Abstract
The functions of two conserved residues, Phe135 and Pro136, located at the apex of the Cys loop of the nicotinic acetylcholine receptor are investigated. Both residues were substituted with natural and unnatural amino acids, focusing on the role of aromaticity at Phe135, backbone conformation at Pro136, side chain polarity and volume, and the specific interaction between the aromatic side chain and the proline. NMR spectroscopy studies of model peptides containing proline and unnatural proline analogues following a Phe show a consistent increase in the population of the cis conformer relative to peptides lacking the Phe. In the receptor, a strong interaction between the Phe and Pro residues is evident, as is a strong preference for aromaticity and hydrophobicity at the Phe site. A similar influence of hydrophobicity is observed at the proline site. In addition, across a simple homologous series of proline analogues, the results reveal a correlation between receptor function and cis bias at the proline backbone. This could suggest a significant role for the cis proline conformer at this site in receptor function.
Keywords: Peptides/Conformation, Receptors/7-Helix Ligand-gated Channels, Receptors/Structure-Function, Ion Channels, Nicotinic Acetylcholine Receptors, Cys Loop, Nicotinic Receptor, Proline, Unnatural Amino Acid Mutagenesis
Introduction
The Cys loop superfamily of neurotransmitter-gated ion channels includes the nicotinic acetylcholine receptor (nAChR),2 the 5-HT3 serotonin receptor, the GABAA and GABAC receptors, and the glycine receptor (1, 2). Together, these receptors mediate both excitatory and inhibitory fast synaptic transmission throughout the central and peripheral nervous systems. They are established targets for potential treatments of Alzheimer disease, Parkinson disease, schizophrenia, attention deficit hyperactivity disorder (ADHD), addiction, and other neurological disorders.
The eponymous Cys loop, a disulfide-linked sequence Cys-Xaa13-Cys, is located at the interface between the extracellular and transmembrane domains of the receptor, and many studies have established that the Cys loop is essential for receptor function. Not part of the agonist binding site, the Cys loop probably plays a key role in receptor gating, transmitting structural changes initiated by agonist binding to the ion channel region of the receptor (3–7).
The intervening residues of the Cys loop show considerable conservation across the family (Fig. 1). Specifically, a completely conserved Phe-Pro motif (followed by Phe or Met) lies at the apex of the Cys loop. (These are residues 135 and 136 in the α1 subunit of the muscle-type nAChR, which is the system studied here.) Proline residues are unique among the 20 natural amino acids in several ways. Of particular interest here is the much greater tendency of prolyl peptide bonds to exist in the cis conformation (8–12). The presence of the Phe in the Phe-Pro motif makes this possibility more enticing. It is well established that an aromatic amino acid N-terminal of a proline enhances the likelihood of a cis conformation, roughly doubling the contribution of the cis peptide in the conformational equilibrium (12). Indeed, previous studies of the analogous motif in the 5-HT3 receptor using conventional mutagenesis led to a postulation that the Pro was in a cis conformation (13).
FIGURE 1.
Sequence alignment of the Cys loop from various subunits of the Cys loop superfamily.
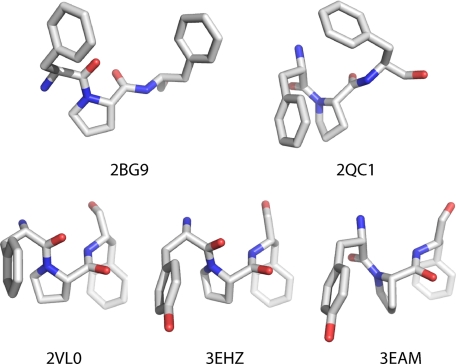
Currently available structural information related to Cys loop receptors adds to the intrigue (Fig. 2). (Note that the acetylcholine-binding protein, arguably the most valuable structural model for the extracellular domain, does not contain a Cys loop and does not contain the Phe-Pro sequence (14).) In the medium resolution electron microscopic structure of the Torpedo nAChR (Protein Data Bank code 2BG9), the proline of the α1 subunit is in the trans conformation, and there is clearly no structural interaction at all between the side chains of Phe135 and Pro136 (15). In contrast, in the high resolution x-ray crystal structure of the mouse muscle nAChR α1-subunit extracellular domain complexed to α-bungarotoxin (Protein Data Bank code 2QC1), the proline is in its cis form, and the Phe-Pro rings are stacked (16). (The Torpedo and mouse muscle receptors show very high sequence identity/similarity throughout their structures). Additionally, an NMR study of the isolated Cys loop of the nAChR found a roughly 1:1 mixture of cis and trans conformers, a ratio that can be modulated by glycosylation (17).
FIGURE 2.
Images of the Phe/Tyr-Pro-Phe unit from five different relevant structures.
In recent years, several pentameric prokaryotic channels that are clearly related to the Cys loop receptors have been discovered and crystallized. Although these bacterial channels lack the cysteines of the Cys loop, they all contain a Phe/Tyr-Pro motif, and x-ray crystal structures confirm that the loop is still clearly in place. In a structure of the ELIC bacterial channel, which is believed to be a closed state (Protein Data Bank code 2VL0), the proline is in the trans conformation, and the Phe-Pro side chains are stacked (18). Very recently, two structures of the GLIC bacterial channel have appeared, and both are thought to be an open state of the channel. Both structures contain a completely stacked Tyr-Pro motif, but in one (Protein Data Bank code 3EAM), the proline is cis (19), and in the other (3EHZ) the proline is trans (20).
Together, the structural data strongly indicate that (i) in the highly conserved Phe-Pro motif at the apex of the Cys loop of Cys loop receptors, both cis and trans conformations around the prolyl amide bond are viable, and (ii) an interaction between the Phe and Pro side chains is possibly involved in the conformational preference. Although it is true that the three-dimensional fold of a protein may influence the cis preference of any given residue, the intrinsic conformational bias of the residue itself can still be expected to play an important role in determining structure and thus function of the protein (12).
The feasibility of both cis and trans conformations at Pro136 presents the tantalizing opportunity that cis-trans isomerization of this conserved proline in the Cys loop, facilitated by the adjacent Phe, might be involved in the receptor gating mechanism. Such a cis-trans isomerization at a different proline has been shown to be essential to channel gating in the 5-HT3 receptor (5).
In the present work, we have used a variety of tools to probe the Phe-Pro motif of the muscle-type nAChR, including unnatural amino acid mutagenesis, electrophysiology, and NMR spectroscopy of model peptides. We find evidence for a strong interaction between the two residues and an important role for the aromatic nature of the Phe. At both sites, side-chain hydrophobicity is favorable to the receptor function. In addition, some preference for cis-biased residues at the Pro site is observed, but the involvement of cis-trans isomerization or the specific role of a cis conformer, if any, remains to be firmly established.
