Abstract
Pannexins (Panx) are a class of integral membrane proteins that have been proposed to exhibit characteristics similar to those of connexin family members. In this study, we utilized Cx43-positive BICR-M1Rk cells to stably express Panx1, Panx3, or Panx1-green fluorescent protein (GFP) to assess their trafficking, cell surface dynamics, and interplay with the cytoskeletal network. Expression of a Sar1 dominant negative mutant revealed that endoplasmic reticulum to Golgi transport of Panx1 and Panx3 was mediated via COPII-dependent vesicles. Distinct from Cx43-GFP, fluorescence recovery after photobleaching studies revealed that both Panx1-GFP and Panx3-GFP remained highly mobile at the cell surface. Unlike Cx43, Panx1-GFP exhibited no detectable interrelationship with microtubules. Conversely, cytochalasin B-induced disruption of microfilaments caused a severe loss of cell surface Panx1-GFP, a reduction in the recoverable fraction of Panx1-GFP that remained at the cell surface, and a decrease in Panx1-GFP vesicular transport. Furthermore, co-immunoprecipitation and co-sedimentation assays revealed actin as a novel binding partner of Panx1. Collectively, we conclude that although Panx1 and Panx3 share a common endoplasmic reticulum to Golgi secretory pathway to Cx43, their ultimate cell surface residency appears to be independent of cell contacts and the need for intact microtubules. Importantly, Panx1 has an interaction with actin microfilaments that regulates its cell surface localization and mobility.
Keywords: Cytoskeleton/Microtubules, Glycoproteins, Membrane/Channels, Protein/Cell Surface, Protein/Molecular Dynamics, Protein/Protein-Protein Interactions, Connexins, Pannexins
Introduction
The pannexin family is a new class of integral membrane glycoproteins that have been identified to share sequence homology with the invertebrate gap junction proteins, innexins (1). Unlike the connexin family that is composed of 21 members (2), both the human and mouse genomes contain only three pannexin-encoding genes (Panx1, Panx2, and Panx3) (1). Although connexins and pannexins exhibit no sequence homology, pannexins are predicted to share similar topology with connexins, which includes four transmembrane domains, two extracellular loops, a cytoplasmic loop, and intracellular amino and carboxyl termini (3–5). Our previous study has shown that ectopically expressed Panx1 and Panx3 are capable of trafficking to normal rat kidney (NRK)2 cell surfaces; however, their distribution profile at cell-cell interfaces is not typically clustered or punctate as seen for Cx43 (5). Consistently, electron micrographs of Panx1 overexpressing Madin-Darby canine kidney cells also revealed dispersed Panx1 localization at the plasma membrane with no evidence of gap junction plaques (4).
Cx43 has a relatively short half-life of ∼1–3 h (6). As a result, Cx43 subunits assembled into connexons within the trans-Golgi network (7) are constantly being transported to and removed from the plasma membrane (8). Recent reports provide evidence that although Panx1 appears to form similar hexameric channel units defined as pannexons (4), they exhibit slower turnover dynamics in comparison with Cx43, as assessed by the use of pharmacological blockers of protein synthesis and protein trafficking (4, 5). Functionally, most studies support the premise that pannexins, in particular Panx1, form single membrane channels (4, 5, 9–12), whereas far less evidence supports the notion that they also can form intercellular channels (13, 14).
The delivery of Cx43 for the assembly of functional channels at the cell surface has been extensively studied using fluorescent protein and epitope tags (15, 16). Cx43-GFP has been shown to exhibit similar distribution and functional characteristics as its untagged counterpart, when assessed by dye permeability and electrical conductance (8, 17). Likewise, GFP tagging of the carboxyl-terminal tail of Panx1 did not significantly alter the localization profile of Panx1 in NRK cells (5). However, a recent electrical conductance assessment provided evidence that mouse Panx1-GFP exhibits reduced channel function when expressed in human embryonic kidney (HEK) 293 cells (18), suggesting that its functional state is somewhat impaired but not completely eliminated by the GFP tag. In other studies, untagged and tetracysteine-tagged Panx1 exhibited comparable capabilities in rescuing the trafficking of the glycosylation-defective mutant of Panx1 in Madin-Darby canine kidney cells (19). Collectively, these studies would suggest that the trafficking and life cycle properties of Panx1 can be assessed by GFP tagging approaches in conjunction with real-time dynamic imaging.
It has been previously established through a number of studies using both untagged and GFP-tagged Cx43 that the delivery and regeneration of Cx43 gap junction plaques is facilitated by microtubules and requires a fully functional secretory pathway (8, 20, 21). Other studies using rapid time lapse imaging of GFP- and tetracysteine-tagged Cx43 have shown that Cx43 is delivered in 100–150-nm vesicles that coalesce laterally into the preexisting gap junction plaques (15, 22). However, the delivery of pannexins to the cell surface, their dynamic organization at specific cell surface microdomains, and their dependence on an intact cytoskeletal network have yet to be investigated.
The life cycle of Cx43 is governed, in part, by many direct and indirect binding partners (23–25), whereas pannexin binding partners are only beginning to be identified with some evidence to support Panx1 interaction with a protein subunit of the voltage-dependent potassium channel (26) and regulatory cross-talk with P2X7 receptors (9, 27). In the present study, we further expand on a very limited number of pannexin-binding proteins by identifying actin as a specific Panx1 binding protein.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
BICR-M1Rk cells originally derived from a rat mammary tumor and HEK 293T, NRK, and REK (rat keratinocyte) cells were cultured in high glucose Dulbecco's modified Eagle's medium (Invitrogen), supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine (all from Invitrogen). B16-BL6 murine melanoma cells were kindly provided by Dr. Moulay A. Alaoui-Jamali-(Department of Medicine and Oncology, McGill University (Montreal, Canada)) and cultured as described previously (28). Trypsin (0.25%, 1 mm EDTA), Opti-MEM I medium, and Lipofectamine 2000 were purchased from Invitrogen. Brefeldin A (BFA), nocodazole, and cytochalasin B were purchased from Sigma.
Expression Constructs of Mouse Panx1, Panx2, and Panx3 and Engineering of the GST-Panx1 Carboxyl Domain
Untagged and GFP-tagged expression constructs encoding full-length mouse Panx1 and Panx3 were previously described (5), and correspond to current NCBI reference sequence encoding 426 amino acids for Panx1 (NP_062355) and 392 amino acids for Panx3 (NP_766042).
As previously described, Panx2 was originally cloned from mouse brain, and the purified PCR product was inserted into the EcoRI-SalI site of pEGFP-N1 vector (Clontech) to generate the untagged Panx2 (29). The sequence was confirmed to encode 677 amino acids according to RefSeq. Furthermore, Panx2 cDNA was amplified using forward primer 5′-CCCAAGCTTATGCACCACCTC to create a HindIII site and 5′-GGCGACCGGTCCAAACTCCACA to create an AgeI site at the 5′- and 3′-ends of Panx2, respectively. PCR products and the vector pEGFP-N1 (Clontech) were digested with HindIII and AgeI and ligated, and clones were selected. GFP was fused in frame to the carboxyl terminus of Panx2 with the addition of a five-amino acid polylinker (GGACCGGTCGCCACC) and validated by sequencing.
Panx1 carboxyl tail primers (forward, 5′-CTAGGATTCCGGCAGAAAACGGAC; reverse, 5′-CGAGTCGACTTAGCAGGACGGATT), with flanking sites for BamHI and SalI amplifying the Panx1 carboxyl tail sequence (corresponding to amino acids 299–426), were created. This sequence was ligated into the pGEX-6P-3 GST vector and transformed into BL21 bacteria. Batch purification using Sepharose 4B was performed as described in the GST Gene Fusion System manual with some modifications (GE Healthcare). A single BL21 clone transformed with either GST alone or GST-Panx1 carboxyl tail was grown to an A600 nm of 1 and was induced by the addition of 0.5 mm isopropyl β-d-1-thiogalactopyranoside. A total of 500 ml of either GST or GST-Panx1 carboxyl tail bacterial culture was shaken overnight at room temperature. The next day, purification was performed using 400 μl of 50% Sepharose slurry, washed in PBS seven times, and stored at 4 °C for the co-sedimentation assays.
Transfection and Engineering of Stable Cell Lines
DsRed-tagged Sar1WT and Sar1H79G cDNA constructs were described previously (8) and used for transfection into BICR-M1Rk cells engineered to express Panx1 or Panx3. Briefly, BICR-M1Rk cells were grown overnight to 50–70% confluence on glass coverslips and transfected in Opti-MEM I medium containing 4 μl of Lipofectamine 2000 and 1 μg of Panx1 or 3 μg of Panx3 plasmid DNA together with 2 μg of Sar1WT or Sar1H79G expression constructs.
The Panx1-GFP cDNA construct was described previously (5) and used to transfect BICR-M1Rk cells stably expressing untagged Panx1 and B16-BL6 cells. Cells were grown overnight to 50–70% confluence on 35-mm glass bottom dishes and transfected in Opti-MEM I medium containing 1.5 μl of Lipofectamine 2000 and 1.5 μg of Panx1-GFP plasmid DNA for 4 h at 37 °C.
For Panx3 and Panx3-GFP co-transfections, BICR-M1Rk cells were grown overnight to 20–30% confluence on 35-mm glass bottom dishes and transfected in Opti-MEM I medium containing 4 μl of Lipofectamine 2000 and 3 μg of Panx3 together with 1.5 μg of Panx3-GFP plasmid DNA. Opti-MEM I medium was replaced with complete culture medium 4 h after transfection at 37 °C.
