Abstract
Transcription activation by androgen receptor (AR), which depends on recruitment of coactivators, is required for the initiation and progression of prostate cancer, yet the mechanisms of how hormone-activated AR interacts with coactivators remain unclear. This is because AR, unlike any other nuclear receptor, prefers its own N-terminal FXXLF motif to the canonical LXXLL motifs of coactivators. Through biochemical and crystallographic studies, we identify that steroid receptor coactivator-3 (SRC3) (also named as amplified in breast cancer-1 or AIB1) interacts strongly with AR via synergistic binding of its first and third LXXLL motifs. Mutagenesis and functional studies confirm that SRC3 is a preferred coactivator for hormone-activated AR. Importantly, AR mutations found in prostate cancer patients correlate with their binding potency to SRC3, corroborating with the emerging role of SRC3 as a prostate cancer oncogene. These results provide a molecular mechanism for the selective utilization of SRC3 by hormone-activated AR, and they link the functional relationship between AR and SRC3 to the development and growth of prostate cancer.
Keywords: Crystal Structure, Nuclear Receptors, Receptor Structure-Function, Steroid Hormone Receptor, Transcription Coactivators
Introduction
The androgen receptor (AR)2 is a hormone-dependent transcriptional factor. In response to the binding of its physiological ligands, testosterone or 5-α-dihydrotestosterone (DHT), AR directs a transcriptional program that is required for normal male development and homeostasis of muscle and skeleton systems (1). In addition, AR plays important roles in the initiation and maintenance of prostate cancer, which has led to increasing research being focused on this receptor (2, 3). AR is an established target of pharmaceutical intervention for prostate cancer, including treatment with antiandrogens such as bicalutamide and flutamide, which bind to the androgen-binding pocket in the C-terminal ligand-binding domain (LBD) and inhibit hormone-dependent activation of AR (4). However, long term therapy with antiandrogens becomes progressively less effective, mostly due to increased expression of AR or its coactivators or to mutations that make AR insensitive to antiandrogens (5–7). Novel strategies to inhibit AR activation, including the disruption of AR/coactivator complexes, are needed for developing the next generation of prostate cancer treatments (8, 9).
As a steroid hormone receptor, AR contains an N-terminal activation function domain (AF1), a central DNA-binding domain, and a C-terminal LBD that also contains a hormone-dependent activation function (AF2). The transcriptional activation of AR is initiated by hormone binding to the LBD, which results in functional recruitment of nuclear receptor coactivators, including steroid receptor coactivator-3 (SRC3, also known as AIB1, RAC3, pCIP, TRAM1, or ACTR). SRC3 is a member of the p160 SRC family that also includes SRC1 and TIF2/GRIP1/SRC2 (referred to as SRC2 hereafter) (10, 11). These coactivators are proposed to interact with steroid receptors in a ligand-dependent fashion and potentiate the activation of the receptor (12, 13).
Among nuclear receptor coactivators, SRC3 has been demonstrated to play particularly important roles in the development of both breast and prostate cancers (14–19). In fact, SRC3 was originally identified as amplified-in-breast cancer 1 (AIB1), which was found overexpressed in breast and ovarian cancers (20). Up-regulation of the SRC3 activity promotes breast cancer growth, invasiveness, and resistance to antiestrogen (21, 22). In parallel, SRC3 is found to be overexpressed in prostate cancer cells (23) and is correlated with human prostate cancer seminal vesicle invasion and lymph node metastasis (24). SRC3 expression is required for prostate cancer proliferation and cell survival (23, 25) in both AR-positive and AR-negative cancer cells. Deletion of SRC3 in mice inhibits initiation and progression of spontaneous prostate cancers, suggesting an important role of SRC3 in the hormone-dependent stage of prostate cancer cell development (17). Interestingly, phosphorylation is required for SRC3 activity, and SRC3 (but not SRC1 and SRC2) is selectively phosphorylated when cells are treated with androgen and estrogen but not with glucocorticoid or progesterone (26). Together, these observations suggest that there is a specific functional interplay between AR and SRC3 in prostate cancer.
