Abstract
Transfer RNA (Gm18) methyltransferase (TrmH) catalyzes methyl transfer from S-adenosyl-l-methionine to a conserved G18 in tRNA. We investigated the recognition mechanism of Thermus thermophilus TrmH for its guanosine target. Thirteen yeast tRNAPhe mutant transcripts were prepared in which the modification site and/or other nucleotides in the D-loop were substituted by dG, inosine, or other nucleotides. We then conducted methyl transfer kinetic studies, gel shift assays, and inhibition experiments using these tRNA variants. Sites of methylation were confirmed with RNA sequencing or primer extension. Although the G18G19 sequence is not essential for methylation by TrmH, disruption of G18G19 severely reduces the efficiency of methyl transfer. There is strict recognition of guanosine by TrmH, in that methylation occurs at the adjacent G19 when the G18 is replaced by dG or adenosine. The fact that TrmH methylates guanosine in D-loops from 4 to 12 nucleotides in length suggests that selection of the position of guanosine within the D-loop is relatively flexible. Our studies also demonstrate that the oxygen 6 atom of the guanine base is a positive determinant for TrmH recognition. The recognition process of TrmH for substrate is inducible and product-inhibited, in that tRNAs containing Gm18 are excluded by TrmH. In contrast, substitution of G18 with dG18 results in the formation of a more stable TrmH-tRNA complex. To address the mechanism, we performed the stopped-flow pre-steady state kinetic analysis. The result clearly showed that the binding of TrmH to tRNA is composed of at least three steps, the first bi-molecular binding and the subsequent two uni-molecular induced-fit processes.
Keywords: Nucleic Acid/Enzymology, Protein/Binding/RNA, RNA/Methylation, RNA/Methyltransferase, RNA/Modification, RNA/Structure, RNA/Transfer RNA
Introduction
Modification of nucleic acids by methylation is a fundamental chemical process that can contribute to structure formation or alter gene expression patterns. For example, tRNA molecules contain abundant methylated nucleotides (1, 2); some stabilize tRNA tertiary structure, whereas others alter molecular recognition of target molecules (3). A conserved guanosine at position 18 (G18) in the D-loop of all tRNAs (1, 2) is often modified to 2′-O-methylguanosine (Gm18). Methylation of G18 to Gm18 promotes formation of the L-shaped three-dimensional structure of tRNA by interacting with pseudouridine 55 (Ψ55)2 (4, 5). In Escherichia coli, the Gm18 modification (in conjunction with Ψ55) has been shown to be involved in translational accuracy; a mutant strain lacking both Gm18 and Ψ55 modifications demonstrated growth defects and an increase in frameshift errors, whereas the strain lacking only the Gm18 modification was normal (6).
Formation of Gm18 is catalyzed by tRNA (Gm18) methyltransferase (7–9) (tRNA (guanosine 2′)-methyltransferase; TrMet (Gm18) (2), EC 2.1.1.34). During the reaction, S-adenosyl-l-methionine (AdoMet) functions as a methyl group donor and is converted to S-adenosyl-l-homocysteine (AdoHcy). The tRNA (Gm18) methyltransferase gene in E. coli (10, 11), Thermus thermophilus (8), and Aquifex aeolicus (9) is named trmH (classical name, spoU), and the homolog in Saccharomyces cerevisiae is trm3 (12). In accordance with this nomenclature, we refer to the bacterial tRNA (Gm18) methyltransferase enzyme as TrmH. AdoMet-dependent methyltransferases are sorted into five classes based on the type of fold present in the catalytic domain (13). The majority of RNA methyltransferases are class I enzymes, which maintain a Rossman fold in the catalytic domain. In contrast, TrmH is a class IV enzyme (13) that maintains a topological (trefoil) knot (14) structure in the catalytic domain (15).
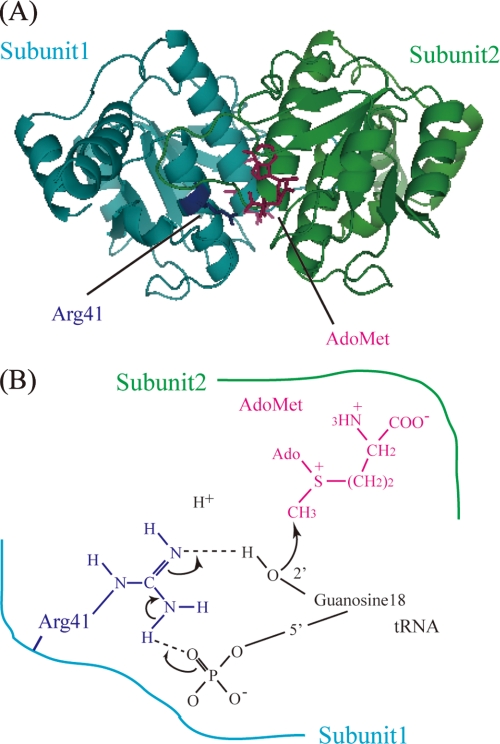
We reported identification of the trmH gene from T. thermophilus in 2002 (8). In extensive further studies, we then solved the crystal structures of apo- and AdoMet-bound forms (Fig. 1A) (15), proposed a hypothetical reaction mechanism (Fig. 1B) (15, 16), clarified roles of the amino acid residues in the conserved motifs (16), and distinguished the first tRNA-binding site and amino acid residues involved in the catalytic mechanism (17). One of strengths of our predicted reaction mechanism (Fig. 1B) is that it explains how the TrmH-tRNA complex might dissociate following methyl transfer: we hypothesize that loss of a positive charge at Arg-41 at the moment of methyl transfer disrupts its interaction with a phosphate. Although there are no discrepancies between this model and our experimental results, the mechanism by which TrmH recognizes the G18 target in tRNA has yet to be resolved.
FIGURE 1.
A, crystal structure of TrmH dimer. Subunit 1 (cyan) acts as the catalytic subunit, although subunit 2 (green) acts as an AdoMet-binding subunit. The catalytic center Arg-41 (dark blue) in subunit 1 and AdoMet (red) in subunit 2 are illustrated using ball-and-stick representations. B, schematic drawing of the proposed catalytic mechanism of TrmH. For binding substrate tRNA, the Arg-41 residue in subunit 1 must come in close proximity to the 5′-phosphate of G18, whereupon the negative charge of the phosphate attracts the guanidino group of Arg-41. Activated Arg-41 displaces or excludes the 2′-OH proton of G18 ribose, and methyl transfer then occurs by nucleophilic attack of the deprotonated oxygen atom of the ribose against the methyl group in AdoMet. The detached proton is depicted in the figure.
The G18 modification site is highly conserved in tRNA, and T. thermophilus TrmH has been shown to modify all tRNA species tested thus far. Because the enzyme catalyzes methyl transfer to various tRNA species with different D-loop nucleotide sequences and sizes, it follows that TrmH should recognize structures common to these tRNAs. In an earlier study, we reported that the D-arm structure of tRNA is essential for recognition by T. thermophilus TrmH and that conserved nucleotides (for example, U8, purine 26, G46, U54, and U55) in the three-dimensional core of L-shaped tRNA affect methylation efficiency (7).
During the past decade, we developed a large scale expression system for T. thermophilus TrmH in E. coli that permitted crystal formation and x-ray structure studies of the enzyme. In this study, we focused on the recognition mechanism between T. thermophilus TrmH and guanosine (G18). We demonstrate that the enzyme recognizes the position of the target guanosine with some flexibility and that the oxygen 6 atom of guanosine is a positive determinant for enzyme recognition. These experimental results suggest that the tRNA binding process consists of at least two steps, recognition of guanine base in tRNA followed by introduction of the guanosine ribose into the TrmH catalytic pocket (from which methylated tRNA is excluded). To confirm the idea, we performed the pre-steady state kinetic study of the binding process between TrmH and tRNA by stopped-flow fluorescence measurements.
EXPERIMENTAL PROCEDURES
Materials
[methyl-14C]AdoMet (1.95 GBq/mmol) and [methyl-3H]AdoMet (2.47 TBq/mmol) were purchased from ICN. Nonradioisotope-labeled AdoMet and AdoHcy were obtained from Sigma, and DE52 was purchased from Whatman. We produced the AdoHcy affinity column by coupling to a HiTrap NHS-activated HP column (Amersham Biosciences) according to the manufacturer's instructions. CM-Toyopearl 650 M was purchased from Tosoh. DNA oligomers were bought from Invitrogen. T4 polynucleotide kinase, calf intestine alkaline phosphatase, and T4 RNA ligase were purchased from Takara. T7 RNA polymerase was from Toyobo. RNase T1 and RNase U2 were from Sankyo. All other chemical reagents were of analytical grade.