For Panx2 and Panx2-GFP expression, BICR-M1Rk cells grown overnight to 50–70% confluence on glass coverslips were transfected in Opti-MEM I medium containing 3 μl of Lipofectamine 2000 and 1.5 μg of Panx2 or 1.5 μg of Panx2-GFP plasmid DNA. For co-transfection of Panx2 with Panx2-GFP, 0.75 μg of each plasmid DNA was used in Opti-MEM I medium containing 3 μl of Lipofectamine 2000.
Full-length constructs encoding mouse Panx1, Panx1-GFP, and Panx3 were inserted into the AP-2 retroviral vector and transfected into the 293GPG packaging cell line as described earlier by Qin et al. (30). Following transfection, replication-defective retroviral supernatants were collected and filtered through a 0.45-μm filter (Pall Gelman Laboratories, Ann Arbor, MI). BICR-M1Rk cells expressing Cx43-GFP were engineered to stably express Panx1 and Panx3 by following a previously described protocol (30). Retrovirus encoding Panx1-GFP was also used to stably express GFP-tagged Panx1 in BICR-M1Rk cells.
Treatments with Pharmacological Reagents
Panx1- and Panx3-overexpressing BICR-M1Rk cells were treated with 5 μg/ml BFA for 19 h at 37 °C, and cell lysates were collected and subjected to immunoblotting. For elucidating the role of cytoskeletal elements in pannexin trafficking, Panx1-GFP-expressing cells were treated with 10 μm nocodazole or 2.5 μg/ml cytochalasin B for 90 min at 37 °C and fixed for immunocytochemistry. For fluorescence recovery after photobleaching (FRAP) studies, Panx1-GFP expressing cells were pretreated with cytoskeletal inhibitors for 90 min prior to imaging up to 3–4 h in presence of these same inhibitors.
Immunocytochemistry
Cells were immunolabeled as previously described (5). Briefly, cells grown on glass coverslips were fixed using ice-cold 80% methanol and 20% acetone for 20 min at 4 °C. Cytochalasin B-treated cells were fixed using 3.7% formaldehyde for 30 min at room temperature and permeabilized for 45 min in a 1% blocking solution (bovine serum albumin (BSA) (Sigma), containing 0.1% Triton X-100). Cells were incubated with a 500-fold dilution of polyclonal anti-Cx43 antibody (Sigma), a 100-fold dilution of polyclonal anti-GPP130 antibody (Convance), polyclonal anti-Panx2 antibody (Zymed Laboratories Inc., San Francisco, CA) or monoclonal anti-tubulin antibody (Convance) for 1 h at room temperature. Affinity-purified polyclonal Panx1 and Panx3 antibodies were used at a concentration of 2 μg/ml. F-actin was localized using a 200-fold dilution of rhodamine phalloidin (Invitrogen). Cells were incubated in goat anti-rabbit antibody conjugated to Texas Red or fluorescein isothiocyanate (1:100; Jackson Laboratories, West Grove, PA) or a goat anti-mouse antibody conjugated to Texas Red (1:100; Jackson Laboratory). Cells were rinsed with PBS, and nuclei were stained with Hoechst 33342 and mounted. Immunolabeled cells were imaged using a ×63 oil objective lens mounted on a Zeiss LSM 510 META system (Zeiss, Toronto, Canada).
Immunoblotting and Co-immunoprecipitation
Cell lysates from BICR-M1Rk cells transiently co-transfected with Sar1 and Panx1 or Panx3 cDNA constructs, and BFA-treated cells were collected using a lysis buffer containing 1% Triton X-100, 10 mm Tris, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.5% Nonidet P-40, 100 mm sodium fluoride, and 100 mm sodium orthovanadate and a protease inhibitor tablet (one tablet/10 ml of buffer; Roche Applied Science), pH 7.4. Protein concentrations were measured using a BCA protein determination kit (Pierce). In total, 20–30 μg of protein was resolved using 10% SDS-PAGE and transferred to nitrocellulose membrane (Pall Life Sciences, NY). Nitrocellulose membranes were blocked in Licor blocking solution (Lincoln, NE) or 3% BSA solution and probed overnight with polyclonal affinity-purified anti-Panx1 or anti-Panx3 antibodies (0.2 μg/ml) at 4 °C. Monoclonal anti-β-actin antibody (1:5000; Sigma) was used to assess gel loading. Detection of primary antibody binding was performed by using mouse IgG IR dye 800 (Rockland Immunochemicals) and rabbit IgG Alexa 680 (Invitrogen) with the Odyssey infrared imaging system (Licor).
For co-immunoprecipitation experiments, 1 mg of protein lysates from WT-, Panx1-, and Panx1-GFP-overexpressing BICR-M1Rk cells was incubated overnight at 4 °C in the lysis buffer (1% Triton X-100, 10 mm Tris, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.5% Nonidet P-40, 1 mm sodium fluoride, and 1 mm sodium orthovanadate) containing 10 μg/ml anti-Panx1 antibody. The antibody complex was pulled down with 30 μl of precleaned protein A-Sepharose beads (in PBS) for 2 h on the rocker at 4 °C. The antibody-bead complex was centrifuged at 4500 rpm at 4 °C for 2 min, and supernatant was aspirated. Unbound nonspecific protein was separated from bound proteins by washing three times with 500 μl of lysis buffer, and the bound complex was detached by boiling for 5 min in 30 μl of 2× Laemmli loading sample buffer containing β–mercaptoethanol. Samples were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes, which were probed with anti-Panx1 and anti-β-actin antibodies.
FRAP Analysis
To assess Panx1 or Panx3 dynamics at the cell surface, BICR-M1Rk cells expressing Panx1-GFP, alone or in combination with the untagged Panx1, or Panx3-GFP together with Panx3 were cultured on 35-mm glass bottom dishes and subjected to FRAP. B16-BL6 cells expressing Panx1-GFP were also subject to FRAP analysis. Rapid time lapse imaging was performed on a Zeiss LSM 510 META system to quantify the movement of Panx1-GFP into the bleached region of interest (ROI), as described previously (16). Briefly, glass bottom dishes were placed in an environmentally controlled chamber, and ROIs representing the various plasma membrane domains were selected and photobleached using scan iterations at 488 nm with 100% laser strength. Images were acquired ∼2–5 s apart for up to 60 s with 0.9% laser strength to avoid further photobleaching. Fluorescence intensities within ROIs were quantified as described previously (16). Briefly, fluorescence recovery was recorded before photobleaching, immediately upon completion of photobleaching (t was set to 0 s), and postbleaching at the following time intervals: 15, 25, 50, and 60 s. Postbleach intensities were corrected and normalized for any residual fluorescence, and the recoverable fraction of Panx1-GFP or Panx3-GFP was calculated using the equation, FNt = (Ft − F0)/(Fi − F0) as previously described (31), where FNt represents normalized fluorescence at a time point t; Ft is fluorescence intensity within ROI at t seconds postphotobleach; F0 is fluorescence intensity upon photobleaching at t = 0; and Fi is fluorescence intensity prior to photobleaching. All FRAP experiments were repeated three times for each experimental set, with each set containing multiple ROIs at t = 15, 25, 50, and 60 s. FNt values from replicates of each experimental set were combined, and a non-linear regression analysis was performed to obtain a curve of best fit using GraphPad Prism software (San Diego, CA). To compare the mobility dynamics of Panx1-GFP or Panx3-GFP in various plasma membrane domains, a one-way analysis of variance followed by Tukey's multiple comparison tests were performed. For comparisons between untreated and nocodazole- and cytochalasin B-treated experimental sets, t tests were performed using GraphPad Prism software.
Vesicle Movement
Panx1-GFP containing vesicles were analyzed by measuring the total distance traveled of vesicles that remained in confocal plane of focus for the duration of the analysis. Vesicles of ∼0.5–0.8 μm in diameter were monitored by 1.8-s interval image scans for a total period of 8.8 s (n = 18–20 over four independent repeats).
Actin Co-sedimentation Assays
Muscle actin (Cytoskeleton Inc., Denver, CO) was resuspended in buffer (5 mm Tris-HCl, pH 8.0, 0.2 mm CaCl2) to a concentration of 1 mg/ml in 250 μl on ice for 30 min. 25 μl of 10× actin polymerization buffer (500 mm KCl, 20 mm MgCl2, 10 mm ATP, 100 mm Tris, pH 7.5) was added to the monomeric actin and incubated at room temperature for 1 h (F-actin stock at 23 μm). Protein samples of BSA, GST, Panx1 carboxyl terminal tail (C-tail), and GST-tagged Panx1 C-tail were centrifuged at 150,000 × g for 1 h at 4 °C. Supernatants were collected and placed on ice. For co-sedimentation assays, a GST fusion protein containing the C-tail of mouse Panx1 was used, either as a fusion protein after elution from the Sepharose beads or after cleavage of the Panx1 C-tail from the fusion protein using PreScission Protease (GE Healthcare) for 16 h at 4 °C, as per the manufacturer's instructions. Following the Cytoskeleton Inc. protocol for actin binding protein assays, 50-μl samples were prepared with F-actin alone, GST protein with or without F-actin, BSA (negative control) plus F-actin, and Panx1 C-tail or GST fusion protein with or without F-actin. Samples were incubated at room temperature for 30 min, followed by centrifugation at 150,000 × g for 1.5 h at 24 °C. Supernatants were carefully removed and mixed with 10 μl of 4× Laemmli reducing sample buffer. The pellets were resuspended in 30 μl of double-distilled water and mixed with 30 μl of 2× Laemmli buffer. Equal volumes of resuspended pellets and supernatants were run in duplicate on 10% SDS-PAGE. Gels containing samples were either stained overnight with Sypro-Ruby Protein gel stain (Invitrogen) or transferred onto nitrocellulose membranes using an iBlot apparatus for dry transfer (Invitrogen). Sypro-Ruby-stained gels were destained and exposed to UV for visualization of the major bands. Nitrocellulose membranes were incubated overnight with primary anti-Panx1 antibodies, washed, and probed with Alexa-680 anti-rabbit secondary for detection of Panx1 bands with a LiCor scanner as described previously (5).
