Abstract
Tumor necrosis factor-related apoptosis-inducing ligand receptors death receptor (DR) 4 and DR5 are potential targets for antibody-based cancer therapy. Activation of the proapoptotic DR5 in various cancer cells triggers the extrinsic and/or intrinsic pathway of apoptosis. It has been shown that there are several functional domains in the DR5 extracellular domain. The cysteine-rich domains of DR5 have a conservative role in tumor necrosis factor-related apoptosis-inducing ligand-DR5-mediated apoptosis, and the pre-ligand assembly domain within the N1-cap contributes to the ligand-independent formation of receptor complexes. However, the role of the N-terminal region (NTR) preceding the N1-cap of DR5 remains unclear. In this study, we demonstrate that NTR could mediate DR5 activation that transmits an apoptotic signal when bound to a specific agonistic monoclonal antibody. A novel epitope in the NTR of DR5 was identified by peptide array. Antibodies against the antigenic determinant showed high affinities for DR5 and triggered caspase activation in a time-dependent manner, suggesting the NTR of DR5 might function as a potential death-inducing region. Moreover, permutation analysis showed that Leu6 was pivotal for the interaction of DR5 and the agonistic antibody. Synthetic wild-type epitopes eliminated the cytotoxicity of all three agonistic monoclonal antibodies, AD5-10, Adie-1, and Adie-2. These results indicate that the NTR of DR5 could be a potential target site for the development of new strategies for cancer immunotherapy. Also, our findings expand the current knowledge about DR5 extracellular functional domains and provide insights into the mechanism of DR5-mediated cell death.
Keywords: Receptors/Structure-Function, Signal Transduction, Tumor/Therapy, Antibodies, Epitope Mapping
Introduction
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)4 is a attractive candidate for cancer therapy because it can triggers the extrinsic and/or intrinsic pathway of apoptosis in a variety of tumor cells by engaging the death receptors DR4 and DR5, while sparing most normal cells (1–5). However, decoy receptors (DcR1, DcR2, and OPG) compete for binding of TRAIL and protect cells from TRAIL-mediated cell death (6–10). In this case, agonistic monoclonal antibodies (mAbs) against DR4/5 have shown encouraging results and potential in the therapeutic aspect of diseases.
There are a number of agonistic mAbs against human DR4 or DR5 reported in the literature (11–13), and they have demonstrated more specific cell death-inducing activities. Most of these mAbs either need a cross-linker to ensure effective killing of tumor cells (14, 15) or compete with TRAIL for binding with DR5 (12, 16), mimicking the apoptosis-inducing mechanism of TRAIL. Guo et al. (17) from our laboratory reported that a novel anti-human DR5 monoclonal antibody AD5-10 induces the apoptosis of various carcinoma cell lines without cross-linking in vitro and exhibits strong tumoricidal activity in vivo. Furthermore, AD5-10 does not induce cell death of normal human hepatocytes or primary peripheral blood lymphocytes, and injection of AD5-10 into mice causes nontoxic reactions in the liver, spleen, and kidney. Unlike other anti-DR5 mAbs, AD5-10 does not compete with TRAIL for binding with DR5. There is also a synergistic effect of TRAIL and AD5-10 on tumoricidal activity. Although both TRAIL and AD5-10 are capable of activating NF-κB, there are differences between the regulation of NF-κB activity by TRAIL and that by AD5-10 in certain cell lines. TRAIL, but not AD5-10, could induce significant activation of caspase-3 and cleavage of c-FLIPL in H460 cells (18). It is notable that AD5-10-induced cell death is different from that induced by TRAIL in terms of ROS aggregation and JNK activation (19). DR5 can signal various downstream messages when activated by different ligands. However, the underlying mechanism is poorly understood.
Given the differences in the cell signaling induced by AD5-10 and TRAIL, we speculate that changes in the conformational structure of DR5 may contribute to this mechanism. Biological conformational changes can be induced by many factors, especially the binding of an agonist. For the ligand-receptor interaction, the binding affinity and the interaction pattern are important. Ligands interacting with DR5 with various affinities initiate a series of signaling events, including classical apoptosis and other programmed cell death. TRAIL and agonistic mAbs with high affinities to DR5 can mediate apoptotic cell death in TRAIL-sensitive cancer cells, predominantly through the sequential activation of caspases (13). In other cases, depending on the external stimuli and specific cell types, TRAIL and anti-DR5 mAbs with relatively low affinities can also induce multiple cellular signaling pathways, such as autophagy (20, 21). Consistent with these findings, on the one hand, the affinity between AD5-10 and DR5 is much higher than that between TRAIL and DR5. This might induce a distinct conformation of activated DR5. On the other hand, interactions between TRAIL and AD5-10 are easily discriminated from each other. TRAIL acts as a death messenger by forming a trimer complex, whereas AD5-10 is a bivalent monoclonal antibody. Therefore, different components might be recruited into the DISC and mediate distinct downstream cascade events when different agonists bind to DR5. However, the variety of DR5 signaling pathways cannot be fully explained by these reasons. The noncompetitive binding between AD5-10 and TRAIL suggests that there may be a previously unreported potential death-triggering binding site on DR5.
To further understand the unique activity of AD5-10 and to illustrate the underlying molecular mechanism, it is important to map the exact AD5-10-binding site on DR5. Determination of a novel death-triggering region on DR5 and demonstration of the interaction between AD5-10 and DR5 will provide an overall view of the complicated signaling pathways mediated by DR5 and could lead to the development of new cancer immunotherapy strategies.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Recombinant human TRAIL was purchased from R & D Systems (Minneapolis, MN). The homemade mouse anti-human DR5 monoclonal antibody AD5-10 was described previously by Guo et al. (17). The QuikChange® II XL site-directed mutagenesis kit was purchased from Stratagene Corp. (La Jolla, CA). Antibodies were sourced as follows: anti-FLAG was from Sigma; anti-phospho-ERK1/2, anti-ERK1/2, anti-phospho-MEK1/2, anti-MEK1/2, anti-FADD, anti-caspase-3, -8, and -9, and anti-cFLIP were from Cell Signaling Technology, (Danvers, MA); anti-DR4, anti-DR5, anti-DcR1 and anti-DcR2 were from Chemicon (Temecula, CA); phycoerythrin (PE)-labeled anti-mouse DR5 was from eBioScience Inc. (San Diego); PE-conjugated mouse IgG2B isotype and anti-human DR5 (clone FAB71908) were from R & D Systems Inc.(Minneapolis, MN); normal mouse IgG3 control and anti-RIP1 were from Santa Cruz Biotechnology (Santa Cruz, CA); horseradish peroxidase (HRP)-conjugated and FITC/TRITC-conjugated secondary antibodies were from ZhongShan Co., Beijing, China).
Cell Lines, Cell Culture, and Transfection
HeLa (human cervix carcinoma), HCT116 (human colon cancer), and HEK293T/17 (human embryonic kidney) cell lines (ATCC, Manassas, VA) were cultured in high glucose Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT) and penicillin/streptomycin (100 μg/ml of each). The Jurkat (human T-lymphoma) and H460 (non-small-cell lung cancer) cell lines were cultured in RMPI 1640 medium (Invitrogen) containing 10% heat-inactivated fetal calf serum (Hyclone) and penicillin/streptomycin. The NIH3T3 (mouse embryonic fibroblast) cell line was cultured in high glucose Dulbecco's modified Eagle's medium supplemented with 10% calf serum (Hyclone) and penicillin/streptomycin (100 μg/ml of each). All cell lines were grown in 5% CO2 at 37 °C. Cells were transfected with appropriate plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Analyses were performed 48 h after transfection unless otherwise noted.
Plasmid Constructs, Retrovirus Packaging, and Transduction
Full-length DR5a cDNA in pCR3.1 (Invitrogen) was inserted into the p3×FLAG-cmv-14a vector (Sigma) at the EcoRI/XbaI restriction site. The extracellular domain (corresponding to 1–128 amino acids) of DR5 was ligated with the transmembrane and intracellular domains (corresponding to 657–1259 amino acids) of the rat ErbB2 receptor to generate the chimeric receptor DR5e/ErbB2i (Der, where Der is death receptor 5 extracellular domain fused with ErbB2 intracellular domain) (Fig. 4A). Briefly, PCR was used to introduce an EcoRI site into the 5′ end of the DR5 cDNA fragment encoding the extracellular domain (hDR5-ECD) and an XhoI site into the 3′ end of the ErbB2 cDNA fragment encoding its transmembrane and intracellular domains. The cDNA fragments were subsequently inserted into the pMSCV-IRES-GFP retroviral vector using the EcoRI/XhoI restriction site to generate the chimeric receptor DR5e/ErbB2i (Der) expression vector. Site-directed mutagenesis was conducted using the QuikChange® II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions to produce DR5e/ErbB2i epitope L6A (Der-L6A) and DR5e/ErbB2i-truncated (Der-Δ) mutant expression vectors. The primer sequences are listed in Table S1. The retroviral vectors encoding the chimeric receptor Der and its mutants, Der-L6A and Der-Δ, were transduced into HEK293T/17 cells to generate viral particles. NIH3T3 cells were incubated with a viral suspension containing Polybrene (8 μg/ml) for 16 h and then washed with PBS and cultured in complete medium before analysis.