TrmH Protein
TrmH was expressed in the E. coli BL21 (DE3) RIL codon plus strain (Takara) and purified as reported previously (8). Protein concentration was measured using the Bio-Rad protein assay kit with bovine serum albumin as the standard. Purified protein was diluted to a concentration of 50% glycerol and stored at −30 °C.
Preparation of Synthetic Wild-type and Variant Yeast tRNAPhe Transcripts
Full-length yeast tRNAPhe transcript was prepared as reported previously (7). Transcripts were purified by electrophoresis in 10% polyacrylamide gels (7 m urea). 5′-half-fragments containing deoxyguanylic acid (dG) or inosine (I) were synthesized by Hokkaido System Science. The sequences were as follows: YF dG18-(1–36), 5′-GCG GAU UUA GCU CAG UUdG GGA GAG CGC CAG ACU GAA-3′; YF dG18dG19A20-(1–36), 5′-GCG GAU UUA GCU CAG UUdG dGAA GAG CGC CAG ACU GAA-3′; YF I18G19G20, 5′-GCG GAU UUA GCU CAG UUI GGA GAG CGC CAG ACU GAA-3′. All oligomers were purified using high pressure liquid chromatography C18 reverse phase column chromatography and 10% PAGE (7 m urea). 3′-Half-fragments were transcribed by T7 RNA polymerase as reported previously (7) and purified using Qiagen plasmid mini kit column chromatography and 10% PAGE (7 m urea). 3′-Fragments were treated with bacterial alkaline phosphatase and then phosphorylated using T4 polynucleotide kinase and ATP. Purified 5′-half (2.5 A260 units) and 3′-half (2.0 A260 units) fragments were mixed together in 100 μl of annealing buffer (10 mm Tris-HCl (pH 7.5), 5 mm MgCl2, and 100 mm NaCl), heated to 80 °C, and then allowed to anneal by cooling to 40 °C for 40 min. For ligation, 5 μl of RNA ligation buffer (500 mm Tris-HCl (pH 7.5), 100 mm MgCl2, 100 mm dithiothreitol, 10 mm ATP, and 0.1% bovine serum albumin) and 50 units of T4 RNA ligase were added directly to each annealed sample and incubated at 14 °C for 24 h. Ligated RNAs were extracted with phenol/chloroform, recovered by ethanol precipitation, and purified by 10% PAGE (7 m urea). RNA sequences were verified from RNase T1 and RNase U2 digestions using the Donis-Keller method (18).
Measurement of Enzymatic Activity
Incorporation of 14C-methyl from AdoMet to yeast tRNAPhe transcripts was used to assay TrmH activity (7). To obtain kinetic parameters of TrmH for each tRNA, concentrations of TrmH and AdoMet were fixed at 0.1 and 37 μm, respectively, whereas incubation times varied from 2 to 30 min depending on the methyl group acceptance activity for each transcript. We used a gel assay described previously to visualize methylated RNAs (19).
Modified Nucleotide Analysis Using Thin Layer Chromatography
Purified protein (1 μg), 3.3 μm tRNA transcript, and 7.8 μm [methyl-14C]AdoMet in 100 μl of buffer A were incubated at 50 °C for 1 h. The RNA was extracted using phenol/chloroform and then recovered by precipitation in ethanol. The RNA pellet was dissolved in 5 μl of 50 mm ammonium acetate (pH 5.0) and digested with 2.5 units of nuclease P1, after which 2 μl of standard ribonucleotides containing 0.05 A260 units each pA, pG, pC, and pU were added. Two microliters of sample was spotted onto a thin layer plate (Merck code 1.05565, cellulose F) and separated using the following solvent systems (20): first dimension, isobutyric acid/concentrated ammonia/water, 66:1:33, v/v; second dimension, isopropyl alcohol/HCl/water, 70:15:15, v/v. Incorporation of 14C-methyl groups was monitored with a Fuji Photo Film BAS2000 image analyzer. The locations of standard nucleotides were detected using 260 nm irradiation.
Primer Extension
Primer extension for detection of methylation sites was performed using 5′-32P-labeled primer, a 625 μm mixture of A, G, C, and T deoxynucleotide triphosphates, and Prime Script reverse transcriptase (Takara). The primer sequence (5′-TGG TGC GAA TTC TGT GGA TCG-3′) was complementary to the T-arm at the CCA end of yeast tRNAPhe. DNA sequencing with an fmol DNA Cycle Sequencing System (Promega) was performed using the same primer.
Gel Mobility Shift Assay
Gel mobility shift assays were performed as described in previous reports (16, 17). In brief, purified protein and 0.05 A260 unit of yeast tRNAPhe transcript were incubated in 20 μl of buffer A (50 mm Tris base, 50 mm acetic acid, and 5 mm Mg(OAc)2) at 37 °C for 20 min. Four microliters of loading solution (0.25% bromphenol blue and 30% glycerol) were then added to each sample, and samples were resolved on a 6% polyacrylamide gel (width, 90 mm; length, 90 mm; thickness, 1 mm) prepared with 1× buffer A. Electrophoresis was performed at room temperature for 1 h at 100 V. To detect protein, the gel was stained with Coomassie Brilliant Blue. Methylene blue was used for RNA detection. RNA and protein were visualized from stain intensity using a Fuji Photo Film BAS2000 imaging analyzer.
Inhibition Experiments
Methyl transfer reaction by TrmH was inhibited with methylated dG18 or dG18dG19A20 tRNA. Initial velocities of the reaction with 80 nm TrmH, 50 μm [3H]AdoMet, and various concentrations (25, 50, 100, 200, 500, and 1000 nm) of wild-type yeast tRNAPhe transcript in 30 μl of buffer B (50 mm Tris-HCl (pH 7.5), 5 mm MgCl2, and 50 mm KCl) at 55 °C were compared in the absence or presence (25 or 50 nm) of methylated dG18 or dG18dG19A20 mutant tRNA transcript.
Stopped-flow Fluorescence Measurements
To investigate the pre-steady state kinetics of the binding process between TrmH and tRNA, we performed stopped-flow fluorescence measurements using an Applied Photophysics SX20 stopped-flow apparatus (Applied Photophysics Ltd., Leatherhead, UK). We monitored changes in the fluorescence intensity of tryptophan residues in TrmH during the complex formation with tRNA. The excitation wavelength was 295 nm, and the fluorescence emission above 320 nm was monitored using a long pass UV cut-off filter. TrmH dissolved in buffer B containing 10 μm AdoMet was rapidly mixed with equimolar yeast tRNAPhe transcript in buffer B at 25 °C. The mixing experiments were carried out at two different TrmH and tRNA concentrations, 1.93 and 7.7 μm, after the mixing. The kinetic data were acquired in a split time-base mode, in which data points were distributed in two linear intervals, one from 0 to 0.1 s and the other from 0.1 to 1.1 s. The scan was repeated 20 times for 7.7 μm TrmH and tRNA and 80 times for 1.925 μm TrmH and tRNA. The dead time of the stopped-flow mixing, determined by the method of Brissette et al. (21), was 3 ms, and the optical path length was 1 mm. To estimate the ratio of methylated tRNA to unmodified tRNA, methyl transfer filter assay was performed with [3H]AdoMet under the same conditions (7.7 μm TrmH and tRNA).
RESULTS
Deoxyribose at G18 Changes the TrmH Modification Site to G19
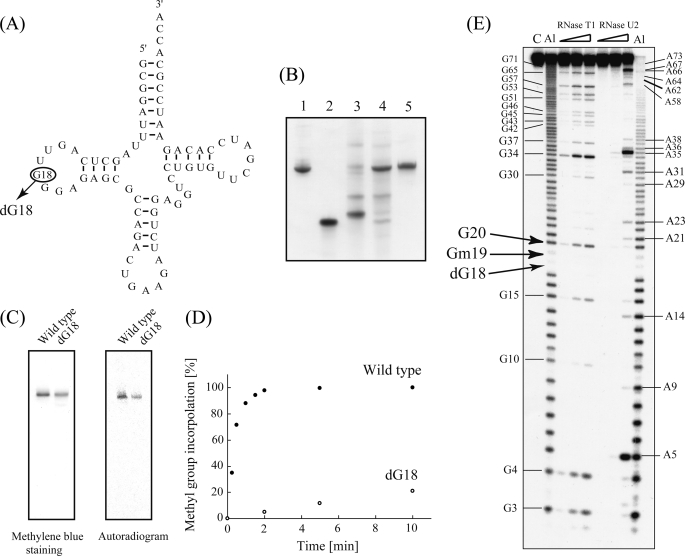
We used the sequence of yeast tRNAPhe to investigate TrmH target recognition because its three-dimensional structure is established (4, 5) and because this tRNA is an effective substrate for T. thermophilus TrmH methylation (7). TrmH is composed of two subunits (Fig. 1A). Subunit 1 generally associates with G18 in the tRNA, positioning the 2′-oxygen atom of the G18 ribose for nucleophilic attack of the AdoMet residue associated with subunit 2 (Fig. 1B). To investigate these interactions, we prepared a synthetic yeast tRNAPhe molecule in which deoxyguanosine 18 (dG18) replaced guanosine 18 (G18) (Fig. 2A). A 5′-half-fragment synthesized with a dG18 substitution was ligated to a 3′-half-fragment, and successful ligation products were purified by gel electrophoresis (Fig. 2B, lane 5).