Biotinylation Assays
BICR-M1Rk cells grown on 100-mm dishes were transiently transfected with 5–10 μg of Panx3-GFP-encoding cDNA constructs, and 48 h post-transfection, cells expressing Panx3-GFP were subjected to biotinylation treatment on ice as described previously (5). Cells were incubated in PBS or with cold PBS containing EZ-link Sulfo-NHS-LC-biotin (0.5 mg/ml; Pierce) for 20 min at 4 °C. Control and biotin-treated cells were washed and incubated in 100 mm glycine buffer for 15 min at 4 °C to quench the biotin. Cells were then lysed with SDS lysis buffer (1% Triton X-100 and 0.1% SDS in PBS), and protein concentrations were measured using a BCA protein determination kit (Pierce). In total, 1000 μg of protein from control and biotin-treated cell lysates were rocked overnight at 4 °C in the presence of 50 μl of neutravidin-agarose beads (Pierce). Beads were washed three times with immunoprecipitation lysis buffer (150 mm NaCl, 10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 0.5% Nonidet P-40, and 1% Triton X-100) containing 1 mm NaF and 1 mm Na3VO4 and once with PBS containing 1 mm NaF and 1 mm Na3VO4. The beads were air-dried and resuspended in 50 μl of 2× Laemmli loading sample buffer containing β-mercaptoethanol before boiling for 5 min. As a lysate control, 40 μg of total protein from control and biotin samples was also resolved by SDS-PAGE and transferred to nitrocellulose membranes for immunoblotting with anti-Panx3 antibody. Glyceraldehyde-3-phosphate dehydrogenase was used as a control to detect any unexpected biotin internalization.
RESULTS
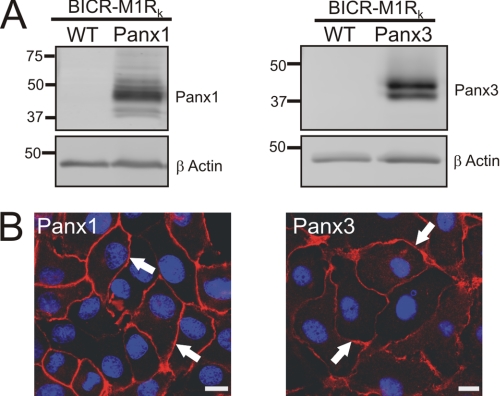
We have previously shown that the Cx43-positive BICR-M1Rk (rat mammary tumor) cell line is an excellent reference model for investigating dynamic delivery events and assembly and turnover mechanisms of Cx43 (16, 32); therefore, we designed our experimental approach to compare Panx1 and Panx3 trafficking dynamics in Cx43-positive BICR-M1Rk cells. We engineered stable cells lines to ectopically express Panx1 or Panx3. Western blots analysis revealed that wild-type BICR-M1Rk cells are negative for Panx1 and Panx3 (Fig. 1A), but when engineered to express pannexins, Panx1 resolved as multiple species ranging from ∼41 to 48 kDa, whereas Panx3 was detected as a doublet at ∼43 kDa (Fig. 1A). These multiple pannexin species have previously been shown to be the result of glycosylation (4, 5). Immunolabeling of both Panx1 and Panx3 revealed that both of these pannexins were capable of trafficking and localizing in a relatively uniform pattern at the apposing cell surface (Fig. 1B). In contrast, detectable Panx2 and Panx2-GFP were mainly localized in the intracellular compartments of not only BICR-M1Rk cells but also HEK 293T, NRK, and rat keratinocytes (supplemental Fig. 1). Not unexpected, the co-expression of Panx2 with Panx2-GFP did not alter the distribution pattern of untagged or tagged Panx2. These studies indicate that Panx2 has a unique distribution when compared with Panx1 and Panx3 even when expressed in the same reference cells.
FIGURE 1.
Panx1 and Panx3 are capable of trafficking to the cell surface in BICR-M1Rk cells. BICR-M1Rk cells were engineered to stably express Panx1 or Panx3. Immunoblotting with affinity-purified antibodies (anti-Panx1 and anti-Panx3) revealed multiple banding profiles of Panx1 (∼41–48 kDa) and Panx3 (∼41–43 kDa) (A). β-Actin was used as a protein loading control (A). Immunolabeling of Panx1 and Panx3 (B) identified that both pannexins trafficked and localized to the cell surface (arrows). Nuclei are stained with Hoechst 33342 (blue). Bars, 10 μm. Results shown are representative of three independent experiments.
Trafficking of Panx1 and Panx3 Is Mediated through Sar1-dependent COPII Vesicles
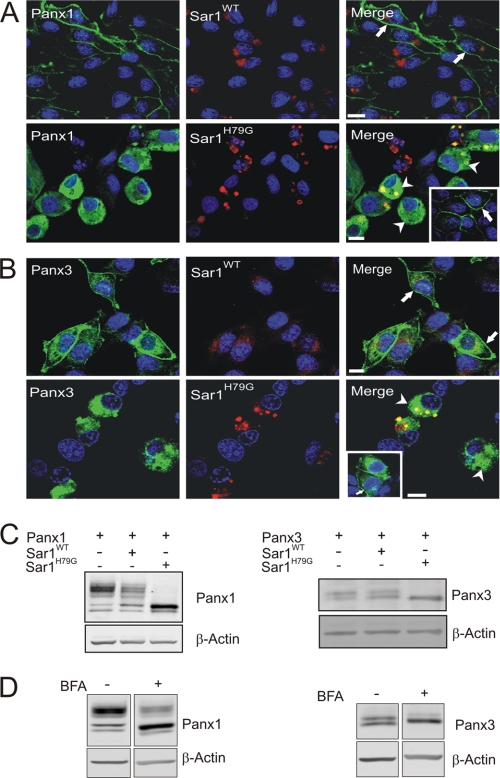
It has been widely documented that Sar1 (secretion-associated and Ras-related) GTPase is critical for COPII (coat protein II) assembly and vesicular transport of newly synthesized proteins from the endoplasmic reticulum (ER) compartment (33, 34). Dominant negative GTP-bound mutant Sar1H79G has previously been shown to block the ER-Golgi transport of newly synthesized Cx43 in BICR-M1Rk cells (8). In order to determine if Panx1 and Panx3 follow the classical ER-Golgi secretory pathway mediated through a COPII-dependent mechanism, we engineered BICR-M1Rk cells to co-express Panx1 or Panx3 along with Sar1WT or Sar1H79G. Co-expression of Panx1 with Sar1WT revealed a typical uniform distribution of Panx1 at the cell surface (Fig. 2A, arrows), whereas Sar1WT was localized to the paranuclear region in an intact Golgi-like compartment (Fig. 2, A and B). In comparison, expression of Sar1H79G resulted in fragmentation of a Golgi-like compartment in the paranuclear region (Fig. 2, A and B) and retention of Panx1 and Panx3 in ER-like patterns with little evidence of cell surface localization (Fig. 2, A and B, arrowheads). In the same culture environments, BICR-M1Rk cells expressing Panx1 or Panx3 but not Sar1H79G revealed a uniform cell surface labeling of Panx1 and Panx3 (Fig. 2, A and B, insets, arrows). Furthermore, as we previously reported (8), the distribution of Cx43 gap junction plaque-like structures was not evident in the presence of Sar1H79G (data not shown).
FIGURE 2.
Trafficking of Panx1 and Panx3 was disrupted in the presence of a dominant-negative Sar1 mutant. BICR-M1Rk cells expressing Panx1 or Panx3 together with Sar1WT or Sar1H79G were immunolabeled for Panx1 (A) or Panx3 (B). Both Panx1 and Panx3 were capable of trafficking and localizing to the cell surface in the presence of Sar1WT (A and B, filled arrows). Expression of Sar1H79G resulted in Panx1 and Panx3 being retained in an ER-like compartment (A and B, arrowheads); however, when cells expressed Panx1 or Panx3 without expressing Sar1H79G in the same cellular environment, both Panx1 and Panx3 trafficked to the plasma membrane (A and B, insets, arrows). Western blotting of Panx1 and Panx3 in the presence of Sar1H79G (C) or after long term BFA treatment (19 h) (D) revealed an accumulation of the high mannose species of Panx1 and Panx3, with a noticeable reduction in the higher molecular weight glycosylation species (C and D). Nuclei are stained with Hoechst 33342 (blue). Bars, 10 μm. Results shown are representative of three independent experiments.
Because Panx1 and Panx3 resolve as multiple bands (Figs. 1A and 2C), we wanted to determine the effect of dominant-negative Sar1 on the different molecular species. When compared with Panx1 alone or with Sar1WT, the presence of Sar1H79G revealed an accumulation of the intermediate species (previously demonstrated to be a high mannose glycosylation species (4)), with a noticeable reduction in the most extensively glycosylated species of Panx1 (Fig. 2C). Similarly, an accumulation of lower molecular weight species (previously reported to be a high mannose glycosylation species (29)) of the Panx3 doublet was also revealed in the presence of Sar1H79G (Fig. 2C), suggesting that the higher molecular weight species of Panx1 and Panx3 is the consequence of additional post-translational processing that occurs upon exiting the ER. Consistently, long term BFA treatment (19 h) (a pharmacological blocker known to inhibit anterograde transport of proteins between the ER and Golgi apparatus (35)) of Panx1- and Panx3-expressing BICR-M1Rk cells caused a detectable increase in the high mannose species of Panx1 and Panx3, with a noticeable reduction in the higher molecular weight species of both Panx1 and Panx3 (Fig. 2D). Collectively, these results suggest that both Panx1 and Panx3 are co-translationally inserted into the ER and transported in a COPII-dependent mechanism to the Golgi apparatus, where they are substrates for further glycosylation and editing.
