Abstract
Expression of gp78, an E3 ubiquitin ligase in endoplasmic reticulum-associated degradation, is associated with tumor malignancy. To study gp78 overexpression in mammary gland development and tumorigenicity, we generated murine mammary tumor virus (MMTV) long terminal repeat-driven gp78 transgenic mice. Embryos carrying the gp78 transgene cassette were implanted in FVB surrogate mothers, and two founders with high copy integration showed elevated gp78 expression at both transcript and protein levels at the virgin stage and at 12 days gestation. Transgenic mammary glands showed increased ductal branching, dense alveolar lobule formation, and secondary terminal end bud development. Bromodeoxyuridine staining showed increased proliferation in hyperplastic ductal regions at the virgin stage and at 12 days gestation compared with wild type mice. Reduced expression of the metastasis suppressor KAI1, a gp78 endoplasmic reticulum-associated degradation substrate, demonstrates that gp78 ubiquitin ligase activity is increased in MMTV-gp78 mammary gland. Similarly, metastatic MDA-435 cells exhibit increased gp78 expression, decreased KAI1 expression, and elevated proliferation compared with nonmetastatic MCF7 cells whose proliferation was enhanced upon knockdown of KAI1. Importantly, stable gp78 knockdown HEK293 cells showed increased KAI1 expression and reduced proliferation that was rescued upon KAI1 knockdown, demonstrating that gp78 regulation of cell proliferation is mediated by KAI1. Mammary tumorigenesis was not observed in repeatedly pregnant MMTV-long terminal repeat-gp78 transgenic mice over a period of 18 months post-birth. Elevated gp78 ubiquitin ligase activity is therefore not sufficient for mammary tumorigenesis. However, the hyperplastic phenotype observed in mammary glands of MMTV-gp78 transgenic mice identifies a novel role for gp78 expression in enhancing mammary epithelial cell proliferation and nontumorigenic ductal outgrowth.
Keywords: Cell/Division, Development Differentiation/Tissue, Diseases/Cancer/Mammary, Proteases/Ubiquitination, Protein/Degradation, Tumor/Suppressor, gp78/Autocrine Motility Factor Receptor
Introduction
gp78, also called autocrine motility factor receptor, is an E3 ubiquitin ligase implicated in endoplasmic reticulum-associated degradation that also has a dual function as a cell surface cytokine receptor for autocrine motility factor/phosphoglucose (1). gp78 overexpression is closely linked to tumor malignancy and human cancer (2) and was identified as one of the 189 most mutated genes in breast and colon cancers (3). gp78 expression correlates with aggressive tumor biology and poor outcome for malignancies of the lung, tongue, esophagus, stomach, colon, rectum, liver, breast, thymus, and skin (2). Notably, in bladder, colorectal, gastric, skin, and esophageal cancers (4–8), gp78 is either not expressed or expressed at significantly reduced levels in adjacent normal tissue. However, whereas overexpression of gp78 in tumors is well documented, it has yet to be determined whether gp78 overexpression contributes to tumor progression.
Substrates of gp78 E3 ubiquitin ligase activity include CD3-δ, the T cell receptor, apoB lipoprotein, hydroxymethylglutaryl-CoA reductase, cystic fibrosis transmembrane conductance regulator, and the metastasis suppressor KAI1 (9–13). KAI1 (CD82) is a tetraspanin whose expression is lost in advanced stages of many human malignancies; however, the mechanism of action of this suppressor remains unknown (14, 15). The recent identification of KAI1 as a gp78 endoplasmic reticulum-associated degradation substrate functionally links gp78 ubiquitin ligase activity in endoplasmic reticulum-associated degradation to its established role in metastasis (16).
In human breast cancer, ∼25% of 370 primary invasive breast carcinomas showed membranous and cytoplasmic gp78 labeling, although gp78 positivity did not correlate with prognosis (17). To further evaluate the role of gp78 in breast cancer progression, we generated a transgenic MMTV3 -gp78 mouse model overexpressing gp78 in the mammary gland. Analysis of gp78 transgenic mammary glands showed that gp78 expression induces a hyperplastic phenotype, increased ductal branching, and dense alveolar lobule formation as well as down-regulation of the KAI1 metastasis suppressor. Using a knockdown approach in vitro, KAI1 was found to mediate gp78 regulation of cell proliferation defining a functional relationship between this E3 ubiquitin ligase and its substrate in cell proliferation.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and Animals
Rat anti-gp78 3F3A monoclonal antibody was as described previously (18) and mouse anti-gp78 (ab54787-100) was purchased from Abcam. Rabbit monoclonal anti-Ki67 (Clon-SP6) was from Medicorp, rabbit anti-KAI1 (sc-1087) and anti-cyclin-D1 were from Santa Cruz, and mouse anti-β-actin and horseradish peroxidase-conjugated anti-rabbit or anti-mouse secondary antibodies were from Sigma. Glucocorticoid analogue dexamethasone and MTT reagent were from Sigma. DIG probe labeling, probing kit, and BrdUrd were from Roche Applied Science. The animals were maintained at the British Columbia Cancer Agency facility following the guidelines and protocols of the University of British Columbia Animal Care Committee. All of the animals in both wild type and transgenic groups were between 8 and 10 weeks of age, and a minimum of four animals from several litters was used for the assessment.
Plasmids and siRNAs
MMTV-LTR plasmid (19–21) was obtained from the H. Lee Moffitt mouse model core facility. Mouse gp78 cDNA (22) was amplified using the following set of primers (5′-GAA TTC ATG CCG CTG CTC TTC CTC GAG CGC-3′ and 5′-GAA TTC CTA GGT TGT CCG TTG CCT CTG AAG-3′). Amplified cDNA was cloned into pCRII (Invitrogen) and subsequently subcloned into the MMTV-LTR plasmid at the EcoRI site in the third exon of the rabbit β-globin gene that ends with the bovine growth hormone (BGH)-poly(A) sequence. The final plasmid was sequenced to verify ligation junctions and cDNA integrity. pcDNA6.2-GW/ +EmGFP-miR was obtained from Invitrogen. Nonspecific, nontargeting control (D-001810-10-05) as well as human CD82 (KAI1) (L-003901-00)-specific siRNAs pools were purchased from Dharmacon.
Cell Lines and Transfection
MCF7 and MDA-435 cells were obtained from ATCC and maintained in RPMI medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamate as described (17). HEK293 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, and 1% l-glutamate. MMTV-LTR-gp78 or pCDNA were transiently transfected into MCF7 cells and cultured for 24–48 h in the absence or presence of dexamethasone (final concentration, 1 mg/ml) prior to harvesting.