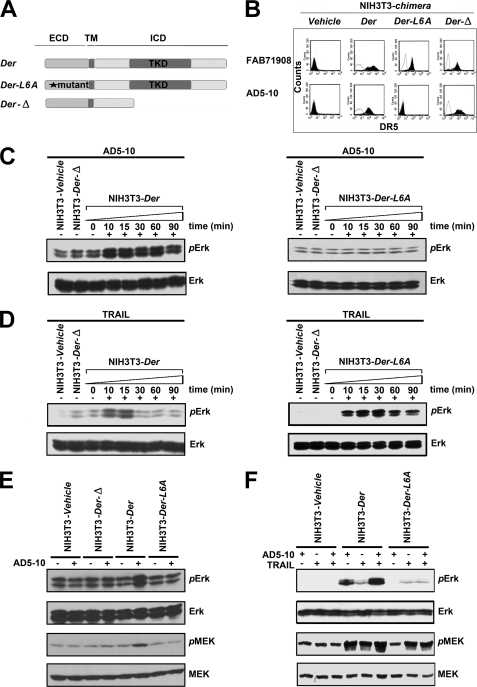
FIGURE 4.
MAPK pathway is investigated in chimera signaling. Before treatment, all cells were starved in serum-free medium for 24 h and then cultured in fresh assay medium. Then NIH-chimera cells were treated with 500 ng/ml AD5-10 or 500 ng/ml rsTRAIL for the indicated times. Lysates were probed for protein phosphorylation by Western blot using the respective specific antibodies. A, schematic diagram depicting DR5e/ErbB2i and derived chimeric receptors. B, expression of surface chimeras in NIH3T3 cells. NIH3T3 cells were transfected with chimeric constructs and stained with PE-labeled anti-DR5 antibody (FAB 71908) or AD5-10 and TRITC-labeled secondary antibody followed by flow cytometry. C and D, immunoblot analysis of the activation of ERK in NIH3T3 chimeras treated with 500 ng/ml AD5-10 (C) or rsTRAIL (D) for the indicated times. E and F, analyses of phosphorylated ERK1/2 and phosphorylated MEK1/2 levels in NIH3T3 wild-type or NIH3T3 chimera-transfected cells with 500 ng/ml AD5-10 (E) and/or rsTRAIL (F) treatment for 15 min. p-, phosphorylated; ICD, intracellular domain. All experiments were repeated at least three times with similar results.
Membrane Protein Extraction
5 × 107 Jurkat cells were harvested and washed with 20 ml of HB buffer (20 mm Tris-HCl (pH 7.5), 4 mm EDTA, 2 mm EGTA, 10% glycerol, 1× protease inhibitor mixture). Then cells were suspended in 4 ml of HB buffer containing 300 mm NaCl and 1% Triton X-100 followed by a 30-min incubation at 4 °C. The lysed cells were centrifuged at 86,000 × g for 45 min at 4 °C. The membrane fraction were collected and diluted in 1× HB buffer containing 150 mm NaCl and 0.5% Triton X-100 with proper concentration.
OPAL and Permutation Array
The OPAL was described by Rodriguez et al. (22). The peptide arrays were generated on cellulose membranes using an Intavis Multipep instrument (Intavis, Germany) and Fmoc (N-(9-fluorenyl)methoxycarbonyl)-based chemistry according to the manufacturer's instructions. The estimated peptide yield at each spot was about 5 nmol. All peptide sequences were based on the primary structure of mature DR5a (NCB accession number NP_671716.1). The sequence of the primary structure was addressed by using an oriented peptide walking library synthesized and printed onto a cellulose membrane. The walking library consisted of a series of overlapping 12-amino acid peptides with a sliding window of two residues. Alanine scanning assay was used to identify the key amino acids in the epitope. Each amino acid residue of the indicated peptide was replaced by alanine. To generate a permutation array, each degenerated residue of the specific sequence was substituted with any of the 20 amino acids. The membrane was first blocked with 5% bovine serum albumin in TBST buffer (0.1 m Tris-HCl (pH 7.4), 150 nm NaCl, and 0.1% Tween 20) for 2 h at room temperature and followed by incubation with AD5-10 or an isotype control antibody of normal mouse IgG3 (0.2 μg/ml) for 4 h at room temperature. The array membrane was then washed three times with TBST buffer and incubated with HRP-conjugated goat anti-mouse IgG for 2 h at room temperature. Subsequently, the membrane was washed again and peptide spots that bound AD5-10 were visualized by enhanced chemiluminescence (Millipore Corp., Danvers, MA). If a peptide in the library pool at any given residue was preferred by the antibody, the peptide would be detected by immunoblotting and would show up as a positive spot.
In Vitro Neutralization Assay
A series of peptides was synthesized, partially corresponding to the amino acid sequence of DR5. Synthetic wild-type epitopes are AQI1TQQDLAPQQRA12 and AI1TQQDLAPQQRA12. The mutant peptides possessed identical sequences, except at position 6 where leucine was replaced with alanine. The sequences of the control peptides were either from death receptor 5 (26CPPGHHISEDGRDC39 and 45GQDYSTHWNDLLFCLRC61) or a random sequence (SQRLHTPCFNKMEA). The purity of the peptides, which was judged to be approximately >95%, was confirmed by high pressure liquid chromatography and mass spectrometry. For the neutralization assay, these peptides were incubated with AD5-10 or rsTRAIL at 37 °C for 2 h. The mixture was then added to the cell culture medium. The final concentrations of the synthesized peptides, AD5-10 (or rsTRAIL), were 10 μm and 500 ng/ml, respectively. After treatment, the cell viability assay was conducted.
Hoechst 33258 Staining and Cell Viability Assay
Hoechst 33258 staining was performed as described previously (23). Briefly, cells were fixed with 4% paraformaldehyde for 30 min at room temperature and then washed once with PBS. Hoechst 33258 (1 μg/ml) was added to the fixed cells, incubated for 15 min at room temperature, and washed with PBS. Cells were mounted and examined by fluorescence microscopy. Apoptotic cells were identified by the condensation and fragmentation of their nuclei. For cell viability assay, cells (104 cells/well in 96-well plates) were incubated with the indicated concentration of AD5-10 or rsTRAIL at 37 °C for the indicated time. Cell viability was assayed using the Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer's specification. Relative cell viability = absorbance of treated sample/absorbance of control sample × 100%.
Enzyme-linked Immunosorbent Assay
Binding of antibodies to synthetic peptides or DR4/5 was carried out by an enzyme-linked immunosorbent assay in 96-well plates. Enzyme-linked immunosorbent assay plates were first coated with synthetic peptides (100 ng/well) or recombinant soluble human DR4/5-ECD (300 ng/well) in 0.1 m sodium phosphate (pH 9.6). The plates were rinsed with PBS containing 0.05% (v/v) Tween 20 (PBS/Tween) and incubated at room temperature for 2 h with 3% bovine serum albumin in PBS/Tween to block nonspecific sites, followed by three more rinses in PBS/Tween to remove excess reagents. The plates were then incubated at 37 °C for 2 h with 100 μl of the indicated concentrations of primary antibodies. Subsequently, the plates were washed three times with PBS/Tween and incubated with 100 μl of HRP-conjugated secondary antibodies at 37 °C for 60 min. The excess of conjugate was removed by washing the plates three times with PBS/Tween. The amount of bound conjugate was determined by adding tetramethylbenzidine substrate solution and incubated for 30 min. The reaction was stopped by adding sulfuric acid, and absorbance was measured at 450 nm on a microplate reader.
Immunofluorescence and Surface Staining
Cells were grown on glass coverslips coated with poly(l-lysine) and collagen, fixed in 4% paraformaldehyde, and blocked with 5% bovine serum albumin in PBS. Primary antibody incubation was done at 4 °C overnight, followed by incubation for 1 h at room temperature with FITC-labeled secondary antibody in the dark. The samples were washed three times with cold PBS, and then the coverslips were mounted on glass slides using mounting media with propidium iodide to allow visualization of the nuclei. Images were captured by confocal microscopy (Zeiss LSM 510, Germany). For the surface expression assay, cells (106) were washed twice with PBS and suspended in 100 μl of flow cytometry staining buffer (1× PBS, 3% fetal bovine serum). For direct staining, PE-conjugated antibody was added to the cell suspension and kept in the dark at 4 °C for 30 min. For indirect staining, cells were incubated with the primary antibody (1:50 to 1:100) at 4 °C for 60 min and then washed with 1 ml of cold staining buffer and incubated with FITC- or TRITC-labeled secondary antibody in the dark at 4 °C for 60 min. After an additional wash with 2 ml of cold PBS, cells were suspended in 500 μl of cold staining buffer, and 104 cells per sample were analyzed by flow cytometry (Pharmingen).
Immunoprecipitation
For TRAIL-DISC analysis, 108 cells were stimulated with 5 μg of FLAG-TRAIL cross-linked with 5 μg of anti-FLAG M2 in 1 ml of medium at 37 °C for the indicated times. For antibody-DISC analysis, 108 cells were stimulated with 5 μg of different anti-DR5 agonistic antibodies in 1 ml of medium at 37 °C for the indicated times, respectively. Cells were then washed with cold PBS and lysed in 1 ml of lysis buffer containing 1% Nonidet P-40, 20 mm Tris-HCl (pH 7.5), 150 mm NaCl, and 10% glycerol. In unstimulated control, 1 μg of AD5-10 was added after lysis to immunoprecipitate death receptor 5. Lysates were pre-cleared with Sepharose 4B (GE Healthcare) and immunoprecipitated overnight at 4 °C with protein G-Sepharose beads (GE Healthcare). In both cases, beads were then washed three times with the lysis buffer before being processed for immunoblotting.
