Abstract
Large DNA viruses, such as herpesvirus and poxvirus, encode proteins that target and exploit the chemokine system of their host. UL146 and UL147 in the cytomegalovirus (CMV) genome encode the two CXC chemokines vCXCL1 and vCXCL2. In this study, vCXCL1 was probed against a panel of the 18 classified human chemokine receptors. In calcium mobilization assays vCXCL1 acted as an agonist on both CXCR1 and CXCR2 but did not activate or block any of the other 16 chemokine receptors. vCXCL1 was characterized and compared with CXCL1/GROα, CXCL2/GROβ, CXCL3/GROγ, CXCL5/ENA-78, CXCL6/GCP-2, CXCL7/NAP-2 and CXCL8/IL-8 in competition binding, calcium mobilization, inositol triphosphate turnover, and chemotaxis assays using CXCR1- and CXCR2-expressing Chinese hamster ovary, 300.19, COS7, and L1.2 cells. The affinities of vCXCL1 for the CXCR1 and CXCR2 receptors were 44 and 5.6 nm, respectively, as determined in competition binding against radioactively labeled CXCL8. In calcium mobilization, phosphatidylinositol turnover, and chemotaxis assays, vCXCL1 acted as a highly efficacious activator of both receptors, with a rather low potency for the CXCR1 receptor but comparable with CXCL5 and CXCL7. It is suggested that CMV uses the UL146 gene product expressed in infected endothelial cells to attract neutrophils by activating their CXCR1 and CXCR2 receptors, whereby neutrophils can act as carriers of the virus to uninfected endothelial cells. In that way a lasting pool of CMV-infected endothelial cells could be maintained.
Keywords: Chemokines, Cytokine, Endothelium, G Protein-coupled Receptors (GPCR), 7-Helix Receptor, Herpesvirus, Neutrophil, CMV, UL146, vCXCL1
Introduction
Cytomegalovirus (CMV)2 or human herpesvirus 5 was identified in 1956 (1). The spectrum of primary CMV infections in the immunocompetent host ranges from subclinical infection in children to a mononucleosis syndrome in young adults. Primary CMV infection in pregnancy can result in congenital infection of the infant with severe neurological sequelae. In immunodeficiency CMV infection can be a serious problem. Thus a human immunodeficiency virus-infected patient with low CD4 cell count can develop retinitis and colitis, whereas the immunosuppressed transplanted patient in addition to colitis can develop pneumonitis and hepatitis (reviewed by Mocarski et al. (2)).
CMV, HHV6, and HHV7 constitute the members of the human β-herpesvirus group, which is characterized by a long reproductive cycle, slow progression in culture, and an ability to enlarge cells. CMV has by far the largest genome of the viruses that infect humans. Its 230-kilobase pair genome encodes more than 160 genes, and several of these encode viral homologs of human proteins (reviewed by Mocarski et al. (2)). CMV belongs, along with Epstein-Barr virus, HHV6, HHV7, Kaposi sarcoma-associated herpesvirus, and the poxviruses molluscum contagiosum, vaccinia and variola, to a group of large human DNA viruses that encode proteins that exploit the human chemokine system. These proteins fall into three groups: chemokine proteins; seven-transmembrane G protein-coupled receptors, some of which have been shown to bind chemokines; and chemokine binding proteins (3–5). Chemokines are 70–80-amino acid proteins with a characteristic three-dimensional fold that are involved in guiding and activating distinct leukocyte subsets. Chemokines can be divided into four subfamilies on the basis of the pattern and number of the conserved cysteine residues located near their N terminus, which are involved in disulfide binding formation: the CC, CXC, CX3C, and XC families, respectively. Chemokines act through seven-transmembrane G protein-coupled receptors, of which we today know 10 CC chemokine receptors (CCR1–10), six CXC receptors (CXCR1–6), one CX3C receptor (CX3CR1), and one XC receptor (XCR1). CXC chemokines can be further subdivided into two groups depending on the presence or absence of an ELR motif adjacent and N-terminal to the CXC motif (Fig. 1). CXCR1 and CXCR2 are exclusively activated by the ELR-positive CXC chemokines. CXCR1 and CXCR2 are the principal chemokine receptors expressed on neutrophils and are considered to play a major role in the acute inflammatory response (reviewed by Murphy et al. (6)).
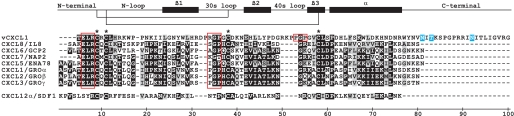
FIGURE 1.
Alignment of the CMV-encoded chemokine vCXCL1 with the human ELR-positive CXC chemokines. Primary structure of vCXCL1 and the human encoded CXC chemokines with the ELR motif aligned using CLUSTALW 1.8. The non-ELR CXC chemokine CXCL12α/SDF-1α was included for comparison. Identical amino acids are shown in white on black, whereas similar amino acids are shown in black on light gray. Conserved cysteines are marked by asterisks. The secondary structures of CXCL1/GROα, CXCL2/GROβ, CXCL7/NAP-2, and CXCL8/IL-8, as determined by NMR and x-ray diffraction crystallography and available in the Protein Data Bank, are indicated by the line above the alignment. The ELR motif and the PGP/SGP motif are marked by red boxes. N-Glycosylated asparagines and a potential O-glycosylated threonine in the C-terminal of vCXCL1 are shown in white on blue.
The virally encoded chemokines have different pharmacological phenotypes, ranging from being very broad spectrum to highly selective and/or behaving as agonists or antagonists (7–16). The sequencing of various wild type CMV strains suggested the presence of additional host interacting genes (17). Two of these genes, UL146 and UL147, encode CXC chemokines. The gene product of UL147 has not yet been characterized, whereas the UL146 gene encodes the protein vCXCL1 with an ELR motif that has been shown to work as a selective CXCR2 agonist (16). In the present study we find that the CMV UL146 gene product encodes a CXC chemokine, which functions as a selective agonist not only for CXCR2 but also for CXCR1, although with lower affinity and potency. The targeting of both CXCR1 and CXCR2 by CMV supports the notion that neutrophils are intimately connected to CMV biology and pathogenesis.
EXPERIMENTAL PROCEDURES
Chemokines and Cytokines
The virally encoded chemokines vCCL1, vCCL2, and vCXCL1 and the human encoded chemokines CCL1, CCL5, CCL11, CCL16, CCL20, CCL21, CCL22, CCL28, CXCL8, CXCL12, CXCL13, CXCL16, CX3CL1, XCL1, and tumor necrosis factor-α were bought from R & D (Minneapolis, MN), whereas CCL25, CXCL1, CXCL2, CXCL3, CXCL5, CXCL6, CXCL7, and CXCL11 were bought from PeproTech (Rocky Hill, NJ). CCL2, CCL7, vCCL4 (amino acid residues 21–113 from GenBankTM 000898) and vCXCL2 (amino acid residues 41–159 from GenBankTM AY446871) were chemically synthesized by Ian Clark-Lewis as described previously (18). The virally encoded chemokines MC148 and vCCL3 were purified from cell medium from COS7 cells transfected with their respective genes as described previously (11, 14). Lipopolysaccharide (L4391), interferon γ (I3265), and IL-1α (I2778) were bought from Sigma-Aldrich.