To generate gp78 knockdown stable cell lines, pre-miRNA sequences for gp78 and nontargeting control miRNAs were cloned into pcDNA6.2-GW/+EmGFP-miR (Invitrogen). The microRNA nucleotide sequence targeting human gp78 (GenBankTM accession number NM_001144.4) corresponds to amino acids 293–300 relative to the initiating methionine codon. HEK293 cells were transfected with either control or gp78 miRNA vectors and selected for ∼2 weeks against 5 μg/ml blasticidin. Green fluorescent protein-positive clones were sorted by fluorescence-activated cell sorter, and gp78 down-regulation was confirmed by Western blotting using 3F3A antibody.
Control and KAI1-specific siRNA smart pools were transiently transfected in to MCF7 or HEK293 stable cell lines using Lipofectamine 2000 (Invitrogen) following the protocol described before (23). KAI1 knockdown was assessed by Western blotting.
DNA Preparation and Microinjection
All of the experiments involving mice were performed following the guidelines and regulations of the University of British Columbia Animal Care Committee. MMTV-LTR-gp78 plasmid was digested with NruI-AatII and trimmed off the residual backbone leaving the MMTV-LTR promoter, gp78 cDNA embedded into β-globin exon/intron, and BGH-poly(A) tail. The released cassette was extracted from agarose gel and purified for microinjection using GeneClean (MP Biomedicals, LLC). The DNA was resuspended to a final concentration of 2.5 ng/μl in TE buffer (10 mm Tris-HCl, pH 7.5, 1 mm EDTA). Transgenic lines were generated by DNA microinjection of superovulated 4-week-old FVB/Ntac zygotes at the British Columbia Cancer Agency transgenic facility. The surviving zygotes were transferred to pseudo-pregnant ICR females that gave birth to pups 20 days post-implantation. Tail snips of pups were collected 2 weeks after birth and used for genomic DNA extraction.
Genotyping by PCR and Southern Blotting
Genomic DNA was extracted from mouse tails following standard protocols. Briefly, the tails were digested overnight at 55 °C in buffer (10 mm Tris-HCl, pH 8.0, 50 mm NaCl, 25 mm EDTA, pH 8.0, and 0.5% SDS) containing 0.5 mg/ml proteinase K. Digested proteins and debris were separated from DNA using 6 m saturated NaCl and centrifugation. Subsequently, ethanol-precipitated DNA from the supernatant was resuspended in 50 μl of sterile TE buffer. Purified DNA (1 μl) was PCR-amplified using a forward primer designed from the MMTV-LTR promoter (5′-TTG CCC AAC CTT GCG GTT CCC AG-3′) and a reverse primer from gp78 cDNA (5′-GCT GAG GCC CGT GTA GGT GCG-3′). Genomic DNA obtained from the wild type FVB/N mouse was used as a negative control, and the MMTV-LTR-gp78 plasmid was used as a positive control. The 1.5-kb PCR product was separated on 1% agarose gels.
Genotyping or verification of copy number integration of MMTV-LTR-gp78-BGH-poly(A) cassette was confirmed by Southern blotting. Briefly 10 μg of genomic DNA isolated from mouse tails was restriction-digested overnight with high concentrate KpnI (New England Biolabs). Restriction-digested DNAs were phenol-extracted, ethanol-precipitated, and resuspended in sterile RNase/DNase-free PCR water (Invitrogen). KpnI-digested genomic DNA fragments were separated on 0.8% agarose gel, depurinated, denatured, neutralized, and capillary-blotted overnight in 10× SSC onto Zeta probe charged nylon membrane following the manufacturer's protocol (Bio-Rad) and then UV cross-linked (UV Stratalinker 2400; Stratagene).
DIG-labeled probe was prepared and optimized using the PCR DIG probe synthesis kit (Roche Applied Science). Briefly, PCR was done using the following set of primers (5′-CCT CTG CTA ACC ATG TTC ATG-3′ and 5′-GCT GAG GCC CGT GTA GGT GCG-3′) and MMTV-LTR-gp78 plasmid as a template to generate a ∼550-bp product. The probe was stored at −20 °C until required.
Prehybridization, hybridization, and immune probing of membrane were performed using the manufacturer's protocol supplied with the DIG hybridization kit (Roche Applied Science). The membrane was exposed to autographic film, and the signal was developed.
Tissue Harvest
The mice were killed by CO2 asphyxiation. The age- and estrous cycle-matched, superovulated female mice were injected with 100 μg/g of body weight BrdUrd in PBS (Sigma) 2 h prior to sacrifice. The right third thoracic and fourth inguinal mammary glands were fixed in 10% formalin for 48–72 h and processed for paraffin embedding. Tissue embedding and sections (5 μm) were prepared by University of British Columbia Hospital Histopathology Core or Wax-IT Histology (Vancouver, Canada) and stained with hematoxylin and eosin. The left third and fourth glands were whole mounted, fixed overnight in Carnoy's fixative (60% absolute alcohol, 30% chloroform and 10% glacial acetic acid), and defatted through three changes of acetone. The specimens were rehydrated and stained overnight in 0.2% carmine, 0.5% aluminum potassium sulfate. The slides were dehydrated to absolute ethanol, cleared in xylene, permanently mounted (CC/Mount; Sigma), and imaged using a dissection microscope.
RT-PCR
Total RNA was isolated from MCF7 cells or mouse mammary gland (pubertal virgin (8 weeks old) or 12-day mid-gestation wild type or transgenic) using TRIzol (Invitrogen) following the protocol supplied with reagent. Integrity of the total RNA was assessed on formaldehyde gel prior to using them. Reverse transcription and semi-quantitative cDNA synthesis was carried out using a Superscript double-stranded cDNA synthesis kit (Invitrogen) following the supplied protocol and the following set of primers: (5′-CCT CTG CTA ACC ATG TTC ATG-3′ and 5′-GCT GAG GCC CGT GTA GGT GCG-3′).
Lysate Preparation and Western Blotting
For Western blotting, the cells were grown to 80% confluency and washed in cold PBS, and the cell lysates were separated on SDS-PAGE, transferred to nitrocellulose membranes, and Western blotted for the desired proteins as described previously (17). For tissues, ∼10 mg of wild type or transgenic mammary glands were manually homogenized in 1× SDS-PAGE loading buffer, boiled for 5 min, and centrifuged at 14,000 rpm for 10 min at 4 °C. The supernatants were collected, and equal protein amounts were loaded on 10% acrylamide gels; the proteins were separated, transferred to nitrocellulose membrane, and Western blotted for target proteins (gp78, KAI1, and β-actin).