Immunoblotting Analysis
After the indicated treatment, the cells (2–3 × 106) were washed twice with cold PBS and lysed in SDS-PAGE loading buffer (50–100 μl). The proteins in the cell lysate were separated by 10% SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (GE Healthcare). The membrane was then probed with the indicated primary antibody at 4 °C overnight, followed by incubation with horseradish peroxidase-conjugated secondary antibody for 1.5 h at room temperature. Immunoreactive proteins were visualized by enhanced chemiluminescence (Millipore Co, Danvers, MA) and autoradiography.
Computational Molecular Simulation
Structures were entered into molecular operating environment 2006.08 (Chemical Computing Group, Montreal, Canada). Briefly, the “VL + VH” fragment of AD5-10 was determined and refined using the molecular operating environment antibody homology modeling module. The interaction between AD5-10 and the core epitope was assessed using the ligand-receptor docking module in the CHARMM27 force field (24). After a series of models was created, a functional model was culled from the candidates by evaluating the fitness and importance of the molecular descriptors that were built interactively. All structure refinements were performed by macromolecular protonation and energy minimization. Output for the final interaction model and predictions was shown after ligand explorer modification.
RESULTS
Novel Death-inducing Region Is Located within the DR5 N-terminal Region
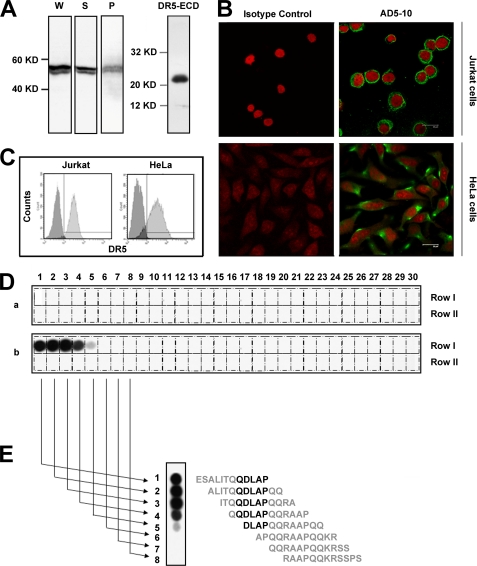
Most antigenic determinants recognized by antibodies can be thought of as special three-dimensional surface features and protein structures of an antigen molecule. Antibody recognition requires a precise fit of these features. To map the epitope in DR5 recognized by AD5-10, we first identified the type of the antigenic determinant, which was considered to be either a sequential epitope or conformational one. DR5+ Jurkat cells were lysed and subjected to denatured SDS-PAGE to eliminate the conformational information of DR5, followed by immunoblotting with the AD5-10 antibody. As shown in Fig. 1A, AD5-10 specifically recognized the two denatured DR5 alternative splicing variants, DR5a and DR5b, implying that both DR5a and DR5b share the same region recognized by AD5-10, and the alternative splicing region in DR5b can be excluded. Moreover, this finding suggested that AD5-10 might recognize a linear epitope instead of a conformational one. To verify this result, immunofluorescence assays were carried out in Jurkat and HeLa cells. As shown in Fig. 1, B and C, AD5-10 bound to Jurkat and HeLa cell membranes specifically (also seen in supplemental Fig. S1). These data indicated that AD5-10 recognizes not only a denatured epitope, but also a native epitope on DR5 that should be a continuous and linear sequence.
FIGURE 1.
Epitope recognized by AD5-10 is localized in the NTR of DR5. A, AD5-10 recognizes denatured endogenous DR5 (both isoform-A and -B) in Jurkat cells. Whole cell lysate (W), soluble fraction (S), and membrane fraction (P) of Jurkat cells were separated by 12% SDS-PAGE, whereas recombinant soluble DR5-ECD was taken as a positive control. The blot was then probed with AD5-10. B, AD5-10 binds to native membrane DR5. Cells were grown on glass coverslips coated with poly(l-lysine) and collagen. After fixation, cells were incubated with AD5-10 or normal mouse IgG3 isotype control at 4 °C for 60 min and then stained with an FITC-labeled secondary antibody. C, AD5-10 binds to native membrane DR5 on Jurkat cells and HeLa cells. Membrane DR5 on Jurkat cells and HeLa cells was determined by indirect immunofluorescence with AD5-10 and FITC-labeled secondary antibody followed by flow cytometry assay. D, OPAL of DR5-ECD is detected by AD5-10. An array of 60 peptides derived from the DR5-ECD was probed for binding to AD5-10 (panel b). Meanwhile, mouse normal IgG3 isotype (panel a) was used as a negative control to monitor the nonspecific binding. All the antibodies were added to the cellulose membrane at a final concentration of 0.2 μg/ml. Bound proteins were identified by an HRP-conjugated goat anti-mouse IgG3 and visualized by ECL. Positive spots indicated definite binding. E, determining the minimal sequence of peptides required for binding to AD5-10 by analyzing the signal strength of the spots. All experiments were repeated at least three times with similar results.
Although biochemical analysis of DR5 identified several ectodomain O-(N-acetylgalactosamine-galactose-sialic acid) structures, the precise O-glycosylation sites in DR5 and how they affected DR5-mediated signaling still remain unelucidated. There are three potential O-glycosylation regions (25). Two of them (19SSPS22 and 75TTTRNTVCQCEEGT88) are in the common ECD sequence of mature human DR5 splice variants, and the third is located within the alternatively spliced DR5 region. However, the effect of O-glycosylation modification on the DR5-AD5-10 complex can be ignored in our study because AD5-10 was generated by immunizing mice with recombinant DR5a expressed in Escherichia coli. Then an unmodified OPAL screening was applied to precisely detect the amino acid residues of the epitope in the extracellular region recognized by AD5-10.
A series of overlapping 12-amino acid peptides covering the major part of the mature human DR5 ectodomain (residues Glu−4 to Lys126 without the signal peptide Met−55 to Ala−5) was synthesized and assembled on a cellulose membrane in an array format with a sliding window of two residues. A total of 60 peptides (Table 1) was generated, and each numbered peptide differed from the adjacent one by two amino acids. The cellulose membrane was immunoblotted with AD5-10 and probed with an HRP-labeled secondary antibody. As shown in Fig. 1D, strong binding spots corresponding to peptides 1–5 were observed, whereas nonspecific binding was excluded using a negative control. Inspection of the peptide sequences corresponding to the positive signals revealed that AD5-10 bound to peptides containing QDLAP residues (Fig. 1E). Due to the appearance of the strongest signal at peptides 2 (−2ALITQQDLAPQQ10) and 3 (1ITQQDLAPQQRA12), we were able to determine that AD5-10 recognizes the epitope 1ITQQDLAPQQ10 with the core sequence of QDLAP in the DR5 N-terminal domain. This finding has not been reported previously.
TABLE 1.
