Abstract
We reveal a set of divergent octamer elements in Drosophila melanogaster (dm) core histone gene promoters. These elements recruit transcription factor POU-domain protein in D. melanogaster 1 (Pdm-1), which along with co-activator dmOct-1 coactivator in S-phase (dmOCA-S), activates transcription from at least the Drosophila histone 2B (dmH2B) and 4 (dmH4) promoters in a fashion similar to the transcription of mammalian histone 2B (H2B) gene activated by octamer binding transcription factor 1 (Oct-1) and Oct-1 coactivator in S-phase (OCA-S). The expression of core histone genes in both kingdoms is coordinated; however, although the expression of mammalian histone genes involves subtype-specific transcription factors and/or co-activator(s), the expression of Drosophila core histone genes is regulated by a common module (Pdm-1/dmOCA-S) in a directly coordinated manner. Finally, dmOCA-S is recruited to the Drosophila histone locus bodies in the S-phase, marking S-phase-specific transcription activation of core histone genes.
Keywords: Cell/Cycle, DNA/Synthesis, Evolution/DNA, Evolution/Protein, Gene/Promoters, Histones
Introduction
Transcription is the foremost critical step for regulating gene activities. Transcription regulation involves interplay among trans-acting activities such as general and gene-specific transcription factors and co-activators in conjunction with RNA polymerase II and cis-acting regulatory elements such as core and proximal promoter elements that are pivotal in recruiting transcription regulators to specify gene expression programs; in turn, changes in transcription (co)factor activities and/or selectivity dictate gene expression outputs, and understanding mechanistic aspects of various gene expression programs often leads to a better understanding of many aspects of physiology (1).
We are interested in characterizing the cis- and trans-regulatory networks of the Drosophila core histone genes, which are among the most conserved eukaryotic genes. The conserved DNA replication-dependent canonical histone genes belong to a multigene family, and the encoded proteins (core histones H2A, H2B, H3, and H4 and linker histone H1) are essential components of nucleosomes, the building blocks of metazoan chromatin. Core histone genes of diverse species have clustered features (2). There are two clusters of histone genes in mammalian cells, with the larger cluster (human chromosome 6, mouse chromosome 13) comprising ∼80% of the genes and the smaller one (human chromosome 1, mouse chromosome 3) containing the remaining (3–5). In Drosophila, multicopied core and linker histone genes are clustered as ∼5-kb repeats on chromosome 2 (6); Xenopus histone genes are similarly organized (7).
Histone biosynthesis occurs almost exclusively in the S-phase (8). For instance, the human H2B (hH2B)3 gene promoter contains an octamer element (ATTTGCAT) that anchors octamer binding transcription factor 1 (Oct-1), which recruits OCA-S to bring about S-phase-specific H2B expression (9). The transcription of mammalian (core) histone genes is mediated by subtype-specific promoter elements and associated transcription (co)factors (10–13); however, the expression of these genes is highly coordinated via a mechanism that is not yet characterized (2, 4, 9–12, 14).
A recent study (15) suggests that the TATA-less Drosophila melanogaster histone H1 (dmH1) gene promoter is selectively regulated by TATA box-binding protein-related factor 2 (TRF2) and that this selective usage versus that of TATA-box-binding protein (TBP) for TATA-containing core histone genes provides a novel mechanism that differentially directs histone gene transcription within the histone gene cluster; however, a detailed molecular description of the mechanism(s) that governs coordinated expression of Drosophila core histone genes is not yet established and requires investigation as one of the critical steps toward understanding Drosophila histone gene regulation pathways.
POU-domain protein in D. melanogaster 1 (Pdm-1, also dubbed dmPOU19 or Nubbin) was first characterized as a protein containing a highly conserved POU domain and is highly expressed during early Drosophila embryo development and expressed at lower levels throughout the rest of development (16). Subsequent studies had identified roles of Pdm-1 in neuronal cell fate specification (17, 18) and in setting up a threshold for the Notch activity in boundary formation in the Drosophila wing (19); however, a potential involvement in the expression of the choline acetyltransferase gene aside (20), little is known about the roles of Pdm-1 as a transcription factor. This is especially true given the roles of its mammalian counterpart Oct-1 in regulating a number of genes including the H2B gene, in which an octamer (ATTTGCAT) element in the gene promoter mediates the S-phase-dependent transcription (10–13).
Characterization of the Drosophila hydei histone genes have revealed no canonical promoter octamer elements (21). Nevertheless, we have focused on the potential transcriptional role(s) of Pdm-1 on Drosophila core histone genes, and this effort identified multiple evolutionarily diversified octamer elements on not only the dmH2B but also dmH4 promoters pivotal for Pdm-1 to function as a transcription factor. Pdm-1 may also act on other Drosophila core histone genes and is absolutely essential for recruiting the D. melanogaster co-activator dmOCA-S, which likely exerts an S-phase transcriptional regulation to directly coordinate the expression of all core histone genes.
EXPERIMENTAL PROCEDURES
RNA Interference (RNAi) in Drosophila Schneider-2 (S2) Cells Using Double-stranded RNAs (dsRNAs)
Cells (1 × 106/ml) in serum-free medium (Invitrogen) were plated in a six-well cell culture dish (Nunc); dsRNA was added directly to the media at different concentrations followed by vigorous agitation. Cells were incubated for 30 min at room temperature followed by the addition of 2 ml of Schneider's Drosophila medium (Invitrogen) containing 5% each of fetal bovine serum and bovine calf serum (JRH Biosciences). The cells were typically incubated for 3 days to allow for completion of RNAi. For dsRNA production, DNA fragments ∼700 bp in length containing coding sequences for the proteins to be knocked down were amplified by PCR using primers containing T7 RNA polymerase promoter (GAATTAATACGACTCACTATAGGGAGA) sequence in front of gene-specific sequences (below). PCR products were used as templates to produce dsRNAs with a MEGAacript T7 transcription kit (Ambion); dsRNAs generated exhibited heterogeneous bands indicating secondary structures that were eliminated by denaturing and re-annealing. Finally, dsRNAs were purified using a MEGAclear kit (Ambion).
Primers
Primer pairs for dsRNA synthesis were: ATGGTTATGTCGGAGCTACGTTGGC and GATTGTTCATGCCCAAGCCAGCT, Pdm-1; ATGGCGGCTAACAAGGAGAGGACTT and CTATTCGTAGATCCAGTCCTTGGCG, Awd; ATGGCCGCCATTAAGGACAGTCTGT and CCTGCTTGTGCAGCTCGTTCCACTT, dmLdh; GTGGCCGTCAACGATCCCTTCAT and CGTTTAGCGAAATGCCAGCCTTG, dmGapdh; ATGGAAGACGCCAAAAACATAAAG and CTACCGGAATGATTTGATTGCCAA, luciferase. Primer pairs for scoring mRNA levels in RT-relative PCR (rPCR) and RT-quantitative PCR (qPCR) (see below) were: GGAAAAGGTGGCAAAGTGAA and TGCAGATGACGCGGAATAA, dmH2A; GAAGGAGAGCTATGCCATCTA and TAGAGCTGGTGTACTTGGTGA, dmH2B; ATGGCTCGTACCAAGCAAA and GAATCGCAAGTCCGTCTTA, dmH3; TGCGTGATAACATCCAAGGTA and TACACAACATCCATGGCTGTA, dmH4; TCGTTGGAGAAGTCCTACGA and GTTGTAGGTGGTCTCGTGGA, dmActin (an internal control for both assays). Primer pairs for detecting the promoter regions in chromatin immunoprecipitation (ChIP) assays (see below) were: ACTTTGCCTTTCCCTTCA and ATTCACTTATCGTAATGTGGG, dmH2B; GTTCACGTTCACTACTTCACG and TATTAT ACACGCACAGCACG, dmH4; AGTACACTCT TCATGGCGA and TCTCTGGATTAGACGAC TGC, dmActin5C.
RT-PCR (RT-rPCR) and Real-time PCR (RT-qPCR)
Total RNA was isolated using the RNeasy kit (Qiagen) and reverse transcribed using SuperScript III (Invitrogen) and random primers. The cDNA levels were scored by rPCR on PTC-200 DNA Engine Peltier thermal cyclers (MJ Research). Semiquantitative evaluations of histone expression levels by RT-rPCR were supported by quantifications using real-time PCR (RT-qPCR) with SYBR Green Core Reagents (ABI).
Luciferase Assays
The test (wild-type or mutant dmH2B and dmH4 or dmActin) promoters were fused to the firefly luciferase gene in pGL3 (Promega). The SV40 promoter sequence in pRL-SV40 (Promega) that contains the Renilla luciferase gene was replaced with the dmActin promoter sequence to generate pRL-dmActin that served as an internal control in the Drosophila system. S2 cells (1 × 106/ml) fed with or without target dsRNAs (37 nm) for 2 or 4 days were transfected (FuGENE 6; Roche Applied Science) with reporter genes: 1 μg of pRL-dmActin with 1 μg of pGL3-dmH2B, pGL3-dmH4, or pGL3-dmActin. After 24 h, cells were harvested, and whole cell lysates were used to measure luciferase activities using Dual-luciferase Reporter Assay System (Promega). All firefly luciferase readings were normalized with the Renilla luciferase readings. For Luciferase assays of wild-type or mutant dmH2B and dmH4 promoters without prior RNAi, the reporters were transfected into cells followed by measuring the luciferase activities at 24 h.