FIGURE 2.
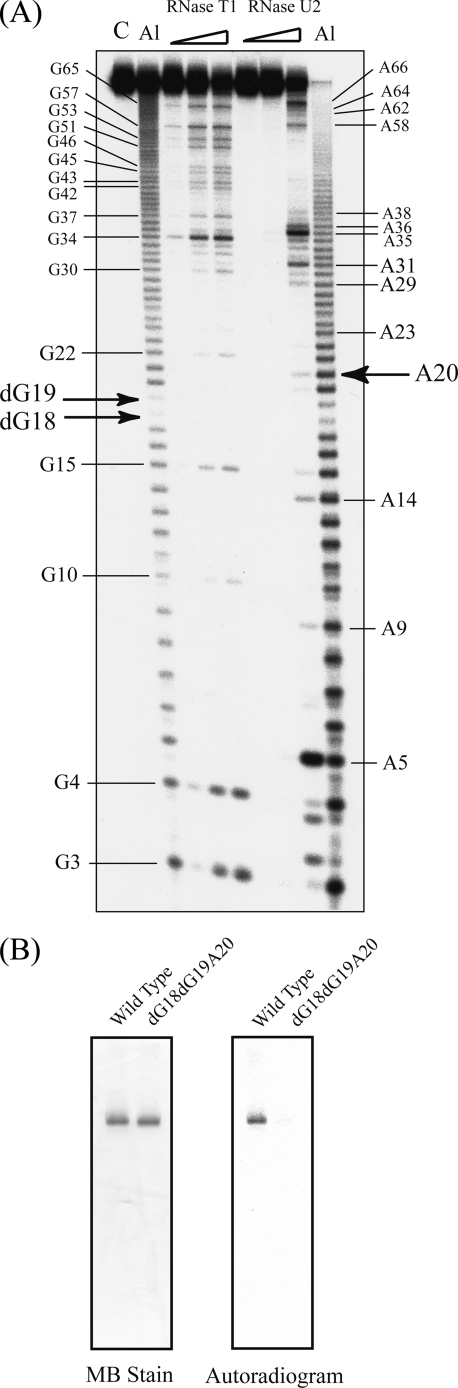
Methylation of a yeast tRNAPhe-dG18 variant. A, cloverleaf structure of wild-type yeast tRNAPhe, with the substitution site G18 enclosed by a circle. B, preparation of yeast tRNAPhe-dG18. RNA fragments were analyzed by electrophoresis in 10% polyacrylamide gels (7 m urea) and then stained with methylene blue. Lane 1, wild-type tRNAPhe (control); lane 2, 5′-half-fragment containing dG18; lane 3, 3′-half-fragment; lane 4, ligation sample of 5′- and 3′-half-fragments; lane 5, purified tRNAPhe-dG18. C, methylation of tRNA-dG18 monitored by 10% PAGE (7 m urea). TrmH was pre-incubated with [14C]AdoMet and then with either wild-type tRNAPhe (left lanes) or tRNAPhe-dG18 (right lanes) prior to analysis by 10% PAGE (7 m urea). The gel was stained with methylene blue (left panel), and an autoradiographic image of the same gel was then captured using a Fuji BAS2000 imaging analyzer (right panel). D, time course experiment of tRNA-dG18 methylation (performed under the same conditions as shown in C). Filled and open circles indicate rates of methylation of wild-type tRNAPhe and tRNA-dG18, respectively. E, identification of the tRNA-dG18 methylation site. Analyses using RNase T1 partial digestion and alkaline-limited hydrolysis reactions are described under “Experimental Procedures.” Lane C, untreated control RNA; lane Al (left), alkaline hydrolysis (95 °C, 2 min); lane RNase T1 included 0.03, 0.1, or 0.3 units of enzyme; lanes RNase U2 included 0.01, 0.03, or 0.1 units of enzyme; lane Al (right), alkaline hydrolysis (95 °C, 4 min). Positions of G and A nucleotides are marked on both sides of the gel, and those of dG18, Gm19, and G20 are indicated with arrows.
Based on our understanding of TrmH catalysis, we predicted that tRNA-dG18 would not be methylated by TrmH. To investigate this hypothesis, TrmH protein pre-incubated with [14C]AdoMet was mixed with wild-type tRNA or tRNA-dG18. Transfer of 14C-methyl to these tRNA molecules was visualized by imaging analysis of RNA products resolved by 10% denaturing PAGE. In contrast to our prediction, we observed 14C-methyl transfer to tRNA-dG18 as well as wild-type tRNA (Fig. 2C). It should be noted that the autoradiogram in Fig. 2C does not depict the marked differences in reaction rates between TrmH and these two substrates. In these reactions, the molar ratio of TrmH dimer to G18 or dG18 tRNAPhe transcripts was 1:1, conditions that promote nearly 100% methylation of yeast tRNAPhe transcripts within 2 min (Fig. 2D).
In a previous study, we calculated the turnover time for one cycle of TrmH methylation of yeast tRNAPhe to be 10 s (17), with methylated tRNA functioning as an inhibitor. In this experiment, our analyses of the kinetic parameters showed that the initial velocity for tRNA-dG18 methylation was ∼400-fold less than that of the wild-type yeast tRNAPhe transcript (Table 1). To analyze methylation sites within the transcripts, we applied alkaline hydrolysis and RNase T1 partial digestion using the Donis-Keller method (see Ref. 18). Both 2′-O-methylation of ribose or removal of a hydroxyl at this site prevent alkaline hydrolysis and RNase T1 digestion, making these sites immune to cleavage. To test for cleavage, tRNA-dG18 was methylated to completion with TrmH and nonradioisotopic AdoMet and its 5′-end radiolabeled using T4 polynucleotide kinase and [γ-32P]ATP. Alkaline cleavage products showed an absence of two specific nucleotides corresponding to positions dG18 and G19 in yeast tRNAPhe (Fig. 2E). In addition, all guanine residues except for G18 and G19 in tRNA-dG18 were seen in the RNase T1 ladder (Fig. 2E).
TABLE 1.
Kinetic parameters of TrmH for tRNA variants
Not detectable means that the methyl transfer could not be detected under the tested conditions.
| Variant | Km | Vmax | Relative Vmax/Km |
|---|---|---|---|
| nm | μmol/mg h | ||
| Wild type tRNAPhe transcript | 82 | 3.1 | 1.0 |
| tRNAPhe-dG18 | 820 | 0.11 | 0.0036 |
| tRNAPhe-dG18dG19A20 | Not detectable | Not detectable | Not detectable |
| tRNAPhe-GGA | 245 | 1.2 | 0.13 |
| tRNAPhe-GAA | 200 | 0.135 | 0.018 |
| tRNAPhe-AGA | 12500 | 0.00675 | 0.000014 |
| tRNAPhe-GUA | 300 | 0.18 | 0.016 |
From this experiment, we verified that the dG18 nucleotide was impervious to hydrolysis by alkali or RNase T1. Curiously, the absence of a band at G19 implies that this residue also remained intact in the presence of either RNase T1 or alkaline hydrolysis and suggests that methyl transfer occurred specifically at G19 in the artificial tRNA-dG18. We conclude that changing the nucleotide at position G18 to dG18 inhibits methylation by TrmH at this site but that the modification also results in less efficient TrmH methyl transfer to the neighboring residue G19.