Stable Cell Lines
The pTej vector (19) was used for generating the following constructs: CCR2 (HindIII-BamHI), CCR5 (HindIII-BamHI), CCR8 (HindIII-EcoRI), CCR9 (EcoRI-BamHI), CCR10 (EcoRI-BamHI), CXCR1 (HindIII-EcoRI), CXCR2 (HindIII-EcoRI), and CXCR6 (EcoRI-BamHI). CCR1 (EcoRV-EcoRV) was inserted into pcDNA1. XCR1 (BamHI-XhoI) inserted into the vector pcDNA3.1 was purchased from the UMR cDNA Resource Center (Rolla, MO). CXCR1 and CXCR2 were transfected into the murine pre-B cell lines L1.2 and 300.19 by electroporation, and stable transfectants were obtained after limiting dilution and chemical selection with G418. Subsequently clones were functionally selected by testing them for calcium responses to CXCL8. Likewise stable clones were generated for CCR2, CCR9, and XCR1 in 300.19 cells and CCR1, CCR5, CCR8, CCR10, and CXCR6 in L1.2 cells. The L1.2 cell lines expressing CCR3 and CCR7 receptors were obtained from ICOS (Seattle, WA) (20). The L1.2 cells stably expressing CCR4, CCR6, and CX3CR1 were a kind gift from Osamu Yoshie (Kinki University, Japan) (21). Kuldeep Neote (Pfizer, Groton, CT) kindly provided 300.19 cells expressing the CXCR3 receptor, which originated from B. Moser (22), and Bernhard Moser (Thedor-Kocher Institute, Bern, Schwitzerland) kindly provided 300.19 cells expressing the CXCR5 receptor (23). Chinese hamster ovary (CHO) cells stably expressing CXCR1, CXCR2, and CXCR4 were provided by Tim Wells (Serono, Geneva, Schwitzerland) (24). L1.2 and CHO cells were grown in 1640 RPMI supplemented with 180 units/ml penicillin, 45 μg/ml streptomycin, and 10 mm glutamine. 300.19 cells were grown in the same medium supplemented with 55 μm mercaptoethanol.
Radioligand Binding Experiments
Whole cell radioligand binding assays (2–5 × 104 cells/well) were performed at 4 °C for 3 h in 0.5 ml of 25 mm Hepes buffer containing 1 mm CaCl2 and 5 mm MgCl2 at pH 7.2, supplemented with 0.5% bovine serum albumin on CHO cells stably transfected with CXCR1 or CXCR2. The incubation was stopped by washing four times with 0.5 ml of the same buffer made 0.5 m in NaCl. Cell-associated radioactivity was determined after extraction of the cells with 8 m urea in 3 m acetic acid supplemented with 1% Nonidet P-40. Nonspecific binding, determined in the presence of the relevant chemokine peptide (0.1 μm), was subtracted. The radioactivity labeled peptide 125I-CXCL8 (IM249) was bought from GE Healthcare.
Phosphatidylinositol Assay
COS-7 cells were transiently transfected as mentioned above. Briefly, 2 × 106 COS-7 cells were transfected with 30 μg of cDNA encoding the promiscuous chimeric G protein, GαΔ6qi4myr, which allows the Gαi-coupled receptor to couple to the Gαq pathway (phospholipase C activation measured as PI turnover) (25), with or without 20 μg of CXCR1 or CXCR2 cDNA. Gqi4myr or GαΔ6qi4myr is a chimeric Gαq subunit in which the six N-terminal amino-acid residues have been deleted, an N-terminal consensus motif for myristoylation has been created, and the four C-terminal residues have been replaced with the corresponding Gαi sequence. After transfection, COS-7 cells (2.5 × 104 cells/well) were incubated for 24 h with 2 μCi of myo-[3H]inositol in 0.4 ml of growth medium/well. The cells were washed twice in 20 mm Hepes, pH 7.4, and supplemented with 140 mm NaCl, 5 mm KCl, 1 mm MgSO4, 1 mm CaCl2, 10 mm glucose, and 0.05% (w/v) bovine serum albumin, and the chemokines were incubated in 0.4 ml of the same buffer supplemented with 10 mm LiCl at 37 °C for 90 min. The cells were extracted by the addition of 1 ml of 10 mm formic acid to each well followed by incubation on ice for 30–60 min. The generated [3H]inositol phosphates were purified on AG1-X8 anion exchange resin (Bio-Rad).
Intracellular Calcium Mobilization Assays
The cells stably transfected with the respective chemokine receptors were loaded with Fura-2AM (Molecular Probes, Eugene, OR) in RPMI with 1% fetal calf serum for 20–30 min, and aliquots of 1 × 106 cells were resuspended in RPMI with 10 mm EGTA. Fluorescence was measured on a Jobin Yvon FlouroMax-2 (Jobin Yvon Spex, Edison, NJ) as the ratio of emission at 490 nm when excited at 340 and 380 nm, respectively.
Chemotaxis Assays
Chemotaxis was measured as the number of migrated cells. 1 × 106 CXCR1/L1.2, CXCR2/L1.2, or naïve L1.2 cells were resuspended in 0.1 ml of chemotaxis buffer and added to the top chamber insert of 24 Transwell polycarbonate 3-μm membranes (Corning Costar, Cambridge, MA). The chemokines were diluted in 0.6 ml of chemotaxis buffer (RPMI medium containing 0.5% bovine serum albumin) and added to the lower chemotaxis chamber. The plates were then incubated for 4 h at 37 °C in a 5% CO2-humidified incubator. Following incubation, the cells from the bottom wells were collected and counted by a fluorescence-activated cell sorter (FACSCalibur; Becton Dickinson, Franklin Lakes, NJ) using the CellQuest software.
RESULTS
vCXCL1 Targets CXCR1 and CXCR2 as an Agonist When Screened against a Panel of the Human Chemokine Receptors
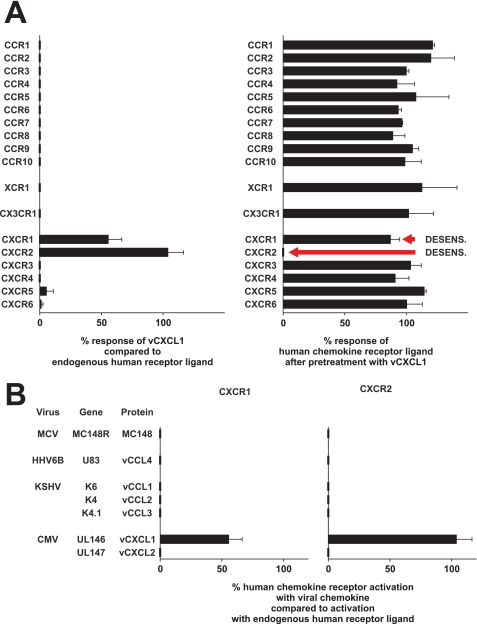
To screen for a possible interaction of the UL146 encoded gene product vCXCL1 with the 18 known human chemokine receptors, vCXCL1 was probed in calcium mobilization assays on a panel of cell lines individually expressing these receptors.
Initially, 100 nm of vCXCL1 was unable to elicit a calcium response through 16 of the 18 human chemokine receptors (Fig. 2A, left panel, for specific cell lines used see legend). However, in L1.2 cells stably transfected with either CXCR1 or CXCR2, vCXCL1 generated a robust calcium response (Fig. 2A). For CXCR2 the response was similar in magnitude to the response generated by CXCL1. For CXCR1 the response was of lower magnitude compared with the response generated by CXCL8, thus indicating a lower potency for vCXCL1 on this receptor. vCXCL1 was not able to induce a calcium response in any of the 10 other stable L1.2 cell lines expressing CCR1, 3, 4, 5, 6, 7, 8, and 10, CXCR6, and CX3CR1 (see legend to Fig. 2A), showing that the effect was mediated through CXCR1 and CXCR2 and not an endogenous receptor in L1.2 cells.
FIGURE 2.