GST Fusion Proteins
Sequences encoding dmPOU-1 and dmOCA-S components were inserted into the GST fusion protein vector pGEX-TK4E, and GST-tagged proteins were bacterially expressed and purified on glutathione-Sepharose beads (GE Healthcare). Purified proteins were dialyzed (in 5 mm NaH2PO4/Na2HPO4, pH 7.4, and 0.15 m NaCl), concentrated, and used for raising antibodies. GST and GST-dmPOU-1 used in electrophoresis mobility shift assays (EMSAs) and DNase I footprinting were eluted from affinity beads with 100 mm glutathione, 0.15 m NaCl, 5 mm NaH2PO4/Na2HPO4, pH 7.4, and 8 m urea and refolded by sequential dialysis against 6, 4, 3, 2, 1, and 0 m urea over 2 days in BC-100 buffer (20 mm Tris-HCl, pH 7.9, 20% glycerol, 0.025% Nonidet P-40, 0.25 mm EDTA, 0.125 mm EGTA, 0.5 mm DTT, 0.25 mm PMSF, and 100 mm KCl). Proteins for the functional analyses were stored at −80 °C.
Antibodies
In-house mice were immunized with the GST fusion proteins with dmPOU-1 and dmGapdh, dmAwd, dmLdh, and stress inducible protein 1 (dmSti1/p60) partial sequences to generate polyclonal antibodies. Preimmune serum was from tail bleeds of non-immunized mice. Mouse antibodies against TBP-associated factor 5 (dmTAF5) were purchased from Abcam, and goat antibodies against dmβ-tubulin and normal mouse IgGs were from Santa Cruz Biotechnologies. Rabbits were immunized with commercially available enzymatic GAPDH to generate polyclonal rabbit antibodies used in confocal analyses. Monoclonal mouse anti-Ser/Thr-ProMPM-2 was purchased from Upstate Biotechnology. Rabbit Anti-Pdm-1 is a generous gift from Dr. Yang Xiaohang, Developmental Neurobiology Research Laboratory, Department of Developmental Biology, Institute of Molecular and Cell Biology.
EMSAs
We synthesized sense and antisense oligonucleotides with the promoter-specific sequences (Fig. 1, D and E) and flanking spacer sequences (total lengths 49 nucleotides) such that the annealed probes had a 5′ T residue overhang at either end allowing Klenow enzyme to fill-in with [α33P]dATP (GE Healthcare). The reaction contained 5–40 ng of GST or GST-tagged dmPOU1 or 10 μg of crude nuclear extract in 2.5 μl of BC-100 buffer, 1 μg of double-stranded polydeoxyinosinic-deoxycytidylic acid (poly[dI-dC]-poly[dI-dC]), and ∼10 of fmol end-labeled probe in 6.25 μl of 2× EMSA buffer (25 mm Hepes, pH 7.9, 62.5 mm KCl, 0.05% Nonidet P-40, 2 mm MgCl2, 8% Ficoll (400), 500 μg/ml bovine serum albumin, protease inhibitors (0.5 mm PMSF, 5 μg/ml each of antipain, leupeptin, aprotinin, chymastatin, and pepstatin A)), and 3.75 μl of H2O. The incubation was for 30 min at room temperature. Cold probes in a 50–100-fold molar excess were included at the time of reactions for competition assays. 10 μl of the reaction was loaded on a 4% non-denaturing polyacrylamide gel (in 6.25 mm Tris, pH 8.2, 50 mm glycine, 0.1 mm EDTA, 1 mm MgCl2, 0.025% Nonidet P-40, and 0.5 mm DTT) that was pre-run for 75 min at 20 mA and 4 °C and was run for another 150 min with the buffer changed once in the middle of the run. The gel was then dried and autoradiographed. All EMSAs were repeated at least three times.
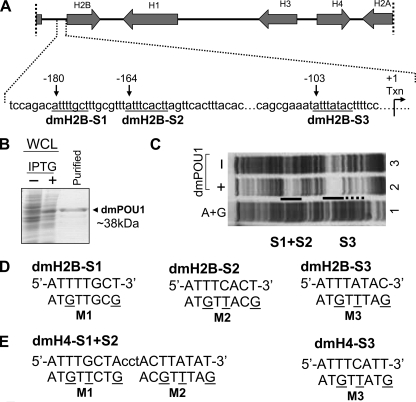
FIGURE 1.
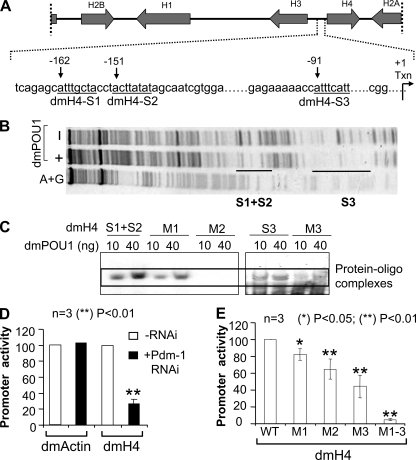
Three pivotal octamer elements for the Pdm-1 anchorage to the dmH2B promoter. A, depicted on top is a D. melanogaster histone genes cluster (∼5 kb) on chromosome 2L, with transcriptional directions of the genes indicated by arrows. Illustrated below are details of the dmH2B promoter, with octamer positions (relative to the transcription start site) indicated with arrows, and sequences (from distal to proximal, dmH2B-S1, dmH2B-S2, and dmH2B-S3) underlined. B, shown is bacterially expressed GST-Pdm-1 POU domain fusion protein (dmPOU1). Whole cell lysates (WCL) of the host XA90 Escherichia coli strain from the control and isopropyl 1-thio-β-d-galactopyranoside-induced samples were assessed for protein production (staining of the SDS-gel-resolved proteins with Coomassie Blue). Lane 3 shows the purified protein. C, shown is a footprint analysis of the promoter region of the dmH2B gene without (lane 3) or with 1 μg dmPOU1 (lane 2). The protected regions that cover dmH2B-S1, -S2, and -S3 sites are indicated. D, sequences of the dmH2B-S1, -S2, and -S3 octamer sites (uppercase) are shown. Oligonucleotides encompassing individual sites were end-labeled with 33P and used in EMSAs. Base-substituted oligonucleotides (M1, M2, and M3) are indicated with mutated residues underlined. E, sequences of the dmH4-S1+S2 and -S3 octamer sites (uppercase) are shown. Base-substituted oligonucleotides (M1, M2, and M3) are indicated with the mutated residues underlined. These oligonucleotides were used in later EMSAs (Fig. 6C).
DNase I Footprinting
The pGL3-dmH2B DNA was cut by XhoI and end-labeled with [γ-33P]ATP using T4 polynucleotide kinase, and the probe was released by a cut with NheI and gel-purified. The DNA-protein binding reaction was carried out by mixing GST-dmPOU1 (1 μg in 5 μl of BC100 (20 mm Tris-HCl, pH 7.9, 20% glycerol, 0.25 mm EDTA, 0.125 mm EGTA, 0.025% Triton X-100, 100 mm KCl, 0.5 mm DTT, 0.25 mm PMSF), ∼10 fmol of probe, and 1 μg of poly(dI-dC)-poly(dI-dC) in 12.5 μl of 2× footprinting buffer (25 mm Hepes, pH 7.9, 62.5 mm KCl, 0.05% Nonidet P-40, 2 mm MgCl2, 1 mm DTT, 16% glycerol, 6% polyethylene glycol 8000, 200 μg/ml bovine serum albumin) and 7.5 μl of H2O. The reaction was for 30 min at room temperature. DNase I (New England Biolabs) freshly diluted in 5 μl of 1× footprinting buffer was added for 1 min, and the digestion was stopped by 100 μl of 0.5 mm NH4Ac, 25 mm EDTA, 0.2% SDS, and 2 mg/ml yeast tRNA and extracted with phenol-chloroform twice. After ethanol precipitation, the footprinting and control samples were analyzed on an 8% polyacrylamide sequencing gel along with an (A+G)-cleavage reaction prepared as earlier described (22).
ChIP Assays
Each ChIP assay used 1 × 106 S2 cells, which was performed following the detailed protocol provided by the ChIP assay kit (Upstate Biotechnology). Immune complexes were collected with 60 μl of protein G-agarose and washed once with 1.4 ml each of the following buffers: low salt wash buffer ((0.1% SDS, 1% Nonidet P-40, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 150 mm NaCl; 4 h); high salt wash buffer (0.1% SDS, 1% Nonidet P-40, 2 mm EDTA, 20 mm Tris-HCl, pH 8.0, and 1500 mm NaCl; overnight); LiCl wash buffer (250 mm LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mm EDTA, and 10 mm Tris-HCl, pH 8.0, 4 h)) and twice with TE (10 mm Tris-HCl, pH 8.0, and 1 mm EDTA) for 10 min each. Immune complexes were then eluted with freshly prepared elution buffer (1% SDS and 0.1 m NaHCO3). Cross-links were then reversed by heating at 65 °C overnight in the presence of NaCl followed by proteinase K treatment. DNA was recovered by QIAquick gel extraction kit spin column (Qiagen) and subjected to PCR or real-time PCR analyses using earlier-specified primers (see “Primers”).
Western Blots
All samples were thoroughly dissolved in the radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 8, 150 mm NaCl, 1.0% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 0.2 mm NaVO4, 10 mm NaF, 0.4 mm EDTA, and 10% glycerol). To ensure an equal loading, protein concentrations were normalized for each sample, which after boiling was resolved on an 8 or 12% SDS-polyacrylamide gel. Resolved proteins were transferred to nitrocellulose membranes (Hybond-C Extra; GE Healthcare) in 25 mm CAPS (pH 11.5) and 20% methanol, and the were membranes blocked by 5% nonfat milk, 0.1% Tween 20 in Tris-buffered saline (20 mm Tris-HCl, pH 7.6, 150 mm NaCl) for 1 h before incubation with primary antibodies (in 1% nonfat milk, 0.1% Tween 20 in Tris-buffered saline) for 1 h. The bound primary antibodies were detected by horseradish peroxidase-conjugated secondary antibodies and visualized by the enhanced chemiluminescence (ECL) kit (GE Healthcare).
