interaction of the FVIII C2 domain and its inhibitor. This is presumably due to the high ionic strength of ammonium sulfate that precludes the protein-inhibitor interaction. Thus, we searched for new crystallization conditions for the FVIII C2 domain.
We produced the recombinant human FVIII C2 domain (amino acids 2,170–2,328) in Pichia pastoris, using a similar method to that reported by Pratt et al. (11). We generated the crystals of this C2 domain under three different crystallization conditions (see Table 1). One crystal form was obtained using sodium chloride as precipitant at two different pH values (6.0 or 8.0), and the other was generated using PEG-MME 2000 as precipitant. The crystal formed crystals in the presence of NaCl at either pH (PDB code 3HNY) have the same crystal packing with identical cell parameters and space group (P212121), whereas the crystal formed in the presence of PEG (PDB code 3HOB) has a different crystal packing (space group of P21). The structures from these crystal forms were determined by molecular replacement using the crystal structure of the FVIII C2 domain determined by Pratt et al. (11; PDB code 1D7P). The structure of the NaCl form (PDB code 3HNY) was refined to an R value of 0.185 and an Rfree value of 0.200 to 1.07 Å, whereas the structure of the PEG-MME 2000 form (PDB code 3HOB) was refined to an R value of 0.213 and an Rfree value of 0.272. All crystal structures have reasonable stereochemistry, as shown by the Ramachandran plots (Table 1).
The crystals formed in the presence of NaCl (PDB code 3HNY) have cell parameters and space groups identical to the ones determined by Pratt et al. (11; r.m.s.d. of 0.59 Å for 544 main chain atoms), despite the different precipitant used in crystallization (Fig. 1). The crystals formed in the presence of PEG (PDB code 3HOB) have different crystal packing and contain two FVIII C2 domain molecules in one crystallographic asymmetric unit. The two molecules are similar to each other with an r.m.s.d. of 0.48 Å. Both are also similar to the crystal formed in the presence of NaCl (r.m.s.d. of 0.46 Å). Our human C2 domain is a recombinant protein produced in yeast (P. pastoris). The overall crystal structure of the current FVIII C2 domain is quite similar to the crystal structures of C2 domains produced in mammalian cells, including the C2 domain structures of the full-length B-domain-deleted FVIII (PDB code 3CDZ; 3), r.m.s.d. 0.97 Å; PDB code 2R7E (4) r.m.s.d. 1.14 Å for main chain atoms), and the inactivated bovine FVai (PDB code 1SDD (17), r.m.s.d. 1.29 Å, see supplemental Fig. S2). The structural resemblance of FVIII C2 domains from different sources and different crystal packing (Fig. 1 and supplemental Fig. S2) suggests that the overall C2 domain structure is quite robust and not perturbed by crystal packing forces.
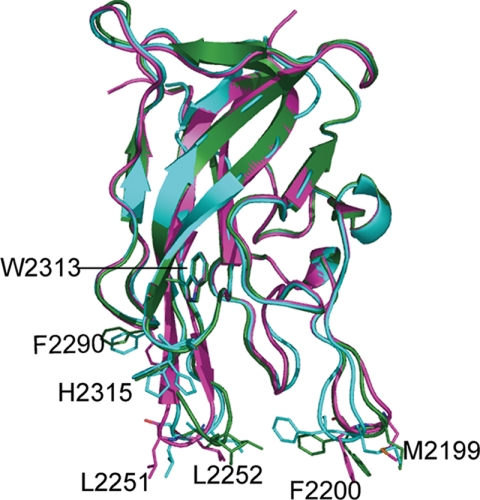
FIGURE 1.
Structural superposition of FVIII C2 domain structures formed under different crystallization conditions. The C2 domain in the protein-inhibitor complex from crystals formed in the presence of NaCl (PDB code 3HNB) is shown in cyan; the C2 domain formed in the presence of ammonium sulfate (PDB code 1D7P) in magenta, the C2 domain from crystals formed in the presence of PEG-MME (PDB code 3HOB) in green. The phenyl ring of residue Phe2200 was modeled in dual conformations. The conformations of Phe2200 are clearly different. The two membrane-binding spikes (2251–2252 and 2199–2200) are more open in 1D7P (magenta) than in the structure of F8C2–005B10 (cyan).