EXPERIMENTAL PROCEDURES
Synthesis of dCA-Amino Acids
The preparations of amino acids coupled to the dinucleotide (dCA) have been described previously (21) with the exception of dCA-Dhp and dCA-Mor. (S)-3-morpholinecarboxylic acid HCl was purchased from Tyger Scientific Inc. (Ewing, NJ), and 3,4-dehydro-l-proline (Dhp) was from Chem-Impex International Inc. (Wood Dale, IL). The amino groups were protected as the O-nitroveratryloxycarbonyl (NVOC) group. NVOC-Cl was purchased from Aldrich. (NVOC)-3,4-Dehydroproline cyanomethyl ester and (NVOC)-morpholine cyanomethyl ester were prepared according to the representative protocol reported in Ref. 22. Products were characterized by NMR spectroscopy. The NMR spectra, both 1H and 13C, are complicated because each compound shows two distinct conformations in the solution. (NVOC)-3,4-Dehydroproline cyanomethyl ester. 1H NMR (500 MHz, CDCl3) δ3.95–4.03 (m, 6H), 4.34–4.43 (m, 2H), 4.69–4.87 (m, 2H), 5.20–5.21 (m, 2H), 5.43–5.67 (m, 2H), 5.76–5.83 (m, 1H), 7.01 (s, 1H), 7.71 (m, 1H). 13C NMR (125 MHz, CDCl3) δ49.20, 49.23, 53.57, 54.20, 56.54, 56.57, 56.87, 64.66, 65.08, 65.92, 66.28, 108.34, 108.37, 110.07, 111.25, 113.96, 113.98, 123.46, 123.71, 127.44, 127.85, 130.37, 130.49, 139.88, 139.92, 148.32, 148.43, 153.39, 153.76, 153.81, 153.96, 168.41, 168.83. High resolution MS analysis (FAB+) calcd for C17H18N3O8 m/z = 392.1094, found 392.1109. (NVOC)-Morpholine cyanomethyl ester. 1H NMR (500 MHz, CDCl3) δ3.24–3.48 (m, 1H), 3.51 (dt, 1H), 3.69–3.75 (m, 1H), 3.83–3.95 (m, 2H), 3.94–3.95 (m, 3H), 3.99–4.02 (m, 3H), 4.33–4.41 (m, 1H), 4.63–4.85 (m, 3H), 5.41 (dd, 1H), 5.69 (dd, 1H), 6.88–6.97 (m, 1H), 7.66–7.70 (m, 1H). 13C NMR (125 MHz, CDCl3) δ41.22, 41.73, 49.39, 49.45, 54.53, 55.05, 56.53, 56.60, 56.81, 64.91, 65.31, 66.29, 66.73, 66.99, 67.34, 108.32, 108.36, 109.91, 111.13, 113.84, 113.89, 126.93, 127.66, 139.78, 140.11, 148.32, 148.53, 153.66, 153.81, 155.14, 156.04, 168.69, 168.89. High resolution MS analysis (FAB+) calcd for C17H20N3O9 m/z = 410.1199, found 410.1180.
Dhp and Mor cyanomethyl esters were coupled to dCA following the protocol in Ref. 22. dCA-Dhp. ES-MS calcd for C34H40N10O20P2 m/z 970.2; found (M − H)− m/z 969.0, (M + Na − 2H)− m/z 991.1, and (M + Na − H)− m/z 992.0. dCA-Mor. ES-MS calcd for C34H42N10O21P2 m/z 988.2; found (M − H)− m/z 987.0, (M + Na − 2H)− m/z 1009.0, and (M + Na − H)− m/z 1010.0.
Molecular Biology
Subunits of embryonic mouse muscle nAChR were in pAMV vectors. The α subunit contains the hemagglutinin epitope tag in the M3-M4 loop. There is no significant shift in EC50 caused by the insertion of the hemagglutinin epitope tag at this location. Site-directed mutagenesis was performed using the Stratagene QuikChange protocol. For single unnatural amino acid incorporation, the site of interest was mutated to an amber stop codon. For double unnatural acid incorporation, the 135 site was mutated to the opal stop codon, and the 136 site was mutated to the amber stop codon. Circular cDNA was linearized with NotI or KpnI. After purification (Qiagen), linearized DNA was used as a template for run-off in vitro transcription using the T7 mMessage mMachine kit (Ambion). The resulting mRNA was purified (RNAeasy minikit, Qiagen) and quantified by UV-visible spectroscopy.
THG73 (23) and TQOpS′ (24, 25) were used as amber suppressor tRNA and opal suppressor tRNA, respectively. Conjugated dCA-amino acid was ligated to 74-nucleotide tRNA as previously reported (21). Crude tRNA-amino acid product was used without desalting, and the product was confirmed by matrix-assisted laser desorption ionization time-of-flight MS on a 3-hydroxypicolinic acid matrix. Deprotection of the NVOC group on tRNA-amino acid was carried out by 5-min photolysis immediately prior to injection.
Microinjection
Stage V-VI Xenopus laevis oocytes were employed. For wild-type receptor and receptors containing conventional mutations, quantified mRNAs of all subunits were mixed in a ratio of α/β/γ/δ = 2:1:1:1 by mass. If an unnatural amino acid was to be incorporated into the α subunit, the mRNA stoichiometry was α/β/γ/δ = 10:1:1:1 by mass. Total amount of injected mRNA was 0.5–5 ng/cell for the wild type, 5–50 ng/cell for conventional mutations, and 25–125 ng/cell for suppression mutations. More mRNA was used in the double mutation experiments and with some mutations that gave abnormally low expression level. Equal volumes of the mRNA mixture and unprotected tRNA-amino acid were mixed prior to injection. Approximately 15 ng of tRNA/cell was used in the single suppression experiments, and 50 ng was used in the double suppression experiments. Each oocyte was injected with 50 nl of RNA solution, and cells were incubated for 18–72 h at 18 °C in culture medium (ND96+ with 5% horse serum). In the case of low expressing mutant receptors, a second injection was required. As a negative control for all suppression experiments, 76-nucleotide tRNA (dCA ligated to 74-nucleotide tRNA) was co-injected with mRNA in the same manner as fully charged tRNA.
Electrophysiology
Acetylcholine chloride and succinylcholine dihydrate were purchased from Sigma. Drug dilutions were prepared from 1 m stock solutions in the calcium-free ND96 buffer.
Ion channel function in oocytes was assayed by current recording in two-electrode voltage clamp mode using the OpusXpress 6000A (Axon Instruments). For dose-response experiments, 1 ml of each drug solution was applied to the cells, and between 12 and 16 concentrations of drug were used. Oocytes were clamped at −60 mV. Cells were perfused in calcium-free ND96 solution at flow rates of 1 ml/min before agonist application, 4 ml/min during agonist application, and 3 ml/min during wash. Drug application was 15 s in duration. Data were sampled at 125 Hz and filtered at 50 Hz.
Data Analysis
All dose-response data were obtained from at least five cells and at least two batches of oocytes. Data were normalized (Imax = 1) and averaged. EC50 and Hill coefficient (nH) were determined by fitting averaged, normalized dose-response relations to the Hill equation. Dose responses of individual oocytes were also examined and used to determine outliers. Individual dose-response data with nH > 2 or nH < 1 were discarded.
The coupling parameter (Ω) between any two mutations at residues 135 and 136 was calculated from Equation 1,
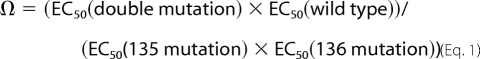 |
Side chain logP values were obtained using the ChemDraw program (CambridgeSoft Corp.).