GFP-tagged Panx1 Mimics the Distribution Profile of Untagged Panx1 and Is Suitable to Investigate the Dynamic Distribution of Panx1
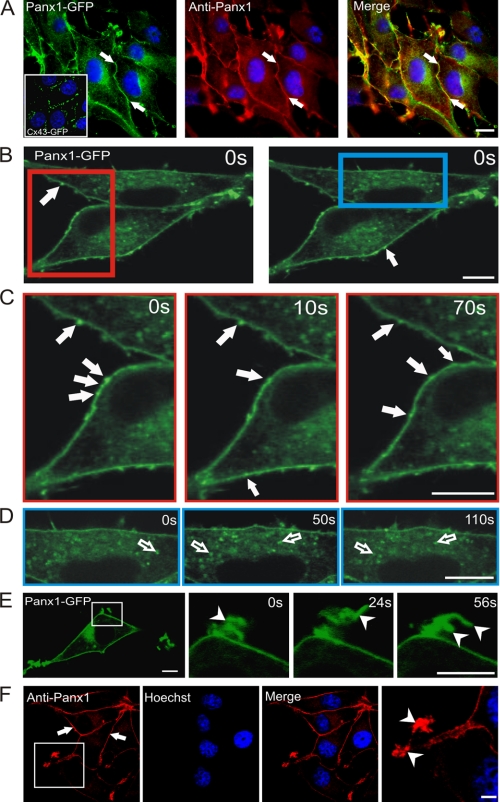
In order to assess the dynamic properties of Panx1, we first stably expressed Panx1-GFP in BICR-M1Rk cells and evaluated its distribution profile with respect to untagged Panx1. Immunofluorescent labeling revealed that Panx1-GFP exhibited a cell surface distribution pattern (Fig. 3A, arrows) similar to that observed for untagged Panx1 (Fig. 3F, arrows), with a notable increase in intracellular fluorescent signal (Fig. 3A). Clearly, the cell surface pattern for Panx1-GFP was distinct from that observed for Cx43-GFP when expressed in the same cell type (Fig. 3A, inset).
FIGURE 3.
Panx1 was localized to multiple sites, compartments, and microdomains. Immunolabeling of Panx1-GFP with an anti-Panx1 antibody revealed its localization at the cell surface (A, filled arrows) in a pattern that was distinct from Cx43-GFP (A, inset). Regions of interest from live BICR-M1Rk cells expressing Panx1-GFP (B, red and blue rectangles) were chosen for live imaging and imaged at t = 0, 10, 50, 70, and 110 s (C and D). Rapid time lapse imaging revealed that Panx1-GFP is distributed primarily in a uniform pattern, whereas mobile bright fluorescent clusters could be identified at the cell surface (B and C, filled arrows) and within the cell (D, unfilled arrows). Panx1-GFP was clearly localized to dynamic plasma membrane protrusions (E, arrowheads) that were evident in BICR-M1Rk cells expressing untagged Panx1 (F, arrowheads). Nuclei in A and F are stained with Hoechst 33342 (blue). Bars, 10 μm. Representative of three independent experiments.
To assess the dynamic activity of Panx1 at the cell surface and within intracellular compartments, we performed rapid time lapse imaging on cells that expressed Panx1-GFP. Panx1-GFP was not only visualized in a relatively uniform pattern at the cell surface (Fig. 3A) but also as mobile bright fluorescent clusters (Fig. 3, B and C). Rapid time lapse imaging revealed that these clusters (suggestive of Panx1-GFP aggregates) were mobile at the plasma membrane and in regions devoid of cell-cell contacts (Fig. 3, B and C, filled arrows; see supplemental Movie 1). In addition, Panx1-GFP was also found in distinct intracellular vesicle-like structures that were highly mobile (Fig. 3D, unfilled arrows, and supplemental Movie 1). Surprisingly, and distinct from that observed for functional Cx43, Panx1-GFP was found in dynamic finger-like projections indicative of membrane protrusion (Fig. 3E, arrowheads, and supplemental Movie 1). This localization to membrane protrusion was also frequently evident in cells expressing untagged Panx1, suggesting that this localization profile is not a consequence of the GFP tag (Fig. 3F, arrowheads).
Panx1-GFP Is Highly Mobile at All Plasma Membrane Domains as Revealed by FRAP Analysis
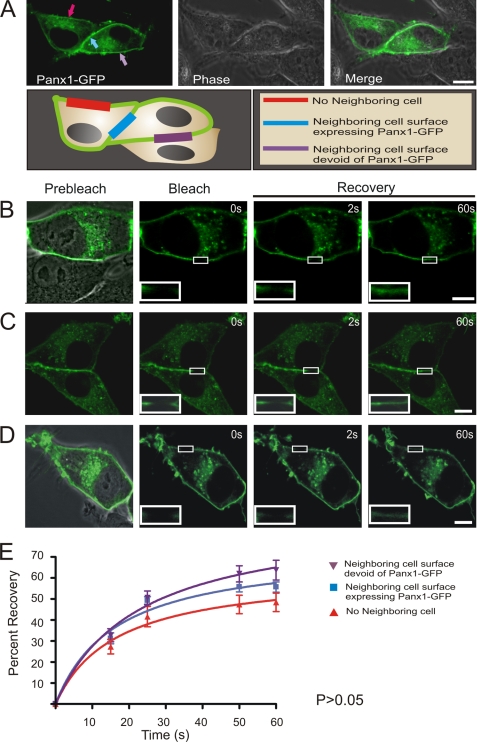
To analyze the dynamic mobility characteristics of Panx1-GFP, we first assessed the ability of Panx1-GFP to traffic and localize at three distinct plasma membrane domains where there was no neighboring cell (Fig. 4A, red arrow), the neighboring cell expressed Panx1-GFP (Fig. 4A, blue arrow), or the neighboring cell was devoid of Panx1-GFP (Fig. 4A, purple arrow). Once it was determined that Panx1-GFP was not differentially distributed to any of these cell surface domains (Fig. 4A), regions of Panx1-GFP located at these distinct domains were selected and subjected to FRAP analysis (Fig. 4, B–E). After initial photobleaching, within 2 s, there was a rapid movement of fluorescent molecules from the outer edges into the photobleached area of all bleached cell surface domains (Fig. 4, B–D, insets). FRAP curve analysis revealed the mobile fluorescent fractions to be 45–60% at all microdomains over a time course of only 60 s (Fig. 4E). Moreover, there was no significant difference in the total recovered fraction of Panx1-GFP within any of the plasma membrane domains, an observation that was similar to when we analyzed the percentage recovery of transiently transfected Panx1-GFP (40–50%) in BICR-M1RK cells stably expressing untagged Panx1 (supplemental Fig. 2), or B16-BL6 cells, identified for the first time to express endogenous Panx1 (supplemental Fig. 3, A and B).
FIGURE 4.
Panx1-GFP is highly mobile at all plasma membrane locations. Panx1-GFP was localized to three distinct plasma membrane domains of BICR-M1Rk cells, as depicted by the schematic diagram (A). Fluorescent images of Panx1-GFP were superimposed with DIC images to highlight the microenvironment surrounding the cell being analyzed (A–D). Selected cell regions where Panx1-GFP was localized at the three distinct plasma membrane domains were photobleached, and fluorescence recovery back into the photobleached areas was assessed and normalized over 60 s (B–D). Panx1-GFP recovery within the photobleached area was not significantly different (p > 0.05) among all three domains (E). Bars, 10 μm. n = 6–9 per plasma membrane domain collected from three independent experiments.
A Subpopulation of GFP-tagged Panx3 Is Evident at the Cell Surface When Expressed Alone or Co-expressed with Panx3 and Reveals Dynamic Localization to the Membrane Protrusions
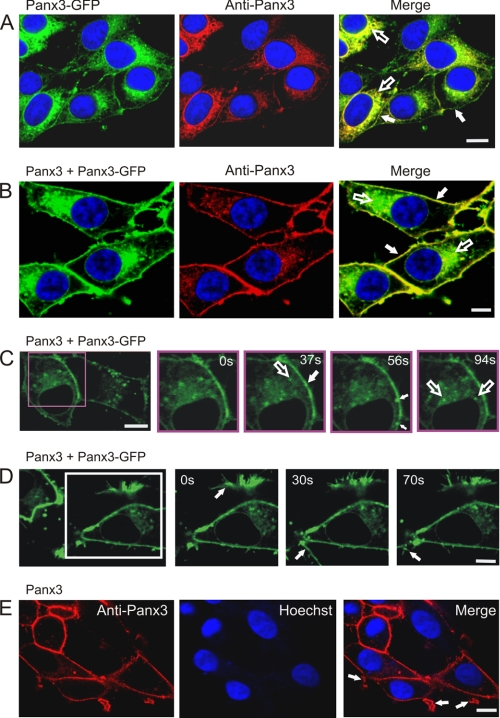
We have previously reported that Panx3-GFP has a substantial trafficking defect, causing it to be retained within the endoplasmic reticulum of NRK cells (5). However, when overexpressed in BICR-M1Rk cells, some evidence of Panx3-GFP localization to the cell surface was detected (Fig. 5A, filled arrows), along with the intracellular ER-like distribution of Panx3-GFP (Fig. 5A, unfilled arrows). To further confirm that a subpopulation of Panx3-GFP can traffic and localize to the cell surface, we performed biotinylation assays in live BICR-M1Rk cells transiently expressing Panx3-GFP. Incubation of Panx3-GFP-expressing cells with biotin followed by pull-downs with neutravidin beads revealed that the ∼70-kDa Panx3-GFP indeed traffics to the cell surface (supplemental Fig. 4). Glyceraldehyde-3-phosphate dehydrogenase was used as a control to confirm that biotin was not internalized (supplemental Fig. 4). It was interesting to note that when Panx3-GFP was co-expressed with Panx3 in BICR-M1Rk cells, there was an apparent increase in the cell surface population of Panx3-GFP (Fig. 5B, filled arrows), with some expected intracellular distribution (Fig. 5B, unfilled arrows). This finding suggests that Panx3-GFP may interact with Panx3 to facilitate its traffic to the plasma membrane.