The Structure of the C2 Domain-Inhibitor Complex and the Inhibitor-binding Sites of FVIII C2
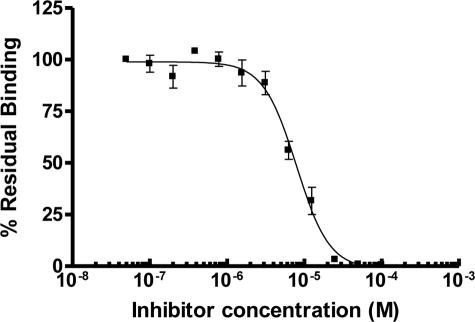
A FVIII C2 domain inhibitor, 005B10, that interferes with C2 domain membrane binding was identified by Segers et al. (15) using a computational approach. This compound inhibited membrane binding of FVIII with an IC50 of 7.8 μm, as measured by surface plasmon resonance (Fig. 2). We obtained crystals of this inhibitor in complex with the FVIII C2 domain using NaCl as a precipitant. The crystals diffracted to 1.15 Å with the synchrotron x-ray source. We determined the crystal structure of this complex and refined it to an R value of 0.185 and an Rfree value of 0.220 at 1.15 Å (Table 1, PDB code 3HNB). The structure had a satisfactory stereochemistry with 84.8% of the residues in the most favored region and 13.8% in the additional allowed region in the Ramachandran plot. The structure of the C2 domain in the complex is similar to the molecular replacement search model (r.m.s.d. of 0.56 Å) and to the other C2 structures in our study (r.m.s.d. of 0.07 Å for 3HNY; r.m.s.d. is 0.46 Å for 3HOB), suggesting that the inhibitor does not perturb the FVIII C2 structure.
FIGURE 2.
Inhibition by compound 005B10 of FVIII binding on immobilized phospholipid vesicles composed of 20% phosphatidylserine and 80% phosphatidylcholine. Inhibitory IC50 were determined to be 7.8 μm.
The small molecular inhibitor was clearly visible from the electron density maps in the structure of the complex (supplemental Fig. S3). The inhibitor binds to residues Trp2313-His2315 of the FVIII C2 domain (Fig. 3). Two hydrogen bonds were formed between the small molecule inhibitor (labeled 005B10 in Fig. 3) and residues Trp2313 and Lys2258 (Fig. 3). Trp2313 seems to be the key residue for inhibitor binding as 73% of its total area (contact area of 55.5 Å2) interacts with the inhibitor. Comparison of the current inhibitor-bound FVIII C2 structure (PDB code 3HNB) with the unbound form (PDB code 1D7P) shows the local conformations of residues Ser2312-Val2314 do not change upon inhibitor binding (r.m.s.d. of 0.14 Å).
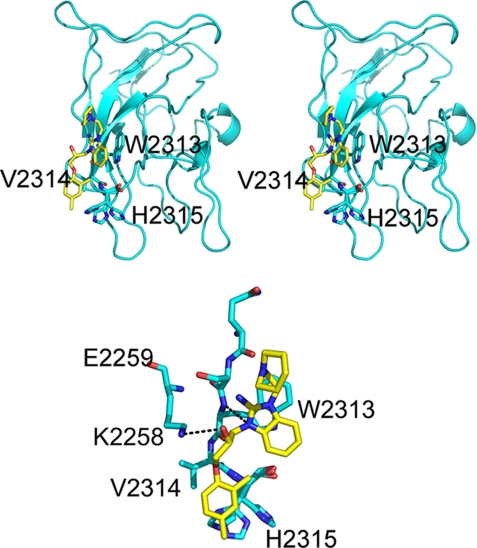
FIGURE 3.