BrdUrd-FACS or FACS Analyses
Cells grown in 6-well plates were treated with BrdUrd(10 μm) for 45–60 min, fixed with 80% cold ethanol, treated with 3 n HCl, washed, and incubated with anti-BrdUrd monoclonal antibody (BD Biosciences) for 60 min. After washing, cells were incubated with Alexa Fluor®488 goat anti-mouse IgG (Invitrogen) for 30 min. These cells or ethanol-fixed cells (for direct FACS assays) were treated with propidium iodide and RNase A for 30 min before FACS analyses.
Protein Alignment
Comparisons of mammalian OCA-S/Oct-1 and D. melanogaster dmOCA-S/Pdm-1 proteins were performed using a Protein Information Resource at the Georgetown University Medical Center, which uses a Smith-Waterman full-length alignments module.
Cell Synchronization
Synchronization of Drosophila S2 cells was conducted as described (23). Briefly, log-phase (2 × 106/ml) cells were first incubated with 0.2 nm ponasterone A for 24 h to obtain G2 cells. Cells were then rinsed with phosphate-buffered saline (PBS) three times, resuspended in fresh Schneider's Drosophila medium (Invitrogen) containing 5% each of fetal bovine serum and bovine calf serum (JRH Biosciences), along with 1.5 mm hydroxyurea (HU), and cultured for 18 h to obtain G1/S cells. Afterward, these cells were rinsed with PBS three times, cultured in the above-specified medium without HU, and harvested at various time points for analyses.
Microscopy
Cells grown on coverslips (Fisher) were washed 2 times with PBS and then fixed with a 1:1 ratio of methanol:acetone for 10 min at room temperature. Fixed cells were blocked with indirect immunofluorescence blocking buffer (1% bovine serum albumin, 0.2% Triton X-100) for 1 h at room temperature. The cells were then immunolabeled with appropriate primary antibodies diluted in fluorescence dilution buffer (PBS with 5% normal goat serum, 5% fetal bovine serum, 2% bovine serum albumin) for 1 h at room temperature. The coverslips were then washed 5 times with 0.2% saponin PBS at 5-min intervals and incubated with properly diluted secondary antibodies in fluorescence dilution buffer at room temperature for 1 h. The coverslips were washed again with 0.2% saponin PBS 5 times at 5-min intervals and twice with PBS before being mounted on microscopic slides with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA). Confocal microscopy was performed with an Olympus Fluoview 1000 confocal microscope (Center Valley, PA). Seventeen sequential planes were acquired at axial (z) spacing of 0.44 μm to form a z-stack image.
Trypan Blue Cell Staining
Cells were harvested at 200 × g. The supernatant was discarded, and cells were stained with trypan blue in 0.2 ml of PBS (0.2% trypan blue in 0.85% PBS). The cells were left at room temperature for 3 min and then placed on ice until counted.
Mutagenesis
Base-substitution mutants (single or combinatorial sites) targeting the dmH2B and dmH4 promoters were generated by site-directed mutagenesis using a PCR-based strategy as provided by the manufacturer (TransformerTM site-directed mutagenesis kit; Clontech). All the mutations were confirmed by sequencing.
Nuclear Extract
Nuclear extract was modified from Roberts et al. (24) Nuclei were isolated from 5 × 15-cm plates, and cells were harvested by centrifugation at 5000 × g for 10 min at 4 °C. Cell pellets were resuspended in PBS and washed twice. The pellet was then resuspended in 3 volumes of ice-cold hypotonic buffer (10 mm Hepes, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 1 mg/ml pepstatin, 0.5 mg/ml leupeptin, and 0.5 mm PMSF) and incubated on ice for 10 min. The cells were lysed by 10 expulsions through a 27-gauge needle and centrifuged at 5000 × g for 20 min at 4 °C. The supernatant was discarded, and the pellet was suspended in 2.5 volumes of S1 buffer (0.25 m sucrose, 10 mm Hepes, pH 7.9, 3.3 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 1 mg/ml pepstatin, 0.5 mg/ml leupeptin, and 0.5 mm PMSF) and S3 buffer (0.88 m sucrose, 10 mm Hepes, pH 7.9, 3.3 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 1 mg/ml pepstatin, 0.5 mg/ml leupeptin, and 0.5 mm PMSF) in a 1:1 ratio followed by centrifugation at 1100 × g for 15 min at 4 °C. The supernatant was again discarded, and the pellet was suspended in BC100 (20 mm Tris-HCl, pH 7.9, 20% glycerol, 0.25 mm EDTA, 0.125 mm EGTA, 0.025% Triton X-100, 100 mm KCl, 0.5 mm DTT, and 0.25 mm PMSF). The suspension was then sonicated on ice using a Misonix sonicator with a microtip at setting 3 and with five 10-s pulses followed by 30-s intervals to release nuclear proteins. The suspension was again centrifuged at 1100 × g for 15 min at 4 °C, and the supernatant was kept and snap-frozen with liquid nitrogen before storage as nuclear extract at −80 °C.
Statistical Analyses
Statistical data in Figs. 2–4 and 6–7 were analyzed by unpaired homoscedastic t test; two-tailed p values less than 0.05 are considered statistically significant, and values less than 0.01 are considered more statistically significant. Accordingly, * corresponds to p values less than 0.05; ** indicates p values less than 0.01. Data without these indicators, or labeled with N.S. (not significant), suggest p values at ≥0.05. Experiments were carried out either three or five times (n = 3 or n = 5), and the data were presented as averages ± S.D. (error bars).
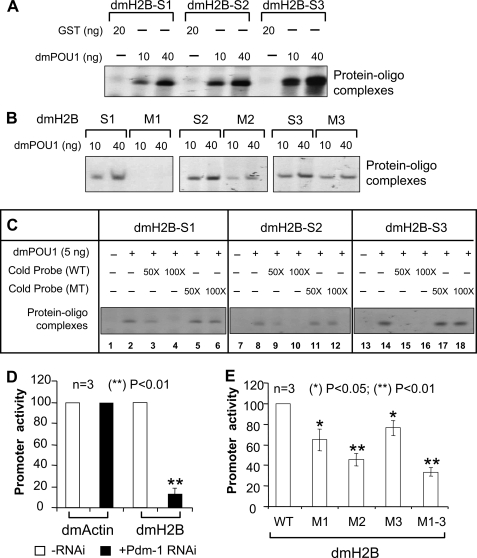
FIGURE 2.
Sequence-specific interaction of Pdm-1 with dmH2B octamer elements dictating dmH2B transcription. A, EMSAs using dmH2B octamer-containing probes with either GST-dmPOU1 or, as a control, GST are shown. B, oligonucleotide probes containing wild-type or mutant (M) dmH2B-S1, -S2, or -S3 site were EMSA-analyzed. C, shown are oligonucleotide probes containing wild-type dmH2B-S1, -S2, or -S3 site were EMSA-analyzed in the absence or presence of a 50- or 100-fold molar excess of non-labeled oligonucleotides that contained either wild-type or mutant (MT) octamer sites. D, luciferase reporter assays are shown of the dmH2B or the dmAct5C promoter in S2 cells treated without or with 37 nm Pdm-1-specific dsRNA for 72 h. E, shown are luciferase reporter assays on the wild-type versus the mutant dmH2B promoters.
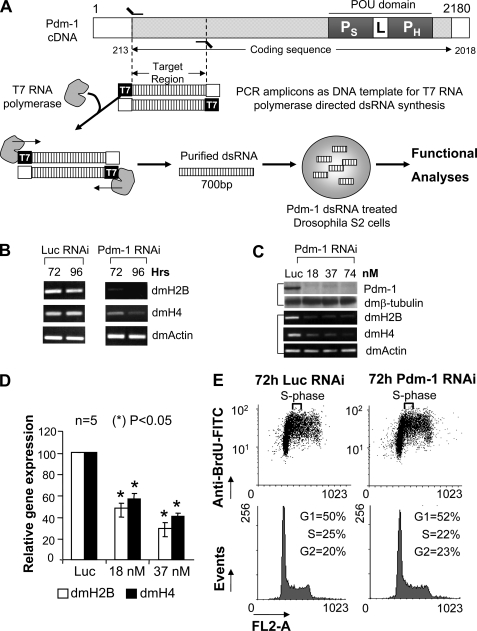
FIGURE 3.
RNAi-mediated silencing of Pdm-1 down-regulates dmH2B expression. A, shown is a dsRNA-mediated Pdm-1 RNAi strategy. Depicted is the Pdm-1 cDNA; the POU domain with POU specific (PS) and POU homeo (PH) subdomains along with the linker (L) region is shown. DNA corresponding to the first 700 bases of the coding sequence was PCR-amplified, which incorporated the T7 promoter on both ends. Then dsRNA was synthesized using T7 RNA polymerase, purified and used to treat Drosophila S2 cells. B, shown is a time course. S2 cells were treated with 37 nm luciferase dsRNA (left panel) or Pdm-1 dsRNA (right panel) for 72 or 96 h. mRNA levels of dmH2B, dmH4, and dmActin were assessed by RT-rPCR. C, shown is a dose response. S2 cells were treated with the indicated doses of Pdm-1 dsRNA or 37 nm luciferase dsRNA for 72 h. Pdm-1 and dmβ-tubulin protein levels were determined by immunoblot (upper panel), and dmH2B, dmH4, and dmActin mRNA levels were scored by RT-PCR (lower panel). D, quantification is shown of dmH2B and dmH4 expression in Pdm-1-silenced S2 cells using RT-qPCR. E, shown are cell cycle profiles of cells treated with luciferase- or Pdm-1-specific dsRNA (37 nm, 72 h). FITC, fluorescein isothiocyanate.