Synthesis of Fmoc-protected Amino Acid
Fmoc-Cl was purchased from Fluka. (S)-3-Morpholinecarboxylic acid HCl was purchased from Tyger Scientific Inc. (Ewing, NJ), (2S,3S)-3-methylpyrrolidine-2-carboxylic acid (3-Me-Pro) was from Acros Organics USA (Morris Plains, NJ), α-methyl-l-proline (2-Me-Pro) was from Fluka, and Dhp was from Chem-Impex International Inc. (Wood Dale, IL). The amino acids were coupled to the Fmoc protecting group using the following protocol.
l-Amino acid (0.06 mmol) was dissolved in 10% Na2CO3 in water (2 ml), resulting in a solution with pH ∼9. To this solution was added Fmoc-Cl (1.5 eq) in dioxane (2 ml) at room temperature. Diisopropylethylamine was added dropwise while the reaction was stirred. Typically, the reaction was complete within 6 h. The reaction mixture was diluted by the addition of brine (20 ml). This was extracted with ether (5 ml) five times. The aqueous layer was acidified with 6 n HCl to pH ∼1 (solution became cloudy) and extracted with ether (5 ml) three times or until the organic layer was clear. The combined organic layers were dried over Na2SO4, and the solvent was removed under reduced pressure. Crude product was dried under vacuum overnight and was used in the next step (solid-phase peptide synthesis) without further purification. N-Fmoc-2-methyl-proline. 1H NMR (500 MHz, CDCl3) δ1.26–1.62 (m, 3H), 1.75–1.98, (m, 3H), 2.15–2.42 (m, 1H), 3.51–3.63 (m, 2H), 4.13–4.56 (m, 3H), 7.27–7.41 (m, 4H), 7.55–7.61 (m, 2H), 7.70–7.77 (m, 2H). 13C NMR (125 MHz, CDCl3) δ22.31, 22.72, 22.83, 23.42, 39.23, 41.11, 47.48, 47.58, 48.28, 48.91, 65.00, 66.28, 67.25, 67.65, 120.12, 120.20, 124.92, 124.95, 125.30, 125.35,127.27, 127.29, 127.30, 127.74, 127.78, 127.93, 141.54, 141.56, 141.60, 141.62, 144.05, 144.17, 144.21, 144.43, 154.86, 155.35, 178.42, 179.54. High resolution MS analysis (FAB+) calcd for C21H22NO4 m/z = 352.1549, found 352.1534. N-Fmoc-3-methyl-proline. 1H NMR (500 MHz, CDCl3) δ1.17–1.28 (m, 3H), 1.49–1.63 (m, 1H), 2.01–2.15 (m, 1H), 2.40–2.49 (m, 1H), 3.50–3.68 (m, 2H), 3.85–3.97 (m, 1H), 4.12–4.28 (m, 1H), 4.33–4.46 (m, 2H), 7.27–7.41 (m, 4H), 7.53–7.62 (m, 2H), 7.69–7.77 (m, 2H). 13C NMR (125 MHz, CDCl3) δ18.65, 18.91, 31.58, 32.50, 38.24, 39.70, 45.92, 46.29, 47.27, 47.30, 65.59, 66.07, 67.72, 67.77, 119.94, 119.97, 120.04, 125.05, 125.11, 125.16, 125.24, 127.10, 127.13, 127.15, 127.68, 127.78, 127.79, 141.27, 141.34, 141.36, 141.39, 143.80, 143.84, 144.07, 144.12, 154.75, 155.46, 176.79, 177.69. ESI MS on an LCQ ion trap mass spectrometer (positive ion mode) calcd for C21H21NO4 m/z = 351.1, found 351.9. N-Fmoc-morpholine. 1H NMR (500 MHz, CDCl3) δ3.04–3.92 (m, 6H), 4.20–4.68 (m, 5H), 7.27–7.34 (m, 2H), 7.36–7.42 (m, 2H), 7.48–7.60 (m, 2H), 7.71–7.77 (m, 2H). 13C NMR (125 MHz, CDCl3) δ41.16, 41.73, 47.27, 54.45, 54.84, 66.37, 66.75, 67.32, 67.72, 67.77, 68.12, 120.11, 120.14, 124.79, 124.89, 125.13, 127.19, 127.25, 127.27, 127.86, 127.90, 141.37, 141.42, 141.45, 141.48, 143.78, 143.82, 143.94, 155.91, 156.58, 174.83, 175.02. ESI MS on an LCQ ion trap mass spectrometer (positive ion mode) calcd for C20H19NO5Na m/z = 376.1, found 376.3.
Solid-phase Peptide Synthesis
All peptides were synthesized by solid-phase methods from Fmoc-protected amino acids using HBTU (Fluka) as a coupling reagent. Fmoc-l-proline was purchased from Sigma, Fmoc-l-pipercolic acid was from Peptech Corp. (Burlington, MA), Fmoc-l-azetidine-2-carboxylic acid was from Fluka, Fmoc-cis-2-fluoro-l-proline was from AnaSpec Inc. (San Jose, CA), N-Fmoc-glycine was from Aldrich, and Fmoc-l-phenylalanine was from Sigma. All chemicals were used as purchased without purification.
PAL resin (Sigma; estimated 0.4–0.8 mmol/g loading, 1% cross-linked with divinylbenzene, 100–200 mesh) was used to afford C-terminal primary amides. For conventional amino acids, couplings were performed with 3 eq of Fmoc amino acid, 3 eq of HBTU, and 6 eq of diisopropylethylamine. For unnatural amino acids, couplings were performed with 2 eq of Fmoc amino acid, 2 eq of HBTU, and 4 eq of diisopropylethylamine. The reaction time for each coupling step was 1–2 h. A Kaiser test was performed to monitor the progress of the reaction. After each coupling step, unreacted free amine was acetylated (5% acetic anhydride and 5% pyridine and 90% DMF) for 8 min, followed by deprotection of Fmoc-protected amine groups (20% piperidine/DMF, 15 min). In the last step, after Fmoc deprotection, the peptides were acetylated at the N termini on the resin using a solution of 5% pyridine, 5% acetic anhydride, and 90% DMF. Peptides were cleaved from the resin by treatment with trifluoroacetic acid and water (95:5) for 2 h. After filtration to collect the filtrate, solvents were removed as much as possible under reduced pressure. Following the addition of 5% acetic acid solution, this solution was lyophilized to dryness. The peptides were purified by preparative scale reversed phase high pressure liquid chromatography with gradient elution using an A-B gradient (buffer A, 0.05% trifluoroacetic acid in water; buffer B, 20% water and 0.05% trifluoroacetic acid in acetonitrile) and a flow rate of 15 ml/min. Peptide identity was characterized by ESI MS on an LCQ ion trap mass spectrometer (positive ion mode). GFProG (M + Na)+ expected 440.2, observed 440.3. GF(2-Me-Pro)G (M + Na)+ expected 454.2, observed 454.3. GF(3-Me-Pro)G (M + Na)+ expected 454.2, observed 454.4. GFflpG (M + Na)+ expected 458.2, observed 458.4. GFAzeG (M + Na)+ expected 426.2, observed 426.2. GFPipG (M + Na)+ expected 454.2, observed 454.3. GFMorG (M + Na)+ expected 456.2, observed 456.2. Note that the synthesis of Gly-Phe-Dhp-Gly peptide did not give the desired product in the first trial, and no further attempt has been made to obtain the product.