Effect of recombinant vCXCL1 on calcium mobilization on a panel of cell lines stably transfected with the human chemokine receptors. A, 100 nm vCXCL1 or vehicle was first added to the cells followed by a submaximal dose of an appropriate endogenous human chemokine. The height of the response induced by the endogenous ligand only, the height of the response induced by vCXCL1 only, and the height of the response induced by endogenous ligand after pretreatment with vCXCL1 were measured. The left panel shows the calcium mobilization response of vCXCL1 compared with the responses of endogenous ligands on 18 different chemokine receptors. The right panel shows the calcium mobilization response via the 18 different receptors to the endogenous ligands in the presence of 100 nm vCXCL1. The data are the means ± S.E. (n = 2). The following cell lines, ligands, and concentrations were used: L1.2/CCR1, 10 nm CCL5/RANTES; 300.19/CCR2, 10 nm CCL2/MCP-1; L1.2/CCR3, 10 nm CCL7/MCP-3; L1.2/CCR4, 10 nm CCL22/MDC; L1.2/CCR5, 10 nm CCL5/RANTES; L1.2/CCR6, 10 nm CCL20/LARC; L1.2/CCR7, 1 nm CCL21/SLC; L1.2/CCR8, 10 nm CCL1/I-309; 300.19/CCR9, 10 nm CCL25/TECK; L1.2/CCR10, 10 nm CCL28; L1.2/CXCR1, 10 nm CXCL8/IL-8; L1.2/CXCR2, 10 nm CXCL1/GROα; 300.19/CXCR3, 1 nm CXCL11/I-TAC; CHO/CXCR4, 10 nm CXCL12/SDF-1α; 300.19/CXCR5, 10 nm CXCL13/BCA-1; L1.2/CXCR6, 10 nm CXCL16; 300.19/XCR1, 100 nm XCL1/Lymphotactin; and L1.2/CX3CR1, 10 nm CX3CL1/Fractalkine. B, the calcium mobilization response to viral chemokine compared with the response of endogenous ligand on cells stably transfected with either CXCR1 or CXCR2. The concentrations of viral chemokine used were 100 nm for MC148, vCCL1, vCCL2, vCCL3, and vCXCL1 and 1 μm for vCCL4 and vCXCL2. The cell lines used were L1.2/CXCR1 cells (vCCL1, vCCL2, vCCL3, vCXCL1, and vCXCL2), 300.19/CXCR1 (MC148), CHO/CXCR1 (vCCL4), L1.2/CXCR2 ((vCCL1, vCCL3, vCXCL1, and vCXCL2), 300.19/CXCR2 (MC148 and vCCL2), and CHO/CXCR2 (vCCL4). The data are the means ± S.E. (n = 2–4).
vCXCL1 did not act as an antagonist on any of the tested human chemokine receptors, because pretreatment of the cell lines with vCXCL1 was unable to inhibit or block the responses to the relevant endogenous chemokines through any of the human chemokine receptors tested (Fig. 2A, right panel). However, in CXCR2-transfected L1.2 cells, no response to CXCL1 was observed after pretreatment with 100 nm vCXCL1 (Fig. 2A, right panel, red arrow). This inhibitory effect is due to specific desensitization. In contrast, pretreatment of the L1.2/CXCR1 cells with vCXCL1 reduced the response of CXCL8 only slightly, if at all. Again indicating that vCXCL1 has a lower potency for this receptor compared with CXCL8. To find out whether this was the case, we did calcium mobilization desensitization experiments on 300.19/CXCR1 and L1.2/CXCR1 cells using 1 μm of vCXCL1 and 1 nm of CXCL8, which resulted in an 81% decrease of the CXCL8 response in 300.19 cells (n = 3; data not shown) and 88% in L1.2 cells (n = 2; data not shown). These results show that vCXCL1 is able to desensitize the response of CXCL8 through the CXCR1 receptor. To show that the activation of CXCR1 and CXCR2 was not an assay-specific phenomenon, we tested a panel of known virally encoded chemokines on these two receptors. Of the seven known chemokines encoded by human viruses, only the UL146 gene product was able to activate CXCR1 and CXCR2 (Fig. 2B). These viral chemokines were able to activate or block their known receptor targets (7, 9–16) (data not shown), establishing that they were biologically active with the exception of vCXCL2, where the receptor target has not yet been identified.
Having established that vCXCL1 targeted CXCR1 and CXCR2 as an agonist, we decided next to characterize this interaction in further detail. Hence, we performed competition binding, additional receptor signaling, and chemotaxis assays. vCXCL1 was tested in parallel with the seven human ELR-positive CXC chemokines (Fig. 1), which are known to target CXCR1 and/or CXCR2.
vCXCL1 Binds to CXCR1 and CXCR2
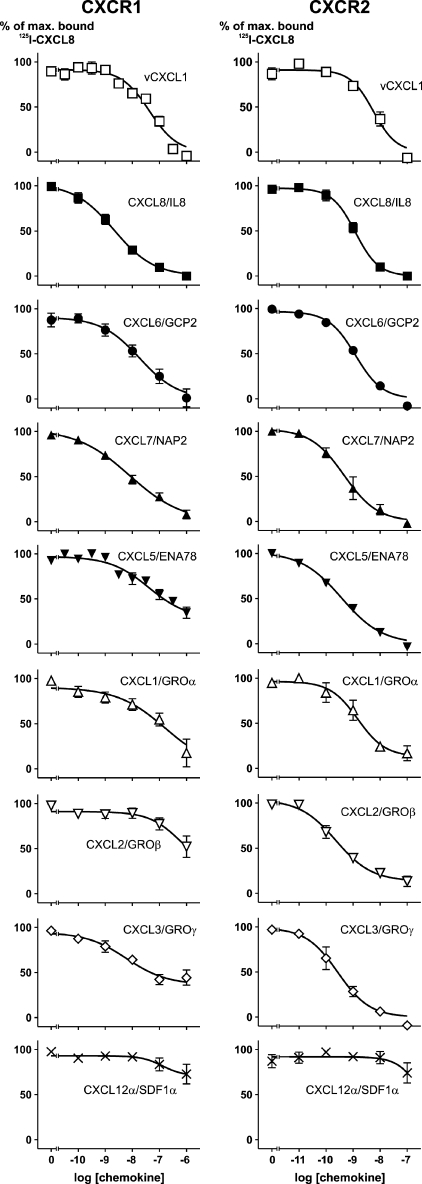
To characterize the affinity of vCXCL1 to both CXCR1 and CXCR2 and compare this affinity to the known endogenous human ELR-positive ligands, we performed binding studies on CHO cells stably transfected with either CXCR1 or CXCR2 using 125I-CXCL8. In these experiments vCXCL1 had an affinity of 44 nm (log Ki −7.37 ± 0.12) for the CXCR1 receptor. By comparison, CXCL6 and CXCL7 had higher affinities for CXCR1 than vCXCL1, but in the same order of magnitude (Fig. 3 and Table 1), whereas CXCL8 bound the receptor with the highest affinity with an IC50 of 2.5 nm (log Kd −8.61 ± 0.26). Only vCXCL1, CXCL6, CXCL7, and CXCL8 were able to displace all bound 125I-CXCL8 from the CXCR1 receptor, whereas CXCL1, CXCL2, CXCL3, and CXCL5 were unable to do so at a concentration of 1 μm (Fig. 3, left panels). By comparison, vCXCL1 and all of the endogenous human ELR-positive ligands bound CXCR2 with affinities in the nanomolar or subnanomolar range (Fig. 3, right panels, and Table 1). CXCL12α/SDF-1α, a non-ELR CXC chemokine included as a negative control, was unable to displace 125I-CXCL8 from either receptor (Fig. 3, bottom panels).