List of OPAL in the peptide-walking array screen
| Row I (DR5 residues 1–70) |
Row II (DR5 residues 61–130) |
||
|---|---|---|---|
| Spot no. | Amino acid sequence | Spot no. | Amino acid sequence |
| 1 | ESALITQQDLAP | 1 | FCLRCTRCDSGE |
| 2 | ALITQQDLAPQQ | 2 | LRCTRCDSGEVE |
| 3 | ITQQDLAPQQRA | 3 | CTRCDSGEVELS |
| 4 | QQDLAPQQRAAP | 4 | RCDSGEVELSPC |
| 5 | DLAPQQRAAPQQ | 5 | DSGEVELSPCTT |
| 6 | APQQRAAPQQKR | 6 | GEVELSPCTTTR |
| 7 | QQRAAPQQKRSS | 7 | VELSPCTTTRNT |
| 8 | RAAPQQKRSSPS | 8 | LSPCTTTRNTVC |
| 9 | APQQKRSSPSEG | 9 | PCTTTRNTVCQC |
| 10 | QQKRSSPSEGLC | 10 | TTTRNTVCQCEE |
| 11 | KRSSPSEGLCPP | 11 | TRNTVCQCEEGT |
| 12 | SSPSEGLCPPGH | 12 | NTVCQCEEGTFR |
| 13 | PSEGLCPPGHHI | 13 | VCQCEEGTFREE |
| 14 | EGLCPPGHHIS.E. | 14 | QCEEGTFREEDS |
| 15 | LCPPGHHISEDG | 15 | EEGTFREEDSPE |
| 16 | PPGHHISEDGRD | 16 | GTFREEDSPEMC |
| 17 | GHHISEDGRDCI | 17 | FREEDSPEMCRK |
| 18 | HISEDGRDCISC | 18 | EEDSPEMCRKCR |
| 19 | SEDGRDCISCKY | 19 | DSPEMCRKCRTG |
| 20 | DGRDCISCKYGQ | 20 | PEMCRKCRTGCP |
| 21 | RDCISCKYGQDY | 21 | MCRKCRTGCPRG |
| 22 | CISCKYGQDYST | 22 | RKCRTGCPRGMV |
| 23 | SCKYGQDYSTHW | 23 | CRTGCPRGMVKV |
| 24 | KYGQDYSTHWND | 24 | TGCPRGMVKVGD |
| 25 | GQDYSTHWNDLL | 25 | CPRGMVKVGDCT |
| 26 | DYSTHWNDLLFC | 26 | RGMVKVGDCTPW |
| 27 | STHWNDLLFCLR | 27 | MVKVGDCTPWSD |
| 28 | HWNDLLFCLRCT | 28 | KVGDCTPWSDIE |
| 29 | NDLLFCLRCTRC | 29 | GDCTPWSDIECV |
| 30 | LLFCLRCTRCDS | 30 | CTPWSDIECVHK |
Leu6 in the Novel Epitope of DR5 Has a Pivotal Role in the Interaction of DR5 and AD5-10
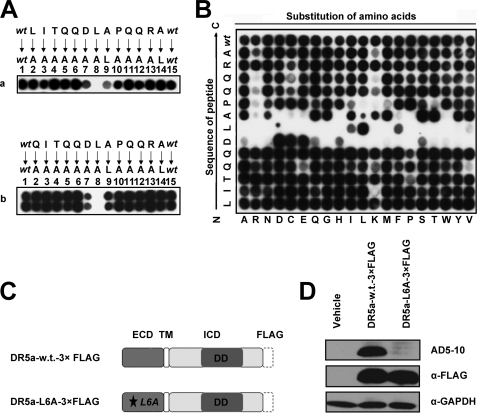
To identify critical residues in the novel epitope of DR5, alanine scanning and permutation arrays were performed. With the purpose of keeping the maximum immunogenicity of the epitope, a 13-amino acid peptide, −1LITQQDLAPQQRA12, was applied as a candidate by combining the sequences of peptides 2 and 3. As shown in Fig. 2A, substitution of any one residue in 5DLA7 led to either complete or partial loss of the peptide binding activity of AD5-10. By contrast, replacement of any other residues outside the “4QDLAP8” by Ala had no significant effect on antibody binding to the epitope. Notably, residue Leu6 played a key part in AD5-10 binding to the epitope suggesting that the hydrophobic amino acid-rich domain around Leu6 is important for antibody binding (Fig. 6). Permutation analysis confirmed the importance of the hydrophobic Leu6 residue in AD5-10 binding to the epitope. As shown in Fig. 2B, substitution of isoleucine or phenylalanine residues with leucine significantly restored the binding ability as compared with the other permutations. Similarly, Asp5 and Ala7 were also vital for epitope binding to AD5-10. However, Leu−1 could be changed to any amino acid without incurring a notable loss in affinity. Basic residues appeared to be disfavored at the pair of glutamines near the C terminus of the epitope, whereas 3QQ4 could be substituted with most of the natural amino acids. Collectively, these data demonstrate at a single amino acid level that the hydrophobic amino acid residue, especially Leu6, has a pivotal role in the interaction between DR5 and AD5-10.
FIGURE 2.
Residues 5DLA7 are essential for the interaction between DR5 and AD5-10. A, alanine-scanning mutagenesis of the epitope. Two alanine-scanning arrays of peptides −1LITQQDLAPQQRA12 (panel a) and QI1TQQDLAPQQRA12 (panel b) were probed with 0.2 μg/ml AD5-10. The sequences of the peptides are shown. The first and the last spot corresponded to the parental peptide, whereas the other spots represented an Ala-substituted analogue of wild-type sequence (wt) as indicated above the individual spots. B, permutation array of peptide −1LITQQDLAPQQRA12. Each residue in this peptide was replaced, one at a time, by a naturally occurring amino acid. The resulting array of 260 (13 × 20) peptides was probed with 0.2 μg/ml of AD5-10. Spots that displayed the “half-moon” pattern were likely to be caused by imperfections in array synthesis. C, FLAG-tagged full-length DR5. A C-terminal FLAG tag was added to the full-length DR5. D, analysis of point mutation in full-length DR5 molecule. Both wild-type and mutant FLAG-tagged DR5 constructs were introduced into HEK293T/17 cells by transient transfection. Cell lysates were probed by immunoblot analysis using AD5-10 or anti-FLAG mAb. All experiments were repeated at least three times with similar results. TM, transmembrane; w.t., wild type; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
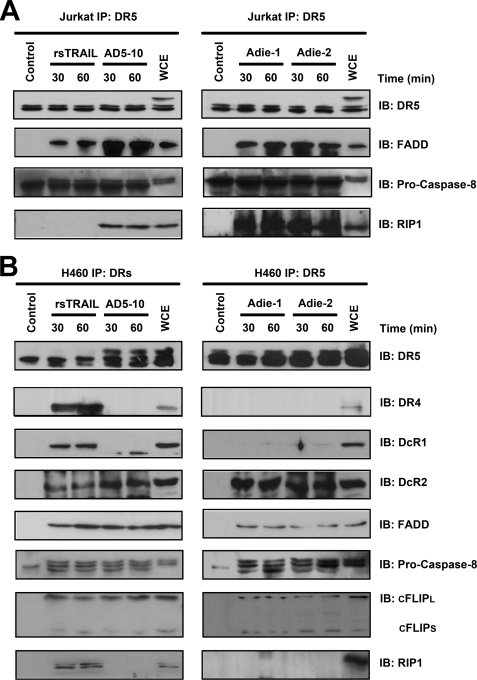
FIGURE 6.
Agonistic anti-DR5 mAbs and TRAIL induce DISC formation in Jurkat cells and H460 cells. A and B, analysis of TRAIL- and agonist anti-DR5 mAbs-induced DISC in Jurkat cells (A) and H460 cells (B). Cells were treated with anti-DR5 mAbs or “TRAIL-FLAG + anti-FLAG” for indicated time before cell lysis, although AD5-10 was added to the cells after cell lysis in unstimulated cells. The samples were pre-cleared with-Sepharose 4B (GE Healthcare) and immunoprecipitated (IP) overnight at 4 °C with protein G-Sepharose beads and analyzed by Western blotting. All experiments were repeated at least three times with similar results. IB, immunoblot; WCE, whole cell extract.
To further explore the exact binding site of AD5-10 in the whole DR5 molecule, a pCMV-FLAG plasmid encoding full-length DR5 fused with a FLAG tag at the C terminus was constructed, and a substitution of Leu6 with Ala was made in DR5. Wild-type and mutant DR5 were expressed in mammalian cells, immunoblotted using AD5-10, and probed with anti-FLAG antibody. As shown in Fig. 2C, both wild-type and mutant FLAG-tagged DR5 were detected, and only the wild-type molecule was recognized by AD5-10. These results reveal a crucial role for Leu6 in the antigen-antibody recognition.
Synthetic DR5 Epitope Eliminates the Cytotoxicity of AD5-10
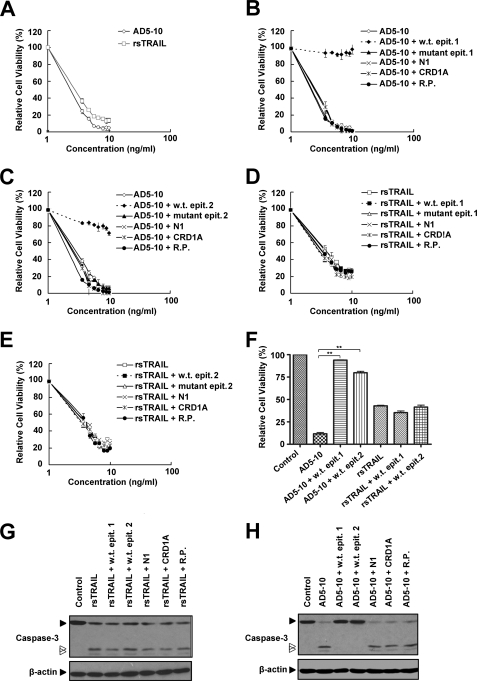
AD5-10 can kill a variety of tumor cells in vitro through binding with DR5. To further confirm the functionality of the epitope recognized by AD5-10, a series of peptides derived from the wild-type epitope was synthesized (Table 2). To increase water solubility of these peptides, Ala or AQ was added to the N terminus of 1ITQQDLAPQQRA12. The mutant peptides possessed identical sequences, except at position 6 where leucine was replaced with alanine. Earlier reports showed that an “50s loop” within the cysteine-rich domain 1 (CRD1) of DR5-ECD is crucial for TRAIL-DR5 interaction (26). An N1-cap (residues 26–39) contains a pre-ligand assembly domain that mediates death receptor-death receptor interactions (27). Thus, two peptides, N1 (26CPPGHHISEDGRDC39) and CRD1A (45GQDYSTHWNDLLFCLRC61), corresponded to the primary structure of the N1-cap, and CRD1A of DR5 were generated as negative controls. A random peptide was created to rule out nonspecific binding of AD5-10. These peptides were preincubated with AD5-10 or rsTRAIL, and the mixture was then added into DR5+ Jurkat T lymphoma cells, followed by a cytotoxicity neutralization assay. As shown in Fig. 3A, both AD5-10 and rsTRAIL triggered significant cell death in Jurkat T lymphoma cells. Wild-type epitopes 1 and 2 completely eliminated AD5-10-induced cell death, although the mutant epitopes failed to do so (Fig. 3, B, C and H). Neither the wild-type peptides nor the mutants of the epitope neutralized rsTRAIL-induced cell death in Jurkat cells (Fig. 3, D, E, and G). Moreover, none of the control peptides eradicated AD5-10 and rsTRAIL cytotoxicity, indicating that AD5-10 did not bind to other functional domains of DR5-ECD, and TRAIL failed to interact with the linear 50s loop. These data provide further evidence that AD5-10 functionally recognizes the epitope.