NMR Spectroscopy of Model Peptides
The peptide samples were dissolved in 5 mm phosphate buffer with 25 mm NaCl in 90% H2O, 10% D2O at pH 5. Samples for NMR experiments were between 2 and 5 mm. NMR spectra were acquired on a Varian 600-MHz spectrometer, and the temperature was set to 298 K. The water signal was suppressed by presaturation. Sequential assignments were achieved using gradient-selected correlated spectroscopy (gCOSY) and total correlation spectroscopy (TOCSY) experiments. Spectra were all internally referenced to 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid sodium salt (∼200 μm final concentration) at 0.0 ppm. The fraction of cis conformer was determined by integrating well resolved peaks in the one-dimensional 1H NMR spectra after base-line correction. NMR data were processed using the MestReNova software version 5.1.0 (Mestrelab Research S.L.).
RESULTS
Mutational Studies at Pro136
A previous study of the muscle-type nAChR in HEK293 cells showed that P136G mutations in the β and γ subunits prevented receptor assembly, whereas analogous mutations in the α or δ subunits prevented trafficking of receptors to the cell surface (26). Similarly, in previous studies of the analogous proline in the homopentameric 5-HT3 receptor, the P136A mutant revealed no surface expression in HEK293 cells (13). In the more permissive Xenopus oocyte expression system, the muscle-type nAChR containing the αP136A mutation produces <10% of the current levels seen for wild type. Surprisingly, this mutant receptor has an ACh EC50 value similar to that of the wild type. As discussed below, this result can be interpreted in several different ways; we therefore anticipated that the more subtle mutations enabled by unnatural amino acid mutagenesis would provide a more revealing analysis of the role of this residue.
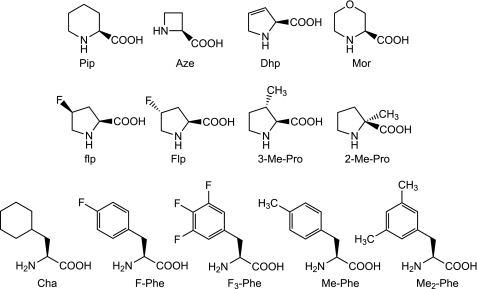
Several unnatural analogues of proline (Fig. 3) (5) were incorporated into the receptor using the in vivo nonsense suppression method. These unnatural proline analogues have varying ring size, side chain substitution, and intrinsic preferences for the cis conformer when probed in model systems (Table 1). The wild-type rescue experiment (i.e. incorporating Pro by nonsense suppression) displays the full phenotype of the wild-type receptor, including ACh EC50 value, Hill coefficient, and current traces. This indicates that the nonsense suppression methodology is viable at the 136 site. Interestingly, Pro analogues at position 136 are generally gain-of-function (lower EC50), the sole exception being 2-Me-Pro, which gives essentially wild-type behavior. Similar to the Ala mutation mentioned above, the current levels from experiments involving 2-Me-Pro are <10% of those seen in comparable experiments with other mutations. Despite the relative subtlety of the mutations, the gain-of-function effects can be substantial, as seen with Pip and 3-Me-Pro, which show 13- and 22-fold decreases in EC50, respectively, relative to wild type.
FIGURE 3.
Structures of unnatural amino acids used in this study.
TABLE 1.
EC50 and Hill constant values of mutant receptors containing unnatural amino acid at α136
| αPro136 | Reported percentage cisa | ACh EC50 | EC50(mut)/EC50(wild type) | Hill constant | n |
|---|---|---|---|---|---|
| % | μm | ||||
| Pro | 5 | 23 ± 0.2 | 1 | 1.5 ± 0.02 | 35 |
| Prob | 5 | 22 ± 0.2 | 1 | 1.6 ± 0.03 | 8 |
| Pip | 12 | 1.8 ± 0.1 | 0.1 | 1.7 ± 0.08 | 7 |
| Aze | 18 | 5.8 ± 0.2 | 0.3 | 1.7 ± 0.07 | 10 |
| Flp | ∼5 | 12 ± 0.2 | 0.5 | 1.5 ± 0.04 | 6 |
| Flp | ∼5 | 10 ± 0.2 | 0.5 | 1.7 ± 0.04 | 7 |
| 3-Me-Pro | ∼5 | 1.0 ± 0.02 | 0.04 | 1.7 ± 0.04 | 7 |
| 2-Me-Pro | 0 | 25 ± 0.7 | 1 | 1.6 ± 0.06 | 10 |
| Dhp | NRc | 18 ± 0.3 | 0.8 | 1.6 ± 0.03 | 10 |
| Mor | NRc | 8.7 ± 0.4 | 0.4 | 1.7 ± 0.05 | 9 |
Correlation between the cis-trans energy gap and the energy of channel activation could be expected if the receptor gating mechanism involves cis-trans isomerization of Pro136. However, no simple correlation is found (Table 1). For example, although both Pip and Aze show a stronger inherent cis preference than Pro and a lower EC50, 3-Me-Pro shows a conformational bias very similar to that of Pro but a greatly diminished EC50. Before analyzing these results in greater detail, however, we must consider the role of Phe135.
Mutational Studies at Phe135
Previous single channel studies have shown that the F135A mutation in the nAChR alters the gating mechanism, leading to two uncoupled open states that produce independent gating reactions from the diliganded closed state (27). In our studies of the nAChR, we found that the F135A mutation nearly obliterates receptor function; only very weak ACh-induced currents are observed despite normal surface expression levels (supplemental Fig. 1).
Seeking a more insightful analysis of the role of this residue, we probed the Phe135 site with an extensive series of Phe analogues. Again, the wild type rescue experiment displays the full characteristics of the wild-type receptor. The Phe135 site is sensitive to even very subtle mutations, as shown in Table 2. Similar to what is observed with Pro136, Phe analogues consistently produce gain-of-function mutants. ACh sensitivity increases with the volume and number of hydrophobic substituents on the aromatic ring. For example, Me-Phe has a lower EC50 than F-Phe, and Me2-Phe has a lower EC50 than Me-Phe. Surprisingly, cyclohexylalanine (Cha), which is similar to Phe in size and shape but is not aromatic (28), produces functional receptors with a small perturbation; EC50 is near the wild-type value. Given that the F135Cha mutant receptor is functional, aromaticity at position 135 is not an absolute requirement for the receptor to function.
TABLE 2.
EC50 and Hill constant values of mutant receptors containing unnatural amino acid at α135
| αPhe135 | ACh |
SuCh |
Efficacya | ||||
|---|---|---|---|---|---|---|---|
| EC50 | Hill constant | n | EC50 | Hill constant | n | ||
| μm | μm | ||||||
| Phe | 23 ± 0.2 | 1.5 ± 0.02 | 35 | 59 ± 1 | 1.3 ± 0.03 | 13 | 0.14 ± 0.01 |
| Pheb | 23 ± 0.4 | 1.5 ± 0.03 | 8 | NAc | NA | NA | NA |
| F-Phe | 2.6 ± 0.03 | 1.6 ± 0.02 | 7 | 32 ± 0.8 | 1.4 ± 0.04 | 9 | 0.54 ± 0.02 |
| F3-Phe | 1.0 ± 0.02 | 1.5 ± 0.05 | 15 | 8.1 ± 0.2 | 1.6 ± 0.05 | 8 | 0.86 ± 0.02 |
| Me-Phe | 1.0 ± 0.02 | 1.6 ± 0.04 | 12 | 11 ± 0.2 | 1.5 ± 0.04 | 11 | 0.82 ± 0.02 |
| Me2-Phe | 0.22 ± 0.01 | 1.6 ± 0.07 | 9 | 1.6 ± 0.06 | 1.5 ± 0.07 | 7 | 0.93 ± 0.04 |
| Cha | 16 ± 0.2 | 1.6 ± 0.02 | 15 | 60 ± 1 | 1.6 ± 0.03 | 9 | 0.10 ± 0.01 |
a Determined by the average of Imax(SuCh)/Imax(ACh).
b Data obtained by suppression mutation.
c NA, data not available.