FIGURE 3.
125I-CXCL8 displacement experiments with vCXCL1 and the human ELR-positive CXC chemokines on CHO cells stably transfected with either CXCR1 or CXCR2. The cells were labeled with 125I-CXCL8 and incubated with increasing amounts of cold chemokine ligand as indicated. ○, vCXCL1; ●, CXCL8/IL-8; ■, CXCL6/GCP-2; ▴, CXCL7/NAP-2; ▾, CXCL5/ENA-78; ▵, CXCL1/GROα; ▿, CXCL2/GROβ; ◇, CXCL3/GROγ; ×, CXCL12/SDF-1α. Nonspecific binding was determined in the presence 0.1 μm of the relevant chemokine peptide. The data are the means ± S.E. (n = 3).
TABLE 1.
Binding affinities for the CMV UL146 gene product vCXCL1 compared with the human ELR-positive CXC chemokines on CXCR1 and CXCR2
Competition binding experiments were performed on CHO cells stably transfected with either CXCR1 or CXCR2. An IC50 value is shown in parentheses if 1 μm of the chemokine was unable to displace all bound radioligand and not shown at all if 1 μm was unable to displace 50% of the bound radioligand.
| Chemokine | CXCR1 |
CXCR2 |
||
|---|---|---|---|---|
| IC50 | IC50 ± S.E. | IC50 | IC50 ± S.E. | |
| nm | log | nm | log | |
| vCXCL1 | 44 | −7.37 ± 0.12 | 5.6 | −8.25 ± 0.09 |
| CXCL8/IL8 | 2.5 | −8.61 ± 0.26 | 1.2 | −8.92 ± 0.12 |
| CXCL6/GCP2 | 15 | −7.84 ± 0.29 | 1.2 | −8.92 ± 0.03 |
| CXCL7/NAP2 | 8.2 | −8.09 ± 0.17 | 0.5 | −9.34 ± 0.27 |
| CXCL5/ENA78 | (238) | (−6.63 ± 0.24) | 0.4 | −9.44 ± 0.12 |
| CXCL1/GROα | (127) | (−6.89 ± 0.48) | 1.5 | −8.83 ± 0.18 |
| CXCL2/GROβ | 0.2 | −9.63 ± 0.12 | ||
| CXCL3/GROγ | (42) | (−7.37 ± 0.41) | 0.3 | −9.56 ± 0.30 |
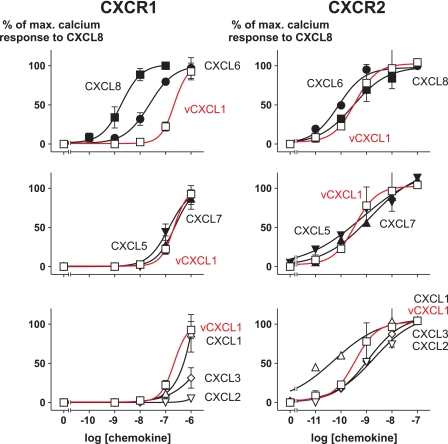
vCXCL1 Induces Calcium Mobilization through CXCR1 and CXCR2 in a Concentration-dependent Manner
To calculate potencies (EC50) we treated 300.19 cells stably transfected with either CXCR1 or CXCR2 with increasing concentrations of vCXCL1 and measured intracellular calcium release. For comparison the seven endogenous ELR-positive CXC chemokines were tested in parallel. As shown in Fig. 4 (red curves), the CMV-encoded gene product of UL146 was able to induce a maximal calcium response through CXCR1 in stably transfected 300.19 cells. Interestingly the different ELR+ CXC chemokines had a wide range of potencies for the CXCR1 receptor (Fig. 4 and Table 2). The potency of vCXCL1 was 217 nm (log EC50 −6.66 ± 0.18), whereas the potencies of CXCL6 and CXCL8 were 1 and 2 orders of magnitude higher, respectively. CXCL1, CXCL5, and CXCL7 had comparable potencies to vCXCL1, whereas CXCL2 and CXCL3 elicited a rather poor calcium response at concentrations of 1 μm. In contrast, vCXCL1 had a potency of 0.4 nm (log EC50 −9.45 ± 0.54), equal to the potency of CXCL8 for the CXCR2 receptor. The other ELR-positive ligands had potencies for the CXCR2 receptor within a range of 1 order of magnitude from vCXCL1 and CXCL8 (Fig. 4 and Table 2). Thus CXCL1 had the highest potency for CXCR2 with an EC50 of 0.05 nm (log EC50 −10.3 ± 0.23), whereas CXCL2 had the lowest potency with an EC50 of 2 nm (log EC50 −8.71 ± 0.17).
FIGURE 4.
Calcium mobilization experiments in 300.19 cells stably expressing either CXCR1 or CXCR2 stimulated with increasing concentrations of vCXCL1 or the endogenous human ELR-positive CXC chemokines. □, vCXCL1; ■, CXCL8/IL-8; ●, CXCL6/GCP-2; ▴, CXCL7/NAP-2; ▾, CXCL5/ENA-78; ▵, CXCL1/GROα; ▿, CXCL2/GROβ; ◇, CXCL3/GROγ. The data are the means ± S.E. (n = 3).
TABLE 2.
Potencies for the CMV UL146 gene product vCXCL1 compared with the human ELR-positive CXC chemokines on CXCR1 and CXCR2
Dose-response calcium mobilization experiments were performed on 300.19 cells stably transfected with either CXCR1 or CXCR2. An EC50 value is not shown if 1 μm was unable to elicit a response 50% of maximum.
| Chemokine | CXCR1 |
CXCR2 |
||
|---|---|---|---|---|
| EC50 | EC50 ± S.E. | EC50 | EC50 ± S.E. | |
| nm | log | nm | log | |
| vCXCL1 | 217 | −6.66 ± 0.18 | 0.4 | −9.45 ± 0.54 |
| CXCL8/IL8 | 1.8 | −8.74 ± 0.21 | 0.3 | −9.51 ± 0.25 |
| CXCL6/GCP2 | 23 | −7.64 ± 0.13 | 0.08 | −10.1 ± 0.07 |
| CXCL7/NAP2 | 219 | −6.66 ± 0.20 | 1.4 | −8.85 ± 0.63 |
| CXCL5/ENA78 | 139 | −6.86 ± 0.29 | 0.7 | −9.17 ± 0.36 |
| CXCL1/GROα | 325 | −6.49 ± 0.30 | 0.05 | −10.3 ± 0.23 |
| CXCL2/GROβ | 2.0 | −8.71 ± 0.17 | ||
| CXCL3/GROγ | 1.2 | −8.93 ± 0.33 | ||
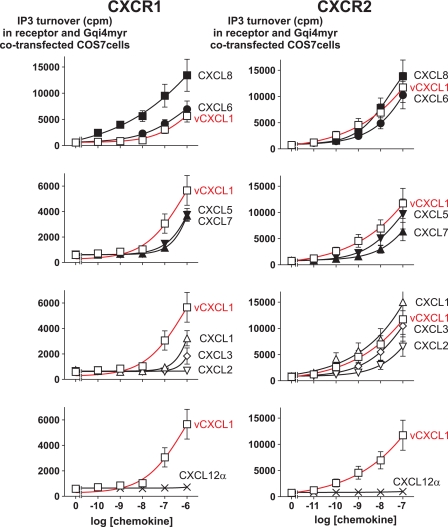
vCXCL1 Induces IP3 Accumulation through CXCR1 and CXCR2 in a Concentration-dependent Manner
To confirm that vCXCL1 activated CXCR1 and CXCR2 in another signal transduction assay, dose-response studies were performed in COS-7 cells co-transfected with a chimeric G protein, the Gqi4myr. Co-transfection of cells with the Gqi4myr gene enables Gαi-coupled receptors, such as CXCR1 and CXCR2, to signal through the Gαq pathway, and IP3 accumulation can be measured (25).