TABLE 2.
List of synthetic peptides used in cytotoxic neutralization assay
| Peptide | Sequence | Mr | Purity |
|---|---|---|---|
| % | |||
| Wild-type epitope 1 | AQITQQDLAPQQRA | 1566.81 | 83.9 |
| Mutant epitope 1 | AQITQQDAAPQQRA | 1524.76 | 98.2 |
| Wild-type epitope 2 | AITQQDLAPQQRA | 1438.75 | 95 |
| Mutant epitope 2 | AITQQDAAPQQRA | 1396.71 | 98.5 |
| N1 | CPPGHHISEDGRDC | 1521.61 | 99 |
| CRD1A | GQDYSTHWNDLLFCLRC | 2069.91 | 100 |
| Random peptide | SQRLHTPCFNKMEA | 1660.78 | 99.7 |
FIGURE 3.
Synthetic wild-type epitopes eliminate tumoricidal activity of AD5-10. A, Jurkat cells were treated with the indicated concentrations of AD5-10 or rsTRAIL for 8 h. AD5-10 (B and C) or rsTRAIL (D and E) was preincubated with 10 μm wild-type or mutant epitope or 10 μm control peptides (N1, CRD1A, and random peptide) at 37 °C for 2 h. Jurkat cells were then treated with these mixtures for 8 h. Both rsTRAIL and AD5-10 concentrations shown in the graphs are in logarithmic scale. F, Jurkat cells were treated with 100 ng/ml AD5-10 or 100 ng/ml rsTRAIL in the absence or presence of synthetic wild-type epitopes. G and H, Western blot analysis of caspase-3 activation in Jurkat cells treated with 100 ng/ml AD5-10 or 100 ng/ml rsTRAIL in the absence or presence of synthetic peptides for 2 h. Cell viability was determined using a CCK-8 assay. Values represent the mean ± S.D. of triplicate samples. **, p < 0.01 versus cells incubated with AD5-10 alone. w.t., wild-type; epit., epitope; R.P., random peptide.
Epitope Modulates a Signaling Pathway upon Stimulation with AD5-10
Most cancerous tissues and cells express high levels of DR5, whose overexpression always induces a caspase cascade and apoptosis. ErbB2 is a receptor tyrosine kinase belonging to the family of epidermal growth factor receptors, which is generally involved in cell differentiation, proliferation, and tumor growth (28). In an attempt to confirm the signaling capacity of the DR5 epitope in cell death, a series of chimeric receptor constructs and mutants was established. Der was a chimeric receptor containing the DR5 extracellular domain fused with the intracellular and transmembrane domains of ErbB2 (Her-2), whereas Der-L6A was the Der epitope mutant with L6A. Der-Δ was the mutant with deletion of the tyrosine kinase domain (Fig. 4A). These constructs were transduced into mouse NIH3T3 cells with null expression of ErbB2 (29) so that the endogenous ErbB2 expression could be avoided and the signaling capacity of the DR5 epitope could be explored. The expression of Der, Der-L6A, and Der-Δ on the cell surface was validated (Fig. 4B). Cells expressing these chimeric receptors were treated with AD5-10 and/or rsTRAIL, and the signaling pathway mediated by Der, Der-L6A, and Der-Δ was investigated. As shown in Fig. 4C, upon stimulation with AD5-10, ERK1/2 was significantly phosphorylated over a 10–90-min time course in cells expressing Der but not in cells expressing Der-L6A. Compared with AD5-10, rsTRAIL can activate ERK phosphorylation in both Der+ and Der-m+ cells (Fig. 4D). It indicated that AD5-10 activated the tyrosine phosphorylation of the chimeric receptor tyrosine kinase by binding to the epitope, whereas the N terminus of the DR5-ECD did not mediate the formation of TRAIL-DR5 complex. Similarly, after treatment with AD5-10, both ERK1/2 and MEK1/2 were markedly phosphorylated in cells expressing Der, but not in cells with Der-L6A or Der-Δ expression (Fig. 4, E and F). Although bound to AD5-10 and/or TRAIL, the deletion mutant Der-Δ failed to activate MAPK. In line with our previous findings, both AD5-10 and rsTRAIL can bind to wild-type DR5-ECD and show synergistic effect on MAPK activation (Fig. 4F). These data indicate that the epitope modulates the signaling pathway in cells stimulated with AD5-10.
Epitope Functions as a Novel Death-inducing Region
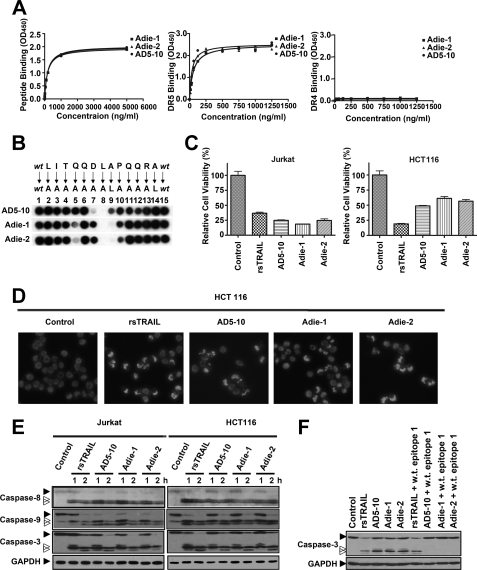
We investigated the biological function of the epitope in the death of tumor cells. The hapten-peptide “CLITQQDLAPQQRA” harboring the epitope core sequence was synthesized as an immunizing peptide. Keyhole limpet hemocyanin was then conjugated to the N-terminal cysteine of the synthetic peptide. The complex was subsequently used to immunize BALB/c mice to generate the monoclonal antibody against the epitope. Two monoclonal antibodies against the death-inducing epitope, Adie-1 and Adie-2, were established with specific high affinity (Kd = 1.153 ± 5.180 nm and Kd = 1.202 ± 6.463 nm, respectively), although not interacting with DR4. Compared with AD5-10, both Adie-1 and Adie-2 specifically bound to DR5-ECD as well (Fig. 5A). Moreover, Leu6 still played a key part in Adie-1/2 recognition pattern (Fig. 5B and supplemental Fig. S2). As shown in Fig. 5C, the viability of Jurkat lymphoma cells and HCT116 colon cancer cells treated with the two antibodies were dramatically decreased. These data suggested these antibodies possess an ideal cell death inducing activity, which was confirmed by Hoechst 33258 staining of HCT116 cells. Significant chromatin condensation and fragmentation were visualized by fluorescence microscope (Fig. 5D). Further Western blot analysis demonstrated that caspase-8, -9, and -3 were activated by the two antibodies in a time-dependent manner within 1 h in Jurkat and HCT-116 cells (Fig. 5E). In agreement with our hypothesis, synthetic wild-type epitope neutralized the tumoricidal activity of all anti-DR5 mAbs but not rsTRAIL (Fig. 5F). These data indicate that the two antibodies against the epitope exert tumoricidal activity and the NTR of DR5 contain a novel death-inducing domain.
FIGURE 5.
Antibodies against the epitope −1LITQQDLAPQQRA12 possess tumoricidal activities. A, binding affinity of Adie-1 and Adie-2 to the DR5 epitope recognized by AD5-10 (left), soluble DR5-ECD (middle), and soluble DR4-ECD (right). Synthetic immunizing peptide or recombinant soluble DR4/5 was immobilized on a 96-well plate. The plate was then incubated with the indicated concentration of AD5-10, Adie-1, and Adie-2. The binding affinities were then measured by enzyme-linked immunosorbent assay. Values represent the mean ± S.D. of triplicate samples. B, alanine-scanning mutagenesis analysis of the recognition pattern of anti-DR5 mAbs. An array of peptide −1LITQQDLAPQQRA12 was probed with 0.2 μg/ml AD5-10, Adie-1, or Adie-2, respectively. The first and the last spot corresponded to the parental peptide, whereas the other spots represented an Ala-substituted analogue of wild-type (wt) sequence as indicated above the individual spots. C, tumoricidal function of Adie-1 and Adie-2 compared with AD5-10. Jurkat or HCT116 cells were incubated with 500 ng/ml anti-DR5 mAbs or rsTRAIL for 8 h. Cell viability was determined by a CCK-8 assay. D, Hoechst 33258 staining. HCT116 cells were cultured with 500 ng/ml anti-DR5 mAbs or rsTRAIL for 4 h. After fixation, cells were stained with Hoechst 33258 (1 μg/ml), and nuclear condensation and fragmentation of chromatin were observed by fluorescence microscopy (20×). The excitation wavelength was 380 nm. E, Jurkat cells and HCT116 cells were incubated with 500 ng/ml anti-DR5 mAbs or rsTRAIL for the indicated times. The activation of caspase-8, -9, and -3 was tested using the respective antibodies. F, Western blot analysis of caspase-3 activation in Jurkat cells treated with 500 ng/ml anti-DR5 mAbs or rsTRAIL in the absence or presence of synthetic wild-type epitope for 2 h. All experiments were repeated at least three times with similar results. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
NTR-mediated DR5 Activation Initiates DISC Formation
To elucidate the molecular components involved in agonistic anti-DR5 mAbs signaling through DR5, we analyzed death-inducing signaling complex (DISC) associated with anti-DR5 mAbs-DR5 complex, compared with FLAG-tagged TRAIL treatment. Jurkat cells express only DR5, but not other death receptors, and can be killed by TRAIL through apoptosis (30, 31). Treatment of Jurkat T cells with TRAIL led to the rapid recruitment of FADD and pro-caspase-8 to the precipitated complexes (Fig. 6A), consistent with the previous results (32–34). Interestingly, agonistic anti-DR5 mAbs, AD5-10 and Adie-1/2, initiated DISC assembly containing DR5, FADD, caspase-8, and RIP1 (Fig. 6A and supplemental Fig. S3).