Top, stereo representation of the overall structure of human FVIII C2–005B10 complex. FVIII C2 is in cyan, and the carbon atoms of the inhibitor, 005B10, is in yellow. Oxygen atoms are colored red, and nitrogen atoms are colored blue. The inhibitor binds to the surface-exposed residues Trp2313-His2315. Bottom, the interaction between the inhibitor 005B10 and the FVIII C2 domain. The FVIII C2 is shown in cyan, and the small molecular inhibitor 005B10 is shown in yellow. There are two hydrogen bonds (black dotted lines) between the inhibitor and FVIII C2. One is between the inhibitor and Trp2313, the other is between the inhibitor and Lys2258.
Local Structural Flexibility of FVIII C2 Domain
Although all the current and previous FVIII C2 domain structures are quite similar to each other in their overall structures, some local changes are significant. His2315 has three conformations of its side chain in our structure of the complex (Fig. 1). Side chain flexibility of residue Phe2200 was also observed in our PEG2000 crystal form (PDB code 3HOB) and was modeled with two side chain conformations. Two putative membrane-binding loops (residues Met2199-Phe2200 and Leu2251-Leu2252, also termed spikes) are further apart in 1D7P than in our NaCl form crystals (PDB 3HNB, Fig. 1). This variation of distance between these two spikes suggests that the FVIII C2 domain has two conformations: an open and a closed form, much like what has been proposed for the C2 domain of FV (Fig. 4) (18).
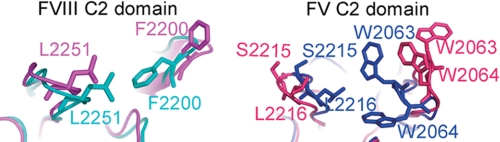
FIGURE 4.
Comparison of the hydrophobic spikes of the C2 domains (view from membrane) of factor VIII (left) and factor V (right). Factor V C2 structures exist in open (red, PDB code 1CZS) and closed (blue, PDB code 1CZV) forms. Factor VIII structures also show conformational flexibility of the spikes (blue for PDB code 3HOB, and magenta for PDB code 1D7P), but the overall structure is more compact and the spikes are less flexible compared with factor V.
DISCUSSION
FV and FVIII C2 domains facilitate the assembly of key coagulation enzymatic complexes, prothrombinase and tenase, onto cell membrane surfaces. This binding interaction was proposed as a target for intervention in blood coagulation (14, 15). Using a computational approach, Segers et al. (15) identified a small molecule (005B10) that had the capability of inhibiting FVIII C2 binding to negatively charged phospholipids in vitro. We determined the structure of the human FVIII C2 domain bound to 005B10. The inhibitor is found to bind to residues 2313–2315 of the FVIII C2 domain. This suggests that residues 2313–2315 are involved in the membrane binding of the FVIII C2 domain. Several other studies support a role for residues 2313–2315 in this function. Three overlapping synthetic peptides encompassing this region (2303 to 2332) were found to inhibit FVIII binding to phosphatidylserine by >90% (9), suggesting that the region between residue 2313–2323 contains a phospholipid binding site. An electron crystallography study revealed the structure of FVIII bound to phospholipid membrane at 15 Å resolution and suggested that four C2 domain loops are involved in phospholipid binding. These loops include the two spike loops (Met2199-Phe2200 and Leu2251-Leu2252), the loop containing Val2223, and the loop containing Trp2313-His2315 (13). Some patients with hemophilia A, who have moderate to severe loss of FVIII function, have a mutation of Trp2313 to Arg (19, 20). A recombinant B-domain-deleted FVIII protein with the mutation W2313A showed normal intracellular protein synthesis and normal binding to von Willebrand factor in a murine hemophilia A model, but defective binding (KD, 28-fold higher) to 4% phosphatidylserine vesicles (21). Sequence alignment of the C2 domain of human coagulation cofactors with their homologs shows that Trp2313 is a conserved residue (supplemental Fig. S4). Taken together, these arguments support that the loop Trp2313-His2315 plays an important role in mediating C2 domain membrane binding.