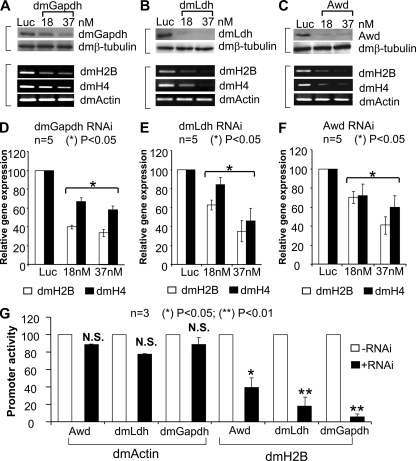
FIGURE 4.
Repressed dmH2B and dmH4 expression by elimination of a dmOCA-S function in vivo. A–C, shown are dmH2B and dmH4 mRNA levels (lower panels) upon RNAi that targeted luciferase or selective dmOCA-S components (upper panels). S2 cells were treated with 37 nm luciferase-specific dsRNA or 18 or 37 nm dmGapdh-specific (A; 5 days), dmLdh-specific (B; 3 days), and Awd-specific (C; 3 days) dsRNA. D–F correspond to A–C lower panels but show dmH2B and dmH4 expression quantification by real-time PCR. G, luciferase reporter assays are shown. S2 cells fed without or with 37 nm dmOCA-S-specific dsRNAs for 48 h (Awd, dmLdh) or 96 h (dmGapdh) were transfected with dmH2B- or dmAct5C-promoter-luciferase reporter genes. After 24 more hours, cell lysates were assayed for luciferase activities. N.S., not significant.
FIGURE 6.
The dmH4 promoter is also a direct target for the Pdm-1/dmOCA-S regulatory module. A, a schematic of dmH4 promoter, illustrated with the octamer positions (relative to the transcription start site) is indicated with arrows and sequences (from distal to proximal, dmH4-S1, dmH4-S2, and dmH4-S3) underlined. B, a footprinting analysis exhibits dmPOU1-protected (from DNase I digestion) regions corresponding to dmH4-S1+S2 and dmH4-S3 sites on the dmH4 promoter. C, shown are EMSAs using the wild-type dmH4-S1+S2 and dmH4-S3 probes and the corresponding dmH4-M1, -M2, -M3 mutants (shown in Fig. 1E). The dmH4-S1 and -S2 sites are very tightly linked (A and B); we did not assay them in separate probes here. This close proximity of the two binding sites might pose a stereo-hindrance for double occupancy by dmPOU1 at least in vitro; nevertheless, both sites along with S3 significantly contribute to the overall promoter activity when individually tested (see E). D, shown are luciferase reporter assays of the dmH4 or the dmAct5C promoter in S2 cells treated without or with 37 nm Pdm-1-specific dsRNA for 72 h. E, shown are luciferase reporter assays on the wild-type versus the mutant dmH4 promoters.
FIGURE 7.
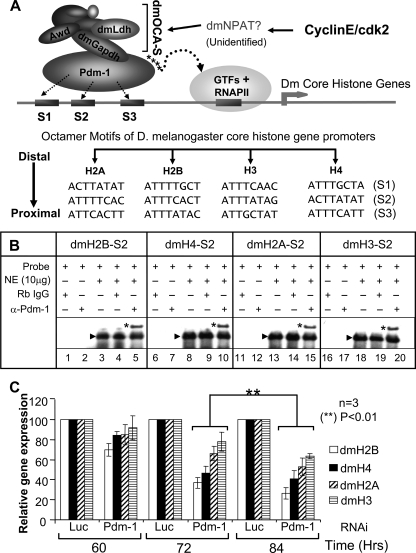
Pdm-1 as a universal transcription factor connecting dmOCA-S to Drosophila core histone gene promoters. A, shown is a transcriptional regulation pathway of D. melanogaster core histone genes. All the core histone genes contain multiple octamer sites in their promoters for recruitment of the common transcription factor Pdm-1, which in turn recruits dmOCA-S that might well be the universal co-activator for the coordinated expression of all Drosophila core histone genes. B, EMSAs analyses are shown using crude nuclear extract (NE) and oligos containing the H2B, H4, H2A, and H3 S2 sites, which formed one major complex (arrowhead; lanes 3, 8, 13, and 18). Naïve rabbit IgG as control did not supershift the complex formed between nuclear extract and probe (lanes 4, 9, 14, and 19); however, rabbit anti-Pdm-1 antibodies produced a supershifted complex (*), hence, demonstrating that the complex contained native endogenous Pdm-1 transcription factor (lanes 5, 10, 15, and 20). Note that, for space considerations the images of the gel portions containing free probes are not shown here as well as in the EMSAs in Figs. 2 and 6. C, core histone gene expression in a time course is shown. Schneider-2 (S2) cells were treated with 37 nm luciferase- or Pdm-1-specific dsRNA and harvested at 60, 72, and 84 h for expression analyses of dmH2A, dmH2B, dmH3, and dmH4 core histone genes using quantitative RT-qPCR.
RESULTS
Three Cryptic Octamer Elements for Anchoring Pdm-1 to the dmH2B Promoter
D. melanogaster contains two histone gene clusters, each cluster with 100–150 tandem repeats of the histone genes per haploid genome (2, 6). Fig. 1A illustrates such a repeat (∼5 kb) with a complete set of core and linker histone genes.
Given the pivotal role of Oct-1 in the hH2B expression (10–13), a similar role of Pdm-1 was explored despite an earlier inability to identify a canonical octamer (ATTTGCAT) in Drosophila histone promoters (21). Thus, as a first step, we used sequence comparison to identify three putative octamer elements (Fig. 1A; dmH2B-S1, -S2, and -S3) on the dmH2B promoter while permitting three mismatches with the canonical octamer in the hH2B gene (Table 1). We then used a DNase I footprinting assay with the dmH2B promoter DNA (Fig. 1A) as a probe and recombinant GST-tagged Pdm-1 POU domain (dmPOU1; Fig. 1B) to assess its binding, reasoning that Pdm-1 may bind to these sites via the conserved bipartite POU-domain, known to be sufficient to confer high affinity sequence-specific DNA binding (25).
TABLE 1.
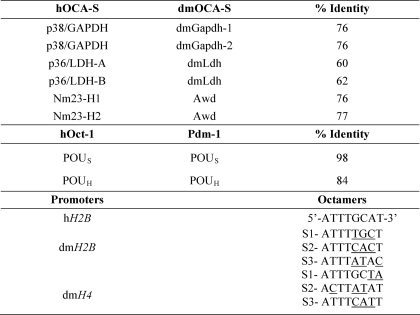
Identities between human (h) and dmOCA-S components (a subset), Oct-1 and Pdm-1 POU domains, and histone promoter octamer motifs
Peptide sequence identities were based upon pairwise alignment using the Smith-Waterman algorithm. GenBankTM accession numbers are: p38/GAPDH (CAA25833.1), p36/LDH-A (NP_005557.1), p36/LDH-B (CAA68701.1), Nm23-H1 (AA085436.1), Nm23-H2 (CAB37870.1), Oct-1 (AAM77920.1), dmGapdh-1 (NP_525108.2), dmGapdh-2 (NP_542445.1), dmLdh (AAB07594.1), Awd (NP_476761.2), Pdm-1 (AAA28829.1). Comparison between Pdm-1 and Oct-1 is limited to the POU domains with the POU-specific (POUS) and POU-homeo (POUH) subdomains. Underlined are the octamer nucleotides, of the dmH2B and dmH4 promoters, which are deviated from the canonical hH2B octamer site.
dmPOU1 created two major footprints corresponding, respectively, to the S1+S2 and S3 on the dmH2B promoter regions (Fig. 1C; lane 2). To prove that the binding of dmPOU1 to the octamer sites was sequence-specific, we generated wild-type (WT) and base-substituted mutant oligonucleotides that encompassed respective dmH2B sites (Fig. 1D) for EMSAs; the WT sites were bound by the recombinant GST-tagged dmPOU1 protein but not the GST tag-only protein (Fig. 2A), and the mutant sites (Fig. 1D) possessed diminished or weakened binding with dmPOU1 (Fig. 2B). In the competition EMSAs, each unlabeled WT dmH2B oligonucleotide, when in molar excess, effectively inhibited complex formation between dmPOU1 and labeled WT probe, whereas unlabeled mutant oligonucleotides failed to do so (Fig. 2C). These results suggest that the dmH2B octamer sites are potential regulatory elements that are bound by Pdm-1 in a sequence-specific manner.
Regulation of the dmH2B Gene by Pdm-1 as a Transcription Factor
To prove that Pdm-1 directly mediates the transcriptional activation of the dmH2B promoter, we employed reporter assays with a dmH2B promoter fragment(s) cloned upstream of the firefly luciferase gene (pGL3). The promoter of the dmActin gene was used as a control. When Pdm-1 expression was silenced by RNA interference (RNAi) using a Pdm-1-specific dsRNA in a Drosophila cell line Schneider-2 (S2) (see Fig. 3A for the RNAi strategy), the dmH2B-promoter-luciferase reporter activity was severely reduced; however, regardless of the Pdm-1 silencing, the dmActin-promoter-luciferase reporter activity remained the same (Fig. 2D). Thus, Pdm-1 might directly dictate dmH2B transcription. To support the idea that a transcriptional function of Pdm-1 is exerted through the octamer sites, we introduced mutated octamer sites into the dmH2B promoter; individual dmH2B promoter mutant at site 1, 2, or 3 (M1, M2, or M3; see Fig. 1D) exhibited significant reduction, whereas a triple-point mutant (M1-M3) showed dramatic reduction of the dmH2B promoter activities (Fig. 2E).