Weak TrmH Recognition of Guanosine at Other Sites in the D-loop
Because introduction of deoxyribose at G18 did not prevent methyl transfer to tRNAPhe, but instead altered the modification site from G18 to G19, we investigated methylation target-site recognition by TrmH with a set of transcripts in which the G18G19G20 trinucleotide in yeast tRNAPhe was changed to GGA, GAA, AGA, or GUA, respectively (Fig. 3A). To our surprise, all four of the mutant transcripts served as substrate for TrmH-catalyzed methyl transfer from [14C]AdoMet (Fig. 3B). The results shown in the figure do not represent the relative initial velocity accurately, because TrmH was present in excess (1:1 tRNA to enzyme). The kinetic parameters for methylation of each transcript are summarized in Table 1.
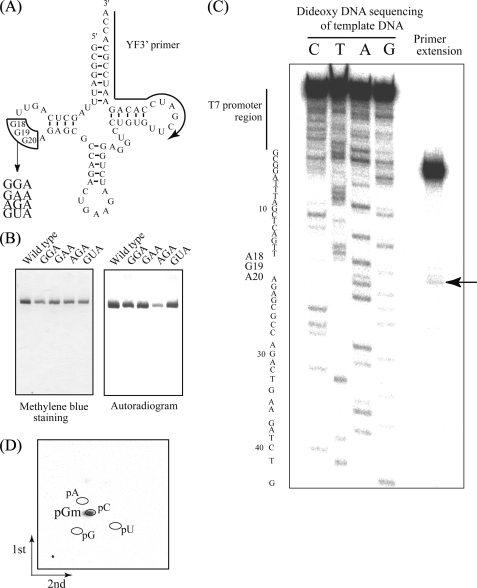
FIGURE 3.
A, substitution sites (G18G19G20) and replaced sequences depicted on a tRNAPhe clover-leaf schematic. The substitution site (G18G19G20) is enclosed by lines, and the region complementary to the primer used for extension (C) is marked with an arrow. B, methylation of tRNAPhe variants. Methyl group acceptance activities of tRNAPhe variants GGA, GAA, AGA, and GUA were analyzed using the same method as Fig. 2C. C, primer extension to locate sites of modification. Following methylation of tRNAPhe AGA using TrmH and AdoMet, primer extension was performed as described under “Experimental Procedures.” To generate the DNA sequence ladder on the left, dideoxy DNA sequencing was performed using the same 5′-32P-labeled primer. The terminated band (marked with an arrow) shows 2′-O-methylation at G19. D, two-dimensional thin layer chromatography analysis of the modified nucleotide. The tRNA AGA variant was methylated using TrmH and [14C]AdoMet, digested with nuclease P1, and then separated on two-dimensional thin layer chromatography plates prior to autoradiography.
In previous studies using purified enzyme from native source (T. thermophilus cells), we reported that methylation of the yeast tRNAPhe AGA variant was less than 0.005 times as frequent as the wild-type tRNAPhe transcript, a value below the detection limit at that time (7). In contrast, recombinant TrmH purified from the E. coli expression system in this study offered a more pure and concentrated source of enzyme. Greater sensitivity of the imaging analyzer system for detecting less frequent 14C-methyl modification products also allowed us to measure the kinetic parameters of weaker TrmH methylation substrates in this study.
Using these improved reagents and tools, we calculated the relative Vmax/Km value for tRNAPhe AGA variant to be 0.000014 times as frequent as the wild-type tRNAPhe transcript. Because complete modification of the tRNAPhe AGA substrate was difficult to attain, we use primer extension, rather than the Donis-Keller method, to identify the modification site (Fig. 3C). Briefly, the tRNAPhe AGA variant was first modified using nonradioisotope-labeled AdoMet and TrmH, and then a reverse transcriptase reaction was performed using a 5′-end 32P-labeled primer. In the presence of a low concentration of dNTPs (625 μm equimolar mixture), there is premature termination of reverse transcriptase extensions at 2′-O-methylated nucleotides. Thus, the faint but clear band observed at nucleotide G19 (Fig. 3C) identifies this nucleotide as the site of methylation in the tRNAPhe AGA variant. The identity of the modified nucleotide as 14C-pGm was verified using two-dimensional thin layer chromatography (Fig. 3D). These experimental results demonstrate clearly that G19, not A18, was the site of modification the tRNAPhe AGA variant. Similar analyses of the three other tRNAPhe variants (GGA, GAA, and GUA) demonstrated modification only at G18 (data not shown). Furthermore, we confirmed that these tRNA variants strongly bind to TrmH (supplemental Fig. S1). Thus, the first binding process occurs in the methylation of these tRNA variants.
Taken together, these experimental results demonstrate that TrmH is most effective at methylating the G18G19 site in the D-loop and that the remaining G base is modified at a slower rate when the G18G19 sequence is disrupted. In addition, when G18 is replaced by dG18 or A18, the methylation target changes to G19 albeit at a decreased initial velocity involving both an increase in Km and a decrease in Vmax.
Flexibility in TrmH Recognition of the D-loop GG Sequence
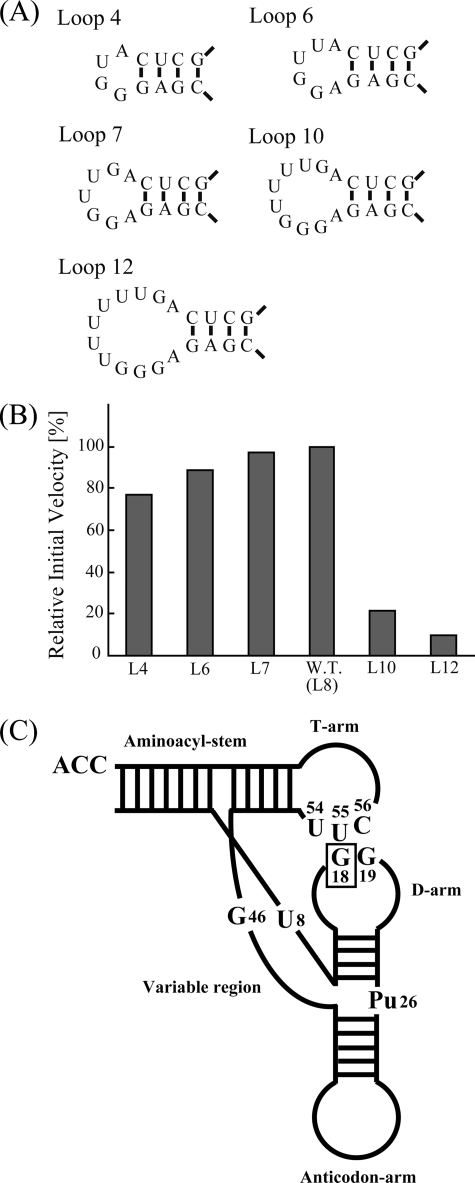
The D-loop regions of different tRNAs vary in size. In the case of T. thermophilus, all tRNA species thus far sequenced are methylated at G18, and D-loop lengths vary from 7 to 9 nucleotides (1). To investigate the effect of D-loop size on the methyl group acceptance activity, we prepared five tRNAPhe variants with loop sizes that varied in length from 4 to 12 nucleotides (Fig. 4A). Surprisingly, all variants of the 8-nucleotide loop (which served as the wild-type control) were methylated. Loops of fewer than 8 nucleotides showed no significant decrease in methyl group acceptance, whereas tRNAs with 10 or 12 nucleotide loops showed a clear decrease in the initial velocity of methylation (Fig. 4B).
FIGURE 4.
Methylation of tRNA D-loop size variants. A, sequence and structure of the D-loop regions (4, 6, 7, 10 and 12 nucleotides in length) in five tRNA variants. B, relative initial velocities for methylation of tRNA D-loop size variants. All tRNAs were methylated, although variants containing 10 and 12 nucleotide D-loops were modified at a significantly slower velocity relative to the wild-type (W.T.) yeast tRNAPhe. C, model of TrmH recognition elements within structured tRNA. In an earlier study (7), we presented a model in which TrmH recognizes the D-arm structure and conserved nucleotides (U8, Pu26, G46, U54, U55, and C56) in tRNA that affect methylation efficiency. The enzyme recognizes the modification site G18 by its position relative to the ribose phosphate backbones in the three-dimensional core of tRNA.
These results, in combination those of a previous report (7), led us to propose the following model for TrmH recognition (Fig. 4C). Because both the D-arm structure and other conserved nucleotides (U8, purine 26, G46, U54, U55, and C56) in the tRNA are components that have been shown to affect methylation efficiency, we hypothesize that TrmH recognition of the G18 modification site is achieved by assessment of the relative distances and angles between ribose phosphate backbones in the three-dimensional core of tRNA. We predict that unusually large D-loops alter the presentation of G18 relative to ribose phosphate backbones in the three-dimensional core. In addition, large D-loops may result in unfavorable interactions between D- and T-loops or between D-loop and TrmH upon complex formation.