Our study also demonstrates the flexibility of the FVIII C2 domain, especially the spike loops. In contrast to the previous 1.5 Å x-ray structure (11), our structures obtained in the presence of NaCl exist in a closed form where the two spikes are close to each other. The structure obtained from the PEG condition showed the presence of both closed and open forms. Wide separation between the spikes was observed in the structure of the FVIII C2 domain in complex with immunoglobulin G4κ (22). In this particular case, the separation of the spikes seems related to the interaction between FVIII C2 domain and the antibody. These observations suggest the spike loops are flexible in nature. Such flexibility was observed in the structures of other C2-containing proteins, including factor V and bovine lactadherin, whose C2 domains are homologous to the FVIII C2 domain (18, 23). The FV C2 domain shows ∼40% sequence similarity to FVIII (24). In the FV C2 domain, two tryptophans (Trp2063-Trp2064) are surface-exposed and are part of a spike loop, corresponding to the spike loop Met2199-Phe2200 in FVIII. FV C2 domain structures are in either the open form (PDB codes 1CZS and 1CZT) or closed form (PDB code 1CZV) (18). In the closed form of FV, the two tryptophans are buried inside the protein, leading to unfavorable interactions with membrane, whereas the open form may be suitable for the interaction with hydrophobic interior of the membrane (18). However, a recent molecular dynamics study concluded that both the closed and open conformations of FV may be equally suitable for membrane binding in terms of binding energy (25). This study, together with previous studies, show that the flexibility of the C2 domain spike loop is a prevalent feature of the C2 domain structure. The current evidence that Trp2313 is involved in membrane binding further supports our previously proposed tenase membrane-binding model (3), where the C2 domain, along with the rest of full-length factor VIII molecule, tilts onto the membrane (supplemental Fig. S5).
In summary, the inhibition of FVIII cofactor function could represent a different but unique approach for antithrombotic intervention. Several small molecule inhibitors that disrupt FVIII/phospholipid binding have been identified by using high throughput screening strategies (14). Segers et al. (15) used a structure-based virtual ligand screening approach to find several small molecules that bind the FV C2 domain and validated their binding to FV or FVIII in vitro by surface plasmon resonance. In the current study, we determined the crystal structure of the FVIII C2 domain in complex with one of the inhibitors. These results provide the first direct structural evidence for the membrane-binding role of the C2 domain Trp2313-His2315 segment. Furthermore, these membrane-binding structural models may facilitate the further design of lead compounds that may be developed into a novel class of anticoagulants.
Acknowledgments
Use of the Advanced Photon Source is supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract W-31-109-Eng-38. We thank the staff of the Argonne Advanced Photon Source Southeast Regional Collaborative Access Team beamline 22ID for help during data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant HL086584. This work was also supported by grants from the Natural Science Foundation of China (30811130467 and 30625011), the Ministry of Science and Technology (2006AA02A313 and 2007CB914304), the Chinese Academy of Sciences (KSCX2-YW-R-082), and the National Science Foundation (NSF-EPSCoR).
The atomic coordinates and structure factors (codes 3HOB, 3HNY, and 3HNB) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- FVIII
- factor VIII
- MES
- 4-morpholineethanesulfonic acid
- PDB
- Protein Data Bank
- r.m.s.d.
- root mean square deviation
- PEG-MME
- polyethylene glycol monomethyl ether.