Gene silencing in Drosophila cells can typically be realized in 3–4 days with 37 nm target-specific dsRNAs (26); when this dose (Fig. 3B) and time-course (Fig. 3C) were used for Pdm-1, the endogenous dmH2B expression was significantly reduced, as opposed to a control (luciferase-specific) dsRNA, in line with the expression of an ectopic H2B gene (Fig. 2D). In a titration experiment, 18 nm Pdm-1-specific dsRNA sufficed to silence the Pdm-1 gene (Fig. 3C, upper panel) and reduce the dmH2B expression at the mRNA level (Fig. 3C, lower panel). Results of semiquantitative RT-rPCR assays (Fig. 3, B and C) were confirmed by RT-qPCR assays (Fig. 3D).
From yeast to humans, the expression of core histone genes is highly coordinated (9, 12, 14, 27), which dictates that repressing the expression of one histone gene leads to expression repression of others. We examined the expression of another core histone gene, dmH4, which was co-repressed in concert with repressed dmH2B expression in Pdm-1-silenced cells with similar kinetics (Fig. 3, B–D). These results support a notion of coordinated expression of (core) histone genes in diverse species.
S-phase histone expression is tightly coupled with DNA-replication and S-phase progression (28); thus, there is a legitimate possibility that histone expression defects (Fig. 3, B–D) as a result of Pdm-1 silencing (Fig. 3C) were not primary but secondary to an S-phase defect that in turn might feed back to coordinately repress overall histone expression. To exclude the possibility, we analyzed cell cycle profiles of cells treated with a Pdm-1-specific dsRNA and those of cells treated with a (control) luciferase-specific dsRNA for 72 h. BrdUrd-FACS (Fig. 3E, upper panel) and propidium iodide FACS assays (Fig. 3E, lower panel) showed similar cell cycle profiles of the two samples, suggesting that the dmH2B expression defects because of Pdm-1 RNAi (Fig. 3, B–D) were primary defects (also see below and “Discussion”).
Transcriptional Regulation of the dmH2B Gene by dmOCA-S
Given that Oct-1 and Pdm-1 share similarities in their evolutionarily conserved POU-domains (Table 1), as do the human OCA-S and putative dmOCA-S components (Table 1; a subset), we proposed a similar trans-regulatory network in the D. melanogaster H2B gene activation pathway in which dmOCA-S abets the role of Pdm-1; thus, silencing individual dmOCA-S subunits may bring about phenotypes similar to those observed in Pdm-1-silenced cells seen in Fig. 3. In particular, glyceraldehyde-3-phosphate dehydrogenase (p38/GAPDH) and lactate dehydrogenase (p36/LDH) were shown to be OCA-S components absolutely required for hH2B transcription (9, 29). We, hence, subjected dmGapdh1/2, dmLdh, and abnormal wing discs (Awd, the non-metastatic protein 23 in human (nm23) homolog) to RNAi. The expression of these proteins was silenced to relative completion (Fig. 4, A–C, upper panels) with 37 nm dsRNA doses, which was accompanied by coordinately repressed expression of dmH2B and dmH4 genes (Fig. 4, A–C, lower panels, RT-rPCR; Fig. 4, D–F, RT-qPCR). The repressed dmH2B expression was attributed to reduced promoter activity of the dmH2B gene, for the activities of the ectopic dmH2B promoter-luciferase reporter were severely reduced upon silencing the protein expression of each of the tested dmOCA-S components (dmGapdh1/2, dmLdh, and Awd; Fig. 4G). Collectively, these results suggest an existence of a dmOCA-S co-activator in Drosophila that comprises of the above-tested proteins and functions in the dmH2B gene regulation pathway by abetting the role of Pdm-1.
Occupancy of the dmH2B and dmH4 Gene Promoters by Pdm-1 and dmOCA-S
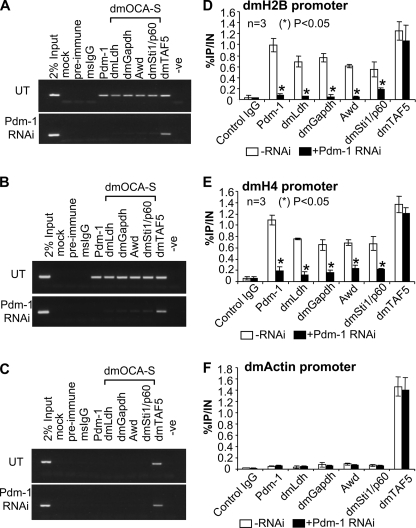
Human OCA-S occupies the hH2B gene via an interaction with promoter-bound Oct-1 (9, 29); we sought to explore whether Pdm-1 and dmOCA-S occupy the dmH2B promoter and whether a dmOCA-S recruitment is Pdm-1-dependent. In particular, we wished to investigate the status of dmH2B promoter occupancy by these transcriptional regulators in living cells. Thus, we used ChIP assays with ChIP-quality antibodies. Pdm-1 and all the tested dmOCA-S components (dmGapdh, dmLdh, Awd, and dmSti1/p60) were found to be engaged to the dmH2B promoter (Fig. 5, A and D) as opposed to non-engagement to the dmActin promoter (Fig. 5, C and F); however, in cells in which the Pdm-1 expression was silenced, the tested dmOCA-S components no longer occupied the dmH2B promoter (Fig. 5, A and D). In sharp contrast, dmTAF5, a subunit of the Drosophila general transcription factor IID (TFIID) complex, occupied all the tested promoters regardless of the Pdm-1 silencing (Fig. 5). Interestingly, the dmH4 promoter, originally employed as an additional negative control, was also occupied by both Pdm-1 and dmOCA-S, and the dmOCA-S occupancy on this promoter diminished in a Pdm-1-knocked down background as well (Fig. 5, B and E).
FIGURE 5.
dmH2B and dmH4 promoter ChIP assays. Assays were performed on S2 cells treated without (UT) or with 37 nm Pdm-1-specific dsRNA using indicated ChIP-quality antibodies. A–C, shown are recovered DNA was used for PCR amplification with primers specific for the dmH2B (A), dmH4 (B), and dmActin (C) promoters. -Ve, negative control for PCR. D–F, ChIP assays were quantified by real-time PCR. The efficacies of respective promoter DNA recovery after immunoprecipitations (IP) are expressed as percentages of the (sheared) input (IN) chromatin. Panels D–F correspond to panels A–C.
The status of the occupancy of dmH2B, dmH4, and dmActin promoters by Pdm-1, dmOCA-S, or dmTAF5 was also quantified using real-time PCR (Fig. 5, D–F) assays, which was in line with the semiquantitative PCR (Fig. 5, A–C) assays. Based on the above results, we conclude that all the dmH2B and dmH4 expression defects in this study are largely primary effects of a missing function of the Pdm-1/dmOCA-S module. That TFIID is recruited to the histone promoters regardless of Pdm-1 silencing (Fig. 5) suggests that Pdm-1/dmOCA-S enhances the potency of histone transcription machineries through a mechanism other than facilitating the TFIID recruitment.
Pdm-1 and dmOCA-S May Be Universal Regulators for Drosophila Core Histone Genes
The observations (Figs. 3–5) prompted a proposition that the Pdm-1/dmOCA-S module is involved in transcriptional activation of diverse Drosophila core histone genes, collectively providing an insight into a mechanism by which the expression of all the Drosophila core histone genes is coordinated in a more direct fashion than the coordinated expression of mammalian core histone genes (see below and “Discussion”).
Sequence comparison allowed us to identify multiple putative octamer sites on the dmH4 promoter (Fig. 6A; also Table 1), raising the possibility that these sites might be anchorages for the Pdm-1/dmOCA-S regulatory module. We used footprinting (Fig. 6B) and EMSAs (Fig. 6C) to confirm that the three putative dmH4 octamer sites were genuine Pdm-1 binding sites and identified base-substituted octamer mutants (Fig. 6C) with much-weakened or diminished octamer-Pdm-1 interaction (Fig. 6C). We next employed reporter assays using the dmH4 promoter-luciferase genes with the wild-type or mutant octamer sites. Silenced expression of Pdm-1 led to reduced dmH4-promoter-luciferase reporter activity, but the dmActin-promoter-luciferase reporter activity was the same regardless of the Pdm-1 silencing (Fig. 6D). Individual dmH4 promoter mutant at dmH4-S1, -S2, or -S3 (M1, M2, M3) exhibited a significant reduction of the dmH4-promoter-luciferase activities, and a dmH4-M1-M3 triple-site mutant led to a more drastic reduction (Fig. 6E). These findings, thus, support the notion that Pdm-1 in conjunction with dmOCA-S directly dictates the transcriptional outputs of not only dmH2B but also dmH4 genes.