TrmH Binds Tightly with a dG18dG19A20 tRNA That Is Not Methylated
To investigate interactions between TrmH and tRNAPhe in the absence of any D-loop methylation targets, we substituted the G18G19G20 sequence with dG18dG19A20. The sequence of the tRNA-dG18dG19A20 variant was verified using the Donis-Keller method following ligation of 5′- and 3′-synthetic fragments (Fig. 5A). Standard methylation assays with TrmH and the tRNA-dG18dG19A20 variant demonstrated near-complete loss of methyl group acceptance activity (Fig. 5B).
FIGURE 5.
tRNA-dG18dG19A20 variant was not methylated by TrmH. A, RNA sequencing of tRNA-dG18dG19A20 variant using the Donis-Keller method. Lane C, untreated control RNA; lane Al (left), alkaline hydrolysis (95 °C, 2 min); lanes RNase T1 included 0.03, 0.1, or 0.3 units of enzyme; lanes RNase U2 included 0.01, 0.03, or 0.1 units of enzyme; Lane Al (right), alkaline hydrolysis (95 °C, 4 min). Positions of G and A nucleotides are marked on both sides of the gel, and those of dG18, Gm19, and G20 are indicated with arrows. B, elimination of the methyl group acceptance activity by substitution of G18G19G20 by dG18dG19A20. Methylation of tRNA-dG18 monitored by 10% PAGE (7 m urea). TrmH was pre-incubated with [14C]AdoMet, and then with either wild-type tRNAPhe (left lanes) or tRNAPhe-dG18dG19A20 (right lanes) prior to analysis by 10% PAGE (7 m urea). The gel was stained with methylene blue (MB, left panel), and an autoradiographic image of the same gel was then captured using a Fuji BAS2000 imaging analyzer (right panel).
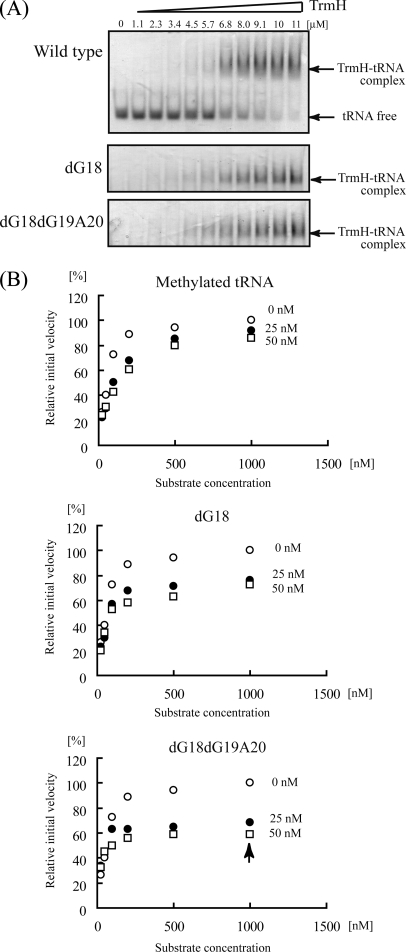
Gel mobility shift assays using tRNAPhe-dG18dG19A20 were used to investigate TrmH binding to this target (Fig. 6A). Wild-type, dG18, or dG18dG19A20 tRNAPhe transcript was incubated with increasing amounts of TrmH protein, and complex formation was assayed by shifts in RNA mobility in 6% native polyacrylamide gels. Gels, stained first with Coomassie Brilliant Blue for detection of protein and then with methylene blue for detection of RNA, clearly showed formation of complexes between TrmH and all three target tRNAs (Fig. 6A). Although formation of TrmH tetramers at high concentrations of dimeric enzyme prevented calculation of precise Kd values (16, 17), these gels clearly demonstrate an affinity between TrmH and the tRNA-dG18dG19A20 variant (Fig. 6A, lower panel) that is comparable with binding with wild-type or dG18 tRNAs.
FIGURE 6.
Gel shift assay and inhibition experiments with tRNA-dG18 and -dG18dG19A20 variants. A, steady-state binding of TrmH to wild-type tRNAPhe (upper panel), tRNA-dG18 (middle panel), and tRNA-dG18dG19A20 (lower panel) variants. Transcripts incubated with increasing concentrations of TrmH protein in a binding reaction were then resolved by 6% native PAGE. Gels were stained sequentially with Coomassie Brilliant Blue and methylene blue for visualization of protein and RNA, respectively. B, inhibition of methyl transfer to wild-type tRNA in the presence of methylated tRNAs containing Gm18 (upper panel), tRNA-dG18 (middle panel), or tRNA-dG18dG19A20 (lower panel). The concentration of TrmH was fixed at 80 nm, whereas the concentrations of the tRNA variant heading each graph are 0 nm (open circles), 25 nm (closed circles), and 50 nm (open squares).
The binding of TrmH to the tRNA-dG18dG19A20 variant was also confirmed in substrate competition experiments (Fig. 6B). Using a wild-type tRNAPhe transcript control that was methylated to completion by TrmH with nonradioisotope-labeled AdoMet, we first confirmed that the pre-methylated tRNA shows no further methyl acceptance in the presence of 80 nm TrmH and 37 μm [14C]AdoMet (data not shown). The upper graph in Fig. 6B demonstrates inhibition of TrmH methyl transfer to unmethylated wild-type tRNAPhe with a pre-methylated form of the same sequence.
Similarly, we performed methyl transfer reactions with TrmH and wild-type tRNAPhe in the presence of increasing concentrations tRNA-dG18 (Fig. 6B, middle panel) or tRNA-dG18dG19A20 (Fig. 6B, lower panel). Unexpectedly, both the tRNA-dG18 (Fig. 6B, middle panel) and tRNA-dG18dG19A20 (Fig. 6B, lower panel) variants inhibited the tRNAPhe methylation by TrmH more effectively than the pre-methylated wild-type tRNA. This was particularly obvious with low concentrations of tRNA-dG18dG19A20 inhibitor, in that a concentration of 25 nm tRNA-dG18dG19A20 (1/40 of the substrate tRNA concentration) resulted in a 35% decrease in the initial velocity of methyl transfer (Fig. 6B, arrow in lower panel).
With the concentration of TrmH dimers fixed at 80 nm, the resulting kinetics of methyl transfer imply that inhibition by tRNA-dG18dG19A20 does not follow a simple competition model. In contrast, these experimental results suggest that TrmH binds tightly with tRNA-dG18dG19A20 and that dissociation of this complex is slow relative to methylated tRNA-TrmH complexes.
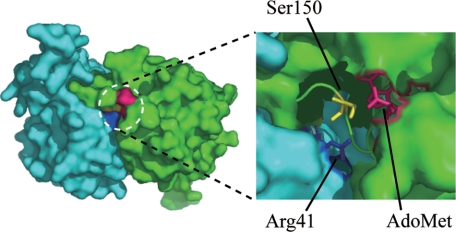
Furthermore, tRNA-dG18 clearly inhibits more effectively for TrmH methyltransferase than wild-type methylated tRNAPhe. These results suggest that although wild-type methylated tRNA is excluded during the induced fit process to TrmH, tRNA-dG18 is bound by TrmH like a normal tRNA with an unmodified G18. The crystal structure of TrmH provides support for exclusion of methylated tRNA. Because the catalytic pocket of TrmH is quite narrow (Fig. 7), the AdoMet would sterically hinder binding of the methylated G18 in the catalytic pocket, thereby excluding methylated tRNA in the induced fit process. In contrast, we hypothesize that the dG nucleotides in tRNA-dG18 or tRNA-dG18dG19A20 are not bulky enough to be excluded from the TrmH catalytic pocket; binding of these variants would therefore result in complexes that cannot accept a transferred methyl group and thereby prevent dissociation of Arg-41 and phosphate and rapid catalytic cycling of enzyme (Fig. 1B).
FIGURE 7.
Catalytic pocket of TrmH. Structure of a TrmH dimer represented by a space-filling model (left). Conserved amino acid residues in the catalytic pocket include Arg-41 (dark blue), Ser-150 (yellow), and AdoMet (red) in the catalytic pocket (right).