Immunohistochemistry
Paraffin sections were rehydrated with PBS and processed using one of the following protocols. The sections to be stained with anti-gp78 antibody (3F3A), anti-KAI1 (Santa Cruz), or anti-Ki67 (Medicorp) were rinsed in distilled H2O and subjected to antigen retrieval in citrate buffer (4.5 ml of 0.1 m citric acid, 20.5 ml of 0.1 m sodium citrate, fill up to 250 ml with distilled H2O, pH 5.6) for ∼30 min at 90–98 °C (24, 25). The slides were cooled for 20 min, rinsed three times with PBS, incubated for 30 min with 0.3% H2O2, and subsequently incubated for 30 min with DAKO protein block reagent. The sections were incubated at 4 °C overnight in primary antibody (3F3A 1/20) prepared in DAKO dilution buffer. The next day the slides were subjected to several washes with PBS-Tween-20 (0.1%) and stained with secondary anti-rat horseradish peroxidase (Vector) antibody (1/100 in DAKO dilution buffer) for 1 h at room temperature and were subsequently washed three times with PBS-Tween-20 on shaker. The slides were incubated with streptavidin-peroxidase conjugate DAKO LSAB + horseradish peroxidase (K0690; Vector) washed and stained with Nova-Red (SK-4800; Vector). The slides were counterstained with Hematoxylin (Gill 1) for 20 s, washed, xylene-cleaned, and mounted. For BrdUrd injected mice, mammary glands were isolated and paraffin-embedded, and sections of 5-μm thickness were used for BrdUrd staining. A 5-bromo-2′-deoxyuridine labeling and detection kit II from Roche Applied Science was used for detecting BrdUrd-labeled proliferating cells. The slides were counterstained with hematoxylin, and the data represent the mean values of total cell nuclei versus BrdUrd-positive cell nuclei from three microscopic fields (10×) in tissue slides from three different animals. All of the slides were imaged using 10×, 20×, and 63× Plan-Apochromat objectives of a Zeiss Axioskop microscope.
MTT Assay
Cell proliferation assays were performed as described previously (23). Briefly, 10,000 cells were seeded on 96-well plates for 48–72 h, prior to adding MTT reagent diluted 1:10 from 5 mg/ml stock in RPMI (MCF7; MDA-435) or Dulbecco's modified Eagle's medium (HEK293) medium, after which the cells were allowed to grow in a CO2 incubator in the dark for 4–5 h. The medium was decanted, precipitates of metabolite (Formazan) were dissolved in 200 μl of Me2SO, and absorbance was measured at 570 nm. Background absorbance in the presence of Me2SO was subtracted, normalized, and subjected to statistical analysis. For siRNA knockdown experiments, the cells were transiently transfected with control and target specific siRNAs in 96-well plates and incubated for 48 h prior to performing the MTT assay.
RESULTS
Preparation of the MMTV-LTR-gp78 Plasmid and in Vitro Validation
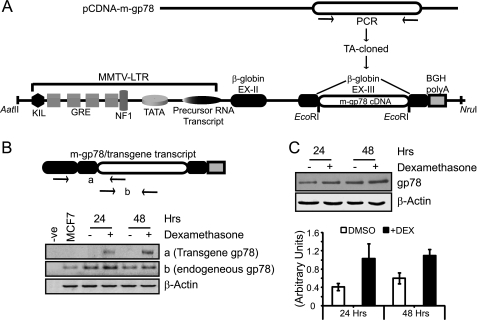
To overexpress gp78 in the mammary gland, we placed gp78 cDNA downstream of the MMTV-LTR promoter gene at the EcoRI site in exon III of the rabbit β-globin gene followed by the BGH-poly(A) sequence that facilitates termination of transcription (Fig. 1A). The MMTV-LTR-gp78 plasmid was validated in vitro prior to generating transgenic animals. The MMTV-LTR promoter carries several glucocorticoid (GC) response elements that can also be turned on by the GC analogue dexamethasone (Fig. 1A). MMTV-LTR-gp78 was transiently transfected into breast carcinoma MCF7 cells that express GC receptors and internalize dexamethasone and treated with Me2SO or 1 mg/ml dexamethasone for 24–48 h. The cells were harvested for RNA as well as protein samples and subjected to RT-PCR or Western blot analysis. As seen in Fig. 1B, the presence of chimeric transcript is selectively observed in dexamethasone-treated cells in both the 24- and 48-h samples and is absent in control Me2SO-treated samples. The total transcript levels are elevated in dexamethasone-treated cells relative to controls. Similarly, when assessed for the gp78 protein level by Western blotting with the 3F3A anti-gp78 antibody (18), dexamethasone-induced cells showed elevated gp78 protein levels (Fig. 1C). These results suggest that in expressing cells, the MMTV-LTR-gp78 plasmid is responsive to the GC analogue dexamethasone and that gp78 overexpression is induced in a controlled fashion.
FIGURE 1.
In vitro validation of the MMTV-LTR-gp78 plasmid. A, schematic diagram showing the design of the MMTV-LTR gp78 construct. Mouse gp78 was PCR-amplified, cloned, and placed at the EcoRI restriction site in β-globin exon III of the MMTV-LTR promoter shown with all elements. GRE represents glucocorticoid response elements. B, schematic presents gp78 transgene transcript with whole mouse gp78 fused to β-globin exons II and III and BGH-poly(A) sequence. a and b labeled on the schematic of the cassette, flanked by arrows (representing primer binding sites), show the areas used for PCR amplification. The bottom panel shows RT-PCR where gp78 transgene transcript is present only in dexamethasone-treated samples, amplified as a, and corresponds to the elevated level of total gp78 transcript, amplified as b, at both 24 and 48 h. Negative controls show no product, and control β-actin transcript levels remain unaltered. C, Western blots probed with 3F3A anti-gp78 rat IgM (1:1000) show elevation in gp78 expression upon dexamethasone treatment at both the 24- and 48-h time points. The graph represents quantification of three individual experiments showing ∼2–3-fold increase in gp78 expression levels relative to β-actin loading control (n = 3; ±S.E.; *, p < 0.05; **, p < 0.001).
Generating gp78-overexpressing Transgenic Mouse
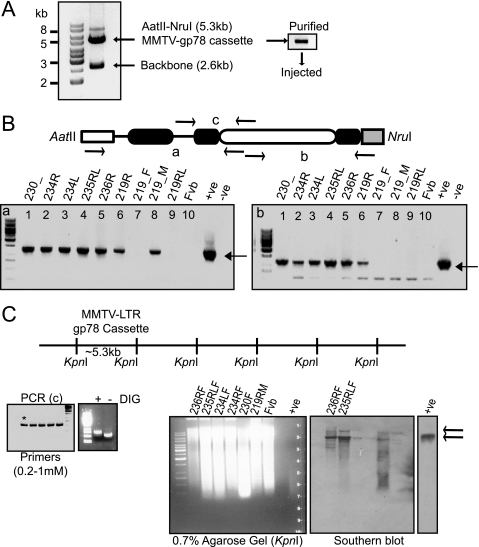
To generate transgenic mice, the cassette of 5.5-kb size containing MMTV-LTR promoter-gp78 cDNA with β-globin exon-intron and BGH-poly(A) was released using AatII-NruI restriction digestion (Fig. 2A). The purified target fragment was microinjected into superovulated 4-week-old FVB/Ntac zygotes and transferred to pseudo-pregnant ICR females. 20-day post-implantation females gave birth to pups, and at 2 weeks of age, tail snips were collected and subjected to genomic DNA extraction and genotyping. For assessing complete cassette integration, we chose two different areas (primer sets a and b) for PCR genotyping (Fig. 2B, schematic). Primer set a identified seven founders (of 40 mice tested; 18% efficiency) containing transgenic DNA (Fig. 2B, left panel). Of these, six founders had intact cassette integration when assessed with the other set (primer set b) of primers (Fig. 2B, right panel), whereas one appeared to have lost part of the DNA during recombination.