REFERENCES
- 1.Toole J. J., Knopf J. L., Wozney J. M., Sultzman L. A., Buecker J. L., Pittman D. D., Kaufman R. J., Brown E., Shoemaker C., Orr E. C. (1984) Nature 312, 342–347 [DOI] [PubMed] [Google Scholar]
- 2.Wood W. I., Capon D. J., Simonsen C. C., Eaton D. L., Gitschier J., Keyt B., Seeburg P. H., Smith D. H., Hollingshead P., Wion K. L. (1984) Nature 312, 330–337 [DOI] [PubMed] [Google Scholar]
- 3.Ngo J. C., Huang M., Roth D. A., Furie B. C., Furie B. (2008) Structure 16, 597–606 [DOI] [PubMed] [Google Scholar]
- 4.Shen B. W., Spiegel P. C., Chang C. H., Huh J. W., Lee J. S., Kim J., Kim Y. H., Stoddard B. L. (2008) Blood 111, 1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fay P. J., Coumans J. V., Walker F. J. (1991) J. Biol. Chem. 266, 2172–2177 [PubMed] [Google Scholar]
- 6.Pittman D. D., Kaufman R. J. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 2429–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Dieijen G., Tans G., Rosing J., Hemker H. C. (1981) J. Biol. Chem. 256, 3433–3442 [PubMed] [Google Scholar]
- 8.Furie B., Furie B. C. (1990) Semin. Hematol. 27, 270–285 [PubMed] [Google Scholar]
- 9.Foster P. A., Fulcher C. A., Houghten R. A., Zimmerman T. S. (1990) Blood 75, 1999–2004 [PubMed] [Google Scholar]
- 10.Saenko E. L., Scandella D. (1995) J. Biol. Chem. 270, 13826–13833 [DOI] [PubMed] [Google Scholar]
- 11.Pratt K. P., Shen B. W., Takeshima K., Davie E. W., Fujikawa K., Stoddard B. L. (1999) Nature 402, 439–442 [DOI] [PubMed] [Google Scholar]
- 12.Gilbert G. E., Kaufman R. J., Arena A. A., Miao H., Pipe S. W. (2002) J. Biol. Chem. 277, 6374–6381 [DOI] [PubMed] [Google Scholar]
- 13.Stoilova-McPhie S., Villoutreix B. O., Mertens K., Kemball-Cook G., Holzenburg A. (2002) Blood 99, 1215–1223 [DOI] [PubMed] [Google Scholar]
- 14.Spiegel P. C., Kaiser S. M., Simon J. A., Stoddard B. L. (2004) Chem. Biol. 11, 1413–1422 [DOI] [PubMed] [Google Scholar]
- 15.Segers K., Sperandio O., Sack M., Fischer R., Miteva M. A., Rosing J., Nicolaes G. A., Villoutreix B. O. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 12697–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vagin A., Teplyako A. T. (1997) J. Appl. Crystallography 30, 1022–1025 [Google Scholar]
- 17.Adams T. E., Hockin M. F., Mann K. G., Everse S. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 8918–8923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Macedo-Ribeiro S., Bode W., Huber R., Quinn-Allen M. A., Kim S. W., Ortel T. L., Bourenkov G. P., Bartunik H. D., Stubbs M. T., Kane W. H., Fuentes-Prior P. (1999) Nature 402, 434–439 [DOI] [PubMed] [Google Scholar]
- 19.Tagariello G., Belvini D., Salviato R., Are A., De Biasi E., Goodeve A., Davoli P. (2000) Haematologica 85, 525–529 [PubMed] [Google Scholar]
- 20.Kemball-Cook G., Tuddenham E. G., Wacey A. I. (1998) Nucleic Acids Res. 26, 216–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schatz S. M., Zimmermann K., Hasslacher M., Kerschbaumer R., Dockal M., Gritsch H., Turecek P. L., Schwarz H. P., Dorner F., Scheiflinger F. (2004) Br. J. Haematol. 125, 629–637 [DOI] [PubMed] [Google Scholar]
- 22.Spiegel P. C., Jr., Jacquemin M., Saint-Remy J. M., Stoddard B. L., Pratt K. P. (2001) Blood 98, 13–19 [DOI] [PubMed] [Google Scholar]
- 23.Lin L., Huai Q., Huang M., Furie B., Furie B. C. (2007) J. Mol. Biol. 371, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kane W. H., Davie E. W. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 6800–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mollica L., Fraternali F., Musco G. (2006) Proteins 64, 363–375 [DOI] [PubMed] [Google Scholar]