Oxygen 6 Atom in Guanine Base Is a Positive Determinant for TrmH
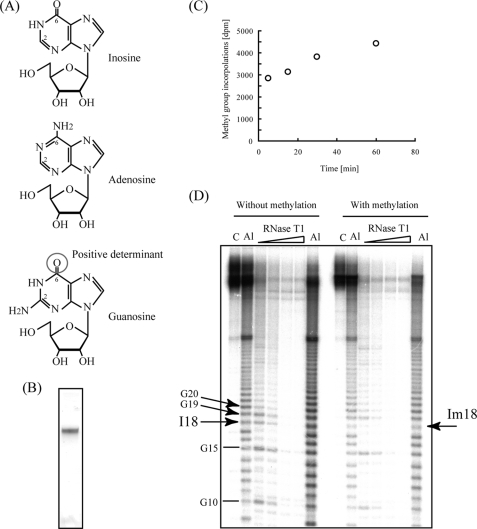
Nucleotide recognition by TrmH is restricted to guanosine in the D-loop, with no demonstrated recognition of adenosine or pyrimidines. To investigate what components within guanosine nucleotides function in recognition, we prepared a mutant tRNAPhe transcript in which G18 was substituted by inosine, which contains a oxygen 6 in its purine ring, similar to guanosine (Fig. 8A). As in the construction of tRNAPhe-dG18, a 5′-fragment containing I18 was synthesized and ligated to a corresponding 3′-fragment (Fig. 8B) to generate synthetic tRNA-I18. Modification of the resulting molecule by TrmH was efficient (Fig. 8C), and the site of modification was identified as I18 using the Donis-Keller method (Fig. 8D). As expected, cleavage of unmodified tRNAPhe-I18 using RNase T1 resulted in a band at the site of the inosine modification (22). In contrast, the intensity of cleavage at I18 in methylated tRNAPhe-I18 decreased in both the RNase T1 and alkaline hydrolysis ladders. These results demonstrate that TrmH is capable of methylating the 2′-O-residue of I18 and lead us to conclude that the corresponding oxygen 6 atom in G18 is a positive determinant for TrmH recognition.
FIGURE 8.
Substitution of G18 by inosine. A, structural representations of inosine (upper panel), adenosine (middle panel), and guanosine (lower panel). The oxygen 6 in guanosine is enclosed by a circle. B, purified tRNA-I18 analyzed by 10% PAGE (7 m urea) and stained with methylene blue. C, time-dependent increase in steady-state levels of methyl group incorporation into tRNA-I18. The time course experiment showed that more than 70% of the variant was methylated within 5 min. D, determination of the modification site of the tRNA-I18 variant using the Donis-Keller method. Unmodified RNA was cleaved by RNase T1 at I18 (left), whereas alkali and RNase T1 ladder rungs corresponding to cleavage at I18 disappeared following methylation by TrmH (right). Amounts of RNase T1 were 0.03, 0.05, 0.07, and 0.1 units per one sample (from left to right). Positions of nucleotides I18, G19, G20, and Im18 are marked with arrows.
Pre-steady State Kinetic Study by Stopped-flow Fluorescence Measurements
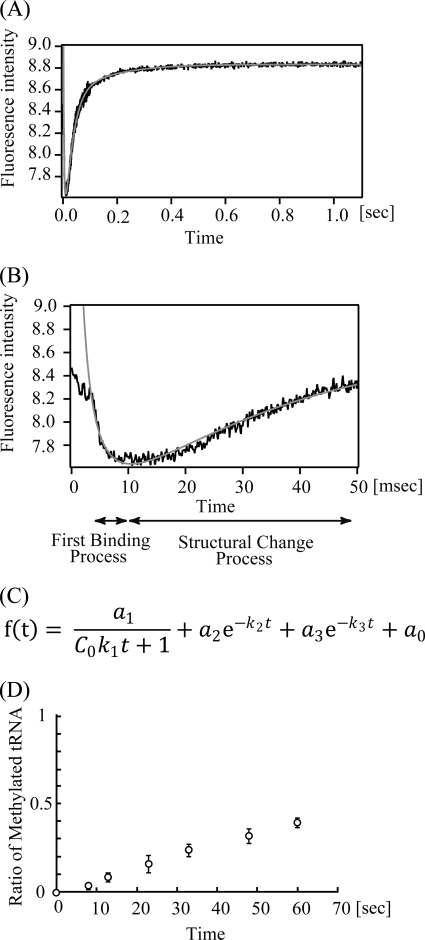
The inhibition experiments suggested that the interaction of TrmH and tRNA is composed of at least two phases, the first binding and the second induced fit processes. This idea is reasonable because the oxygen 6 atom of G18 is embedded in the three-dimensional core of tRNA. For recognition of the oxygen 6 atom of G18 by TrmH, G18 and U55 (or Ψ55) base pair should be disrupted. To investigate the interaction of TrmH and tRNA, we performed pre-steady state kinetic analysis of the interaction by stopped-flow analysis (23, 24). In general, the stopped-flow analysis of tRNA methyltransferase and tRNA may not be so easy because tRNA methyltransferase does not have a chromophore. Fortunately, however, TrmH has three tryptophan residues (Trp-73, Trp-126, and Trp-191) per one subunit, which allowed us to do the stopped-flow experiments using the fluorescence derived from these residues. Because tRNA has an absorption at 295 nm (excitation wavelength), an excess amount of tRNA could not be used. Therefore, we used an equimolar amount of TrmH and tRNA in two experiments. TrmH and tRNA (1.93 or 7.70 μm) were rapidly mixed in a stopped-flow apparatus, and the fluorescence above 320 nm was monitored. Fig. 9A shows a typical kinetic reaction curve with 7.70 μm TrmH and tRNA, and the first 50 ms is highlighted in Fig. 9B. As shown in Fig. 9, A and B, a very fast decrease of fluorescence was observed within the first 10 ms, and then relatively slow increases were observed from 10 ms to 0.5 s. In general, the decrease of tryptophan fluorescence intensity suggests an increase in accessibility of the residues to solvent water if the tryptophan fluorescence is not quenched in the initial state. The obtained data could be fitted to the equation shown in Fig. 9C, where the first term represents the bi-molecular binding reaction of TrmH and tRNA, and the second and third terms represent conformational changes. The kinetic data after 3 ms were used for calculating the kinetic parameters (relative amplitudes a1, a2, and a3 and apparent rate constants k1, k2, and k3), and they are given in Table 2. As shown in Table 2, the first decrease of fluorescence can be well fitted to a second-order reaction with a rate constant k1 of (3–27) × 107 m−1 s−1. The very large k1 value of an order of 107–108 means that the complex formation of TrmH and tRNA is caused by a diffusion-limited association (23, 24). In contrast, the subsequent slow increase of fluorescence can be fitted as first-order reactions, in which two phases with different rate constants (k2 and k3) exist. Thus, the increase of fluorescence is derived from structural changes of the TrmH and tRNA complex, suggesting that these phases may be involved in induced fit processes. To unambiguously implicate complex formation in the structural changes observed by stopped-flow fluorescence, we performed the methyl transfer assay under the same condition (7.70 μm TrmH and tRNA) (Fig. 9D). As shown in Fig. 9D, methyl transfer reaction at 25 °C was very slow. Because tRNA concentration is not in excess, it is difficult to explain the mechanism correctly. However, it should be mentioned that the slow methyl transfer was not caused by slow binding of TrmH to tRNA because the stopped-flow analysis showed that the majority of TrmH bound to tRNA within 10 ms (Fig. 9B). Thus, the majority of TrmH did not transfer the methyl group to tRNA for first 8 s (Fig. 9D), suggesting that the methyl transfer reaction had scarcely occurred for first 1.1 s in the stopped-flow analysis (Fig. 9, A and B). Taking these experimental results together, we concluded that the complex formation of TrmH and tRNA is composed of at least three phases, the very rapid first binding process and the relatively slow structural change process. Induced fitting (disruption of tRNA L-shaped structure, guanine base recognition, and introduction of ribose into the catalytic pocket) should be included in the structural change processes.
FIGURE 9.
Pre-steady state kinetic study by stopped-flow fluorescence measurements. Changes of fluorescence intensity derived from tryptophan residues in TrmH with tRNA binding were monitored (A). This figure shows the average of 20 scans of 7.7 μm TrmH and tRNA complex formation. The first 50 ms is highlighted (B). The dead time of the measurement was 3 ms. The data could be fitted by an equation (C) using IGOR Pro 6.10 (WaveMetrics, Inc.). The abbreviations used are as follows: a0, the fluorescence intensity when t = ∞; a1–a3, each amplitude; C0, the initial concentration of TrmH and tRNA (m); k1, second-order rate constant (m−1s−1); k2–k3, each first-order rate constant (s−1); t, time (s). The theoretical fitting curves are indicated in gray (A and B). The calculated parameters are given in Table 2. D, ratio of the methylated tRNA was monitored by filter assay with [3H]AdoMet.