To further explore the possible role of Phe135 in receptor gating, wild-type and mutant receptors were probed with the partial agonist succinylcholine (SuCh). Compared with ACh, SuCh produces only 14% of the maximal current under saturating drug concentrations in the wild-type receptor (Table 2). This indicates that upon receptor activation by SuCh, the channel open-closed equilibrium is shifted toward the open state, but to a much lesser extent relative to ACh activation. If a mutation produces a gain-of-function effect as a result of enhanced receptor gating, one could expect the mutation to improve the efficacy of a partial agonist like SuCh.
The EC50 trend of SuCh (Table 2) parallels that of ACh, implying that the mutants respond to both drugs in the same way. As anticipated, all of the Phe analogues that show a lowered EC50 do increase the relative efficacy of SuCh with respect to ACh. This suggests that mutations at position 135 primarily affect receptor gating. Note that the non-aromatic analogue Cha shows essentially wild-type EC50 for both ACh and SuCh and that this mutation has no strong effect on the relative efficacy.
Interaction between Phe135 and Pro136
There is considerable evidence supporting a specific interaction in a Phe-Pro sequence that stabilizes the cis form of the Pro. This could possibly involve a polar-π interaction in which polarized C–H bonds (Cδ−–Hδ+) on the proline interact favorably with the negative electrostatic potential on the face of the Phe side chain stacked on the Pro (9, 11). We investigated the possibility of a Phe-Pro interaction in this system by testing double mutant receptors in which Phe135 was substituted with the non-aromatic Cha and Pro136 was substituted with either Pip or 3-Me-Pro, the two mutations that cause the largest EC50 shifts. These experiments required consecutive incorporation of two different unnatural amino acids, an unprecedented experiment for receptors expressed in a living cell that was made possible by recent advances in tRNA design (24, 25). The resulting current signals (1–4 μA) were quite sufficient for quantitative analysis.
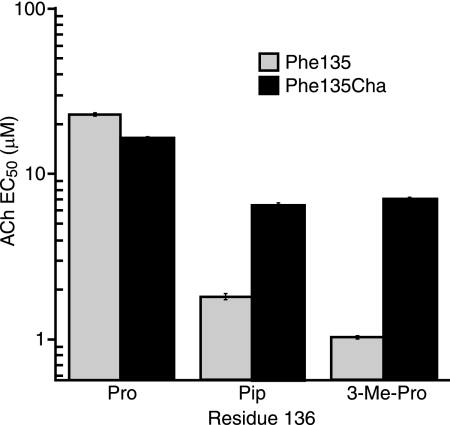
The F135Cha mutation substantially diminishes the large effects of the mutations at Pro136. As shown in Fig. 4, the 13- and 22-fold drops in EC50 for Pip and 3-Me-Pro, respectively, seen in a wild-type Phe background fall to ∼2.5-fold in the presence of F135Cha. A standard evaluation of double mutants employs a mutant cycle analysis, which has been used successfully with EC50 values for Cys loop receptors in several instances (29–32). For the interaction of F135Cha with P136Pip and P136(3-Me-Pro), we find coupling parameters (Ω) of 5 and 10, respectively, which correspond to coupling energies (RTln(Ω)) of 1.0 and 1.3 kcal/mol, respectively. These energies are significant for such subtle mutations and are indicative of a strong interaction between these two residues.
FIGURE 4.
ACh EC50 results from single and double mutation experiments at residues 135 and 136 in comparison with the wild-type value. For F135Cha/P136Pip, EC50 = 6.5 ± 0.2 μm, Hill constant = 1.7 ± 0.07, and n = 9. For F135Cha/P135(3-Me-Pro), EC50 = 6.9 ± 0.2 μm, Hill constant = 1.5 ± 0.06, and n = 12.
Having established a strong interaction between Phe135 and Pro136, we considered whether the intrinsic cis-trans equilibrium reported for proline and the proline analogues would be altered because of the preceding Phe. This would indicate that the percentage cis values used previously (5) and reported in Table 1 may not be appropriate for the present system because no aromatic amino acid was involved. As such, we set out to determine percentage cis values that are more appropriate to the Phe-Pro motif.
Determination of Inherent cis Preferences of Model Peptides Containing Proline Analogues Preceded by Phe
In order to determine whether the data in Table 1 reflect the innate cis preference of residue Pro136, it is necessary to measure the cis-trans energy gap (ΔG(c-t)) for each unnatural amino acid substituted at this site, taking into account the aromatic-proline interaction. In fact, ΔG(c-t) for the Gly-Phe-Pro-Gly and Gly-Phe-Pip-Gly peptides have been reported (33). Using a similar solution NMR technique, it should be possible to determine ΔG(c-t) values for our series of unnatural analogues of proline following a Phe residue in model peptides.
Model peptides Gly-Phe-XPro-Gly, where XPro represents Pro, Pip, Aze, flp, Mor, 3-Me-Pro, and 2-Me-Pro, were synthesized via standard solid-phase peptide synthesis methods. These peptides were then subjected to solution NMR experiments similar to those in Ref. 33. Protons were assigned by two-dimensional gCOSY and/or TOCSY experiments. The proportion of each of the two conformers in solution was measured by integration of a corresponding, well resolved peak after base-line correction. (Representative sample spectra are shown in supplemental Fig. 2). Conformational assignments were based on known chemical shifts of the Gly-Phe-Pip-Gly peptide reported in Ref. 33.
The results from the solution NMR experiments (Table 3) show that the cis preferences are indeed higher than the reported values in model peptides lacking the aromatic residue (Table 1). Note that for the Gly-Phe-(2-Me-Pro)-Gly peptide, the cis form was not observed. The model peptide containing Mor has a very high cis propensity; nearly 50% of the peptide is in the cis form. Moreover, one of the protons attached to the Cβ of the Mor ring displays a large upfield shift in the cis peptide compared with that of the trans peptide (supplemental Table 1), as has also been reported with the structurally similar Pip (33). In the Pip-containing peptide, the chemical shifts of Cβ protons are 1.72 and 2.15 ppm in the trans conformation and 0.35 and 1.90 ppm in the cis conformation. Likewise, for the Mor-containing peptide, the chemical shifts change from 3.74 and 4.37 ppm in the trans conformation to 2.10 and 3.74 ppm in the cis conformation. Most importantly, the Phe residue can alter the trends in cis-trans preferences, as shown for the simple homologous series Aze, Pro, Pip (Table 1 compared with Table 3).