FIGURE 2.
gp78-overexpressing transgenic mice. A, the MMTV-LTR gp78 cassette (∼5.3 kb) was released from plasmid using a combination of AatII-NruI restriction enzymes, and the backbone was removed (∼2.6 kb). Cassette DNA was gel-purified, dissolved in TE at 2.5 ng/μl, and injected into superovulating 4-week-old FVB/Ntac mice. B, schematic showing released cassette with area flanked by arrows marked a, b, and c that were used for PCR genotyping. Tail genomic DNA of the obtained seven founders was assessed by PCR for transgene presence (bottom panels), and only six had complete integration of the cassette (right panel compared with left panel). Negative controls show no product, whereas a positive control using plasmid as template DNA showed amplified product of the correct size. C, schematic shows the unique restriction site KpnI used for copy number integration of the gp78 transgene cassette. Digestion produces ∼5.3-kb repeat fragments if integration of the cassette is head-to-tail. The left panel shows PCR optimization to prepare DIG-labeled PCR probe encompassing area designated as c in B, and the asterisk shows the selected condition. Labeled probe runs slower compared with nonlabeled product on agarose gel. The right panels show Southern blot genotyping where ∼10 μg of tail-genomic DNA from six founders was digested with KpnI and separated on 0.7% agarose gel along with controls (FVB genomic and MMTV-LTR-gp78 DNAs), capillary-transferred on Zeta probe (Bio-Rad) nylon membrane, and subjected to hybridization and development. The top arrow indicates genomic DNA-integrated KpnI-released cassette plus 5′ or 3′ genomic DNA extension, whereas the bottom arrow indicates an actual cassette fragment released with KpnI providing a measure of copy number of integrated cassettes. Founder lines 236RF and 235RLF show elevated levels of KpnI restriction product at the position corresponding to the positive MMTV-LTR-gp78 DNA that was absent in negative control, FVB genomic DNA, and weakly present in the rest of the founder lines.
Although MMTV-LTR promoter is driven by GC, some reports have described an impact of copy number integration on protein expression (26). We therefore assessed which of the six positive founders had an intermediate to high copy number of transgene cassettes integrated using a PCR-generated DIG-labeled primer corresponding to primer set c (Fig. 2B) that was optimized and found to run heavier on gel compared with nonlabeled probe (Fig. 2C, left panel). To check head-to-tail multi-copy integration of cassette, the unique KpnI site in the cassette was used for restriction digestion (Fig. 2C, schematic) of genomic DNAs of six founders and FVB as control. Founder lines 236RF (lane 1) and 235RLF (lane 2) carried higher copy numbers of the transgene cassette compared with other founder lines (lanes 3–6) (Fig. 2C, right panels). Negative control FVB did not show the presence of any band, whereas the positive control MMTV-LTR-gp78 cassette showed hybridization to a band of ∼5.3 kb.
Transgene gp78 Overexpression and Its Impact on KAI1 Expression
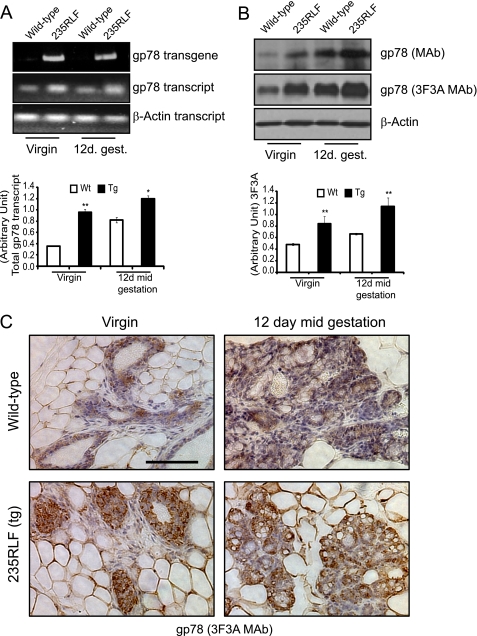
The 235RLF founder line was chosen for further analysis. To assess transgene expression, total RNA was isolated from age- and estrous cycle-matched, superovulated pubertal virgin (8 weeks old) or mid-gestation pregnant (12 days) wild type or transgenic mammary glands. Using primer sets for regions A and B (Fig. 1B, schematic), semi-quantitative RT-PCR was performed on total cDNA generated from extracted RNAs. This facilitated identification of the expression of transgene gp78 (primer set A) as well as total gp78 (primer set B) in both wild type and transgenic mice. Transgene gp78 was expressed only in the 235RLF founder line and, as expected, was absent in wild type virgin or mid-gestation samples. This was further confirmed when the total transcript level assessed at both virgin and mid-gestation time points showed elevated levels of message in 235RLF compared with wild type mice (Fig. 3A).
FIGURE 3.
gp78 overexpression in mammary gland of mouse MMTV-gp78 transgenics. A, RT-PCR analysis of mammary glands from age and estrous cycle matched wild type (Wt) and gp78 transgenic (235RLF) mice. Total RNA prepared from the pubertal virgin or 12-day mid-gestation mammary glands was subjected to cDNA synthesis and assessed for transgene and endogenous transcript levels. gp78 transgene transcript is present only in 235RLF transgenic mice (top panel). Elevated levels of total gp78 message are observed in 235RLF mammary glands compared with wild type. β-Actin transcript levels are unaltered. Quantification of total gp78 transcript normalized to β-actin is presented as a bar graph (n = 3; ±S.E.). B, 10 mg of tissue from mammary glands of wild type and gp78 transgenic mice was homogenized in 1× SDS-PAGE loading buffer, boiled for 5 min, and centrifuged, and 15 μl of lysates were loaded on 10% gel and subjected to Western blotting. gp78 expression levels probed with anti-gp78 monoclonal antibody or anti-gp78 3F3A rat IgM are increased in both the pubertal virgin and 12-day mid-gestation mammary glands of 235RLF transgenic mouse compared with age-matched wild type. Loading control β-actin shows equal loading. Quantification of three individual experiments showed 2–3-fold elevation in expression level of gp78 in the transgenic line probed with 3F3A (n = 3; ±S.E.). C, immunohistochemistry analysis of mouse mammary gland. The panels show 60× magnification of wild type as well as gp78 transgenic mouse mammary glands from pubertal virgin and 12-day mid-gestation time points stained for gp78 with 3F3A rat-IgM antibody. Increased gp78 staining of ducts and alveolar structures in transgenic glands is visible at both time points. Scale bars, 100 μm (*, p < 0.05; **, p < 0.001).
gp78 Western blotting of lysates prepared from the second right mammary gland showed an approximately 2-fold increase at both the virgin and 12-day mid-gestation time points in the 235RLF founder line compared with wild type mice (Fig. 3B). This was further verified by immunohistochemistry of mammary gland frozen sections with the anti-gp78 3F3A monoclonal antibody to examine transgene expression within mammary glands of wild type and transgenic mice (Fig. 3C). Elevated levels of gp78 were observed in mammary glands of pubertal virgin or mid-gestation pregnant 235RLF transgenic mice compared with age-matched wild type mice.