FIGURE 5.
Delivery of Panx3-GFP to the cell surface. Wild-type BICR-M1Rk cells engineered to express Panx3-GFP (A) or both Panx3 and Panx3-GFP (B) were immunolabeled with anti-Panx3 antibody. Panx3-GFP was retained mainly in an ER-like pattern (A and B, unfilled arrows) with some evidence of a cell surface distribution (A and B, filled arrows), whereas co-expression of Panx3 appeared to increase the cell surface population of Panx3-GFP (B). Rapid time lapse imaging of live BICR-M1Rk cells co-expressing Panx3-GFP and Panx3 revealed that Panx3-GFP was distributed primarily in a uniform pattern with notable mobile fluorescent clusters at the cell surface (C, filled arrows) and within the cell (C, unfilled arrow). Localization of Panx3-GFP to plasma membrane protrusions (D, arrows) was similar to that found in cells expressing only Panx3 (E, arrows). Nuclei in A, B, and E are stained with Hoechst 33342 (blue). Bars, 10 μm. Results shown are representative of three independent experiments.
To assess the dynamic activity of Panx3-GFP at the cell surface and within intracellular compartments, we performed rapid time-lapse imaging on cells co-expressing Panx3 and Panx3-GFP. Panx3-GFP was localized at the cell surface in a relatively uniform pattern and as bright clusters either approaching or at the cell surface (Fig. 5C, filled arrows). In addition, Panx3-GFP was also visualized in distinct vesicle-like structures (Fig. 5C, unfilled arrows) and observed in cell surface protrusions (Fig. 5D, arrows) that were also occasionally identified in cells expressing only Panx3 (Fig. 5E, arrows).
Panx3-GFP Is Highly Dynamic at All Cell Surface Domains
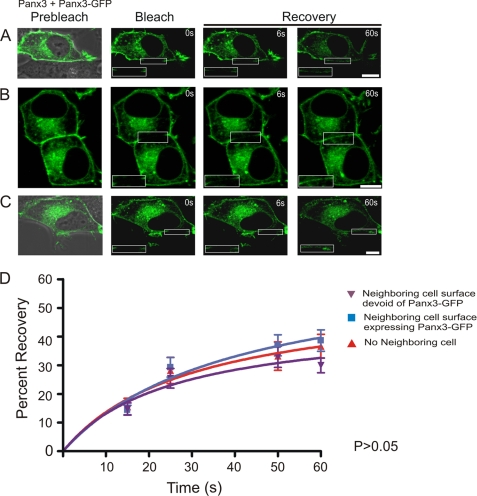
We co-expressed Panx3 with Panx3-GFP and used GFP-tagged Panx3 as a tracer to assess and quantify the mobile fraction in various plasma membrane domains where the neighboring cell surface was devoid of Panx3-GFP (Fig. 6A), the neighboring cell expressed Panx3-GFP (Fig. 6B and supplemental Movie 2), or there was no neighboring cell (Fig. 6C). FRAP analysis of selected regions expressing Panx3-GFP revealed that within 60 s after photobleaching, there was a rapid reentry of GFP-tagged Panx3 molecules into the photobleached area at all microdomains (Fig. 6, A–C, insets), and the mobile fraction was calculated to be ∼30–40%. However, no significant difference was noticed in the total recovered fraction of Panx3-GFP within any of the plasma membrane domains (Fig. 6D).
FIGURE 6.
Panx3-GFP is highly mobile at all plasma membrane domains. Panx3-GFP was localized to three distinct plasma membrane domains of BICR-M1Rk cells co-expressing Panx3 (A–C). Selected cell surface regions containing Panx3-GFP were photobleached, and fluorescence recovery back into the photobleached areas was assessed and normalized over the time course of 60 s. The percentage of Panx3-GFP recoverable fraction was not found to be significantly different among all three plasma membrane domains examined (p > 0.05) (D). Bars, 10 μm. n = 12–25 per plasma membrane domain collected from three independent experiments.
Cell Surface Population of Panx1-GFP and Panx3 Is Insensitive to Nocodazole Treatment
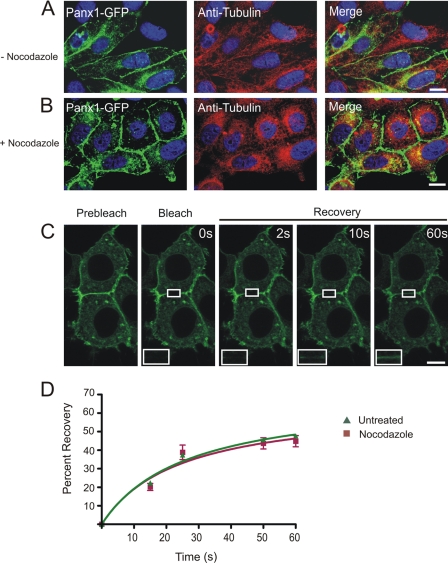
It has been documented that nocodazole disruption of microtubules impairs the continuous trafficking and regeneration of Cx43-GFP at the cell surface (8). To investigate if trafficking and recovery of Panx1-GFP into the photobleached area is dependent on microtubules, Panx1-GFP-expressing BICR-M1Rk cells were exposed to nocodazole for 90 min. Nocodazole treatment resulted in characteristic morphological changes in the cells from spindle-shaped to more cuboidal with a notable disruption of the microtubule architecture (Fig. 7, A–C). However, the distribution of Panx1-GFP (Fig. 7B) and Panx3 (supplemental Fig. 5B) at the cell surface remained relatively unaffected by the nocodazole treatment. Furthermore, FRAP studies revealed that Panx1-GFP migrated into the photobleached area similarly in both untreated and nocodazole-treated cells (Fig. 7, C and D). Interestingly, the inward progression of fluorescent recovery from the edges of the photobleached area toward the center was not detected until 10 s postbleaching (Fig. 7C, inset). Furthermore, FRAP analysis of Panx1-GFP in the presence of nocodazole treatment revealed no significant difference in the recovered fraction when compared with the untreated cells (Fig. 7D). Thus, nocodazole treatment does not visually affect the cell surface localization of Panx1-GFP or the dynamics of recovery into the photobleached area at the cell surface.
FIGURE 7.
The cell surface population of Panx1-GFP is insensitive to nocodazole treatment. Untreated (A) or nocodazole-treated (B) Panx1-GFP-expressing BICR-M1Rk cells were immunolabeled for tubulin. As expected, nocodazole treatment collapsed tubulin into paranuclear regions (A and B); however, the distribution profile of Panx1-GFP at the cell surface and in the intracellular compartments (A and B) remained relatively unchanged with collapsed tubulin (B). FRAP analysis in presence of nocodazole revealed that Panx1-GFP was able to recover into the photobleached area, and the percentage of recoverable fraction was not significantly different from the untreated cells (C and D). Bars, 10 μm. n = 5–10 per plasma membrane domains, data collected over four independent repeats.
The Cell Surface Stability of Panx1-GFP and Panx3 Is Sensitive to Cytochalasin B Treatment, whereas the Mobility of Panx1-GFP Transport Vesicles Is Perturbed in the Absence of Intact Microfilaments
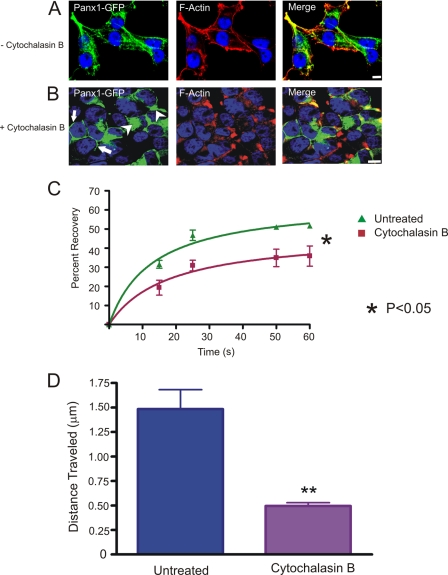
We next wanted to assess the role of actin microfilaments in stabilizing Panx1 and Panx3 at the cell surface as well as evaluating their cell surface dynamic properties and transport of Panx1-GFP. Treatment of cytochalasin B (90 min) caused a redistribution of F-actin from the cell periphery (Fig. 8A and supplemental Fig. 5A) to the paranuclear region (Fig. 8B and supplemental Fig. 5A), with a subsequent change in cell morphology. In cytochalasin B-treated cells, Panx1-GFP and Panx3 were mainly localized in intracellular compartments (Fig. 8B and supplemental Fig. 5A, arrowheads), whereas a small population remained evident at the cell surface (Fig. 8B and supplemental Fig. 5A, arrows). These findings suggest that actin may play a crucial role in the cell surface stability of Panx1-GFP and Panx3. FRAP analysis of the remaining and detectable cell surface population of Panx1-GFP after cytochalasin B treatment (supplemental Movie 3) revealed that Panx1-GFP was mobile, but the recovery rate was slower and the amount of the recovered fraction was significantly less (∼15–20%) when compared with the untreated cells (Fig. 8C). Furthermore, rapid time lapse imaging on the same field of cells visualized before (supplemental Movie 4) and after (supplemental Movie 5) cytochalasin B treatment revealed that Panx1-GFP-carrying vesicles were freely able to move and travel in the untreated cells; however, this movement was greatly perturbed when cells were treated with cytochalasin B with a ∼60% decrease in vesicle movement over a fixed interval of time (8.8 s) (Fig. 8D). Collectively, these studies suggest that microfilaments play a multifaceted role in pannexin stabilization at the cell surface and vesicular transport.