TABLE 3.
ΔΔG(c-t) calculated from the percentage of cis results of solution NMR experiments for each amino acid and the ΔΔG(EC50) calculated from electrophysiology results of mutant receptors containing the corresponding amino acid at α136
ΔΔG(c-t) = RTln(% cis(Pro analogue)/% cis(Pro)). ΔΔG(EC50) = −RTln(EC50(Pro analogue)/EC50(Pro)).
| XPro | Percentage cis | ΔΔG(c-t) | ΔΔG(EC50) |
|---|---|---|---|
| % | kcal/mol | kcal/mol | |
| Pro | 17 | 0 | 0 |
| Pip | 39 | 0.65 | 1.5 |
| Aze | 30 | 0.42 | 0.81 |
| Flp | 32 | 0.49 | 0.38 |
| 3-Me-Pro | 12 | −0.24 | 1.8 |
| 2-Me-Pro | 0 | −0.056 | |
| Mor | 48 | 0.87 | 0.57 |
DISCUSSION
Cys loop neurotransmitter-gated ion channels are remarkable molecular machines. In response to the binding of a small molecule ligand, these large proteins undergo a global conformational change, opening a selective ion channel and thereby converting a chemical event (i.e. ligand binding) to an electrical signal. The precise mechanism of this process is a central issue in molecular neurobiology. Recently, chemical scale studies have provided valuable insights into the structure and function of these receptors, yet significant challenges still remain.
Here we have evaluated the highly conserved and structurally intriguing Phe135-Pro136 motif of the prototypic Cys loop receptor, the nAChR. Proline is well appreciated to display novel conformational behaviors compared with all other natural amino acids. Additionally, it has been proposed that prolines might play a key role in the conformational changes that are essential to the function of many types of receptors (34). Several lines of evidence establish that local amino acids flanking proline can influence proline conformational preferences (9, 11, 12, 35). In particular, an aromatic residue preceding the proline is found to enhance the fraction of the cis isomer for peptides in solution (12). As shown in Fig. 2, Pro136 can exist in both cis and trans conformations, and the two crystal structures with a cis peptide bond (2QC1 and 3EAM) show stacking of the Phe-Pro side chains. Given the complete conservation of the Phe-Pro motif and the available structural data, it seemed reasonable to speculate that the cis conformer of Pro136 could be involved in receptor function.
Our primary measure of receptor function is EC50, the effective concentration of agonist required to achieve half-maximal response. Agonist binding to a receptor induces step-by-step conformational changes that lead to opening of the ion channel; therefore, EC50 is a value that reflects the composite effect of the agonist binding affinity and the sequential gating events. The Phe-Pro motif is remote from the agonist binding site, and the Cys loop is firmly established to play an essential role in gating (36). In addition, we find that a number of mutations at residue 135 greatly increase the efficacy of the partial agonist SuCh, supporting the notion that this residue participates in the gating mechanism. As such, we interpret changes in EC50 to reflect primarily, if not exclusively, changes in receptor gating.
The involvement of the Phe-Pro motif in gating is further supported by a previous single channel study on the F135A mutation, which indicated that the gating mechanism is modified as a result of this mutation (27). The new mechanism appears to be much less efficient at coupling agonist binding to channel opening, consistent with our macroscopic observations of greatly reduced current for this mutant.
Conventional mutations at Pro136 also have strong effects on the receptor. When expressed in HEK293 cells, both Gly mutants in the nAChR subunits and an Ala mutant in the related 5-HT3 receptor (13) gave receptors that were substantially impaired in the ability to assemble and/or traffic to the surface. In the Xenopus oocyte system, we find that the P136A mutant gave <10% of the current levels seen from wild type, again suggesting a disruption of assembly and/or trafficking or a disruption of gating.
Similarly, in an earlier study of Pro308 in the M2-M3 loop of the 5-HT3 receptor, in which a compelling correlation between cis propensity of incorporated proline analogues and receptor function was demonstrated, structural disruption by conventional mutagenesis produced ambiguous results (5). In that study, Ala, Cys, Gly, Lys, Val, and Gln conventional mutants gave nonfunctional receptors. More recently, studies of an orthologous 5-HT3 receptor showed that His and Trp mutants did give functional receptors (37). We note that aromatic amino acids, such as His and Trp, are more than twice as likely to be in a cis conformation as other non-proline natural amino acids (38). Again, the implications of the conventional mutagenesis results are open to debate.
Using conventional mutagenesis to probe the role of the cis conformation of a highly conserved proline is, in our view, unlikely to produce compelling results. Such studies frequently assume that simply seeing a functional receptor with a non-proline natural amino acid incorporated rules out a role for the cis conformer. However, previous studies have demonstrated that in some cases, when a cis proline is mutated to an alanine, the main-chain cis bond is preserved, presumably because the three-dimensional structure favors the cis conformation (11). In such cases, the Pro to Ala mutation often reduces the stability of the protein, which could manifest as lower expression levels, as we see with the P136A mutant. In addition, as with Pro, the presence of an aromatic amino acid (such as Phe) N-terminal to an aliphatic residue (such as Ala) doubles the probability of a cis conformation (38). Alternatively, replacement of a proline with another natural amino acid could produce functional receptors via a different gating path that has become more energetically accessible, parallel to what is seen with the F135A mutation (27).
When studying such a structurally distinctive motif as Phe-Pro, the benefits of unnatural amino acid mutagenesis are amplified. The subtle perturbations allow one to maintain the essential motif while probing its intrinsic features. We have used unnatural amino acids to probe several aspects of the Phe-Pro motif, including the importance of Phe aromaticity, the roles of side chain hydrophobicity and volume, and the possibility of cis-trans isomerization at the proline backbone.
Several intriguing observations emerge from the unnatural amino acid mutagenesis studies. Considering Pro136, subtle mutations produce noticeable changes in EC50. For example, simply adding a methyl group (3-Me-Pro) can lower EC50 22-fold, and adding a single CH2 group to the ring (Pip) can lower EC50 13-fold. Mutations are generally gain-of-function; EC50 decreases. The only residue that is not gain-of-function but instead gives nearly wild-type EC50 is 2-Me-Pro. Similar to Ala, 2-Me-Pro also produces much smaller whole cell currents.
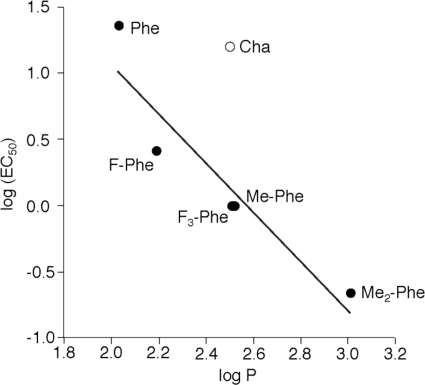
As with the proline, subtle mutations of Phe135 can produce substantial changes in EC50; a 100-fold shift arises from just the addition of two methyl groups fairly remote from the protein backbone (Me2-Phe). Paralleling the proline results, all of the unnatural amino acid mutants are gain-of-function. Moreover, an interesting trend is evident; Fig. 5 shows a plot of log(EC50) for the receptor versus the side chain logP, a measure of its hydrophobicity. Although the cyclohexyl compound (Cha) is clearly an outlier, a significant correlation is seen among the aromatic side chains. These results indicate that hydrophobicity is an important determinant at position 135, with an increase in hydrophobicity making the channel easier to open. This is consistent with a molecular dynamics simulation of the α7 nAChR that places Phe135 in a hydrophobic pocket in an open state (39). In addition, the logP analysis (Fig. 5) highlights the role of aromaticity at residue 135 because Cha has essentially the same hydrophobicity as both Me-Phe and F3-Phe but a much higher EC50. As such, the F135Cha mutant, having being more hydrophobic than the wild-type Phe but lacking the aromaticity, appears to have a nearly wild-type ACh EC50. From these data, we conclude that both hydrophobicity and aromaticity at position 135 are important in receptor function.