Non-small cell lung cancer cells, even though highly expressing DR5, demonstrated more sensitivity to TRAIL than to AD5-10 (18). Cleavage of c-FLIPL was observed in H460 treated with TRAIL, and inhibition of endogenous c-FLIPL expression significantly enhanced AD5-10-induced H460 cell death. All the above findings indicated the involvement of c-FLIP in DISC formation in H460 cells, which has been described everywhere (35). As shown in Fig. 6B, anti-DR5 mAbs DISC assembly in H460 cells involved the agonistic receptor DR5, decoy receptor DcR2, the adaptor protein FADD, caspase-8, and c-FLIP. The same was true for TRAIL DISC formation, except that in addition the endogenous DR4 and DcR1 coimmunoprecipitated with DR5 in a ligand-dependent manner. RIP1 was detected in TRAIL-induced DISC in H460 cells. These data reveal differential composition of DISC in the TRAIL signaling complex and the agonistic anti-DR5 mAbs signaling complex.
DISCUSSION
TRAIL attracts great research interest for its selective induction of apoptosis in malignant cells via its receptors, DR5 and DR4. Potential clinical applications of rsTRAIL or various agonistic DR5 monoclonal antibodies are being tested in various clinical stages (36–38).
Previous reports showed that there are two disulfide bridges within the mature DR5 extracellular region. These loops divide soluble DR5 into three modular compositions as follows: the N1-cap (residues 26–39), the cysteine-rich domain 1 (residues 42–82), and the cysteine-rich domain 2 (CRD2, residues 84–123) (39). The N-terminal cap contains a partial cysteine knot with a single noncanonical disulfide (40). Both cysteine-rich regions have “A + B” modules, which are composed of 12–17 and 21–24 amino acids, respectively. They are defined by the consensus sequences C1-X2GX(Y/F)XX4–9-C2 and C1-X2-C2-X3–6X5-C3-TX2–5NTV-C4 (here n in Xn is the number of intervening amino acids), respectively. The A module exhibits a single disulfide bond between C1 and C2 (C1:C2), although the B module has two disulfide bonds (C1:C3 and C2:C4). The N1 module has only one disulfide bond but is structurally related to the B module. Residues 26–32 of the N1 module, which is structurally homologous to the B module, are involved in tight backbone interactions with the A module in CRD1 of DR5 (39). Structural analysis of the TRAIL-DR5 complex suggests that the cysteine-rich regions, especially the 50s loop (residues 50–64) and the “90s loop” (residues 90–103), on the receptor have a conservative and functional role in mediating receptor function (26). It has been proposed that the molecular mechanism shared by agonistic anti-DR5 antibodies involves mimicking of the binding of TRAIL to these two sites, with subsequent receptor aggregation and apoptotic signaling cascades. This idea is supported by the evidence that binding sites for those agonists and the endogenous DR5 ligand TRAIL on DR5 are overlapping (41, 42) Other regions of the DR5 extracellular domain, however, may also have their own characteristics. For instance, the N-terminal pre-ligand assembly domain is located within the partial N1-cap, which contributes to the ligand-independent formation of receptor complexes and plays a crucial role in the function and signaling of death receptors (27). This leaves us to question whether there are any other undefined regions in DR5 that can function as death triggers.
We previously described a novel anti-human DR5 agonistic monoclonal antibody, AD5-10, which synergically complements the tumoricidal activity of rsTRAIL and yet shows undetectable cytotoxicity toward normal cells. Unlike other DR5 agonistic antibodies reported in the literature, where bivalency is required to trigger cell death, the monovalent scFv of AD5-10 alone induces cell death without cross-linking (43), raising the question of how AD5-10 activates DR5 and transmits the cell death signal.
In this study, we used a synthetic peptide array to identify a novel death-inducing epitope within the NTR of DR5. The NTR (position 1–23) preceding the N1-cap of DR5 used to be considered structurally flexible, and its function has not been clarified yet, because the first 23 residues are disordered and not expected to have regular secondary structure (26, 39). Interestingly, our experiments show that the linear and highly hydrophobic sequence “4QDLAP8” within this region is recognized by the agonistic antibody AD5-10. Fine mapping suggests that the Leu residue at position 6 (Leu6) is vital for the antibody recognition. Replacement of Leu6 with Ala in a synthetic peptide results in a complete loss of antibody binding activity.
In the chimeric receptor assay, the DR5 extracellular domain was linked to the MAPK signaling pathway. The activated chimeric receptor results in the downstream phosphorylation of ERK and MEK as long as the intracellular tyrosine kinase domain is kept intact. However, the mutant chimeric receptor lacking the hydrophobic profile at position 6 (Ala6) failed to bind to AD5-10 and prime the kinase activation of MAPK. However, the mutant chimeric receptor could still interact with TRAIL and be functional, given that the 50s and 90s loop existed. Therefore, we imagine that DR5 is simultaneously able to bind to AD5-10 and TRAIL. This might explain why there is a synergistic effect between TRAIL and AD5-10 with respect to death inducing activity.
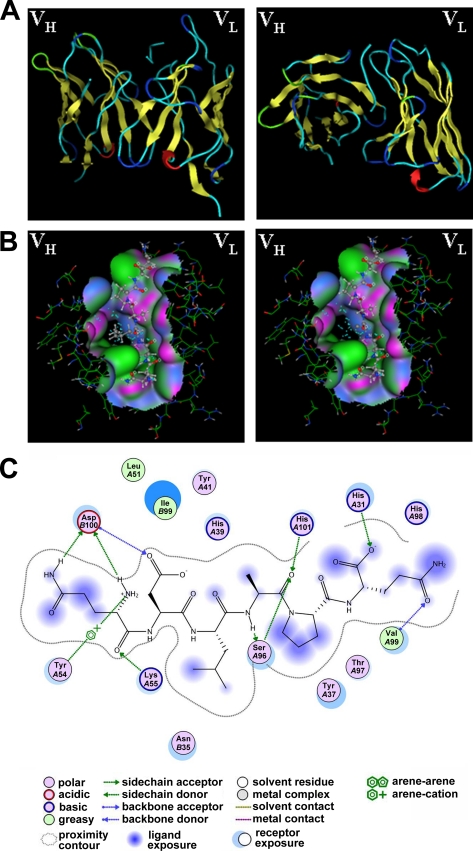
Because there is a lack of electron density in crystal structure determination, the first N-terminal 28 residues are structure-disordered and even have an irregular secondary structure (26, 39). Thus, it is difficult to include this confusing part when modeling the DR5 extracellular structure. Also, it is more complicated to estimate the overall conformational switch when DR5 binds to its agonistic antibody. We showed that a linear epitope within the complicated fraction is functional when recognized by AD5-10. Together with the findings of Shi et al. (43), the core epitope of DR5 and the scFv-like fragment (VL + VH) of AD5-10 can be used to model the epitope recognition pattern. By antibody homology modeling of AD5-10, we found that the complementary-determining regions (CDRs) of the light and heavy chain variable domain formed a hydrophobic binding pocket to accommodate the “4QDLAPQ9” appropriately (Fig. 7A). The core sequence of the determinant is also highly hydrophobic. Moreover, Leu6 primarily contributes to the hydrophobicity, because the side chain of Leu6 stretches deeply into the binding pocket. When Leu6 was replaced with Ala, the hydrophobic profile of the epitope was dramatically impaired, which may explain why the mutant epitope cannot be “accepted” by AD5-10 (Fig. 7B). Consistent with the results of the peptide array, the remarkable hydrophobic side chain composed of the Leu6 residue is required, because substitution of an isoleucine or phenylalanine residue partially restored the binding ability. After modeling the ligand-receptor complex, a prediction interaction map was drawn to predict the possible interactions in the active site. As shown in Fig. 7C, besides the dominant hydrophobic interaction, other actions such as electrostatic interactions could be involved in the stabilization of the “antibody-DR5” complex.
FIGURE 7.
Computational simulation of AD5-10 epitope recognition. A, overall structure of AD5-10-epitope complex, side view (left), and top view (right). B, comparison of the wild-type core epitope (QDLAP, left) and the mutant epitope (QDAAP, right) in the binding pocket of AD5-10. C, ligand interaction plot for core epitope bound to AD5-10. All the diagrams were generated using the molecular operating environment 2006.08 (Chemical Computing Group, Montreal) Ligand Interactions Module.