Multiple putative octamer sites were also spotted within the promoters of the dmH2A and dmH3 genes (Fig. 7A), thus supporting that Pdm-1/dmOCA-S coordinately regulates transcription of all Drosophila core histone genes. To test the functional relevance of these sites, we chose the S2 sites as representatives and used the dmH2B-S2-containing oligo as the backbone and replaced the dmH2B-S2 octamer sequence with that of dmH4, dmH2A, or dmH3-S2 sites (Fig. 7A, lower panel). Probes containing each site were bound, sequence-specifically, by recombinant dmPOU-1 (data not shown). To ensure that these sites could recruit native Pdm-1, we used crude Drosophila nuclear extracts in EMSAs. The dmH2B, dmH4, dmH2A, and dmH3 S2 sites each produced a major oligo-protein complex (Fig. 7B, lanes 3, 8, 13, and 18) that was comprised of endogenous native Pdm-1 because they allowed a supershift by anti-Pdm-1 antibodies (Fig. 7B, lanes 5, 10, 15, and 20; i.e. bands with asterisks).
Therefore, the expression of all Drosophila core histone genes might be coordinately and directly repressed by a missing function of Pdm-1/dmOCA-S, prompting us to use a Pdm-1 minus background to investigate this possibility. Indeed, in a time course, the expression of all the core histone genes was repressed concertedly with similar kinetics upon RNAi-mediated Pdm-1 elimination; the significant histone expression defects manifested at 72 h (Fig. 7C), at which cell cycle profiles (Fig. 3E) and viability (Fig. 8C) of cells treated with Pdm-1-specific dsRNAs were similar to those of cells treated with the luciferase-specific dsRNAs, thus suggesting that the coordinated histone expression defects are primary (also see below and “Discussion”).
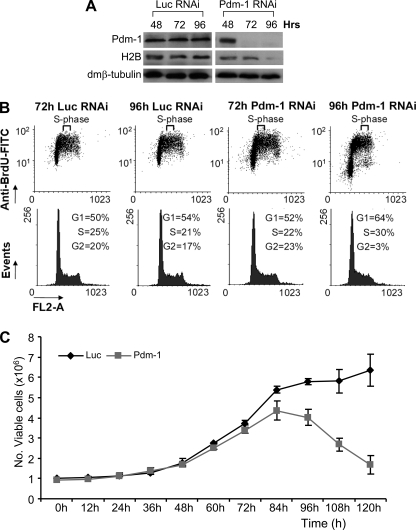
FIGURE 8.
Physiological effects of Pdm-1 RNAi cells. A, shown are Western analyses of Pdm-1 RNAi-treated cells showing time-dependent down-regulation of Pdm-1 and dmH2B protein levels. The lagged deficiency of dmH2B protein expression (96 h) as compared with that of dmH2B mRNA expression (72 h; Fig. 7C) might suggest increased stabilities of preexisting histone proteins in dmOCA-S-deficient cells. B, cell cycle profiles at 72 and 96 h post-Pdm-1 and control RNAi, showing prominent cell cycle defects at 96 h in Pdm-1-silenced cells. The obvious sharp reduction of G2-phase cells is in principle a function of disallowance of significant number of S-phase cells to exit into G2-phase, presumably attributed to a shortage of histone proteins beyond 72 h (see A). Thus, a significant number of cells are retarded in the S-phase; there is ongoing DNA replication, thus BrdUrd incorporation, but it must be with a much-reduced rate in concert with histone proteins shortage. On the other hand, cells already in G2-phase before manifestation of histone expression defects would progress into the G1-phase. These cells, however, would have difficulty to enter S-phase due to a shortage of histone proteins, thus, the increase in the number/percentage of G1-phase cells. FITC, fluorescein isothiocyanate. C, the trypan blue exclusion assays indicated that cell viability was reduced at 84 h, which became more prominent when monitored from 96 h and beyond.
Histone biosynthesis is essential for cell viability; thus, physiological effects manifested from the loss of Pdm-1 were also evaluated up to 120 h post-Pdm-1 RNAi. The mRNA levels of dmH2B were already significantly decreased at 72 h (Figs. 3, B–D, and 7C); however, decreased dmH2B protein levels were observed to become prominent only at 96 h (Fig. 8A). Together with other lines of evidence from cell cycle profiles exhibiting cell cycle defects (Fig. 8B) and also decreased cell viability that manifested from 96-h post-Pdm-1-RNAi (Fig. 8C), these results suggest that the physiological defects due to the Pdm-1 loss-of-function from 96 h and beyond were secondary to the defects monitored at the 72-h time point (Figs. 3, B–D, and 7C).
dmOCA-S Is a Novel Component of the Drosophila Histone Locus Bodies
In mammalian cells, Oct-1 binds the H2B promoter throughout the interphase, whereas OCA-S, represented by p38/GAPDH, is recruited to the promoter in an S-phase-specific fashion (9). We revealed a Pdm-1/dmOCA-S module that regulates the expression of the cell cycle-dependent core histone genes; recently, Drosophila HLB were characterized (30, 31). In S-phase, HLB are labeled with a monoclonal antibody (MPM-2) that recognizes a cyclinE/cyclin-dependent kinase 2 (cdk2)-dependent phospho-epitope shared by diverse species (32); HLB are nuclear suborganelles in which histone expression takes place (30).
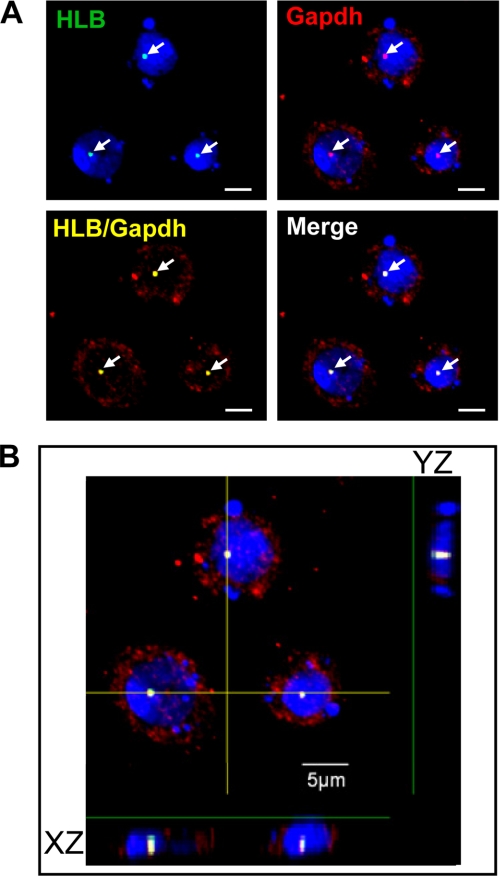
We sought to investigate an S-phase dmOCA-S function or location; in particular, we wished to link dmOCA-S to HLB by tracking dmGapdh, which is an essential dmOCA-S component (Fig. 4), during S-phase progression. To this end we first used HU to synchronize Drosophila S2 cells at a very early S-phase. Intriguingly, these cells showed strong foci colocalization of nuclear dmGapdh and HLB (Fig. 9A). When viewed through a deconvoluted image, i.e. through the z axial plane, we further affirmed colocalizing of the HLB with dmGapdh in the nucleus. Apparently, there exists a nuclear translocation of abundant cytoplasmic dmGapdh upon S-phase entry/progression, the mechanism of which requires further investigation.
FIGURE 9.
Confocal immunofluorescence analysis of HLB foci in cells synchronized at the early S-phase. A, double staining for dmGapdh (red) and HLB (green) is shown. Cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, blue). B, the image represents the z-stack projection of 17 confocal sections (stack z-spacing, 0.44 μm), showing both XZ and YZ sections. Nuclear dmGapdh was co-localized with HLB (arrowheads). Scale bar, 5 μm.
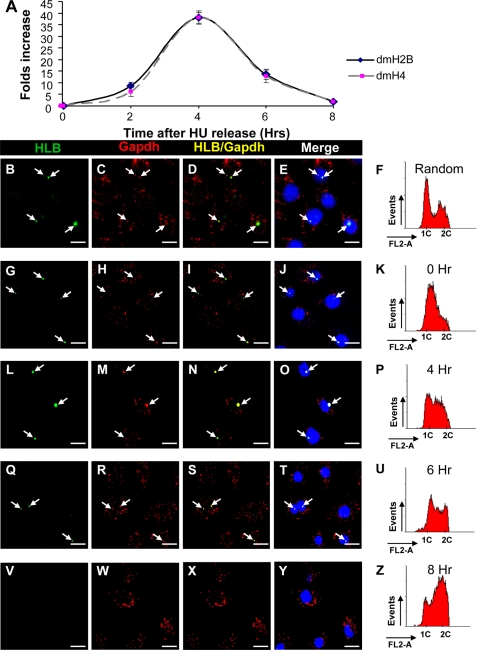
We then used HU-synchronized cells (Figs. 9A and 10K) and released the cells, allowing them to progress into the mid-S (Fig. 10P) and late S (Fig. 10U) phase and G2-phase (Fig. 10Z). Both the dmH2B and dmH4 mRNA levels increased as cells entered the S-phase; their steady-state levels peaked at the 4-h time point and decreased to base line by the 8-h time point (Fig. 10A).