FIGURE 8.
Effect of cytochalasin B on Panx1-GFP. Untreated (A) or cytochalasin B-treated (B) Panx1-GFP-expressing BICR-M1Rk cells were labeled with phalloidin for F-actin localization. As expected, cytochalasin B caused the redistribution of F-actin from the cell surface (A) to the paranuclear region (B). The collapse of F-actin microfilaments coincided with the intracellular accumulation of Panx1-GFP (B, arrowheads), whereas a small population of Panx1-GFP remained evident at the cell surface (B, arrows). FRAP analysis in the presence of cytochalasin B treatment revealed that the cell surface population of Panx1-GFP was significantly impaired from entering the photobleached area (p < 0.05) (C) n = 3. Quantification of the total distance traveled by Panx1-GFP carrying vesicles within the same field of cells analyzed before and after the cytochalasin B treatment indicated a significant (p < 0.05) reduction in vesicle mobility in cytochalasin B-treated cells (D). Bars, 10 μm. Results shown are representative of five independent experiments.
F-actin Directly Binds Panx1 at the Carboxyl Terminus
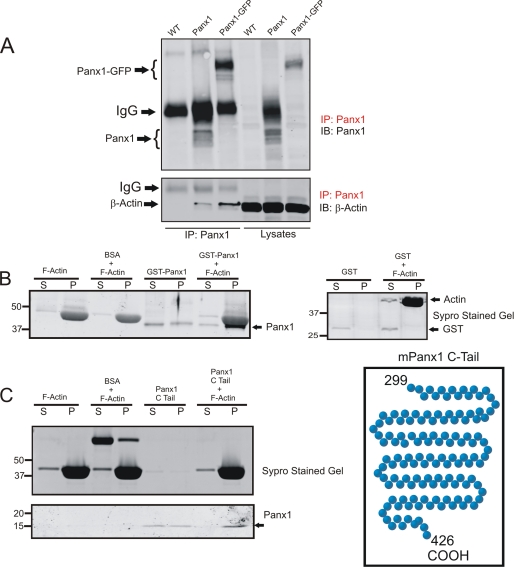
To further examine the possible interaction of Panx1 with actin, lysates of wild-type BICR-M1Rk cells engineered to express Panx1 or Panx1-GFP were subjected to immunoprecipitation of Panx1 prior to immunoblotting for Panx1 or β-actin. As expected, multiple glycosylated species of Panx1 (resolved below the IgG band) and Panx1-GFP were detected in the immunoprecipitates and cell lysates of Panx1-overexpressing cells but not wild-type BICR-M1Rk cells (Fig. 9A, top). Interestingly, β-actin was found to co-immunoprecipitate with Panx1 from both Panx1- and Panx1-GFP-expressing BICR-M1Rk cells (Fig. 9A, bottom). To further validate the interaction of actin with Panx1, we conducted co-sedimentation assays where polymerized actin was mixed with GST fusion protein containing the C-tail of Panx1. As observed by Sypro-stained gel, once polymerized, F-actin typically sediments in the pellet fraction. In the absence of polymerized actin, GST-Panx1 C-tail was found in both the soluble and pellet fractions (Fig. 9B). However, when combined with F-actin, GST-Panx1 C-tail sediments preferentially in the pellet fraction (Fig. 9B). As a control, GST alone did not sediment in the pellet fraction with or without F-actin (Fig. 9B). Because the Panx1 C-tail fusion protein detection at ∼43 kDa was partially masked by actin, we cleaved the C-tail of Panx1 from the GST and performed a similar co-sedimentation assay. Further Panx1 immunoblots revealed the Panx1 C-tail at ∼15 kDa in the pellet fraction (Fig. 9C). These results suggest that F-actin binds directly to the carboxyl terminus of Panx1.
FIGURE 9.
F-actin binds Panx1 at the carboxyl terminus. Wild type (WT) or Panx1- or Panx1-GFP-expressing BICR-M1Rk cells were lysed and subjected to immunoprecipitation (IP) for Panx1 prior to immunoblotting (IB) the immunoprecipitates and cell lysates for Panx1 or β-actin. β-Actin co-immunoprecipitated with Panx1 and Panx1-GFP (A). Monomeric actin was polymerized into F-actin, incubated with either GST fusion protein containing the carboxyl-terminal tail of Panx1 (B) or the carboxyl-terminal tail of Panx1 alone (C) and separated into supernatant or pellet fractions (denoted by S and P, respectively) prior to immunoblotting for Panx1. Panx1 was found to co-sediment with F-actin in the pellet fractions (B and C). Parallel gels were stained with Sypro gel stain, and BSA and GST were used as controls in the co-sedimentation assays. Results shown are representative of three independent experiments.
DISCUSSION
With the recent discovery of pannexins as a new family of channel- or conduit-forming proteins, there has been a growing interest in elucidating their biochemical properties, life cycle, and cellular roles. Previous reports have linked Panx1 channels to the release of ATP in neurons (36) and astrocytes (37) and in cellular response to pathological insults, such as initiation of inflammatory action (10, 38), ischemia-induced death of neurons (39), and tumor suppression (13). Our understanding of Panx3 is even more rudimentary. Although Panx3 has been shown to be a cell surface glycoprotein that forms conduits capable of dye uptake (5, 40), its cellular function remains largely unknown. It was our hypothesis that, being integral membrane proteins with sequence relationships to invertebrate innexin gap junction proteins, Panx1 and Panx3 would exhibit characteristics similar to those of the well studied Cx43 gap junction protein in terms of the secretory pathway governing their trafficking, dynamic properties within the plasma membrane, and interplay with the cytoskeletal network.
Trafficking of Panx1 and Panx3 to the Cell Surface
It has been well established that Cx43 is co-translationally inserted into the ER and oligomerizes into connexons in the trans-Golgi network before trafficking to the plasma membrane (25). In the case of Panx1, treatment with endoglycosidase H and peptide:N-glycosidase F enzymes have revealed that the lower molecular weight species constitutes the core protein that gets glycosylated to the high mannose form in the ER before further glycosylation and editing in the Golgi and delivery to the cell surface (4, 5). Although recent studies have extensively focused on correlating glycosylation status with the delivery of Panx1 to the cell surface (4, 19), molecular mechanisms underlying the secretory pathway taken by Panx1 and Panx3 were largely unknown. To address if the transport of Panx1 and Panx3 from the ER to the Golgi apparatus is mediated through COPII vesicles, we transiently co-expressed Panx1 or Panx3 with a dominant-negative GTP-bound mutant Sar1H79F. Stabilization of Sar1 in the dominant negative GTP-bound state has been previously shown to efficiently block COPII-mediated ER transport of proteins to the cell surface (41). Our data demonstrated that Sar1H79F expression severely inhibited the cell surface localization of Panx1 and Panx3, thus suggesting that COPII vesicular trafficking of these pannexin family members is required prior to their eventual delivery to the cell surface. These data also indicate that efficient GTP hydrolysis of Sar1 is crucial for regulating their transport from the ER compartment because restriction of GTP hydrolysis of Sar1H79G to GDP has been previously shown to arrest the cargo-containing vesicles at the ER exit sites (34). Consistent with our findings, we previously showed that Sar1 function was necessary for delivery of Cx43 to the cell surface (8). Our data support the premise that the post-ER pool of Panx1 and Panx3 is correlated with pannexin processing to the highly complex glycosylation species, whereas the accumulation of the high mannose intermediate species of Panx1 and Panx3 is consistent with retention of Panx1 and Panx3 in the ER. Similar Sar1 GTPase dependence was observed for KATP channels, where expression of dominant negative mutants Sar1H79G (mutant incapable of hydrolyzing GTP) and Sar1T39N (mutant restricted in its ability to exchange GDP for GTP) prevented proper channel processing and the cell surface expression of the channel (42).
ER to Golgi transport of Panx1 and Panx3, as well as their state of glycosylation, was further confirmed by the use of BFA, which is known to inactivate Arf1, thereby inhibiting ER to Golgi transport (43). Consistent with a previous study (19), we noticed a dramatic increase in the high mannose form of Panx1 in BFA-treated cells. Interestingly, we also observed an accumulation of the high mannose species of Panx3 in response to BFA treatment, consistent with the inhibition of both Panx1 and Panx3 being delivered to the Golgi apparatus for further processing. Vesicular trafficking of Panx1 and Panx3 would also strongly suggest that, like Cx43, both proteins are integral transmembrane proteins that get transported from ER membranes in COPII vesicles.