When bound to the TRAIL trimer complex, DR5 changes its conformation so as to initiate death signaling (44, 45). Like TRAIL and other agonistic anti-death receptor antibodies, AD5-10 binds to DR5 with great affinity, and the latter may curve to an ideal shape for subsequent signal transduction. Notably, several lines of evidence have shown that an oligomeric mAb-DR5 complex forms on the cell membrane at the initiative stage (41). We propose that the agonist antibody against the NTR of DR5 may induce DR5 clustering in a conformational way, permitting the optimal recruitment of DISC components or other distinct adaptor molecules.
DISC analysis showed that agnostic mAbs against NTR of DR5 triggers tumor cell death through recruitment of FADD and caspase-8 to the anti-DR5 mAbs-DR5 complex. One interesting finding was that in H460 cells, DcR2 was coimmunoprecipitated with DR5. It was in line with the recent findings indicating the spontaneous ligand-independent interaction of DcR2 with DR5 through the pre-ligand associated domain (27, 31). Another attractive finding was that in Jurkat cells, RIP1 participated in the AD5-10 signaling but not TRAIL signaling, whereas in H460 cells the result is quite the reverse. One possible explanation for this discrepancy could be that RIP1 is associated with the sensitivity of tumor cells to various treatments. Earlier findings indicate that RIP1 is a short lived anti-apoptotic protein that plays a pivotal role in TRAIL-induced JNK-NF-κB activation and protects cells against TRAIL-induced apoptosis (46, 47). Inhibition of RIP1 enhances TRAIL-induced apoptosis in pancreatic cancer cells (48). In contrast, recent data demonstrate that RIP1 is also required for necrotic death induced by tumor necrosis factor and TRAIL (49). Caspase inhibitor enhances the recruitment of RIP1 to the TRAIL DISC (50), and RIP1 can interact with RIP3 to initiate cellular necrotic response (51–53). Taken together, these results indicate the role of RIP1 is determined by the nature of the stimulus and certain cell type. In this study, Jurkat cells are more sensitive to AD5-10 than to TRAIL, whereas H460 cells perform in the opposite way (Fig. 3 and supplemental Fig. S4). RIP1 may have a potential role in modulating cell death type. This hypothesis, however, remains to be tested.
Although there is a range of agonistic DR5 monoclonal antibodies, the interactive patterns between each antibody and DR5 are shared, and the conformational determinants recognized by the antibodies are confined to the cysteine-rich region of DR5. It requires great effort to subsequently generate an identical antibody through special three-dimensional epitope immunization. Our study, on the one hand, might facilitate the generation of therapeutic antibodies against DR5. As we report here, tumoricidal anti-DR5 antibodies can be produced in a relatively easy manner because of the linear epitope. The N terminus of DR5 can function as a deadly trigger once activated by the specific agonistic antibody, which may offer an alternative therapeutic strategy in clinical medicine to artificially activate the DR5 receptor. On the other hand, one of the major limitations in the clinical application of mouse antibody-based therapies is the human anti-mouse antibody reaction, which makes long term use infeasible in patients with chronic progressive conditions, such as cancer. Identification of the exact epitope of AD5-10 provides critical information that could allow the development of therapeutic anti-DR5 antibodies by combining the unique features of the AD5-10 antibody (high tumoricidal activity and low cytotoxicity in normal cells) with humanization. For example, we are currently screening a humanized antibody library using a synthetic peptide containing the epitope information as a hapten. Furthermore, small molecular compounds targeting this region with high affinity and tumoricidal activity might be developed in the near future through high throughput screening approaches.
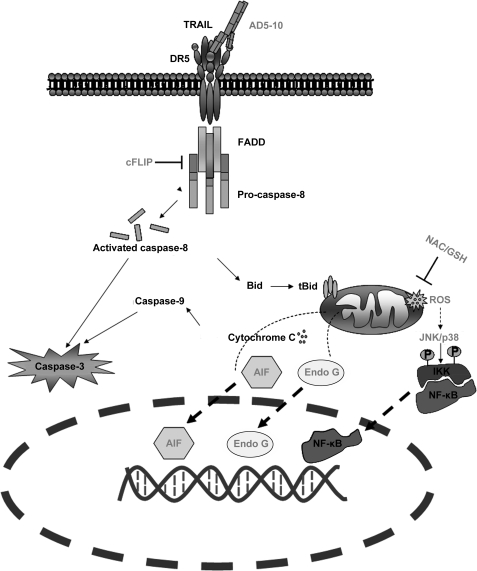
We previously reported that the signal transduction cascade induced by AD5-10 only partially overlaps with that stimulated by the rsTRAIL-receptor interaction. Although both TRAIL and AD5-10 are capable of activating NF-κB, there are differences between the regulation of NF-κB activity by TRAIL and that by AD5-10 in certain cell lines. Even the downstream active signaling in mitochondria differs in rsTRAIL- and AD5-10-treated Jurkat T leukemia cells. Significant ROS accumulation and secondary activation of JNK were observed in AD5-10-treated cells but not in rsTRAIL-treated cells (summarized in Fig. 8). Here, we demonstrated that AD5-10 binds to a core epitope sequence that is different from any reported sites. One possibility is that the binding of AD5-10 induces a specific conformational change in DR5 that is distinct from that induced by TRAIL binding, resulting in the activation of additional signal transduction pathways that contribute to tumor killing activity. Efforts to identify the downstream players after antibody recognition are actively being pursued. This may shed light on how DR5 transfers extracellular structural changes to its intracellular function and subsequently allows development of new strategies by targeting the novel site of DR5 for cancer immunotherapy.
FIGURE 8.
Differences between TRAIL- and AD5-10-induced signaling pathways. In the initiation stage of the caspase cascade, FADD and caspase-8 are recruited to DR5 (and possibly DR4) DISC. c-FLIP is involved in DISC formation in non-small cell lung cancer. Moreover, unlike TRAIL and other reported agonistic antibodies for DR5, AD5-10 particularly induces ROS accumulation and time-dependent reduction of GSH and GSSG content in the intrinsic mitochondrial pathway of Jurkat leukemia cells. ROS scavengers, antioxidant (N-acetyl-l-cysteine (NAC)) and GSH, suppress AD5-10-triggered cell death by inhibiting the release of apoptosis-inducing factor (AIF) and endonuclease G (EndoG), which contribute to the fragmentation of the nuclear DNA. Under ROS stress, JNK was triggered simultaneously and a subsequent secondary activation of JNK was observed; NF-κB was also activated during this time course.
Supplementary Material
This work was supported in part by Natural Science Foundation of China Grants 30623009, 30721063, 30772495, and 30972684 and State Key Basic Research Program of China Grants 2007CB507404 and 2006CB504200.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- rsTRAIL
- recombinant soluble TRAIL
- mAb
- monoclonal antibody
- DR5-ECD
- DR5 extracellular domain
- MAPK
- mitogen-activated protein kinase
- Der-L6A
- Der epitope mutant L6A
- Der-Δ
- Der-truncated without tyrosine kinase domain
- ERK
- extracellular signal-regulated kinase 1/2
- MEK
- mitogen-activated protein kinase (MAPK) kinase
- PBS
- phosphate-buffered saline
- HRP
- horseradish peroxidase
- FITC
- fluorescein isothiocyanate
- TRITC
- tetramethylrhodamine isothiocyanate
- NTR
- N-terminal region
- PE
- phycoerythrin
- JNK
- c-Jun N-terminal kinase
- OPAL
- oriented peptide array library
- ROS
- reactive oxygen species.