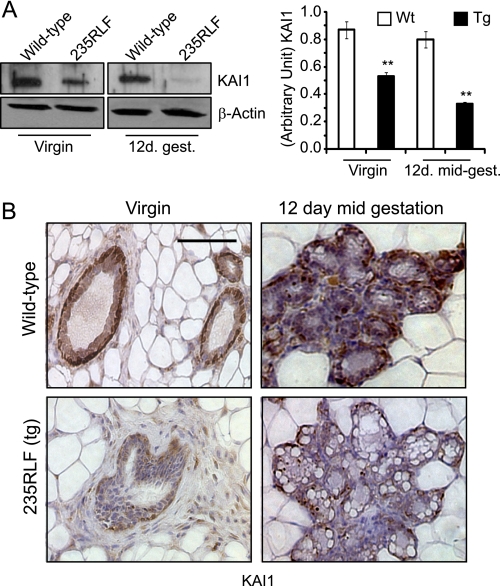
A recent report identified the metastatic suppressor protein KAI1 as a potential gp78 substrate for ubiquitylation and degradation (16). As can be seen in Fig. 4A (left panel), KAI1 levels decrease upon overexpression of gp78 at the pubertal virgin stage and are almost abolished at the 12-day mid-gestation time point in the 235RLF transgenic line, whereas the KAI1 expression levels remain unaltered in wild type mice. This result was further confirmed by immunohistochemistry (Fig. 4B), where KAI1 expression showed a similar reduction in transgenic mammary tissue. Reduced expression of KAI1 provides functional validation of increased gp78 ubiquitin ligase activity in the mammary tissue of the MMTV-LTR-gp78 transgenic mouse.
FIGURE 4.
Reduced KAI1 expression in mammary glands of MMTV-gp78 transgenic mice. A, age-matched mammary gland lysates from wild type (Wt) or 235RLF transgenic mice from virgin or 12-day mid-gestation time points were separated on SDS-PAGE, Western blotted, and probed for KAI1. KAI1 expression levels are significantly reduced in gp78 transgenic 235RLF mice at both time points relative to wild type. Graph presents densitometric quantification (n = 3; ±S.E.). B, immunohistochemistry for KAI1 on mammary glands obtained from wild type and gp78 transgenic mouse is shown at 60× magnification. gp78 transgenic 235RLF shows a reduced level of KAI1 in ducts of transgenic mammary glands at both the virgin and 12-day mid-gestation time points compared with age-matched wild type controls. Scale bars, 100 μm (**, p < 0.001).
gp78 Overexpression Increases Cell Proliferation and Induces Nonclassical Hyperplasia
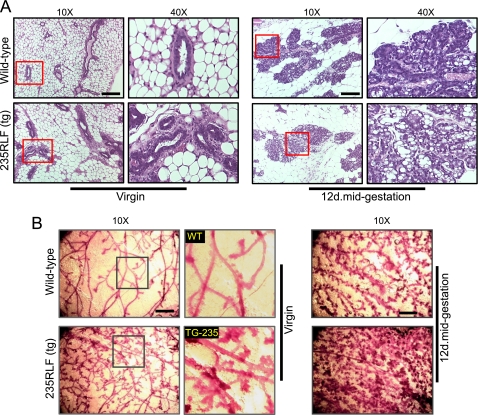
Hematoxylin and eosin staining of mammary glands from both wild type and gp78 transgenic mice showed hyperplasia in ducts and lobules. As shown in Fig. 5A, the wild type mammary gland, at both the virgin and 12-day mid-gestation time points, showed smooth and nearly single layered ductal or alveolar epithelium (top panels). gp78 transgenic mammary gland, at both virgin and 12-day mid-gestation time points, presented multi-layered, diffuse hyperplasia in both ducts and alveolar structures as well as luminal protrusions (bottom panels). Interestingly, mid-gestation alveolar lobules in gp78 transgenic mammary gland also presented numerous vacuoles in addition to the hyperplastic phenotype.
FIGURE 5.
gp78 overexpression induces hyperplasia and increases ductal number and network branching in mouse mammary glands. A, hematoxylin- and eosin-stained sections (10× and 40×) show the mammary gland histology of age-matched wild type and gp78 235RLF transgenic mice. Wild type mammary glands show predominantly adipose tissue and simple epithelia in ductal or alveolar structures, whereas gp78 transgenic mammary glands show ductal alveolar hyperplasia appearing as a multi-layered epithelia protruding into the lumen, with an increased presence of vacuoles. Scale bars, 100 μm (**, p < 0.001). B, mammary ductal structure in carmine alum-stained whole mounts of age- and estrous cycle-matched wild type and transgenic mouse mammary glands shows hyperplastic, differentiated, and densely formed networks of tubules in gp78 transgenic 235RLF line compared with wild type at both the virgin and 12-day mid-gestation time points. The boxed area shows zoomed region. Similarly, 12-day mid-gestation whole mounts show significantly increased branching and alveolar development in gp78 transgenic mouse. Scale bars, 400 μm.
This observation was further confirmed when the third and fourth mammary glands were carnoy-fixed and whole mounted, showing hyperplasia of individual ducts. Age-matched wild type mammary glands from virgin or 12-day mid-gestation mice show smooth normal duct development, whereas gp78 transgenic whole mounts (Fig. 5B, bottom panels) show ductal hyperplasia (i.e. an increase in the number of small ducts) and/or proliferation of ductal epithelium intralumenally (i.e. an increase in the number of epithelial cell layers within a duct as shown in Fig. 5A, bottom panels), resulting in a dense network of ducts at both virgin and 12-day mid-gestation time points that is clearly evident in the zoomed image of pubertal virgin mammary glands (Fig. 5B, boxes).