FIGURE 10.
dmOCA-S is recruited to Drosophila histone locus bodies during S-phase. A, shown is a time course graph following dmH2B and dmH4 levels of S2 cells from early S-phase to G2-phase. Cells were harvested at 0, 2, 4, 6, and 8 h after release from synchronization and assayed for dmH2B and dmH4 mRNA levels by quantitative RT-qPCR. B–E, random cells images are shown. F, shown are cell cycle profiles of random cells; ∼20% of the cells were in the S-phase. A similar percentage was obtained when counting 100 randomly picked cells using HLB-foci-staining as a criterion. G–J, early S-phase cells images show weak HLB foci and co-localization with nuclear dmGapdh. K, early S-phase cell cycle profiles are shown. L–O, mid-S-phase cells images show prominent HLB foci with strong co-localization with nuclear dmGapdh foci. P, mid-S-phase cell cycle profiles are shown. Q–T, late S- and early G2-phase cells images show decreased nuclear dmGapdh and HLB foci in size and number. U, late S- and early G2-phase cell cycle profiles are shown. V–Y, shown are G2-phase cells, with no HLB and dmGapdh nuclear foci. Z, G2-phase cell cycle profiles are shown. When appropriate, arrows indicate HLB (B, G, L, Q, and V) and nuclear dmGapdh (C, H, M, R, and W) foci and their nuclear co-localization (D, I, N, S, and X), which was confirmed by superimposing to 4′,6-diamidino-2-phenylindole (DAPI) nuclei-staining images (E, J, O, T, and Y). HLB foci were stained with the MPM-2 antibody; nuclear dmGapdh foci were stained by anti-p38/GAPDH antibodies. The images and cell cycle profiles of cells at the 2-h time point were similar to those at the 0 h (G–K) and were not shown for space considerations; we reason that cells need certain time to recover from the replication stress imposed by HU. Bar, 10 μm.
In random cells, nuclear dmGapdh co-localized with the discrete HLB spherical bodies at varying degrees in the nuclei of a fraction of cells (Fig. 10, B–D); the percentage of foci-positive cells coincided well with the percentage of the S-phase cells as judged by the cell cycle profile (Fig. 10F and legends). We then focused on the recruitment of dmOCA-S (represented by nuclear dmGapdh) in relation to the appearance of HLB foci representing the cyclinE/cdk2 signaling (30). Therefore, both indirect immunofluorescence confocal and cell cycle profile analyses were performed on cells synchronized at early S (Fig. 10, G–K), mid-S (Fig. 10, L–P), late S (Fig. 10, Q–U), and G2 (Fig. 10, V–Z) phases. In early S-phase cells, HLB foci appeared in ∼90% of the cells, ∼80% of which exhibited dmOCA-S co-localization. The HLB-dmOCA-S co-localization persisted until the end of the S-phase, with the largest degree of co-localization (both in size and number) at the mid-S-phase (Fig. 10, L–P) coinciding with the dmH2B and dmH4 mRNA expression peak (Fig. 10A). Taken together, these data suggest that the recruitment of dmOCA-S to the HLB is S-phase-specific and is downstream of the activation of an unidentified cyclinE/cdk2 substrate recognized by the MPM-2 antibody, linking the dmOCA-S function/nuclear translocation to S-phase-specific histone expression.
DISCUSSION
Although more than 800 million years apart in evolution (33), Drosophila and human share similarities in histone expression pathways, albeit with species-specific features. In Drosophila, core histone gene promoters contain multiple evolutionarily diversified Pdm-1 binding sites (Fig. 7, A and B), contributing to the optimal histone expression (e.g. Figs. 2E and 7C); on the other hand, a single prototype octamer ATTTGCAT mediates the hH2B transcription, and Oct-1 association with this element, which is conserved among vertebrates, is kept under strict sequence requirement (11). This strategy may be also employed by other Oct-1-dependent genes and their cognate coactivators (34–37). These species-specific features could imply significance for metazoan evolution.
Histone biosynthesis and DNA replication are coupled and essential for cell viability. Thus, impeding the function of Pdm-1/dmOCA-S would lead to S-phase defects and likely be detrimental to cell viability. In the case of dmGapdh and dmLdh, their roles in glycolysis and their moonlighting nuclear functions abetting the role of Pdm-1 also dictate negative consequences on the cellular ability to progress through the S-phase in a loss-of-function situation.
Efficient RNAi-mediated protein knockdown requires mRNA destruction and decay of the preexisting protein, which was realized at 72 h for Pdm-1 (Figs. 3C and 8A) with coordinately repressed expression of core histone genes (Fig. 7C); however, the cell cycle profiles were not affected or only marginally affected at this point, which was statistically insignificant (Figs. 3E and 8B). Thus, the histone expression defects at 72 h (Figs. 3, C and D, and 7C) were primary defects. We reason that a most likely scenario is that despite depriving cells of histone transcription at 72 h by a Pdm-1 deficiency, pre-existing histone protein levels are above a threshold that supports DNA replication. Indeed, H2B protein levels at 72 h were largely normal or slightly reduced (Fig. 8A). Beyond 72 h, however, Pdm-1-silenced cells also exhibited a prominent S-phase defect at 96 h (Fig. 8B), and cell viability defects manifested at 84 h with a more drastic viability decrease at 96 h (Fig. 8C).
The cell viability defects might be related to a mechanism coupling histone expression to S-phase progression. The more drastic histone expression defects at 84 h as compared with that at 72 h (Fig. 7C) could be due to additive effects of a primary Pdm-1 loss-of-function and secondary cell viability and S-phase defects, which fed back. Histone transcription defects as a result of the Pdm-1/dmOCA-S deficiency would deprive cells of histone proteins in a long run, thus providing insufficient histone protein levels for chromatin assembly, which ultimately leads to DNA replication, S-phase, and cell viability defects. We propose that although histone transcription defects at 72 h were primary defects, the later-manifested cell viability and S-phase defects were secondary defects due to reduced histone protein levels as exemplified by that of H2B beyond 72 h (96 h; Fig. 8A).
Coordinated histone expression in diverse species (9, 12, 14, 27) is needed to maintain balanced expression of core histone genes, known to sustain genome integrity (38). In mammalian cells, eliminating OCA-S function by silencing p38/GAPDH expression led to H2B expression defect, and a lagged histone H4 expression defect manifested after a severe cell cycle arrest (9); eliminating H2B expression by gene deletion in yeast also led to cell cycle arrest and subsequent expression defects of other core histone genes (27). These observations led to a thought that coordinated histone expression was regulated through an S-phase feedback mechanism (9), which was later revised (14).
In Drosophila cells, the RNAi-mediated silencing of Pdm-1 or dmOCA-S components led to concerted and directly coordinated expression defects of all core histone genes (e.g. Fig. 7C) and as primary transcription effects due to a missing Pdm-1/dmOCA-S function (see above). In mammalian cells, the core histone genes employ distinct promoter elements and associated (co)factors (12), but their expression remains highly coordinated through an uncharacterized mechanism that is indirect but still does not involve S-phase feedback (14). Distinct histone expression coordination mechanisms in fly and mammalian cells might be of crucial relevance in metazoan evolution.
The expression of metazoan-specific linker histone H1 gene is largely S-phase-specific, but the H1 expression output is not tightly coupled with that of core histone genes (15). The TATA-less Drosophila H1 gene utilizes a TRF-containing complex but not the prototype TBP-containing TFIID complex for its expression (15). We found that Drosophila core histone genes, at least that of dmH2B and dmH4, contain TFIID (Fig. 5) in line with recruitment of TBP to dmH3 and dmH4 promoters (15) and the idea that the basal transcription machineries of Drosophila core histone genes use the prototype TFIID. TRF-containing complexes have not been known to function in a cell cycle-dependent manner (15); it is of interest to investigate if the S-phase-specific Drosophila H1 expression is conferred upon by S-phase-specific factors that ought to be distinct from Pdm-1/dmOCA-S because the Drosophila H1 expression was not coordinately repressed with that of core histone genes in Pdm-1-deficient cells (data not shown).
Mammalian histone genes are organized into Cajal bodies in a process facilitated by nuclear protein, ataxia-telangiectasia locus (NPAT), a cyclinE/cdk2 substrate that conveys the cyclinE/cdk2 signaling to histone transcription machineries (28, 31, 39, 40). On the other hand, Drosophila cells contain distinct nuclear domains dubbed HLB, which host all the histone genes and contain a cyclinE/cdk2-dependent phospho-epitope recognized by the MPM-2 monoclonal antibody in S-phase (30, 31). Drosophila HLB are often in close proximity to, but never overlapped with Drosophila Cajal bodies (31, 41).
That cyclinE/cdk2 signaling is conserved from Drosophila to human, and that the HLB foci are associated with nascent histone transcripts (28, 30) prompted an investigation of S-phase-specific recruitment of dmOCA-S (represented by nuclear dmGapdh) to HLB. A higher percentage of HLB (∼90%) foci as compared with dmOCA-S (∼70%) foci in the early S-phase, i.e. ∼80% of HLB foci are nuclear dmGapdh-positive (Fig. 10, G–K), suggests that the dmOCA-S function (dmGapdh-HLB nuclear co-localization) is likely downstream of cyclinE/cdk2 signaling (Fig. 7A). The increase in the foci size and maximal degree of HLB-dmOCA-S co-localization (Fig. 10, L–P) coincided with the peak of dmH2B and dmH4 mRNA levels in the mid-S-phase (Fig. 10A), in line with the notion that the HLB foci size is proportional to histone expression levels (30, 42).
None of dmOCA-S components possesses a consensus sequence(s) for cdk (data not shown); the fly cyclinE/cdk2 signaling might be conveyed to histone genes via an unidentified molecule(s) (dmNPAT; Fig. 7A), for which the phospho-epitope-containing protein recognized by MPM-2 monoclonal antibody (30) is a potential candidate.
Efforts to find dmNPAT have been fruitless; a dmNPAT gene might likely reside in so-far-unsequenced heterochromatic domains or possess sequences drastically divergent from vertebrate NPATs. Alternatively, an NPAT function may not have been acquired during insect evolution, necessitating histone genes to be organized into HLB and compelling cognate promoters to be regulated by diversified octamer elements and a Pdm-1/dmOCA-S module to ensure the directly coordinated expression of histone genes (Fig. 7A).