Despite the functional relationship of AR and SRC3 in prostate cancer, the molecular basis of how hormone-activated AR recruits SRC3 remains unclear. Numerous studies have demonstrated that coactivator binding affinity and selectivity of nuclear receptors are primarily determined by the core LXXLL motifs, which are presented in multiple copies in coactivators (27, 28). However, unlike other nuclear receptors, AR has been considered to interact strongly with FXXLF motifs such as those found in the AR N terminus but weakly with the canonical LXXLL motifs found in members of the SRC family (29–32). In this study, we identify SRC3 as a strong coactivator of hormone-activated AR through synergistic binding of the first and the third LXXLL motifs of SRC3 to AR, and our results link the functional relationship between AR and SRC3 in the development and progression of prostate cancer.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The human AR LBD fragment (amino acids 666–919 with C669S mutation) was expressed as a His6-GST fusion protein from the expression vector pET24a in BL21(DE3) Escherichia coli cells using the same procedures as for the glucocorticoid receptor (GR) LBD (33). A concentration of 10 μm of the AR ligand DHT was used during protein expression and purification. The supernatant of the bacterial lysate was loaded on a GST column, washed with 20 mm Tris-HCl, pH 8.0, 1 m NaCl, 10% glycerol, 0.1% Triton-X100, and 10 μm DHT. The His6-GST-tagged AR LBD was eluted by using a buffer of 20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 10% glycerol, 10 μm DHT, and 20 mm glutathione and then was concentrated to 1 mm for the binding assay. For crystallization, the fusion protein was purified from a nickel column. The His6-GST tag was removed by thrombin (1/1000 ratio), and the AR LBD was further purified on an SP-Sepharose column followed by a gel filtration column. The purified AR LBD was complexed with 1.2 molar excess of peptide motifs and concentrated to ∼10 mg/ml for crystallization.
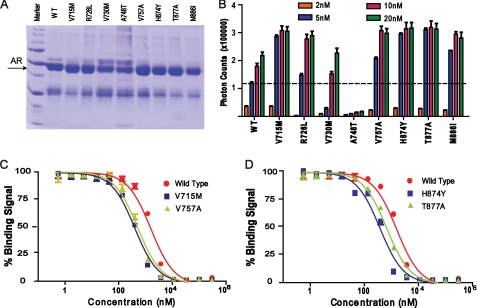
The mutated AR LBD proteins were prepared with the same procedures above. Fourteen AR single mutations, V715M, A721T, L722F, R726L, V730M, W741C, A748T, V757A, H874Y, T877A, M886I, Q902R, G909E, and K910R, were made using a QuikChange site-directed mutagenesis kit (Stratagene). The eight mutations that yielded soluble proteins are indicated in Fig. 6A.
FIGURE 6.
Correlation of AR mutations with SRC3 binding. A, a protein gel showing the purified, mutated LBD of AR for coactivator binding assays. WT, wild type. B, coactivator binding activity of the wild-type and mutated AR proteins as measured by their ability to interact with the biotinylated SRC3-1 motif. Error bars show S.D. C and D, IC50 values of the wild-type and mutated AR to SRC3-1as determined by full-dose competition curves using unlabeled SRC3-1. Error bars represent S.D. from triplicate experiments.
The wild-type and mutated SRC3 fragment (residues 615–746) are expressed as a His6-SUMO fusion protein from the expression vector pSUMO (LifeSensors) in BL21 (DE3) cells. The bacterial lysate supernatant was added onto a 25-ml nickel-nitrilotriacetic acid fast flow column (Amersham Biosciences), which was washed with 600 ml of 20 mm Tris, pH 8.0, 100 mm NaCl, 50 mm imidazole, and 10% glycerol. The His6-SUMO was eluted using buffer A (20 mm Tris, pH 8.0, 100 mm NaCl, and 10% glycerol) supplemented with 250 mm imidazole, cleaved overnight with 1/2000 SUMO protease at 4 °C while dialyzed against 20 mm Tris, pH 8.0, 100 mm NaCl, and 10% glycerol. The protein was then loaded onto a 5-ml nickel-nitrilotriacetic acid chelating Sepharose column (Amersham Biosciences) and eluted at 10% buffer B (10 mm Tris, pH 8.0, 1 m NaCl, 10% glycerol, 50 mm imidazole). Imidazole was removed from the protein by extensive dialysis against 20 mm Tris, pH 8.0, 100 mm NaCl, and 10% glycerol. The large fragments of SRC1 (residues 627–757) and SRC2 (residues 635–753) that contain all three LXXLL motifs were expressed and purified in a manner identical to the SRC3 fragment.