TABLE 2.
Kinetic parameters of pre-steady state kinetics
| TrmH | a0 | a1 | a2 | a3 | k1 | k2 | k3 |
|---|---|---|---|---|---|---|---|
| μm | m−1s−1 | s−1 | s−1 | ||||
| 1.93 | 4.98 ± 0.00 | 2.24 ± 0.10 | −0.560 ± 0.043 | −0.368 ± 0.018 | 2.67 × 108 ± 0.42 × 108 | 70.2 ± 2.1 | 13.9 ± 0.3 |
| 7.70 | 8.79 ± 0.01 | 4.74 ± 0.08 | −2.84 ± 0.12 | −0.782 ± 0.059 | 3.06 × 107 ± 0.31 × 107 | 43.8 ± 0.9 | 8.69 ± 0.39 |
DISCUSSION
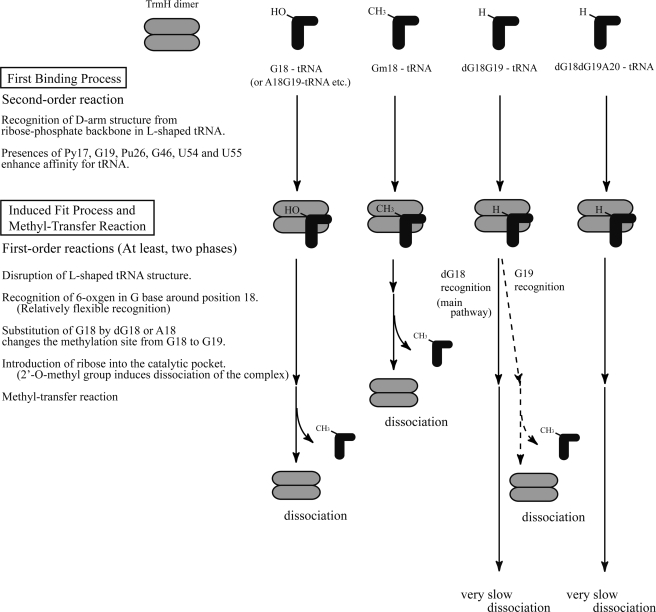
The results of our study, as depicted in the model in Fig. 10, suggest that tRNA recognition by TrmH consists of at least two steps prior to methyl transfer as follows: binding of TrmH to tRNA first, followed by a process of fitting a G nucleotide into the catalytic site of TrmH. A model for the initial binding process was reported in an earlier study (7), where we found that conserved and semi-conserved tRNA nucleotides, such as Py17, G19, Pu26, G46, U54, and U55, enhance TrmH affinity for substrate tRNA. The L-shaped tRNA structure is required for efficient methylation, although the enzyme can methylate a 5′-half-fragment of tRNA (25). In the tRNA structures, D-arm structure is essentially required (7). Because the G18 is embedded in the three-dimensional core of tRNA and oxygen 6 atom forms a hydrogen bond with U55 (or Ψ55), disruption of the tertiary bases pairs between D- and T-loops is required for interaction of the G18 and TrmH. This structural change enables the enzyme to access guanine bases (e.g. G19) around G18 in the D-loop.
FIGURE 10.
Model for TrmH recognition of wild-type and variant tRNA molecules. An outline of the initial binding process is reiterated from a previous study (7). The second process in the model, which we refer to as :induced fit,“ involves at least three steps as follows: disruption of L-shaped tRNA structure, recognition of oxygen 6 in the G base, and introduction of ribose into the TrmH catalytic pocket. Although recognition of the oxygen 6 in guanine is essential, the position of the target guanine within D-loop is somewhat flexible. tRNA methylated at G18 (Gm18) is excluded from TrmH binding in the induced fit process. Mechanistic details of the proposed model are discussed in the text.
In this study, we found that substitution of G18 by dG18 or A18 changes the methylation target site from G18 to G19. Furthermore, we found that the G18G19 dinucleotide sequence is not essential for methylation by TrmH but that methyl transfer efficiency decreases severely in sequence variants due to both an increase in Km and a decrease in Vmax values. The kinetic study suggests that dG18 or A18 substitutions affect both initial binding and the subsequent induced fit process. The pre-steady state kinetic study by stopped-flow fluorescence measurement clearly shows that the first binding process is very rapid as follows: within 10 ms the process completes at 25 °C.
In the case of the tRNAPhe-A18G19A20 variant, 2′-O-methylation did not occur at A18, and only Gm19 resulted, demonstrating the specificity of TrmH for guanosine in the tRNA D-loop. Specificity for guanosine and exclusion of adenosine and pyrimidines would be predicted for the induced fit process, because G18 base is embedded in the elbow region of the three-dimensional core of L-shaped tRNA, and the oxygen 6 atom in G18 forms a hydrogen bond with Ψ55 (4, 5). The position selection of the target guanosine in the D-loop is a relatively flexible process, because TrmH methylates guanosine irrespective of D-loop size (4–12 nucleotides). During the fitting process, the oxygen 6 atom of guanine functions as a positive recognition site for TrmH. To clarify which amino acid residues may be responsible for guanosine recognition, we placed a guanosine into the TrmH crystal structure. However, only structures with substantial steric hindrance could be modeled (data not shown), suggesting that the local structure around the catalytic pocket of the TrmH changes in the presence of tRNA. The structural change processes were observed in the stopped-flow fluorescent measurements.
In the induced fit process, tRNA containing Gm18 is excluded by a proposed steric hindrance between the methyl groups of AdoMet and 2′-O-methyl ribose. At the present level of sensitivity, we detect no modification of Gm18Gm19 tRNAs by TrmH (8, 26). In fact, tRNA containing Gm18Gm19 modification has not been reported (1). Although these facts suggest that the exclusion of the tRNA containing Gm18 occurs prior to guanine base recognition, the precise mechanism for exclusion will likely require x-ray analysis of tRNA-TrmH co-crystals.
In living T. thermophilus cells, exclusion of methylated tRNAs has the effect of enhancing the efficiency of TrmH methylation of acceptable substrates. Initial velocities for the tRNAPhe-dG18 transcript suggest that TrmH recognition of G19 occurs 30-fold less often than dG18 (Table 1). Thus, the majority of tRNA-dG18G19 forms a tight tRNA-TrmH complex, similar to tRNA-dG18dG19A20, in which methyl transfer does not occur. Inhibition experiments reveal that dissociation of TrmH from these ineffective methyl group acceptor molecules is very slow.
The TrmH protein shares a topological knot structure with members of the class IV AdoMet-dependent methyltransferases, the majority of which have been verified as enzymatically active RNA methyltransferases (27, 28). Although numerous crystal structures of the class IV enzymes have been reported in the past 7 years (14, 15, 29–37), information concerning interactions between these enzymes and RNA is limited. E. coli tRNA (m1G37) methyltransferase (TrmD) strongly recognizes the G36 in the G36G37 sequence (38–40), whereas A. aeolicus TrmD recognizes the G36 in both G36G37 and G36A37 sequences (41). Hou and co-workers (42, 44) have reported that the TrmD recognizes the G36 from the anticodon stem side, and this mechanism is completely different from archaeal tRNA (m1G37) methyltransferase (Trm5), which belongs to the class I enzymes (43). Furthermore, archaeal tRNA 2′-O-methylcytidine 56 (Cm56) methyltransferase (aTrm56) recognizes the C56 in the T-loop (37, 45). Moreover, tRNA (Cm32/Um32) methyltransferase (TrmJ) recognizes the C or U at position 32 in the anticodon (46). However, the target nucleotide recognition by these enzymes is still unknown. In this study, we determined that TrmH recognizes target guanosines in the D-loop and that the inducible fit process consists of multiple steps that minimally include recognition of oxygen 6 in guanosine and exclusion of 2′-O-methylated ribose. Although target RNA species, modification sites, and resulting modified nucleotides differ among class IV enzymes, we are optimistic that some of these findings may contribute to structural studies of other class IV AdoMet-dependent methyltransferases.
Supplementary Material
Acknowledgments
We thank members of Ehime University, Prof. Yaeta Endo for use of laboratory facilities, and Drs Akira Hirata and Kazunori Watanabe for valuable discussions.