In cells co-transfected with CXCR1 and Gqi4myr, vCXCL1 stimulated IP3 accumulation in a dose-dependent manner (Fig. 5, red curves). Compared with vCXCL1, higher concentrations of CXCL1, CXCL3, CXCL5, and CXCL7 were needed to induce an IP3 response, whereas CXCL2 was unable to stimulate IP3 accumulation in this assay. CXCL8, as expected, was able to induce IP3 production in a subnanomolar range and with greater efficacy than all the other ligands. The rank order of ligands for CXCR1 was CXCL8 ≫ CXCL6 > vCXCL1 > CXCL7 ∼ CXCL5 ∼ CXCL1 > CXCL3, whereas CXCL2 was unable to induce a response (Fig. 5). Moreover, these results are in agreement with the rank order of potencies from the calcium mobilization assays, except that vCXCL1 was more potent than CXCL5 and CXCL7 in the IP3 assay.
FIGURE 5.
vCXCL1 or ELR-positive CXC chemokines induced IP3 turnover in COS7 cells transiently transfected with CXCR1 or CXCR2 and Gqi4myr. □, vCXCL1; ■, CXCL8/IL-8; ●, CXCL6/GCP-2; ▴, CXCL7/NAP-2; ▾, CXCL5/ENA-78; ▵, CXCL1/GROα; ▿, CXCL2/GROβ; ◇, CXCL3/GROγ; ×, CXCL12/SDF-1α. The data are the means ± S.E. (n = 3).
In cells co-transfected with CXCR2 and Gqi4myr, vCXCL1 was able to induce IP3 accumulation in a dose-dependent manner in the nanomolar range. The rank order of ligands was CXCL1 > vCXCL1 ∼ CXCL8 > CXCL3 ∼ CXCL6 ∼ CXCL5 > CXCL2 ∼ CXCL7 (Fig. 5). These results were in agreement with the EC50 values derived from the calcium mobilization experiments (Table 2), except that CXCL6 was as potent as CXCL1 in the calcium mobilization assay. CXCL12α, a non-ELR CXC chemokine, was included as a negative control and was not able to generate IP3 through either CXCR1 or CXCR2 (Fig. 5, bottom panels). In control experiments with COS-7 cells transfected with Gqi4myr alone, no effect of vCXCL1 was observed, demonstrating that the effect was mediated through CXCR1 and CXCR2 and not through an endogenous receptor expressed in COS-7 cells (n = 2; data not shown).
Surprisingly, in contrast to the calcium mobilization experiments, no Emax values for any of the tested ligands on CXCR1 and CXCR2 could be reached, thus making it difficult to calculate reliable EC50 values for these ligands. However, the results were still useful because the minimal concentrations eliciting an IP3 response could be ranked and compared with the EC50 values in the calcium mobilization assays (Table 1).
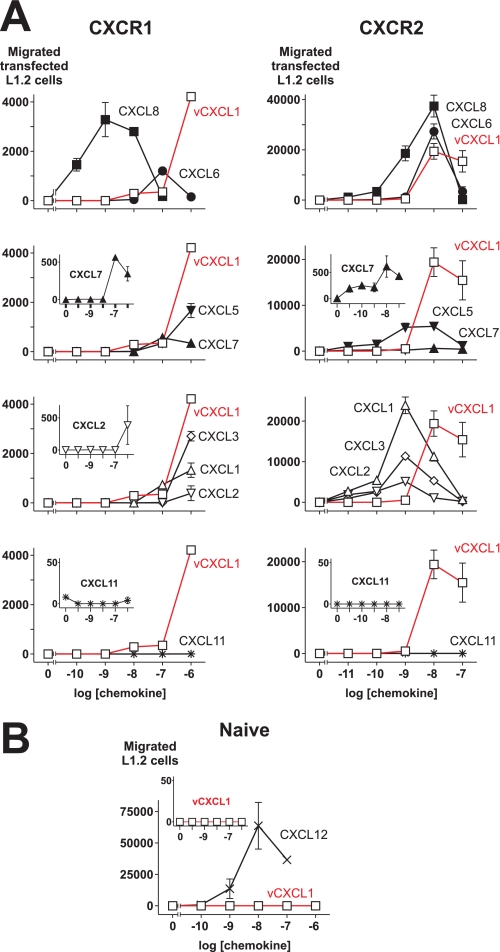
vCXCL1 Induces Migration of CXCR1 and CXCR2-expressing Cells
Having established that vCXCL1 binds to, induces calcium mobilization in, and is able to generate IP3 through the CXCR1 and CXCR2 receptors, we wanted to see whether this viral chemokine could generate a chemotactic response through these receptors. Hence, we explored the ability of vCXCL1 to induce chemotaxis in the murine pre-B lymphocyte cell line L1.2 stably transfected with either CXCR1 (Fig. 6A, left panels) or CXCR2 (Fig. 6A, right panels). We compared the chemotactic activity of the vCXCL1 protein with that of the endogenous human ELR-positive chemokines. As shown in Fig. 6A (left panels), the endogenous chemokines demonstrated different patterns with respect to both efficacy and potency in inducing chemotaxis via the CXCR1 receptor. For instance, CXCL8 was highly potent and efficacious on CXCR1, whereas CXCL1, CXCL3, CXCL5, and CXCL6 showed high efficacies but rather low potencies on CXCR1. In contrast, both CXCL2 and CXCL7 showed low efficacies and potencies. In fact, it looked like CXCL7 was unable to induce a chemotactic response through CXCR1, when the response was compared with the one induced by vCXCL1 (Fig. 6A, left panel, second from top). However (as seen in Fig. 6A, left panel, second from top, inset), CXCL7 was able to induce a chemotactic response with a high chemotactic index through this receptor, although the efficacy compared with the other ligands was low. CXCL6, CXCL7, and CXCL8 elicited classical bell-shaped dose-response curves typical for chemotaxis assays, whereas concentrations higher than 1 μm would be necessary for the rest of the endogenous ligands to evoke the downward slope of the curve (Fig. 6A, left panels).
FIGURE 6.
vCXCL1 or ELR-positive CXC chemokines induced chemotaxis of L1.2 cells stably expressing either CXCR1 or CXCR2, but not of naïve L1.2 cells. A, chemotactic activities of L1.2 cells stably transfected with CXCR1 or CXCR2 toward vCXCL1 or different endogenous ELR-positive or negative chemokines. □, vCXCL1; ■, CXCL8/IL-8; ●, CXCL6/GCP-2; ▴, CXCL7/NAP-2; ▾, CXCL5/ENA-78; ▵, CXCL1/GROα; ▿, CXCL2/GROβ; ◇, CXCL3/GROγ; *, CXCL11/I-TAC. The insets show the chemotactic activities of low efficacy ligands and the negative control on more appropriate scales. The figure shows one representative assay with S.E. as indicated (n = 3 for vCXCL1 and CXCL8 and n = 2 for the rest of the ligands). B, chemotactic activities of naïve L1.2 cells toward vCXCL1 (□) and CXCL12/SDF-1α (×). The inset shows the chemotactic activity of vCXCL1 (□) on a scale from 0 to 50. The data show one representative assay with S.E. as indicated (n = 2).
For the CXCR2 receptor, all of the endogenous human ligands showed the classical bell-shaped curve with potencies between 1 and 10 nm and with high efficacies for CXCL1, CXCL6, and CXCL8, intermediate efficacies for CXCL2, CXCL3, and CXCL5 and a rather low efficacy for CXCL7 (Fig. 6A, right panels) but with a high chemotactic index (Fig. 6A, right panel, second from top, inset).