Mobility Dynamics of Panx1 and Panx3
In our study, we used Panx1-GFP and Panx3-GFP as tracer probes to elucidate the distribution profile and cell surface dynamics of Panx1 and Panx3. The distribution of Panx1-GFP at the plasma membrane appeared to be uniform with occasional Panx1-enriched domains, consistent with previous findings using the untagged Panx1 (5). In contrast, Panx3-GFP alone revealed an increased intracellular profile, as reported earlier (5), with some clear evidence of its localization to the cell surface that was confirmed by cell surface biotinylation assays. Since adequate delivery of Panx3-GFP to the cell surface was required to investigate its dynamics at the plasma membrane, we co-expressed it with untagged Panx3 and found an increased cell surface expression of Panx3-GFP. It is possible that Panx3 co-oligomerized with Panx3-GFP to facilitate its delivery to the plasma membrane; if this holds true, it would also suggest that having a GFP tag at the carboxyl-terminal tail of Panx3 does not interfere with the intermixing of Panx3 subunits. A similar mechanism has been proposed for the cell surface rescue of a trafficking-defective, glycosylation-deficient mutant of Panx1 when co-expressed with either wild-type Panx1 or tetracysteine-tagged Panx1, further indicating that tagging of pannexins does not impair its ability to assemble together with its untagged counterpart (19).
In our study, delivery of both Panx1-GFP and Panx3-GFP to the cell surface appeared to occur at multiple plasma membrane domains via intracellular vesicle-like structures that formed bright clusters upon apparent fusion with the plasma membrane. These clusters were found to be quite mobile and displaced laterally within the cell surface membrane, thus supporting a model of untargeted delivery of Panx1 and Panx3 to all cell surface microdomains. In contrast, Cx43-GFP, typically known to localize in punctate-like structures at the cell surface, has been documented to have both an arbitrary delivery to all plasma membrane domains (8, 16) as well as a preferred microtubule-dependent delivery to adherens junctions that reside in close proximity to the preexisting gap junctions (44). Interestingly, our data support the premise that Panx1 and Panx3 are enriched in membrane protrusions at areas that are devoid of contacting cells, a situation not typically observed for Cx43 unless non-functional Cx43 mutant studies are performed (16). Localization of pannexins in the finger-like membrane protrusions could suggest that it may play a role in cell migration because the process of cell motility typically involves actin polymerization and entails formation of fanlike or pointed projections (lamellipodium and filopodia, respectively) at the leading edge (45). Previously, the absence of Panx1 from the leading edge of a corneal epithelium wound in P2X7−/− mice was correlated with delayed corneal re-epithelialization and compromised wound healing (46). Other channel-forming proteins, such as aquaporin-1, have been implicated in increased cell migration by localizing to the lamellipodia (47), whereas migration of lymphocytes (48) and embryonic nerve cells (49) has been correlated with voltage-dependent K+ channels, thus supporting the role of channel-forming proteins in regulating cell motility.
Our FRAP assessments of Panx1 and Panx3 mobility identified that, similar to Cx43 (16), lateral movement of Panx1-GFP and Panx3-GFP occurred from the outer edge to the center of the photobleached areas. It is notable that although the recovery of fluorescence into the photobleached area is probably the result of lateral movement of pannexins due to the time course being examined, there is also probably a contribution from newly delivery fluorescent protein-tagged pannexins to the cell surface. The relative rate of Panx1-GFP and Panx3-GFP recovery into the photobleached area was quite comparable in any of the examined plasma membrane microdomains, thus suggesting that the assembly state of these pannexin family members remains relatively unchanged with respect to its subcellular location within the plasma membrane. In our study, Panx1-GFP (and to a slightly lesser extent Panx3-GFP) exhibited over twice the mobility of Cx43-GFP molecules, which are typically arranged in gap junction-like clusters (16). Slow recovery of Cx43-GFP was also reported in HeLa cells (50). Given the relatively uniform cell surface distribution of Panx1 and Panx3 that appear to be untargeted to specific microdomains, we speculate that these pannexins are not likely to be packaged into dense crystalline-like structures as reported for Cx43 (51). Thus, the distribution and mobility of these pannexins are more in line with other channels and receptors, such as Na+ channels (52) and acetylcholine receptors (53).
The percentage of fluorescence recovery after photobleaching for both Panx1-GFP and Panx3-GFP (representing the mobile fraction) reached a plateau between 40 and 60%, with the remaining component representing the immobile fraction. Typically, the size of the immobile fraction is dependent on the nature of the protein and the membrane microenvironment being assessed. For instance, the immobile fraction of sodium channels ranges from ∼10% in the cell body to ∼40% in the neurite terminals (52). Likewise, the mobile fraction of glycine receptors ranges from ∼50% in the neuronal cell body to ∼70% in the processes (54). In addition, the viscosity of the membrane microdomain (55), tethering of proteins with scaffolds/binding partners, and interaction with the cytoskeleton can all contribute to the size of the immobile fraction (56).
Cytoskeletal Dependence of Pannexin Trafficking and Mobility
In order to assess the role of the cytoskeleton in pannexin trafficking and cell surface mobility dynamics, we used nocodazole and cytochalasin B to disrupt microtubules and microfilaments, respectively. Nocodazole-induced disruption of microtubules did not significantly alter the cell surface distribution of either Panx3 or Panx1-GFP, which may not be totally unexpected given the predicted long half-life of Panx1 (4, 5). This finding is quite distinct from Cx43, where enhanced growth of gap junctions (20) and Cx43 molecular movement into the photobleached gap junctions was minimal in nocodazole-treated cells (8), suggesting that Cx43 is much more dependent on microtubules than Panx1. In contrast, disruption of microfilaments revealed concomitant accumulation of paranuclear Panx1-GFP and Panx3 with collapsed actin microfilaments, whereas only a subpopulation of Panx1-GFP or Panx3 remained at the cell surface. The longer turnover dynamics of pannexins and the rapid intracellular accumulation upon short term cytochalasin B treatment support the premise that actin provides stability to the cell surface population of Panx3 and Panx1. On the other hand, Cx43 gap junctions have been documented to remain considerably more independent to the assembly state of microfilaments (57). Mobility assessment of the remaining cytochalasin B-insensitive subpopulation of Panx1-GFP at the cell surface revealed a smaller mobile fraction of ∼35% in the absence of intact microfilaments. This surprising finding was somewhat distinct from Na,K-ATPase and reggie-1/flotillin-2, where the disruption of microfilaments caused an increase in the mobile fraction (58, 59). The smaller mobile fraction of Panx1-GFP noticed in our study may be explained by the fact that transport vesicles carrying Panx1-GFP either to or from the plasma membrane were less mobile, and this decrease may mechanistically account for the reduced fluorescent recovery into the photobleached area. The relative speeds of Panx1-GFP-carrying vesicles in untreated cells (∼0.2 μm/s) and cytochalasin B-treated cells (∼0.07 μm/s) were also quite different. Although credited to trafficking on microtubules tracks, vesicles containing Cx43-GFP showed comparable average speeds of ∼0.5 μm/s in untreated HeLa cells (21).
Interaction of Panx1 with Actin
Given the finding that actin microfilaments regulated the distribution and cell surface mobility of Panx1, we speculated that a direct interaction between Panx1 and actin may exist. Because we were best equipped to address this question for Panx1, we first demonstrated that actin does in fact co-immunoprecipitate with Panx1. To further assess 1) if Panx1 binds to monomeric or filamentous actin, 2) if Panx1 interaction to actin is direct, and 3) which domain of Panx1 might be responsible for interaction, we conducted a co-sedimentation assay with a GST-Panx1 C-tail fusion protein. Here we show that Panx1 sediments preferentially in the F-actin fraction, the interaction appears to be direct, and it is the carboxyl-terminal tail of Panx1 that seems responsible for interacting with actin. Comparatively, actin binding to Cx43 is thought to be facilitated via its interaction with zonula occludens-1, which is a known Cx43 binding partner (60). Future studies will be needed to elucidate the Panx1 motif responsible for actin binding at the C-tail and whether actin also binds to Panx3.
In summary, trafficking and assembly of pannexins is a precisely regulated process. Our study is the first to report that Panx1 and Panx3 transport is dependent on Sar1-mediated COPII vesicles, cell surface Panx1 and Panx3 have dynamic mobility properties, and Panx1 directly interacts with F-actin.
Supplementary Material
Acknowledgment
We acknowledge Jamie Simek for providing expertise in optimizing the FRAP technique.
This work was supported by a Canadian Institutes of Health Research operating grant (to D. W. L.) and a National Sciences and Engineering Council of Canada studentship (to R. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–5 and Movies 1–5.
- NRK
- normal rat kidney
- HEK
- human embryonic kidney
- GFP
- green fluorescent protein
- BFA
- brefeldin A
- FRAP
- fluorescence recovery after photobleaching
- BSA
- bovine serum albumin
- PBS
- phosphate-buffered saline
- ROI
- region of interest
- ER
- endoplasmic reticulum
- C-tail
- carboxyl-terminal tail.