This work was supported in part by Research Fellowship for Young Scientists 21-10011 (to A. O.), Grant-in-aid for Science Research on Priority Areas 20034041 (to H. H.), and Grant-in-aid for Science Research 19350087 (to H. H.) from the Japan Society for the Promotion of Science.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- Ψ55
- pseudouridine 55
- AdoMet
- S-adenosyl-l-methionine
- AdoHcy
- S-adenosyl-l-homocysteine.
REFERENCES
- 1.Rozenski J., Crain P. F., McCloskey J. A. (1999) Nucleic Acids Res. 27, 196–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunin-Horkawicz S., Czerwoniec A., Gajda M. J., Feder M., Grosjean H., Bujnicki J. M. (2006) Nucleic Acids Res. 34, D145–D149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia G., R., Goodenough-Lashhua D. M. (1998) in Modification and Editing of RNA (Grosjean H., Benne R. eds) pp. 555–560, American Society for Microbiology, Washington, D. C. [Google Scholar]
- 4.Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. (1974) Nature 250, 546–551 [DOI] [PubMed] [Google Scholar]
- 5.Kim S. H., Sussman J. L., Suddath F. L., Quigley G. J., McPherson A., Wang A. H., Seeman N. C., Rich A. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4970–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urbonavicius J., Durand J. M., Björk G. R. (2002) J. Bacteriol. 184, 5348–5357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hori H., Yamazaki N., Matsumoto T., Watanabe Y., Ueda T., Nishikawa K., Kumagai I., Watanabe K. (1998) J. Biol. Chem. 273, 25721–25727 [DOI] [PubMed] [Google Scholar]
- 8.Hori H., Suzuki T., Sugawara K., Inoue Y., Shibata T., Kuramitsu S., Yokoyama S., Oshima T., Watanabe K. (2002) Genes Cells 7, 259–272 [DOI] [PubMed] [Google Scholar]
- 9.Hori H., Kubota S., Watanabe K., Kim J. M., Ogasawara T., Sawasaki T., Endo Y. (2003) J. Biol. Chem. 278, 25081–25090 [DOI] [PubMed] [Google Scholar]
- 10.Persson B. C., Jager G., Gustafsson C. (1997) Nucleic Acids Res. 25, 4093–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gustafsson C., Reid R., Greene P. J., Santi D. V. (1996) Nucleic Acids Res. 24, 3756–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavaillé J., Chetouani F., Bachellerie J. P. (1999) RNA 5, 66–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schubert H. L., Blumenthal R. M., Cheng X. (2003) Trends Biochem. Sci. 28, 329–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nureki O., Shirouzu M., Hashimoto K., Ishitani R., Terada T., Tamakoshi M., Oshima T., Chijimatsu M., Takio K., Vassylyev D. G., Shibata T., Inoue Y., Kuramitsu S., Yokoyama S. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1129–1137 [DOI] [PubMed] [Google Scholar]
- 15.Nureki O, Watanabe K., Fukai S., Ishii R., Endo Y., Hori H., Yokoyama S. (2004) Structure 12, 593–602 [DOI] [PubMed] [Google Scholar]
- 16.Watanabe K., Nureki O., Fukai S., Ishii R., Okamoto H., Yokoyama S., Endo Y., Hori H. (2005) J. Biol. Chem. 280, 10368–10377 [DOI] [PubMed] [Google Scholar]
- 17.Watanabe K., Nureki O., Fukai S., Endo Y., Hori H. (2006) J. Biol. Chem. 281, 34630–34639 [DOI] [PubMed] [Google Scholar]
- 18.Donis-Keller H., Maxam A. M., Gilbert W. (1977) Nucleic Acids Res. 4, 2527–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamoto H., Watanabe K., Ikeuchi Y., Suzuki T., Endo Y., Hori H. (2004) J. Biol. Chem. 279, 49151–49159 [DOI] [PubMed] [Google Scholar]
- 20.Keith G. (1995) Biochimie 77, 142–144 [DOI] [PubMed] [Google Scholar]
- 21.Brissette P., Ballou D. P., Massey V. (1989) Anal. Biochem. 181, 234–238 [DOI] [PubMed] [Google Scholar]
- 22.Jo Chitester B., Walz F. G., Jr. (2002) Arch. Biochem. Biophys. 406, 73–77 [DOI] [PubMed] [Google Scholar]
- 23.Koren R., Hammes G. G. (1976) Biochemistry 15, 1165–1171 [DOI] [PubMed] [Google Scholar]
- 24.Alsallaq R., Zhou H. X. (2008) Proteins 71, 320–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hori H., Saneyoshi M., Kumagai I., Miura K., Watanabe K. (1989) J. Biochem. 106, 798–802 [DOI] [PubMed] [Google Scholar]
- 26.Kumagai I., Watanabe K., Oshima T. (1982) J. Biol. Chem. 257, 7388–7395 [PubMed] [Google Scholar]
- 27.Tkaczuk K. L., Dunin-Horkawicz S., Purta E., Bujnicki J. M. (2007) BMC Bioinformatics 8, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anantharaman V., Koonin E. V., Aravind L. (2002) J. Mol. Microbiol. Biotechnol. 4, 71–75 [PubMed] [Google Scholar]
- 29.Zarembinski T. I., Kim Y., Peterson K., Christendat D., Dharamsi A., Arrowsmith C. H., Edwards A. M., Joachimiak A. (2003) Proteins 50, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim K., Zhang H., Tempczyk A., Krajewski W., Bonander N., Toedt J., Howard A., Eisenstein E., Herzberg O. (2003) Proteins 51, 56–67 [DOI] [PubMed] [Google Scholar]
- 31.Forouhar F., Shen J., Xiao R., Acton T. B., Montelione G. T., Tong L. (2003) Proteins 53, 329–332 [DOI] [PubMed] [Google Scholar]
- 32.Mosbacher T. G., Bechthold A., Schulz G. E. (2005) J. Mol. Biol. 345, 535–545 [DOI] [PubMed] [Google Scholar]
- 33.Pleshe E., Truesdell J., Batey R. T. (2005) Acta Crystallogr. Sect. F Struct. Biol. Crystalliz. Comm. 61, 722–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahn H. J., Kim H. W., Yoon H. J., Lee B. I., Suh S. W., Yang J. K. (2003) EMBO J. 22, 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elkins P. A., Watts J. M., Zalacain M., van Thiel A., Vitazka P. R., Redlak M., Andraos-Selim C., Rastinejad F., Holmes W. M. (2003) J. Mol. Biol. 333, 931–949 [DOI] [PubMed] [Google Scholar]
- 36.Liu J., Wang W., Shin D. H., Yokota H., Kim R., Kim S. H. (2003) Proteins 53, 326–328 [DOI] [PubMed] [Google Scholar]
- 37.Kuratani M., Bessho Y., Nishimoto M., Grosjean H., Yokoyama S. (2008) J. Mol. Biol. 375, 1064–1075 [DOI] [PubMed] [Google Scholar]
- 38.Byström A. S., Björk G. R. (1982) Mol. Gen. Genet. 188, 440–446 [DOI] [PubMed] [Google Scholar]
- 39.Holmes W. M., Andraos-Selim C., Roberts I., Wahab S. Z. (1992) J. Biol. Chem. 267, 13440–13445 [PubMed] [Google Scholar]
- 40.Redlak M., Andraos-Selim C., Giege R., Florentz C., Holmes W. M. (1997) Biochemistry 36, 8699–8709 [DOI] [PubMed] [Google Scholar]
- 41.Takeda H., Toyooka T., Ikeuchi Y., Yokobori S., Okadome K., Takano F., Oshima T., Suzuki T., Endo Y., Hori H. (2006) Genes Cells 11, 1353–1365 [DOI] [PubMed] [Google Scholar]
- 42.Christian T., Evilia C., Williams S., Hou Y. M. (2004) J. Mol. Biol. 339, 707–719 [DOI] [PubMed] [Google Scholar]
- 43.Goto-Ito S., Ito T., Ishii R., Muto Y., Bessho Y., Yokoyama S. (2008) Proteins 72, 1274–1289 [DOI] [PubMed] [Google Scholar]
- 44.Christian T., Hou Y. M. (2007) J. Mol. Biol. 373, 623–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Renalier M. H., Joseph N., Gaspin C., Thebault P., Mougin A. (2005) RNA 11, 1051–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purta E., van Vliet F., Tkaczuk K. L., Dunin-Horkawicz S., Mori H., Droogmans L., Bujnicki J. M. (2006) BMC Mol. Biol. 18, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.