The mechanistic aspects of co-activation by dmOCA-S seem to be similar to that of human OCA-S, in line with nuclear moonlighting transcription functions of metabolic enzyme conservation in metazoans. Conversely, some non-transcriptional functions of transcription factors or co-factors have been documented: e.g. in the cytoplasm, an isoform of co-activator OCA-B plays an essential non-transcriptional role for B-cell signaling (43).
Pdm-1 is a ubiquitous transcription factor for ubiquitous histone expression; the identified cell-specific roles of Pdm-1 in neuronal cell fate specification (17, 18) and Notch signaling (19) might be due to cell-specific non-transcriptional functions of Pdm-1. Alternatively, cell-specific Pdm-1 phenotypes might be attributed to missing links between mutant pdm-1 alleles and alleles encoding cell-specific co-activators, especially in view of the fact that a given POU-domain is rich in separable surfaces that provide contacts for distinct ubiquitous or tissue-specific co-activators (35). Non-transcriptional development regulators may also interact with partners through different surfaces. Thus, it is not surprising that mutant alleles of even a ubiquitously expressed gene, be it specifying a non-transcription or transcription function, may lead to tissue-specific phenotypes.
The work toward dissecting cis- and trans-regulatory networks of the Drosophila histone transcription regulation pathway(s) will provide a new paradigm for studying transcriptional regulator changes implied in evolution and development as well as permit a broader range of questions to be asked about a cohort of genes involved in histone expression and related DNA replication-dependent transcription and its tight coupling with S-phase progression and about roles of the individual dmOCA-S components as novel S-phase-specific players in above processes in a genetically tractable organism.
Acknowledgments
We thank Dr. Xiaohang Yang, Dr. Edwin Cheung, Dr. Permeen Yusoff, and Dr. Shing-Leng Chan for helpful discussions and encouragement and Dr. Ruping Dai and Dr. Lei Zheng who critiqued an earlier manuscript draft.
This work was supported by the Agency for Science, Technology, and Research, Singapore.
- hH2B
- human H2B
- dm
- D. melanogaster
- Pdm-1
- POU-domain protein in D. melanogaster 1
- Oct-1
- octamer binding factor 1
- OCA-S
- Oct-1 co-activator in S-phase
- GAPDH or Gapdh
- glyceraldehyde-3-phosphate dehydrogenase
- LDH or Ldh
- lactate dehydrogenase
- Awd
- abnormal wing disc
- NPAT
- nuclear protein, ataxia-telangiectasia locus
- Sti1
- stress-inducible protein 1
- ChIP
- chromatin immunoprecipitation
- dsRNA
- double-stranded RNA
- EMSAs
- electrophoresis mobility shift assays
- GST
- glutathione S-transferase
- RT-qPCR
- reverse transcription-quantitative PCR
- RT-rPCR
- RT-relative PCR
- WT
- wild type
- FACS
- fluorescent-activated cell sorting
- BrdUrd
- bromodeoxyuridine
- TBP
- TATA-binding protein
- TFIID
- general transcription factor IID
- TAF
- TBP-associated factor
- dmTAF5
- TBP-associated factor 5
- TRF
- TBP-related factor
- HU
- hydroxyurea
- HLB
- histone locus bodies
- cdk
- cyclin-dependent kinase
- DTT
- dithiothreitol
- PMSF
- phenylmethylsulfonyl fluoride
- CAPS
- 3-(cyclohexylamino)propanesulfonic acid
- PBS
- phosphate-buffered saline
- WT
- wild type.
REFERENCES
- 1.Roeder R. G. (2003) Nat. Med. 9, 1239–1244 [DOI] [PubMed] [Google Scholar]
- 2.Hentschel C. C., Birnstiel M. L. (1981) Cell 25, 301–313 [DOI] [PubMed] [Google Scholar]
- 3.Wang Z. F., Krasikov T., Frey M. R., Wang J., Matera A. G., Marzluff W. F. (1996) Genome Res. 6, 688–701 [DOI] [PubMed] [Google Scholar]
- 4.Marzluff W. F., Duronio R. J. (2002) Curr. Opin. Cell Biol. 14, 692–699 [DOI] [PubMed] [Google Scholar]
- 5.Marzluff W. F., Gongidi P., Woods K. R., Jin J., Maltais L. J. (2002) Genomics 80, 487–498 [PubMed] [Google Scholar]
- 6.Lifton R. P., Goldberg M. L., Karp R. W., Hogness D. S. (1978) Cold Spring Harbor Symp. Quant. Biol. 42, 1047–1051 [DOI] [PubMed] [Google Scholar]
- 7.Perry M., Thomsen G. H., Roeder R. G. (1985) J. Mol. Biol. 185, 479–499 [DOI] [PubMed] [Google Scholar]
- 8.Heintz N. (1991) Biochim. Biophys. Acta 1088, 327–339 [DOI] [PubMed] [Google Scholar]
- 9.Zheng L., Roeder R. G., Luo Y. (2003) Cell 114, 255–266 [DOI] [PubMed] [Google Scholar]
- 10.Fletcher C., Heintz N., Roeder R. G. (1987) Cell 51, 773–781 [DOI] [PubMed] [Google Scholar]
- 11.Luo Y., Roeder R. G. (1999) Cold Spring Harbor Symp. Quant. Biol. 64, 119–131 [DOI] [PubMed] [Google Scholar]
- 12.Osley M. A. (1991) Annu. Rev. Biochem. 60, 827–861 [DOI] [PubMed] [Google Scholar]
- 13.Segil N., Roberts S. B., Heintz N. (1991) Science 254, 1814–1816 [DOI] [PubMed] [Google Scholar]
- 14.Yu F. X., Dai R. P., Goh S. R., Zheng L., Luo Y. (2009) Cell Cycle 8, 773–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isogai Y., Keles S., Prestel M., Hochheimer A., Tjian R. (2007) Genes Dev. 21, 2936–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lloyd A., Sakonju S. (1991) Mech. Dev. 36, 87–102 [DOI] [PubMed] [Google Scholar]
- 17.Bhat K. M., Poole S. J., Schedl P. (1995) Mol. Cell. Biol. 15, 4052–4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yeo S. L., Lloyd A., Kozak K., Dinh A., Dick T., Yang X., Sakonju S., Chia W. (1995) Genes Dev. 9, 1223–1236 [DOI] [PubMed] [Google Scholar]
- 19.Neumann C. J., Cohen S. M. (1998) Science 281, 409–413 [DOI] [PubMed] [Google Scholar]
- 20.Kitamoto T., Ikeda K., Salvaterra P. M. (1995) J. Neurobiol. 28, 70–81 [DOI] [PubMed] [Google Scholar]
- 21.Kremer H., Hennig W. (1990) Nucleic Acids Res. 18, 1573–1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maxam A. M., Gilbert W. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 560–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacAlpine D. M., Rodríguez H. K., Bell S. P. (2004) Genes Dev. 18, 3094–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roberts T. G., Sturm N. R., Yee B. K., Yu M. C., Hartshorne T., Agabian N., Campbell D. A. (1998) Mol. Cell. Biol. 18, 4409–4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herr W., Cleary M. A. (1995) Genes Dev. 9, 1679–1693 [DOI] [PubMed] [Google Scholar]
- 26.Clemens J. C., Worby C. A., Simonson-Leff N., Muda M., Maehama T., Hemmings B. A., Dixon J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6499–6503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han M., Chang M., Kim U. J., Grunstein M. (1987) Cell 48, 589–597 [DOI] [PubMed] [Google Scholar]
- 28.Ewen M. E. (2000) Genes Dev. 14, 2265–2270 [DOI] [PubMed] [Google Scholar]
- 29.Dai R. P., Yu F. X., Goh S. R., Chng H. W., Tan Y. L., Fu J. L., Zheng L., Luo Y. (2008) J. Biol. Chem. 283, 26894–26901 [DOI] [PubMed] [Google Scholar]
- 30.White A. E., Leslie M. E., Calvi B. R., Marzluff W. F., Duronio R. J. (2007) Mol. Biol. Cell 18, 2491–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J. L., Murphy C., Buszczak M., Clatterbuck S., Goodman R., Gall J. G. (2006) J. Cell Biol. 172, 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis F. M., Tsao T. Y., Fowler S. K., Rao P. N. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 2926–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hedges S. B., Shah P. (2003) BMC Bioinformatics 4, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauter P., Matthias P. (1998) Mol. Cell. Biol. 18, 7397–7409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babb R., Cleary M. A., Herr W. (1997) Mol. Cell. Biol. 17, 7295–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cepek K. L., Chasman D. I., Sharp P. A. (1996) Genes Dev. 10, 2079–2088 [DOI] [PubMed] [Google Scholar]
- 37.Luo Y., Roeder R. G. (1995) Mol. Cell. Biol. 15, 4115–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meeks-Wagner D., Hartwell L. H. (1986) Cell 44, 43–52 [DOI] [PubMed] [Google Scholar]
- 39.Ma T., Van Tine B. A., Wei Y., Garrett M. D., Nelson D., Adams P. D., Wang J., Qin J., Chow L. T., Harper J. W. (2000) Genes Dev. 14, 2298–2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J., Kennedy B. K., Lawrence B. D., Barbie D. A., Matera A. G., Fletcher J. A., Harlow E. (2000) Genes Dev. 14, 2283–2297 [PMC free article] [PubMed] [Google Scholar]
- 41.Matera A. G. (2006) J. Cell Biol. 172, 791–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith A. V., King J. A., Orr-Weaver T. L. (1993) Genetics 135, 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siegel R., Kim U., Patke A., Yu X., Ren X., Tarakhovsky A., Roeder R. G. (2006) Cell 125, 761–774 [DOI] [PubMed] [Google Scholar]