The efficacy of vCXCL1 in evoking chemotaxis of CXCR1 receptor-expressing cells was comparable with CXCL6 (Fig. 6A, top left panel) and higher than the rest of the endogenous human ELR+ ligands (Fig. 6A, left panels). However, CXCL8 was more potent and efficacious than vCXCL1 in inducing chemotaxis via CXCR1 (Fig. 6A, top left panel). vCXCL1 induced a chemotactic response with a potency and efficacy comparable with CXCL6 and CXCL8 in CXCR2 (Fig. 6A, top right panel). Summarized, these results show that vCXCL1 can generate chemotactic responses with high efficacies through both the CXCR1 and CXCR2 receptors.
Because the L1.2 cell line is known to express CXCR4, CXCL11/I-TAC and not CXCL12α was chosen as the non-ELR CXC chemokine to be used as a negative control. CXCL11 was not able to generate chemotactic responses through either CXCR1 or CXCR2, showing that the responses were specific for vCXCL1 and the endogenous human ELR-positive ligands (Fig. 6A, bottom panels and insets). vCXCL1 did not induce chemotaxis of naïve L1.2 cells (Fig. 6B and inset), showing that the chemotactic activities were mediated through CXCR1 and CXCR2 and not an endogenous receptor in L1.2 cells.
DISCUSSION
Through a systematic analysis using 18 cloned and categorized human chemokine receptors, we here find that vCXCL1, the UL146 gene product from CMV, targets CXCR1 and CXCR2 and no other chemokine receptor. Moreover, we demonstrate that vCXCL1 binds to both of these receptors and is highly efficacious in activating both CXCR1 and CXCR2 in three different signal transduction assays including calcium mobilization and chemotaxis. CXCR1 and CXCR2 are highly expressed in neutrophils, and this virally encoded chemokine could have a role in the infectious life cycle of CMV by attracting neutrophils to sites of viral infection to use this cell type as a vehicle for dissemination.
Endogenous Human CXCR1 and CXCR2 Ligands
Most binding and activation studies of CXC chemokines belonging to the ELR group have been performed on neutrophils. Results from these studies suggested the existence of two CXCL8 receptors, which led to the identification of CXCR1 and CXCR2 in 1991 (26, 27). Almost all affinities of the ELR-positive group of chemokines on cells expressing either CXCR1 or CXCR2 have been determined in heterologous competition binding experiments. Such assays, however, can sometimes be misleading, because a high affinity ligand may not always be able to compete for another high affinity ligand (28). In fact this was the case for CXCR2, as shown in a comprehensive study by Ahuja and Murphy (29). This group reported that several ELR-positive CXC chemokines had a lower affinity for the CXCR2 receptor in heterologous binding experiments compared with the affinity obtained in homologous binding experiments. For example, CXCL5 and CXCL7 had affinities between 500 and 125 nm in heterologous binding experiments compared with 15 and 1 nm in homologous binding experiments. In contrast to Ahuja et al. (29), however, we found that by performing heterologous competition binding experiments, the affinities of these two chemokines for CXCR2 were in the subnanomolar range (Table 1). We believe that the differences between their results (lower binding affinities) and our results arise in to the different arrangements of our assays. Ahuja et al. used cells in suspension and separated bound from unbound radioligand by pelleting cells through a sucrose cushion. In contrast, we used adherent cells, which were washed four times with a chemokine buffer with 0.5 m NaCl.
Because CXCR1 is 77% identical to CXCR2 at the amino acid level, one has to consider that the affinities of the ELR-positive chemokines for CXCR1 obtained from heterologous competition binding experiments can be misleadingly low, as was the case for CXCR2 receptor. However, this cannot be tested in competition binding experiments, because CXCL8 is the only ligand with sufficient affinity to be used as a radioligand for CXCR1. Therefore, potencies from receptor activation studies, and not just affinities from competition binding experiments, are crucial when deciding whether a ligand targets a receptor or not.
Importantly, the affinities of the endogenous human ELR-positive CXC chemokines for both the CXCR1 and CXCR2 receptors (Table 1) were supported by the potencies obtained in calcium mobilization experiments (Table 2) and the results from two other receptor activation studies. CXCL7 was an exception, because it had an affinity for CXCR1 in the nanomolar range but a potency in the submicromolar range. We are unable to explain this finding, but it is unlikely to be due to the use of a heterologous binding assay compared with the use of a homologous binding assay. If that were the case, we would expect lower affinities rather than lower potencies.
Furthermore the results obtained by calcium mobilization experiments by other research groups are in agreement with the results presented above (Fig. 4) (27, 29–33). In addition, the reported chemotactic activities of the endogenous ELR-positive ligands for the CXCR1 and CXCR2 receptors are also compatible with the results presented above (Fig. 6) (31–35). Chemotactic responses for CXCL2 and CXCL3 on CXCR1- and CXCR2-expressing cells have not been reported before.
The Role of CXCR1 and CXCR2 in Inflammation
CXCR1 and CXCR2 have been found to be co-expressed in great numbers on neutrophils and also on other immune cells such as monocytes, CD8+ T cells, and NK cells (6). No in vivo data are available on the differential roles of CXCR1 and CXCR2 in the immune system. However, two recent independent studies using inflammatory lung models in lethally irradiated CXCR2 knock-out mice reconstituted with bone marrow from wild type mice show that these mice have a surprisingly large defect in their ability to recruit neutrophils (36) or mast cell progenitors (37). These results suggest that CXCR2 expression in pulmonary endothelial cells is important for transcellular migration of effector cells. Other studies have found expression of CXCR2 only on microvascular endothelial cells (36, 38–40) in contrast to macrovascular endovascular cells (41–44).
vCXCL1, a Dual CXCR1 and CXCR2 Agonist
In 1996 sequencing of CMV wild type isolates identified 22 additional open reading frames of the CMV genome that had been deleted from laboratory strains during passage (17). Based on the deduced amino acid sequence of one of these open reading frames, UL146 was predicted to encode a CXC chemokine with an ELR epitope (Fig. 1). Our finding that the protein product of UL146 targets CXCR1 and CXCR2 and none of the other known chemokine receptors is consistent with the fact that all human encoded CXC chemokines with the ELR motif exclusively target one of or both of these two receptors. When the amino acid sequence of vCXCL1 is aligned with those of the human ELR-positive CXC chemokines, some differences are noted. First, the viral chemokine has an extended C terminus, which is glycosylated (Fig. 1) (16) like the extended C terminus of lymphotactin. The C terminus lacks the glycosaminoglycan-binding epitopes, BBXB (B being a basic residue), that is prominent in the extended tails of CCL21/SLC, CCL28, CXCL9/MIG, and CXCL12γ. Second, it has extended “30s-loop” and “40s-loop” regions (Fig. 1). The conserved (P)GP motif in the 30s-loop of ELR-positive CXC chemokines has been shown to be important for receptor binding (45), this motif is preserved in the CMV UL146 gene product, and interestingly, a second PGP motif is found in the extended 40s-loop of the CMV chemokine (Fig. 1, red box).