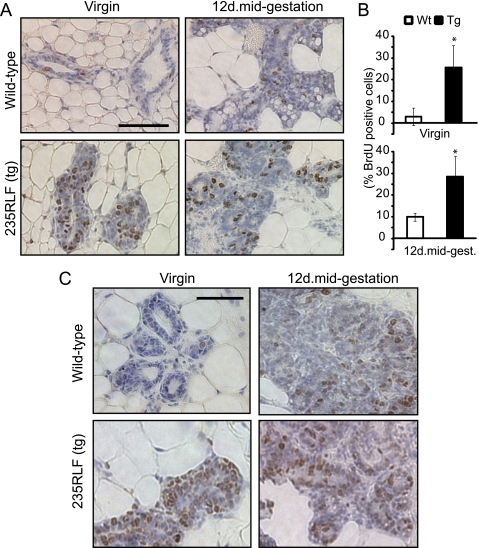
To assess proliferative activities in hyperplastic regions of the transgenic mammary gland, the mice were injected BrdUrd 2 h prior to asphyxiation. Immunohistological staining for BrdUrd-positive cells showed increased levels of BrdUrd incorporation in ductal and alveolar epithelia of gp78 transgenic mammary glands compared with wild type (Fig. 6A). Quantification showed a ∼3–5-fold increase in BrdUrd-positive cells at pubertal virgin and 12-day mid-gestational time points, respectively (Fig. 6B). This was further confirmed by staining with Ki-67, a proliferative marker that showed a markedly increased number of positive cells in ducts and alveolar of gp78 transgenic mammary gland compared with wild type (Fig. 6C). However, there was no significant increase in the expression of the cell cycle marker cyclin D1 at either stage (data not shown). Therefore, overexpression of gp78 induces mammary gland hyperplasia, resulting in increased duct number and network density. However, we did not observe tumor formation in repeatedly pregnant MMTV-LTR-gp78 transgenic mice over a period of ∼18 months after giving birth.
FIGURE 6.
Increased cell proliferation in gp78-overexpressing mammary glands. A, mammary glands of BrdUrd-injected age-matched wild type (Wt) and gp78 235RLF transgenic mice were isolated and paraffin-embedded, and 5-μm sections were stained for BrdUrd to detect proliferating cells. B, bar graph shows quantification (means ±S.E.) of three randomly chosen fields from mammary glands of three individual transgenic or wild type mice scored for number of nuclei presenting BrdUrd incorporation relative to total nuclei. Significantly higher numbers of BrdUrd incorporated nuclei are visible in ducts and alveolar structures of 235RLF gp78 transgenic mice at the pubertal virgin and 12-day mid-gestation time points compared with wild type mice. C, ductal and alveolar epithelium of mammary glands of age-matched gp78 transgenic or wild type mice were stained with the proliferation marker Ki67. Increased Ki67 staining is present in ductal epithelia and alveolae of transgenic mammary glands compared with wild type. Scale bars, A and C, 100 μm.
KAI1 Mediates gp78 Regulation of Cell Proliferation
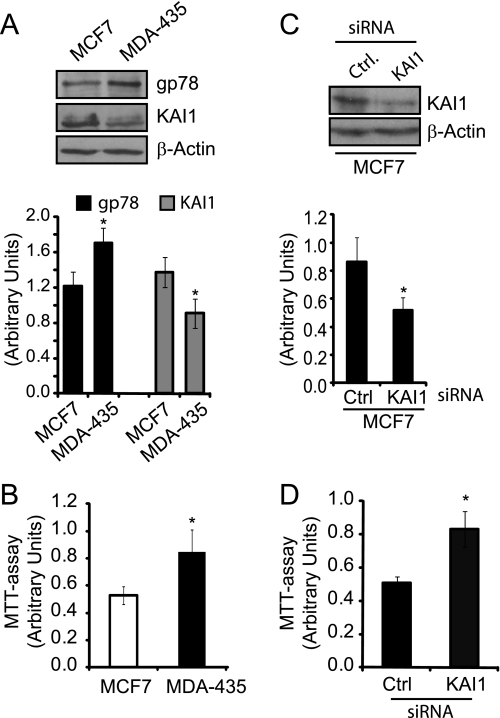
Metastatic MDA-435 cells exhibit elevated gp78 expression levels relative to nonmetastatic MCF7 breast carcinoma cells (17). Interestingly, MDA-435 cells showed reduced KAI1 expression and significantly increased cell proliferation relative to MCF7 (Fig. 7, A and B). siRNA-mediated knockdown of KAI1 in MCF7 cells resulted in a significant increase in proliferation compared with control siRNA transfected MCF7 cells (Fig. 7, C and D), defining a role for KAI1 in the regulation of cell proliferation of these breast tumor cells.
FIGURE 7.
KAI1 regulates cell proliferation in breast carcinoma cells. A, nonmetastatic MCF7 and metastatic MDA-435 breast carcinoma cell lines were Western blotted for gp78 (3F3A rat IgM), KAI1, and β-actin. Quantification of gp78 and KAI1 relative to β-actin is presented as a bar graph (n = 3; ±S.E.). B, cell proliferation of MCF7 and MDA-435 cells was determined using an MTT assay (±S.E.; n = 3; *, p < 0.05). C, MCF7 cells were transfected with KAI1-specific and control (Ctrl) siRNA and KAI1 knockdown validated by Western blot of KAI1 and β-actin. Quantification of KAI1 relative to β-actin is presented as a bar graph (n = 3; ±S.E.). D, MTT assay was performed on control and KAI1 siRNA transfected MCF7 cells (±S.E.; n = 3; *, p < 0.05).
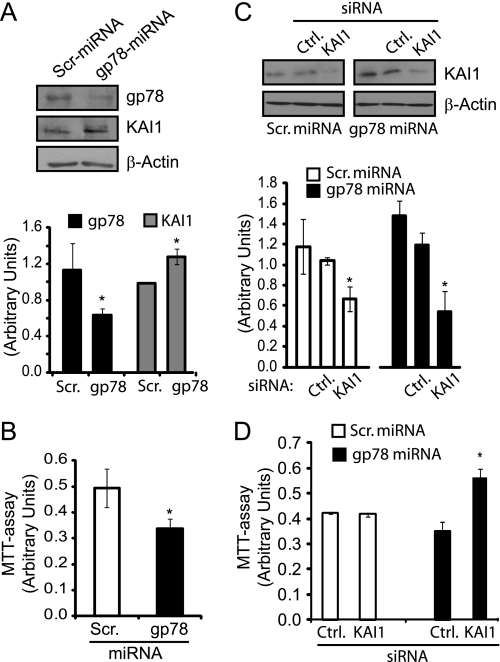
To better define the role of gp78 as a regulator of cell proliferation, we established stable gp78 knockdown HEK293 cell lines using a miRNA-based strategy. Scrambled miRNA and specific gp78 targeted miRNA plasmids were transfected into HEK293 cells, and stable lines were generated. By Western blot, gp78 miRNA lines but not scrambled miRNA lines showed reduced expression of gp78. Stable gp78 knockdown cells showed elevated levels of KAI1 as well as reduced proliferation relative to the scrambled miRNA line (Fig. 8, A and B). Interestingly, when these stable cell lines were subjected to transient KAI1 knockdown by siRNA, only the gp78 knockdown line showed statistically significant elevation in cell proliferation (Fig. 8, C and D). gp78 therefore promotes cell proliferation via degradation of KAI1, defining a mechanistic link between this E3 ubiquitin ligase and its substrate in controlling cell proliferation both in vitro and in vivo in the mammary glands of the MMTV-LTR-gp78 transgenic mice.