REFERENCES
- 1.Wiley S. R., Schooley K., Smolak P. J., Din W. S., Huang C. P., Nicholl J. K., Sutherland G. R., Smith T. D., Rauch C., Smith C. A., et al. (1995) Immunity 3, 673–682 [DOI] [PubMed] [Google Scholar]
- 2.Pan G., O'Rourke K., Chinnaiyan A. M., Gentz R., Ebner R., Ni J., Dixit V. M. (1997) Science 276, 111–113 [DOI] [PubMed] [Google Scholar]
- 3.Walczak H., Degli-Esposti M. A., Johnson R. S., Smolak P. J., Waugh J. Y., Boiani N., Timour M. S., Gerhart M. J., Schooley K. A., Smith C. A., Goodwin R. G., Rauch C. T. (1997) EMBO J. 16, 5386–5397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Falschlehner C., Emmerich C. H., Gerlach B., Walczak H. (2007) Int. J. Biochem. Cell Biol. 39, 1462–1475 [DOI] [PubMed] [Google Scholar]
- 5.Yagita H., Takeda K., Hayakawa Y., Smyth M. J., Okumura K. (2004) Cancer Sci. 95, 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degli-Esposti M. A., Smolak P. J., Walczak H., Waugh J., Huang C. P., DuBose R. F., Goodwin R. G., Smith C. A. (1997) J. Exp. Med. 186, 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pan G., Ni J., Wei Y. F., Yu G., Gentz R., Dixit V. M. (1997) Science 277, 815–818 [DOI] [PubMed] [Google Scholar]
- 8.Degli-Esposti M. A., Dougall W. C., Smolak P. J., Waugh J. Y., Smith C. A., Goodwin R. G. (1997) Immunity 7, 813–820 [DOI] [PubMed] [Google Scholar]
- 9.Miyashita T., Kawakami A., Nakashima T., Yamasaki S., Tamai M., Tanaka F., Kamachi M., Ida H., Migita K., Origuchi T., Nakao K., Eguchi K. (2004) Clin. Exp. Immunol. 137, 430–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashkenazi A. (2002) Nat. Rev. Cancer 2, 420–430 [DOI] [PubMed] [Google Scholar]
- 11.Chuntharapai A., Dodge K., Grimmer K., Schroeder K., Marsters S. A., Koeppen H., Ashkenazi A., Kim K. J. (2001) J. Immunol. 166, 4891–4898 [DOI] [PubMed] [Google Scholar]
- 12.Ichikawa K., Liu W., Zhao L., Wang Z., Liu D., Ohtsuka T., Zhang H., Mountz J. D., Koopman W. J., Kimberly R. P., Zhou T. (2001) Nat. Med. 7, 954–960 [DOI] [PubMed] [Google Scholar]
- 13.Marini P., Denzinger S., Schiller D., Kauder S., Welz S., Humphreys R., Daniel P. T., Jendrossek V., Budach W., Belka C. (2006) Oncogene 25, 5145–5154 [DOI] [PubMed] [Google Scholar]
- 14.Takeda K., Yamaguchi N., Akiba H., Kojima Y., Hayakawa Y., Tanner J. E., Sayers T. J., Seki N., Okumura K., Yagita H., Smyth M. J. (2004) J. Exp. Med. 199, 437–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yada A., Yazawa M., Ishida S., Yoshida H., Ichikawa K., Kurakata S., Fujiwara K. (2008) Ann. Oncol. 19, 1060–1067 [DOI] [PubMed] [Google Scholar]
- 16.Rowinsky E. K. (2005) J. Clin. Oncol. 23, 9394–9407 [DOI] [PubMed] [Google Scholar]
- 17.Guo Y., Chen C., Zheng Y., Zhang J., Tao X., Liu S., Zheng D., Liu Y. (2005) J. Biol. Chem. 280, 41940–41952 [DOI] [PubMed] [Google Scholar]
- 18.Chen F., Guo J., Zhang Y., Zhao Y., Zhou N., Liu S., Liu Y., Zheng D. (2009) Cancer Sci. 100, 940–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C., Liu Y., Zheng D. (2009) Cell Res. 19, 984–995 [DOI] [PubMed] [Google Scholar]
- 20.Han J., Hou W., Goldstein L. A., Lu C., Stolz D. B., Yin X. M., Rabinowich H. (2008) J. Biol. Chem. 283, 19665–19677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park K. J., Lee S. H., Kim T. I., Lee H. W., Lee C. H., Kim E. H., Jang J. Y., Choi K. S., Kwon M. H., Kim Y. S. (2007) Cancer Res. 67, 7327–7334 [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez M., Li S. S., Harper J. W., Songyang Z. (2004) J. Biol. Chem. 279, 8802–8807 [DOI] [PubMed] [Google Scholar]
- 23.Kim M. R., Lee J. Y., Park M. T., Chun Y. J., Jang Y. J., Kang C. M., Kim H. S., Cho C. K., Lee Y. S., Jeong H. Y., Lee S. J. (2001) FEBS Lett. 505, 179–184 [DOI] [PubMed] [Google Scholar]
- 24.Mayaan E., Moser A., MacKerell A. D., Jr., York D. M. (2007) J. Comput. Chem. 28, 495–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wagner K. W., Punnoose E. A., Januario T., Lawrence D. A., Pitti R. M., Lancaster K., Lee D., von Goetz M., Yee S. F., Totpal K., Huw L., Katta V., Cavet G., Hymowitz S. G., Amler L., Ashkenazi A. (2007) Nat. Med. 13, 1070–1077 [DOI] [PubMed] [Google Scholar]
- 26.Hymowitz S. G., Christinger H. W., Fuh G., Ultsch M., O'Connell M., Kelley R. F., Ashkenazi A., de Vos A. M. (1999) Mol. Cell 4, 563–571 [DOI] [PubMed] [Google Scholar]
- 27.Clancy L., Mruk K., Archer K., Woelfel M., Mongkolsapaya J., Screaton G., Lenardo M. J., Chan F. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18099–18104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoffmann I., Eugène E., Nassif X., Couraud P. O., Bourdoulous S. (2001) J. Cell Biol. 155, 133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuda S., Kadowaki Y., Ichino M., Akiyama T., Toyoshima K., Yamamoto T. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10803–10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sprick M. R., Weigand M. A., Rieser E., Rauch C. T., Juo P., Blenis J., Krammer P. H., Walczak H. (2000) Immunity 12, 599–609 [DOI] [PubMed] [Google Scholar]
- 31.Mérino D., Lalaoui N., Morizot A., Schneider P., Solary E., Micheau O. (2006) Mol. Cell. Biol. 26, 7046–7055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuang A. A., Diehl G. E., Zhang J., Winoto A. (2000) J. Biol. Chem. 275, 25065–25068 [DOI] [PubMed] [Google Scholar]
- 33.Bodmer J. L., Holler N., Reynard S., Vinciguerra P., Schneider P., Juo P., Blenis J., Tschopp J. (2000) Nat. Cell Biol. 2, 241–243 [DOI] [PubMed] [Google Scholar]
- 34.MacFarlane M., Kohlhaas S. L., Sutcliffe M. J., Dyer M. J., Cohen G. M. (2005) Cancer Res. 65, 11265–11270 [DOI] [PubMed] [Google Scholar]
- 35.Wang X., Chen W., Zeng W., Bai L., Tesfaigzi Y., Belinsky S. A., Lin Y. (2008) Mol. Cancer Ther. 7, 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hotte S. J., Hirte H. W., Chen E. X., Siu L. L., Le L. H., Corey A., Iacobucci A., MacLean M., Lo L., Fox N. L., Oza A. M. (2008) Clin. Cancer Res. 14, 3450–3455 [DOI] [PubMed] [Google Scholar]
- 37.Plummer R., Attard G., Pacey S., Li L., Razak A., Perrett R., Barrett M., Judson I., Kaye S., Fox N. L., Halpern W., Corey A., Calvert H., de Bono J. (2007) Clin. Cancer Res. 13, 6187–6194 [DOI] [PubMed] [Google Scholar]
- 38.Johnstone R. W., Frew A. J., Smyth M. J. (2008) Nat. Rev. Cancer 8, 782–798 [DOI] [PubMed] [Google Scholar]
- 39.Cha S. S., Sung B. J., Kim Y. A., Song Y. L., Kim H. J., Kim S., Lee M. S., Oh B. H. (2000) J. Biol. Chem. 275, 31171–31177 [DOI] [PubMed] [Google Scholar]
- 40.Mongkolsapaya J., Grimes J. M., Chen N., Xu X. N., Stuart D. I., Jones E. Y., Screaton G. R. (1999) Nat. Struct. Biol. 6, 1048–1053 [DOI] [PubMed] [Google Scholar]
- 41.Adams C., Totpal K., Lawrence D., Marsters S., Pitti R., Yee S., Ross S., Deforge L., Koeppen H., Sagolla M., Compaan D., Lowman H., Hymowitz S., Ashkenazi A. (2008) Cell Death Differ. 15, 751–761 [DOI] [PubMed] [Google Scholar]
- 42.Li B., Russell S. J., Compaan D. M., Totpal K., Marsters S. A., Ashkenazi A., Cochran A. G., Hymowitz S. G., Sidhu S. S. (2006) J. Mol. Biol. 361, 522–536 [DOI] [PubMed] [Google Scholar]
- 43.Shi J., Liu Y., Zheng Y., Guo Y., Zhang J., Cheung P. T., Xu R., Zheng D. (2006) Cancer Res. 66, 11946–11953 [DOI] [PubMed] [Google Scholar]
- 44.Yan N., Shi Y. (2005) Annu. Rev. Cell Dev. Biol. 21, 35–56 [DOI] [PubMed] [Google Scholar]
- 45.Wassenaar T. A., Quax W. J., Mark A. E. (2008) Proteins 70, 333–343 [DOI] [PubMed] [Google Scholar]
- 46.Lin Y., Devin A., Cook A., Keane M. M., Kelliher M., Lipkowitz S., Liu Z. G. (2000) Mol. Cell. Biol. 20, 6638–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mühlethaler-Mottet A., Bourloud K. B., Auderset K., Joseph J. M., Gross N. (2004) Oncogene 23, 5415–5425 [DOI] [PubMed] [Google Scholar]
- 48.Wang P., Zhang J., Bellail A., Jiang W., Hugh J., Kneteman N. M., Hao C. (2007) Cell. Signal. 19, 2237–2246 [DOI] [PubMed] [Google Scholar]
- 49.Holler N., Zaru R., Micheau O., Thome M., Attinger A., Valitutti S., Bodmer J. L., Schneider P., Seed B., Tschopp J. (2000) Nat. Immunol. 1, 489–495 [DOI] [PubMed] [Google Scholar]
- 50.Harper N., Farrow S. N., Kaptein A., Cohen G. M., MacFarlane M. (2001) J. Biol. Chem. 276, 34743–34752 [DOI] [PubMed] [Google Scholar]
- 51.He S., Wang L., Miao L., Wang T., Du F., Zhao L., Wang X. (2009) Cell 137, 1100–1111 [DOI] [PubMed] [Google Scholar]
- 52.Cho Y. S., Challa S., Moquin D., Genga R., Ray T. D., Guildford M., Chan F. K. (2009) Cell 137, 1112–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang D. W., Shao J., Lin J., Zhang N., Lu B. J., Lin S. C., Dong M. Q., Han J. (2009) Science 325, 332–336 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.