In a previous report Penfold et al. (16) found that the UL146 CMV gene product activated and induced chemotaxis of neutrophils, which express both CXCR1 and CXCR2. On the basis of binding studies only, on stable cell lines expressing either CXCR1 or CXCR2, they concluded that vCXCL1 is a selective CXCR2 agonist that does not bind CXCR1. In contrast, here we were able to demonstrate that vCXCL1 (44 nm) binds to CXCR1 with equal affinities as CXCL6 (15 nm) and CXCL7 (8.2 nm) (Fig. 3, left panels, and Table 1). Importantly, we were able to confirm our binding results on the CXCR1 receptor with three different receptor activation assays using three different cell lines, which showed that the UL146 gene product indeed was able to activate the CXCR1 receptor at concentrations of 10–100 nm with an EC50 in the submicromolar range (217 nm) (Figs. 2, 4, 5, and 6 and Table 2). Although the potency of vCXCL1 for CXCR1 was low, it was a surprisingly efficacious ligand. Despite its high efficacy on CXCR1, the question arises as to whether the low potency of vCXCL1 is physiologically relevant for the activation of this receptor. It could very well be the case that vCXCL1 is expressed in CMV-infected cells with the sole purpose of activating CXCR2 and not CXCR1. However, before one dismisses the activation of CXCR1 with vCXCL1 as biologically irrelevant, one has to consider some lines of evidence that could point in the opposite direction.
First, it must be emphasized that it is not uncommon for viral cytokine homologs to have lower affinities for a receptor than their endogenous mammalian counterparts. For instance, this has been shown to be the case for the Epstein-Barr virus-encoded IL-10 homolog, which has a 1000-fold lower affinity to the human IL-10 receptor than endogenous IL-10 (46). Likewise, the HHV8-encoded IL-6 homolog has a 10-fold lower affinity to the human IL-6 receptor than endogenous IL-6 (47). Similarly, poxvirus homologs of the epidermal growth factor (EGF) have 10–1000-fold lower affinities to the EGF receptor than endogenous EGF (48). Importantly, in the latter study the viral EGF homologs were shown to be more potent mitogens than their mammalian counterparts because of attenuation of receptor degradation leading to a sustained signal transduction. These results suggest that viral homologs of human gene products can be significantly different from their human counterparts.
Second, 177 sequences of the UL146 gene have been deposited in GenBankTM, and they display considerable variations in the amino acid sequences, although the ELR motif seems to be preserved (49–52). Thus it is possible that variant UL146 genes encode proteins with different affinities and potencies toward CXCR1 and CXCR2. Finally, vCXCL1 has three glycosylation sites in its extended C-terminal tail (Fig. 1) (16), suggesting that the in vivo form of this viral chemokine may exert an increased biological activity compared with the Escherichia coli recombinant form, as has been demonstrated for the chemokine lymphotactin (53).
One can only speculate as to why the CMV virus has incorporated a gene encoding a CXCR1 and CXCR2 agonist in its genome. One could envision that CMV-infected endothelial cells express vCXCL1 to attract neutrophils, so they can act as passive carriers for CMV (Fig. 7). In that way the virus could maintain a pool of CMV-infected endothelial cells.
FIGURE 7.
A hypothetical function for the UL146 gene product vCXCL1 in CMV biology. A CMV-infected endothelial cell expressing vCXCL1 recruits primary effector cells such as neutrophils and infect them. After infection, the neutrophils carry the virus to and infect endothelial cells at other sites, thus ensuring a constant pool of CMV-infected endothelial cells. For simplicity only CXCR1 and CXCR2 and no other chemokine receptors are shown on the neutrophils. Green circles, CMV-encoded vCXCL1; red circles, human ELR-positive chemokines exemplified by CXCL8/IL-8.
Acknowledgments
We thank Kirsten Culmsee for excellent technical assistance, Professor Thue Schwartz for office and laboratory space, Professor Ulrik Gether for access to the spectroflourometer, and Maria Waldhoer for proofreading the manuscript.
This work was supported by grants from the Danish Medical Research Council, the Foundation of A. P. Møller and Chastine McKinney Møller, the Foundation of Arvid Nilsson, the Augustinus Foundation, the Beckett-Foundation, the Foundation of Bent Bøgh and Wife, the Foundation of Carl and Ellen Hertz, the Foundation of Christian Larsen and Ellen Larsen, the Foundation of Frode V. Nyegaard and Wife, the Foundation of E. Danielsen and Wife, the Foundation of Einar Hansen and Wife, the Foundation of Michael Hermann Nielsen, the Foundation of Else and Mogens Wedell-Wedellsborg, the Foundation of Karl G Andersen, the Harboe-Foundation, the Foundation of Holger Hjortenberg and Wife, the Illum Foundation, the Foundation of Johan Boserup and Lise Boserup, the Foundation of Niels Hansen and Wife, the Foundation of Werner Richter and Wife, the Foundation of Ove William Buhl Olesen and Wife, the Foundation of Meta and Håkon Bagger, and the Foundation of Jakob Madsen and Wife.
- CMV
- cytomegalovirus
- HHV
- human herpesvirus
- CHO
- Chinese hamster ovary
- IP3
- inositol triphosphate
- IL
- interleukin
- EGF
- epidermal growth factor.
REFERENCES
- 1.Smith M. G. (1956) Proc. Soc. Exp. Biol. Med. 92, 424–430 [DOI] [PubMed] [Google Scholar]
- 2.Mocarski E. S., Shenk T., Pass R. T. (2007) in Fields Virology (Knipe D. M., Howley P. M. eds) pp. 2701–2772, Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3.Lindow M., Lüttichau H. R., Schwartz T. W. (2003) Trends Pharmacol. Sci. 24, 126–130 [DOI] [PubMed] [Google Scholar]
- 4.Rosenkilde M. M., Waldhoer M., Lüttichau H. R., Schwartz T. W. (2001) Oncogene 20, 1582–1593 [DOI] [PubMed] [Google Scholar]
- 5.Seet B. T., Johnston J. B., Brunetti C. R., Barrett J. W., Everett H., Cameron C., Sypula J., Nazarian S. H., Lucas A., McFadden G. (2003) Annu. Rev. Immunol. 21, 377–423 [DOI] [PubMed] [Google Scholar]
- 6.Murphy P. M., Baggiolini M., Charo I. F., Hébert C. A., Horuk R., Matsushima K., Miller L. H., Oppenheim J. J., Power C. A. (2000) Pharmacol. Rev. 52, 145–176 [PubMed] [Google Scholar]
- 7.Dairaghi D. J., Fan R. A., McMaster B. E., Hanley M. R., Schall T. J. (1999) J. Biol. Chem. 274, 21569–21574 [DOI] [PubMed] [Google Scholar]
- 8.Dewin D. R., Catusse J., Gompels U. A. (2006) J. Immunol. 176, 544–556 [DOI] [PubMed] [Google Scholar]