FIGURE 8.
KAI1 mediates gp78 regulation of cell proliferation. A, HEK293 stable cell lines expressing control (Scr, Scrambled) and gp78 targeting miRNA were Western blotted for gp78 (3F3A rat IgM), KAI1, and β-actin. Quantification of gp78 and KAI1 levels relative to β-actin is presented as a bar graph (n = 3; ±S.E.). gp78 knockdown results in increased KAI1 expression level but has no effect on β-actin expression levels. B, using the MTT assay, the stable gp78 knockdown cell line shows significantly reduced proliferation compared with control (Ctrl) cell line (±S.E.; n = 3; *, p < 0.05). C, HEK293 stable cell lines expressing control and gp78 targeting miRNA were transfected with control or KAI1-specific siRNA and probed by Western blot for KAI1 and β-actin. D, MTT assay was performed on HEK293 stable cell lines expressing control or gp78 targeting miRNA transfected with either control or KAI1-specific siRNA. KAI1 siRNA increased cell proliferation only in gp78 miRNA stable HEK293 knockdown cells (±S.E.; n = 3; *, p < 0.05).
DISCUSSION
The ability of gp78 overexpression to induce a hyperplastic phenotype upon overexpression in mammary tissue defines a novel potential role for this protein in the early stages of cellular transformation. Hyperplastic morphological patterns in mouse show alveolar outgrowth with lobulo-alveolar morphology, whereas the lesser known nonclassical hyperplasia is associated with growth of small ducts and many tertiary branches that represent nontumorigenic ductal outgrowths (27). The gp78 transgenic mammary gland whole mount shows this nonclassical preneoplastic hyperplasia that was further supported by observations of multi-layered epithelium in ductal and alveolar structures. In the MMTV-p53 (Arg172 to Leu, “gain of function” mutant) transgenic, intraluminal epithelial proliferation results in focal hyperplasia that progresses through DCIS to invasive cancer (27, 28). However, gp78 transgenics, repeatedly pregnant and observed for >18 months after giving birth, did not form tumors. This suggests that the gp78-induced phenotype corresponds to nontumorigenic ductal outgrowth involving development of small ducts and plenty of tertiary branches (27).
In human breast cancer, gp78 expression correlates significantly with pAkt (17), and the gp78 ligand, autocrine motility factor/phosphoglucose isomerase (AMF/PGI) has been shown to induce pAkt expression in various cellular models (17, 29) functionally linking AMF/PGI signaling via gp78 to pAkt in human breast cancer. Autocrine AMF/PGI signaling has long been associated with tumor cell motility and metastasis (30), and AMF/PGI has also been shown to have growth stimulatory and anti-apoptotic activity via pAkt (29, 31). It is therefore possible that increased gp78 cell surface cytokine receptor function enables AMF/PGI growth stimulatory activity promoting mammary cell proliferation. However, we did not observe increases in levels of either AMF or pAkt in gp78-overexpressing mammary tissue (data not shown), suggesting that gp78 overexpression alone is not sufficient to promote AMF/PGI pAkt signaling.
gp78 is a major E3 ubiquitin ligase in endoplasmic reticulum-associated degradation (1, 32). The reduction of KAI1 levels in gp78-overexpressing mammary gland confirms enhanced gp78 ubiquitin ligase activity in these mammary epithelial cells and provides in vivo confirmation of the described previously inverse relationship between gp78 and KAI1 expression in sarcoma (16). KAI1 has been characterized as a metastasis suppressor; however, its mechanism of action remains unknown (14, 33). Down-regulation of KAI1 expression has been widely reported in tumor promotion and distant metastases in various cancers supporting its proposed anti-metastatic function. Reduced or abrogated expression of KAI1 is linked to elevated tumor cell migration, invasion, and proliferation in various carcinomas and tumors (34–36). In pancreatic cancer cells, KAI1 overexpression inhibits cell proliferation and colony formation and abrogates anchorage-independent growth through cell cycle arrest (37). KAI1 physically interacts with the endothelial cell surface protein, DARC (also known as gp-Fy) and promotes inhibition of cell proliferation in vascular endothelial cells (38). Although gp78 overexpression is likely associated with reduced expression of other proteins, these reports are consistent with the association between the reduction of KAI1 and increased proliferation in hyperplastic regions of mammary glands in gp78-overexpressing transgenic mice. A similar relationship between gp78 overexpression, reduced KAI1 expression, and increased cell proliferation was also observed in breast carcinoma cells in vitro and, importantly, upon gp78 knockdown in HEK293 cells. The ability to rescue the reduced proliferation of gp78 knockdown cells, but not gp78-expressing cells, by reducing KAI1 levels with siRNA provides direct evidence that gp78-mediated degradation of KAI1 regulates cell proliferation.
gp78 overexpression in the mammary gland is therefore associated with the induction of hyperplasia defining a novel role for this E3 ubiquitin ligase in the early stages of cellular transformation and potentially promoting enhanced susceptibility to tumorigenesis induced by other agents. These studies further identify KAI1 as a critical mediator of gp78 regulation of cell proliferation. Reduced KAI1 expression in hyperplastic gp78-overexpressing tissue provides novel insight into KAI1 function in tumor progression. Reduced expression of KAI1 caused by gp78 overexpression may lead to preneoplastic hyperplasia as well as predispose these cells to metastasis upon subsequent transformation and tumor formation.
This work was supported by Canadian Institutes of Health Research Grants CIHR MOP-64333 and MT-15132.
- MMTV
- murine mammary tumor virus
- LTR
- long terminal repeat
- BrdUrd
- bromodeoxyuridine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- siRNA
- small interfering RNA
- miRNA
- microRNA
- PBS
- phosphate-buffered saline
- RT
- reverse transcription
- GC
- glucocorticoid
- BGH
- bovine growth hormone
- DIG
- digoxigenin
- AMF/PGI
- autocrine motility factor/phosphoglucose isomerase.