FIGURE 5.
Correlation between EC50 and logP for mutations at Phe135. Note that the Cha point was not included in the linear fit.
The results of our double mutant studies have confirmed an important interaction between residues 135 and 136; the large effects caused by mutation at Pro136 are attenuated when Phe135 is simultaneously mutated to the non-aromatic Cha (Fig. 4). Mutant cycle analysis shows significant coupling energies between residues 135 and 136.
We noted above the intriguing possibility that cis-trans isomerization at Pro136 is involved in receptor gating. In the present work, we did not see a simple correlation between EC50 and previously reported innate percentage cis values of the Pro analogues. However, there is ample precedent showing a deviation of percentage cis from the innate value when the preceding residue is aromatic (12). To probe the impact of the Phe residue on the present system, we used NMR spectroscopy to evaluate the cis-trans preference in the model peptides Gly-Phe-XPro-Gly, where XPro represents Pro, Pip, Aze, flp, Mor, 3-Me-Pro, and 2-Me-Pro. Because the cis form of the Gly-Phe(2-Me-Pro)-Gly peptide was not observed, we cannot comment on the role of Phe in this system. In all other cases, comparisons are possible, and the Phe does increase the percentage cis at the adjacent Pro analogue. The substantial upfield chemical shift of the Cβ proton in the cis conformer supports the existence of the putative interaction between the proline ring and the aromatic ring of the phenylalanine residue (supplemental Table 1).
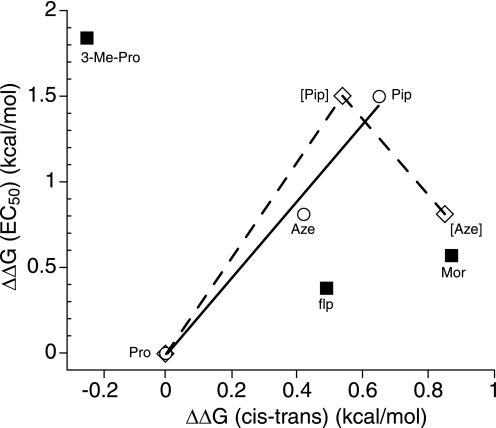
In Table 3, we report ΔΔG(c-t), the extent to which the proline analogue shows an increased bias for the cis form relative to proline. To facilitate comparisons, we also convert each EC50 shift into an energy term, ΔΔG(EC50). We first considered the homologous series of unsubstituted rings Aze, Pro, and Pip, in which the ring size expands from 4 to 5 to 6. The percentage cis and EC50 values track each other; EC50 is Pip < Aze < Pro, whereas percentage cis is Pip > Aze > Pro (Fig. 6, solid line). Note that in this simple series, the Phe substituent is critical because the inherent percentage cis sequence absent the Phe is Aze > Pip > Pro (Fig. 6, dotted line). Having an aromatic residue adjacent to the proline alters the cis bias differentially across this homologous series, and the EC50 values for the receptor mirror this effect. These data suggest that proline cis-trans isomerization at this site may play a role in receptor gating.
FIGURE 6.
Relationship between EC50 values and cis-trans preferences for Pro and analogues at position 136. All values are relative to Pro. Solid line and open circles, Pro, Aze, and Pip using cis-trans values determined in the present study for the Gly-Phe-Xaa-Gly sequence (Table 3). Dashed line and open diamonds, Pro, Aze, and Pip using cis-trans values previously determined for sequences that do not have an aromatic N-terminal to the Pro analogue. Solid squares, data points (flp, Mor, and 3-Me-Pro) that deviate from the trend set by the solid line.
Concerning the more dramatic proline mutations, a simple percentage cis correlation is not evident. It is clear from the Phe135 mutational studies that receptor function is highly sensitive to side chain polarity at the 135 site (Fig. 5), with increased side chain hydrophobicity lowering EC50. It seems reasonable to expect a similar effect at the adjacent Pro136 because Phe and Pro interact, as shown by the mutant cycle analysis. Indeed, our results suggest a preference for side chain hydrophobicity at the Pro136 site as well. Mor is structurally very similar to Pip, but it does not fit into the Aze-Pro-Pip correlation. We propose that EC50 for Mor is anomalously high because of the increased polarity relative to Pip. Similarly, flp has a significantly higher percentage cis than Pro but only a modest decrease in EC50, apparently due to the increased polarity of the fluorine substituent. In fact, a second linear correlation can be seen in Fig. 6 involving the Pro-flp-Mor series, although the structural variation across this series is less consistent than in the Aze-Pro-Pip trio. 3-Me-Pro shows a smaller percentage cis than Pro but the lowest EC50 among the amino acids at the 136 sites. Interestingly, adding a single CH3 group to Pro136 has the same effect on EC50 as adding a single CH3 group to Phe135 (3-Me-Pro and Me-Phe show the same EC50). Inspection of simple molecular models leads to an observation that the two CH3 groups could point into nearly the same region of the receptor when the proline is in the cis form. Perhaps each CH3 fits into a hydrophobic pocket, stabilizing the open state of the receptor and lowering EC50.
As shown Fig. 6, in the most conservative structural series (Pro, Pip, and Aze), we do find a trend that is suggestive of cis-trans isomerization at Pro136. Importantly, this trend is seen only when the perturbing effect of the Phe residue is included, justifying the consideration of the Phe-Pro unit as a single motif. Residues that involve more complex changes do not fit the correlation, but generally the deviation is consistent with the notion that increasing side chain hydrophobicity lowers EC50. From our data, we propose that both cis propensity and side-chain hydrophobicity at Pro136 simultaneously are determinants of nAChR function. Moreover, the possibility of cis-trans isomerization at Pro136 being involved in gating cannot be ruled out.
Summary
The subtle mutations enabled by unnatural amino acid mutagenesis have allowed a detailed study of the Phe-Pro motif in the Cys loop of a Cys loop receptor. Mutant cycle analysis reveals a strong interaction between the two residues and a strong preference for an aromatic residue at position 135. In addition, a clear trend is evident whereby increasing hydrophobicity at either Phe135 or Pro136 lowers EC50. Although the analysis of residue Pro136 is complex, the data provide evidence supporting a role for the cis conformer in receptor function.
Supplementary Material
Acknowledgments
We thank Dr. Scott A. Ross for help with the NMR experiments and Professor Sarah C. R. Lummis for helpful discussion.