- 9.Endres M. J., Garlisi C. G., Xiao H., Shan L., Hedrick J. A. (1999) J. Exp. Med. 189, 1993–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kledal T. N., Rosenkilde M. M., Coulin F., Simmons G., Johnsen A. H., Alouani S., Power C. A., Lüttichau H. R., Gerstoft J., Clapham P. R., Clark-Lewis I., Wells T. N., Schwartz T. W. (1997) Science 277, 1656–1659 [DOI] [PubMed] [Google Scholar]
- 11.Lüttichau H. R., Stine J., Boesen T. P., Johnsen A. H., Chantry D., Gerstoft J., Schwartz T. W. (2000) J. Exp. Med. 191, 171–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lüttichau H. R., Lewis I. C., Gerstoft J., Schwartz T. W. (2001) Eur. J. Immunol. 31, 1217–1220 [DOI] [PubMed] [Google Scholar]
- 13.Lüttichau H. R., Clark-Lewis I., Jensen P. Ø., Moser C., Gerstoft J., Schwartz T. W. (2003) J. Biol. Chem. 278, 10928–10933 [DOI] [PubMed] [Google Scholar]
- 14.Lüttichau H. R., Johnsen A. H., Jurlander J., Rosenkilde M. M., Schwartz T. W. (2007) J. Biol. Chem. 282, 17794–17805 [DOI] [PubMed] [Google Scholar]
- 15.Lüttichau H. R. (2008) Virol. J. 5, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Penfold M. E., Dairaghi D. J., Duke G. M., Saederup N., Mocarski E. S., Kemble G. W., Schall T. J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 9839–9844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cha T. A., Tom E., Kemble G. W., Duke G. M., Mocarski E. S., Spaete R. R. (1996) J. Virol. 70, 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark-Lewis I., Vo L., Owen P., Anderson J. (1997) Methods Enzymol. 287, 233–250 [DOI] [PubMed] [Google Scholar]
- 19.Johansen T. E., Schøller M. S., Tolstoy S., Schwartz T. W. (1990) FEBS Lett. 267, 289–294 [DOI] [PubMed] [Google Scholar]
- 20.Stine J. T., Chantry D., Gray P. (1999) Chem. Immunol. 72, 161–180 [DOI] [PubMed] [Google Scholar]
- 21.Imai T., Baba M., Nishimura M., Kakizaki M., Takagi S., Yoshie O. (1997) J. Biol. Chem. 272, 15036–15042 [DOI] [PubMed] [Google Scholar]
- 22.Loetscher M., Gerber B., Loetscher P., Jones S. A., Piali L., Clark-Lewis I., Baggiolini M., Moser B. (1996) J. Exp. Med. 184, 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legler D. F., Loetscher M., Roos R. S., Clark-Lewis I., Baggiolini M., Moser B. (1998) J. Exp. Med. 187, 655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coulin F., Power C. A., Alouani S., Peitsch M. C., Schroeder J. M., Moshizuki M., Clark-Lewis I., Wells T. N. (1997) Eur. J. Biochem. 248, 507–515 [DOI] [PubMed] [Google Scholar]
- 25.Kostenis E. (2001) Trends Pharmacol. Sci. 22, 560–564 [DOI] [PubMed] [Google Scholar]
- 26.Holmes W. E., Lee J., Kuang W. J., Rice G. C., Wood W. I. (1991) Science 253, 1278–12801840701 [Google Scholar]
- 27.Murphy P. M., Tiffany H. L. (1991) Science 253, 1280–12831891716 [Google Scholar]
- 28.Maggi C. A., Schwartz T. W. (1997) Trends Pharmacol. Sci. 18, 351–355 [DOI] [PubMed] [Google Scholar]
- 29.Ahuja S. K., Murphy P. M. (1996) J. Biol. Chem. 271, 20545–20550 [DOI] [PubMed] [Google Scholar]
- 30.Ahuja S. K., Lee J. C., Murphy P. M. (1996) J. Biol. Chem. 271, 225–232 [DOI] [PubMed] [Google Scholar]
- 31.Loetscher P., Seitz M., Clark-Lewis I., Baggiolini M., Moser B. (1994) FEBS Lett. 341, 187–192 [DOI] [PubMed] [Google Scholar]
- 32.Wolf M., Delgado M. B., Jones S. A., Dewald B., Clark-Lewis I., Baggiolini M. (1998) Eur. J. Immunol. 28, 164–170 [DOI] [PubMed] [Google Scholar]
- 33.Wuyts A., Proost P., Lenaerts J. P., Ben-Baruch A., Van Damme J., Wang J. M. (1998) Eur. J. Biochem. 255, 67–73 [DOI] [PubMed] [Google Scholar]
- 34.Ben-Baruch A., Bengali K., Tani K., Xu L., Oppenheim J. J., Wang J. M. (1997) Cytokine 9, 37–45 [DOI] [PubMed] [Google Scholar]
- 35.Yang W., Schraw W. P., Mueller S. G., Richmond A. (1997) Biochemistry 36, 15193–15200 [DOI] [PubMed] [Google Scholar]
- 36.Reutershan J., Morris M. A., Burcin T. L., Smith D. F., Chang D., Saprito M. S., Ley K. (2006) J. Clin. Invest. 116, 695–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hallgren J., Jones T. G., Abonia J. P., Xing W., Humbles A., Austen K. F., Gurish M. F. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20478–20483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Addison C. L., Daniel T. O., Burdick M. D., Liu H., Ehlert J. E., Xue Y. Y., Buechi L., Walz A., Richmond A., Strieter R. M. (2000) J. Immunol. 165, 5269–5277 [DOI] [PubMed] [Google Scholar]
- 39.Heidemann J., Ogawa H., Dwinell M. B., Rafiee P., Maaser C., Gockel H. R., Otterson M. F., Ota D. M., Lugering N., Domschke W., Binion D. G. (2003) J. Biol. Chem. 278, 8508–8515 [DOI] [PubMed] [Google Scholar]
- 40.Schraufstatter I. U., Chung J., Burger M. (2001) Am. J. Physiol. Lung Cell Mol. Physiol. 280, L1094–L1103 [DOI] [PubMed] [Google Scholar]
- 41.Gupta S. K., Lysko P. G., Pillarisetti K., Ohlstein E., Stadel J. M. (1998) J. Biol. Chem. 273, 4282–4287 [DOI] [PubMed] [Google Scholar]
- 42.Hristov M., Zernecke A., Bidzhekov K., Liehn E. A., Shagdarsuren E., Ludwig A., Weber C. (2007) Circ. Res. 100, 590–597 [DOI] [PubMed] [Google Scholar]
- 43.Petzelbauer P., Watson C. A., Pfau S. E., Pober J. S. (1995) Cytokine 7, 267–272 [DOI] [PubMed] [Google Scholar]
- 44.Schönbeck U., Brandt E., Petersen F., Flad H. D., Loppnow H. (1995) J. Immunol. 154, 2375–2383 [PubMed] [Google Scholar]
- 45.Clark-Lewis I., Dewald B., Loetscher M., Moser B., Baggiolini M. (1994) J. Biol. Chem. 269, 16075–16081 [PubMed] [Google Scholar]
- 46.Liu Y., de Waal M. R., Briere F., Parham C., Bridon J. M., Banchereau J., Moore K. W., Xu J. (1997) J. Immunol. 158, 604–613 [PubMed] [Google Scholar]
- 47.Osborne J., Moore P. S., Chang Y. (1999) Hum. Immunol. 60, 921–927 [DOI] [PubMed] [Google Scholar]
- 48.Tzahar E., Moyer J. D., Waterman H., Barbacci E. G., Bao J., Levkowitz G., Shelly M., Strano S., Pinkas-Kramarski R., Pierce J. H., Andrews G. C., Yarden Y. (1998) EMBO J. 17, 5948–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arav-Boger R., Foster C. B., Zong J. C., Pass R. F. (2006) J. Infect. Dis. 193, 788–791 [DOI] [PubMed] [Google Scholar]
- 50.Hassan-Walker A. F., Okwuadi S., Lee L., Griffiths P. D., Emery V. C. (2004) J. Med. Virol. 74, 573–579 [DOI] [PubMed] [Google Scholar]
- 51.He R., Ruan Q., Qi Y., Ma Y. P., Huang Y. J., Sun Z. R., Ji Y. H. (2006) Intervirology 49, 215–223 [DOI] [PubMed] [Google Scholar]
- 52.Lurain N. S., Fox A. M., Lichy H. M., Bhorade S. M., Ware C. F., Huang D. D., Kwan S. P., Garrity E. R., Chou S. (2006) Virol. J. 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dong C., Chua A., Ganguly B., Krensky A. M., Clayberger C. (2005) J. Immunol. Methods 302, 136–144 [DOI] [PubMed] [Google Scholar]