REFERENCES
- 1.Fairbank M., St-Pierre P., Nabi I. R. (2009) Mol. Biosyst. 5, 793–801 [DOI] [PubMed] [Google Scholar]
- 2.Chiu C. G., St-Pierre P., Nabi I. R., Wiseman S. M. (2008) Expert. Rev. Anticancer Ther. 8, 207–217 [DOI] [PubMed] [Google Scholar]
- 3.Sjöblom T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., Willis J., Dawson D., Willson J. K., Gazdar A. F., Hartigan J., Wu L., Liu C., Parmigiani G., Park B. H., Bachman K. E., Papadopoulos N., Vogelstein B., Kinzler K. W., Velculescu V. E. (2006) Science 314, 268–274 [DOI] [PubMed] [Google Scholar]
- 4.Hirono Y., Fushida S., Yonemura Y., Yamamoto H., Watanabe H., Raz A. (1996) Br. J. Cancer 74, 2003–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nakamori S., Watanabe H., Kameyama M., Imaoka S., Furukawa H., Ishikawa O., Sasaki Y., Kabuto T., Raz A. (1994) Cancer 74, 1855–1862 [DOI] [PubMed] [Google Scholar]
- 6.Otto T., Birchmeier W., Schmidt U., Hinke A., Schipper J., Rübben H., Raz A. (1994) Cancer Res. 54, 3120–3123 [PubMed] [Google Scholar]
- 7.Maruyama K., Watanabe H., Shiozaki H., Takayama T., Gofuku J., Yano H., Inoue M., Tamura S., Raz A., Monden M. (1995) Int. J. Cancer 64, 316–321 [DOI] [PubMed] [Google Scholar]
- 8.Nagai Y., Ishikawa O., Miyachi Y., Watanabe H. (1996) Dermatology 192, 8–11 [DOI] [PubMed] [Google Scholar]
- 9.Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang J. S., Kim T., Fang S., Yamaguchi J., Weissman A. M., Fisher E. A., Ginsberg H. N. (2003) J. Biol. Chem. 278, 23984–23988 [DOI] [PubMed] [Google Scholar]
- 11.Song B. L., Sever N., DeBose-Boyd R. A. (2005) Mol. Cell 19, 829–840 [DOI] [PubMed] [Google Scholar]
- 12.Zhong X., Shen Y., Ballar P., Apostolou A., Agami R., Fang S. (2004) J. Biol. Chem. 279, 45676–45684 [DOI] [PubMed] [Google Scholar]
- 13.Morito D., Hirao K., Oda Y., Hosokawa N., Tokunaga F., Cyr D. M., Tanaka K., Iwai K., Nagata K. (2008) Mol. Biol. Cell 19, 1328–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson P., Marreiros A., Russell P. J. (2005) Int. J. Biochem. Cell Biol. 37, 530–534 [DOI] [PubMed] [Google Scholar]
- 15.Sridhar S. C., Miranti C. K. (2006) Oncogene 25, 2367–2378 [DOI] [PubMed] [Google Scholar]
- 16.Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S. M., Helman L. J., Khanna C., Weissman A. M. (2007) Nat. Med. 13, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 17.Kojic L. D., Joshi B., Lajoie P., Le P. U., Cox M. E., Turbin D. A., Wiseman S. M., Nabi I. R. (2007) J. Biol. Chem. 282, 29305–29313 [DOI] [PubMed] [Google Scholar]
- 18.Nabi I. R., Watanabe H., Raz A. (1990) Cancer Res. 50, 409–414 [PubMed] [Google Scholar]
- 19.Matsui Y., Halter S. A., Holt J. T., Hogan B. L., Coffey R. J. (1990) Cell 61, 1147–1155 [DOI] [PubMed] [Google Scholar]
- 20.Guy C. T., Cardiff R. D., Muller W. J. (1992) Mol. Cell Biol. 12, 954–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guy C. T., Webster M. A., Schaller M., Parsons T. J., Cardiff R. D., Muller W. J. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 10578–10582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Registre M., Goetz J. G., St Pierre P., Pang H., Lagacé M., Bouvier M., Le P. U., Nabi I. R. (2004) Biochem. Biophys. Res. Commun. 320, 1316–1322 [DOI] [PubMed] [Google Scholar]
- 23.Joshi B., Strugnell S. S., Goetz J. G., Kojic L. D., Cox M. E., Griffith O. L., Chan S. K., Jones S. J., Leung S. P., Masoudi H., Leung S., Wiseman S. M., Nabi I. R. (2008) Cancer Res. 68, 8210–8220 [DOI] [PubMed] [Google Scholar]
- 24.Janssen P. J., Brinkmann A. O., Boersma W. J., Van der Kwast T. H. (1994) J. Histochem. Cytochem. 42, 1169–1175 [DOI] [PubMed] [Google Scholar]
- 25.Shi S. R., Key M. E., Kalra K. L. (1991) J. Histochem. Cytochem. 39, 741–748 [DOI] [PubMed] [Google Scholar]
- 26.van Rossum A. G., van Bragt M. P., Schuuring-Scholtes E., van der Ploeg J. C., van Krieken J. H., Kluin P. M., Schuuring E. (2006) BMC Cancer 6, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medina D. (2000) J. Mammary Gland Biol. Neoplasia 5, 393–407 [DOI] [PubMed] [Google Scholar]
- 28.Li B., Murphy K. L., Laucirica R., Kittrell F., Medina D., Rosen J. M. (1998) Oncogene 16, 997–1007 [DOI] [PubMed] [Google Scholar]
- 29.Tsutsumi S., Hogan V., Nabi I. R., Raz A. (2003) Cancer Res. 63, 242–249 [PubMed] [Google Scholar]
- 30.Liotta L. A., Mandler R., Murano G., Katz D. A., Gordon R. K., Chiang P. K., Schiffmann E. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 3302–3306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silletti S., Raz A. (1993) Biochem. Biophys. Res. Commun. 194, 446–457 [DOI] [PubMed] [Google Scholar]
- 32.Fang S., Lorick K. L., Jensen J. P., Weissman A. M. (2003) Semin. Cancer Biol. 13, 5–14 [DOI] [PubMed] [Google Scholar]
- 33.Miranti C. K. (2009) Cell Signal. 21, 196–211 [DOI] [PubMed] [Google Scholar]
- 34.Ruseva Z., Geiger P. X., Hutzler P., Kotzsch M., Luber B., Schmitt M., Gross E., Reuning U. (2009) Exp. Cell Res. 315, 1759–1771 [DOI] [PubMed] [Google Scholar]
- 35.Choi U. J., Jee B. K., Lim Y., Lee K. H. (2009) Cell Biochem. Funct. 27, 40–47 [DOI] [PubMed] [Google Scholar]
- 36.Christgen M., Bruchhardt H., Ballmaier M., Krech T., Länger F., Kreipe H., Lehmann U. (2008) Int. J. Cancer 123, 2239–2246 [DOI] [PubMed] [Google Scholar]
- 37.Guo X. Z., Xu J. H., Liu M. P., Kleeff J., Ho C. K., Ren L. N., Li H. Y., Köninger J., Cui Z. M., Wang D., Wu C. Y., Zhao J. J., Friess H. (2005) Oncol. Rep. 14, 59–63 [PubMed] [Google Scholar]
- 38.Bandyopadhyay S., Zhan R., Chaudhuri A., Watabe M., Pai S. K., Hirota S., Hosobe S., Tsukada T., Miura K., Takano Y., Saito K., Pauza M. E., Hayashi S., Wang Y., Mohinta S., Mashimo T., Iiizumi M., Furuta E., Watabe K. (2006) Nat. Med. 12, 933–938 [DOI] [PubMed] [Google Scholar]