This work was supported, in whole or in part, by National Institutes of Health Grants NS-34407 and NS-11756.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- nAChR
- nicotinic acetylcholine receptor
- ACh
- acetylcholine
- SuCh
- succinylcholine
- Pip
- pipecolic acid
- Aze
- azetedine-2-carboxylic acid
- Dhp
- 3,4-dehydroproline
- Mor
- morpholine-3-carboxylic acid
- flp
- cis 4-fluoro-proline
- Flp
- trans 4-fluoro-proline
- 3-Me-Pro
- trans 3-methyl-proline
- 2-Me-Pro
- 2-methyl-proline
- Cha
- cyclohexylalanine
- F-Phe
- 4-fluorophenylalanine
- F3-Phe
- 3,4,5-trifluorophenylalanine
- Me-Pro
- 4-Me-phenylalainine
- Me2-Phe
- 3,5-dimethyl-phenylalanine
- Fmoc
- N-(9- fluorenyl)methoxycarbonyl
- NVOC
- O-nitroveratryloxycarbonyl
- MS
- mass spectrometry.
REFERENCES
- 1.Corringer P. J., Le Novère N., Changeux J. P. (2000) Annu. Rev. Pharmacol. Toxicol. 40, 431–458 [DOI] [PubMed] [Google Scholar]
- 2.Grutter T., Changeux J. P. (2001) Trends Biochem. Sci. 26, 459–463 [DOI] [PubMed] [Google Scholar]
- 3.Bouzat C., Gumilar F., Spitzmaul G., Wang H. L., Rayes D., Hansen S. B., Taylor P., Sine S. M. (2004) Nature 430, 896–900 [DOI] [PubMed] [Google Scholar]
- 4.Karlin A. (2002) Nat. Rev. Neurosci. 3, 102–114 [DOI] [PubMed] [Google Scholar]
- 5.Lummis S. C., Beene D. L., Lee L. W., Lester H. A., Broadhurst R. W., Dougherty D. A. (2005) Nature 438, 248–252 [DOI] [PubMed] [Google Scholar]
- 6.Millar N. S. (2003) Biochem. Soc. Trans. 31, 869–874 [DOI] [PubMed] [Google Scholar]
- 7.Jha A., Cadugan D. J., Purohit P., Auerbach A. (2007) J. Gen. Physiol. 130, 547–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dugave C., Demange L. (2003) Chem. Rev. 103, 2475–2532 [DOI] [PubMed] [Google Scholar]
- 9.Bhattacharyya R., Chakrabarti P. (2003) J. Mol. Biol. 331, 925–940 [DOI] [PubMed] [Google Scholar]
- 10.MacArthur M. W., Thornton J. M. (1991) J. Mol. Biol. 218, 397–412 [DOI] [PubMed] [Google Scholar]
- 11.Pal D., Chakrabarti P. (1999) J. Mol. Biol. 294, 271–288 [DOI] [PubMed] [Google Scholar]
- 12.Reimer U., Scherer G., Drewello M., Kruber S., Schutkowski M., Fischer G. (1998) J. Mol. Biol. 279, 449–460 [DOI] [PubMed] [Google Scholar]
- 13.Deane C. M., Lummis S. C. (2001) J. Biol. Chem. 276, 37962–37966 [DOI] [PubMed] [Google Scholar]
- 14.Brejc K., van Dijk W. J., Klaassen R. V., Schuurmans M., van Der Oost J., Smit A. B., Sixma T. K. (2001) Nature 411, 269–276 [DOI] [PubMed] [Google Scholar]
- 15.Unwin N. (2005) J. Mol. Biol. 346, 967–989 [DOI] [PubMed] [Google Scholar]
- 16.Dellisanti C. D., Yao Y., Stroud J. C., Wang Z. Z., Chen L. (2007) Nat. Neurosci. 10, 953–962 [DOI] [PubMed] [Google Scholar]
- 17.Rickert K. W., Imperiali B. (1995) Chem. Biol. 2, 751–759 [DOI] [PubMed] [Google Scholar]
- 18.Hilf R. J., Dutzler R. (2008) Nature 452, 375–379 [DOI] [PubMed] [Google Scholar]
- 19.Bocquet N., Nury H., Baaden M., Le Poupon C., Changeux J. P., Delarue M., Corringer P. J. (2009) Nature 457, 111–114 [DOI] [PubMed] [Google Scholar]
- 20.Hilf R. J., Dutzler R. (2009) Nature 457, 115–118 [DOI] [PubMed] [Google Scholar]
- 21.Nowak M. W., Gallivan J. P., Silverman S. K., Labarca C. G., Dougherty D. A., Lester H. A. (1998) Methods Enzymol. 293, 504–529 [DOI] [PubMed] [Google Scholar]
- 22.Cashin A. L., Petersson E. J., Lester H. A., Dougherty D. A. (2005) J. Am. Chem. Soc. 127, 350–356 [DOI] [PubMed] [Google Scholar]
- 23.Saks M. E., Sampson J. R., Nowak M. W., Kearney P. C., Du F., Abelson J. N., Lester H. A., Dougherty D. A. (1996) J. Biol. Chem. 271, 23169–23175 [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez E. A., Lester H. A., Dougherty D. A. (2007) RNA 13, 1703–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodriguez E. A., Lester H. A., Dougherty D. A. (2007) RNA 13, 1715–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu D. X., Sine S. M. (1996) J. Biol. Chem. 271, 31479–31484 [DOI] [PubMed] [Google Scholar]
- 27.Chakrapani S., Bailey T. D., Auerbach A. (2004) J. Gen. Physiol. 123, 341–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMenimen K. A., Dougherty D. A., Lester H. A., Petersson E. J. (2006) ACS Chem. Biol. 1, 227–234 [DOI] [PubMed] [Google Scholar]
- 29.Kash T. L., Jenkins A., Kelley J. C., Trudell J. R., Harrison N. L. (2003) Nature 421, 272–275 [DOI] [PubMed] [Google Scholar]
- 30.Price K. L., Millen K. S., Lummis S. C. (2007) J. Biol. Chem. 282, 25623–25630 [DOI] [PubMed] [Google Scholar]
- 31.Venkatachalan S. P., Czajkowski C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13604–13609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gleitsman K. R., Kedrowski S. M., Lester H. A., Dougherty D. A. (2008) J. Biol. Chem. 283, 35638–35643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu W. J., Raleigh D. P. (1998) J. Org. Chem. 63, 6689–6698 [Google Scholar]
- 34.Sansom M. S., Weinstein H. (2000) Trends Pharmacol. Sci. 21, 445–451 [DOI] [PubMed] [Google Scholar]
- 35.Thomas K. M., Naduthambi D., Zondlo N. J. (2006) J. Am. Chem. Soc. 128, 2216–2217 [DOI] [PubMed] [Google Scholar]
- 36.Sine S. M., Engel A. G. (2006) Nature 440, 448–455 [DOI] [PubMed] [Google Scholar]
- 37.Paulsen I. M., Martin I. L., Dunn S. M. J. (2009) J. Neurochem. 110, 870–878 [DOI] [PubMed] [Google Scholar]
- 38.Jabs A., Weiss M. S., Hilgenfeld R. (1999) J. Mol. Biol. 286, 291–304 [DOI] [PubMed] [Google Scholar]
- 39.Cheng X., Ivanov I., Wang H., Sine S. M., McCammon J. A. (2007) Biophys. J. 93, 2622–2634 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.