REFERENCES
- 1.Panchin Y., Kelmanson I., Matz M., Lukyanov K., Usman N., Lukyanov S. (2000) Curr. Biol. 10, R473–R474 [DOI] [PubMed] [Google Scholar]
- 2.Willecke K., Eiberger J., Degen J., Eckardt D., Romualdi A., Güldenagel M., Deutsch U., Söhl G. (2002) Biol. Chem. 383, 725–737 [DOI] [PubMed] [Google Scholar]
- 3.Baranova A., Ivanov D., Petrash N., Pestova A., Skoblov M., Kelmanson I., Shagin D., Nazarenko S., Geraymovych E., Litvin O., Tiunova A., Born T. L., Usman N., Staroverov D., Lukyanov S., Panchin Y. (2004) Genomics 83, 706–716 [DOI] [PubMed] [Google Scholar]
- 4.Boassa D., Ambrosi C., Qiu F., Dahl G., Gaietta G., Sosinsky G. (2007) J. Biol. Chem. 282, 31733–31743 [DOI] [PubMed] [Google Scholar]
- 5.Penuela S., Bhalla R., Gong X. Q., Cowan K. N., Celetti S. J., Cowan B. J., Bai D., Shao Q., Laird D. W. (2007) J. Cell Sci. 120, 3772–3783 [DOI] [PubMed] [Google Scholar]
- 6.Laird D. W., Puranam K. L., Revel J. P. (1991) Biochem. J. 273, 67–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Musil L. S., Goodenough D. A. (1993) Cell 74, 1065–1077 [DOI] [PubMed] [Google Scholar]
- 8.Thomas T., Jordan K., Simek J., Shao Q., Jedeszko C., Walton P., Laird D. W. (2005) J. Cell Sci. 118, 4451–4462 [DOI] [PubMed] [Google Scholar]
- 9.Iglesias R., Locovei S., Roque A., Alberto A. P., Dahl G., Spray D. C., Scemes E. (2008) Am. J. Physiol. Cell Physiol. 295, C752–C760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pelegrin P., Surprenant A. (2007) J. Biol. Chem. 282, 2386–2394 [DOI] [PubMed] [Google Scholar]
- 11.Dahl G., Locovei S. (2006) IUBMB Life 58, 409–419 [DOI] [PubMed] [Google Scholar]
- 12.Barbe M. T., Monyer H., Bruzzone R. (2006) Physiology 21, 103–114 [DOI] [PubMed] [Google Scholar]
- 13.Lai C. P., Bechberger J. F., Thompson R. J., MacVicar B. A., Bruzzone R., Naus C. C. (2007) Cancer Res. 67, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 14.Bruzzone R., Hormuzdi S. G., Barbe M. T., Herb A., Monyer H. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 13644–13649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaietta G., Deerinck T. J., Adams S. R., Bouwer J., Tour O., Laird D. W., Sosinsky G. E., Tsien R. Y., Ellisman M. H. (2002) Science 296, 503–507 [DOI] [PubMed] [Google Scholar]
- 16.Simek J., Churko J., Shao Q., Laird D. W. (2009) J. Cell Sci. 122, 554–562 [DOI] [PubMed] [Google Scholar]
- 17.Bukauskas F. F., Jordan K., Bukauskiene A., Bennett M. V., Lampe P. D., Laird D. W., Verselis V. K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma W., Hui H., Pelegrin P., Surprenant A. (2009) J. Pharmacol. Exp. Ther. 328, 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boassa D., Qiu F., Dahl G., Sosinsky G. (2008) Cell. Commun. Adhes. 15, 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson R. G., Meyer R. A., Li X. R., Preus D. M., Tan L., Grunenwald H., Paulson A. F., Laird D. W., Sheridan J. D. (2002) Exp. Cell Res. 275, 67–80 [DOI] [PubMed] [Google Scholar]
- 21.Lauf U., Giepmans B. N., Lopez P., Braconnot S., Chen S. C., Falk M. M. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10446–10451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jordan K., Solan J. L., Dominguez M., Sia M., Hand A., Lampe P. D., Laird D. W. (1999) Mol. Biol. Cell 10, 2033–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giepmans B. N., Verlaan I., Hengeveld T., Janssen H., Calafat J., Falk M. M., Moolenaar W. H. (2001) Curr. Biol. 11, 1364–1368 [DOI] [PubMed] [Google Scholar]
- 24.Hervé J. C., Bourmeyster N., Sarrouilhe D., Duffy H. S. (2007) Prog. Biophys. Mol. Biol. 94, 29–65 [DOI] [PubMed] [Google Scholar]
- 25.Laird D. W. (2006) Biochem. J. 394, 527–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bunse S., Haghika A., Zoidl G., Dermietzel R. (2005) Cell. Commun. Adhes. 12, 231–236 [DOI] [PubMed] [Google Scholar]
- 27.Pelegrin P., Surprenant A. (2006) EMBO J. 25, 5071–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan D., Liaw A., Denkins Y. M., Collins J. H., Van Arsdall M., Chang J. L., Chakrabarty S., Nguyen D., Kruzel E., Fidler I. J. (2002) J. Exp. Ther. Oncol. 2, 286–297 [DOI] [PubMed] [Google Scholar]
- 29.Penuela S., Bhalla R., Nag K., Laird D. W. (2009) Mol. Biol. Cell 20, 4313–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qin H., Shao Q., Curtis H., Galipeau J., Belliveau D. J., Wang T., Alaoui-Jamali M. A., Laird D. W. (2002) J. Biol. Chem. 277, 29132–29138 [DOI] [PubMed] [Google Scholar]
- 31.Lippincott-Schwartz J., Snapp E., Kenworthy A. (2001) Nat. Rev. Mol. Cell Biol. 2, 444–456 [DOI] [PubMed] [Google Scholar]
- 32.Laird D. W., Castillo M., Kasprzak L. (1995) J. Cell Biol. 131, 1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee M. C., Miller E. A. (2007) Semin. Cell Dev. Biol. 18, 424–434 [DOI] [PubMed] [Google Scholar]
- 34.Dong C., Zhou F., Fugetta E. K., Filipeanu C. M., Wu G. (2008) Cell. Signal. 20, 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lippincott-Schwartz J., Yuan L. C., Bonifacino J. S., Klausner R. D. (1989) Cell 56, 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson R. J., Jackson M. F., Olah M. E., Rungta R. L., Hines D. J., Beazely M. A., MacDonald J. F., MacVicar B. A. (2008) Science 322, 1555–1559 [DOI] [PubMed] [Google Scholar]
- 37.Scemes E., Suadicani S. O., Dahl G., Spray D. C. (2007) Neuron. Glia Biol. 3, 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanneganti T. D., Lamkanfi M., Kim Y. G., Chen G., Park J. H., Franchi L., Vandenabeele P., Núñez G. (2007) Immunity 26, 433–443 [DOI] [PubMed] [Google Scholar]
- 39.Thompson R. J., Zhou N., MacVicar B. A. (2006) Science 312, 924–927 [DOI] [PubMed] [Google Scholar]
- 40.Penuela S., Celetti S. J., Bhalla R., Shao Q., Laird D. W. (2008) Cell. Commun. Adhes. 15, 133–142 [DOI] [PubMed] [Google Scholar]
- 41.Sato K. (2004) J. Biochem. 136, 755–760 [DOI] [PubMed] [Google Scholar]
- 42.Taneja T. K., Mankouri J., Karnik R., Kannan S., Smith A. J., Munsey T., Christesen H. B., Beech D. J., Sivaprasadarao A. (2009) Hum. Mol. Genet. 18, 2400–2413 [DOI] [PubMed] [Google Scholar]
- 43.Donaldson J. G., Finazzi D., Klausner R. D. (1992) Nature 360, 350–352 [DOI] [PubMed] [Google Scholar]
- 44.Shaw R. M., Fay A. J., Puthenveedu M. A., von Zastrow M., Jan Y. N., Jan L. Y. (2007) Cell 128, 547–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rorth P. (2009) Annu. Rev. Cell Dev. Biol. 25, 407–429 [DOI] [PubMed] [Google Scholar]
- 46.Mayo C., Ren R., Rich C., Stepp M. A., Trinkaus-Randall V. (2008) Invest. Ophthalmol. Vis. Sci. 49, 4384–4391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saadoun S., Papadopoulos M. C., Hara-Chikuma M., Verkman A. S. (2005) Nature 434, 786–792 [DOI] [PubMed] [Google Scholar]
- 48.Levite M., Cahalon L., Peretz A., Hershkoviz R., Sobko A., Ariel A., Desai R., Attali B., Lider O. (2000) J. Exp. Med. 191, 1167–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hendriks R., Morest D. K., Kaczmarek L. K. (1999) J. Neurosci. Res. 58, 805–814 [DOI] [PubMed] [Google Scholar]
- 50.Falk M. M., Baker S. M., Gumpert A. M., Segretain D., Buckheit R. W., 3rd (2009) Mol. Biol. Cell 20, 3342–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Unger V. M., Kumar N. M., Gilula N. B., Yeager M. (1999) Novartis Found. Symp. 219, 22–30; discussion 31–43 [DOI] [PubMed] [Google Scholar]
- 52.Angelides K. J., Elmer L. W., Loftus D., Elson E. (1988) J. Cell Biol. 106, 1911–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Poo M. (1982) Nature 295, 332–334 [DOI] [PubMed] [Google Scholar]
- 54.Srinivasan Y., Guzikowski A. P., Haugland R. P., Angelides K. J. (1990) J. Neurosci. 10, 985–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klausner R. D., Kleinfeld A. M., Hoover R. L., Karnovsky M. J. (1980) J. Biol. Chem. 255, 1286–1295 [PubMed] [Google Scholar]
- 56.Nigg E. A., Cherry R. J. (1980) Proc. Natl. Acad. Sci. U.S.A. 77, 4702–4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Y., Rose B. (1995) J. Cell Sci. 108, 3501–3508 [DOI] [PubMed] [Google Scholar]
- 58.Langhorst M. F., Solis G. P., Hannbeck S., Plattner H., Stuermer C. A. (2007) FEBS Lett. 581, 4697–4703 [DOI] [PubMed] [Google Scholar]
- 59.Paller M. S. (1994) J. Membr. Biol. 142, 127–135 [DOI] [PubMed] [Google Scholar]
- 60.Giepmans B. N., Moolenaar W. H. (1998) Curr. Biol. 8, 931–934 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.