Biochemical Binding Assay
AR/coactivator binding affinities were assessed by fluorescence-based AlphaScreen technology that we have used extensively for coactivator interactions of a number of nuclear receptors (34–37). Briefly, a streptavidin-coated donor bead is brought into proximity with a nickel-chelated acceptor bead by interaction between the biotinylated peptide and the His6-tagged AR LBD. Excited by a laser beam of 680 nm, the donor bead emits singlet oxygen that activates the fluorophore in the acceptor beads, which releases photons of 520–620 nm as the binding signal between the receptor and the coactivator peptide. The experiments were conducted with 5–20 nm AR LBD and 20 nm of biotinylated SRC2-3 peptide (QEPVSPKKKENALLRYLLDKDDTKD) or biotinylated SRC3-1 peptide (SKGHKKLLQLLTCSS) in the presence of 5 μg/ml donor and acceptor beads in a buffer of 50 mm MOPS, pH 7.4, 50 mm NaF, 50 mm CHAPS, and 0.1 mg/ml bovine serum albumin. The rank order of binding affinities was determined by using unlabeled peptides at 500 nm to compete with the binding of biotinylated SRC2-3 to the AR LBD. IC50 values for various peptide motifs were determined from a nonlinear least squares fit of the data based on an average of three repeated experiments, with standard errors typically less than 10% of the measurements. The sequences of the unlabeled peptides have been reported previously (33, 37, 38). The IC50 values for all peptides are in micromole ranges, which are much higher than the AR protein or the biotinylated peptide (5–20 nm) in the experiments. Under these conditions, the IC50 values closely approximate the binding affinities (dissociation constants) (39).
Cell-based Assay for AR Activation
A vector of Gal4 DNA-binding domain fused to the AR LBD was constructed by inserting the AR LBD into the vector pBIND (Promega), and the luciferase reporter vector pG5-Luc containing five GAL4-binding sites (Promega) was used for AR coactivation by SRC coactivators. A full-length AR plasmid containing a CMV promoter and an MMTV reporter vector (pHHLuc from ATCC) were used for coactivation of full-length AR (40). All SRC coactivators and their mutants used in the assays were full-length protein plasmids containing CMV promoter. The assays were performed with COS-7 cells that were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum, 2 mm glutamine, and 15 units/ml penicillin/streptomycin. Cells were plated at 50,000/well in a 24-well plate 24 h prior to transfections. Cells were transfected in Opti-MEM I with 200 ng of reporter plasmid, 0.5 ng of control plasmid phRL-CMV (constitutive expression of Renilla luciferase; Promega), 50 ng of gal4-AR LBD or full-length AR, and wild-type/mutant SRC coactivator plasmids (or empty vector control) by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were induced with 10 nm DHT at 16–18 h after transfection. Twenty-four hours after induction, cells were harvested, and the firefly and Renilla luciferase activities were measured with the Dual Luciferase assay kit from Promega. Luciferase data were normalized to Renilla luciferase as an internal control. All assays were performed in triplicate.
Crystallization, Data Collection, and Structure Determination
The AR LBD/DHT/peptide crystals were grown at room temperature in hanging drops containing 1.0 μl of the protein complex solution and 1.0 μl of well solution containing 1.7–2.0 m ammonium sulfate, 100 mm HEPES, pH 7.5, and 10–20% sorbitol or sucrose as cryoprotectant. Crystals were flash-frozen in liquid nitrogen for data collection.
Both the AR/DHT/SRC3-1 and the AR/DHT/SRC3-3 complex crystals formed in the space group of P212121. Each asymmetric unit cell contained one AR LBD/DHT complex with 47% solvent content. The 180° data sets were collected from a single crystal by use of 1° oscillation of a MAR CCD detector at the ID line of Sector 5 (DuPont-Northwestern-Dow Collaborative Access Team) and Sector 21 (Life Sciences Collaborative Access Team) at the Advanced Photon Source. The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package (41). The structure was determined by molecular replacement using the AR/R1881 structure (Protein Data Bank (PDB) code: 1E3G) as the initial search model with the AMoRe program (42, 43). Manual model building was carried out with QUANTA (Accelrys Inc.), and structure refinement proceeded with CNS (44) using the maximum likelihood target.
RESULTS
Selective Binding of AR to the SRC3-1 LXXLL Motif
Ligand-dependent transcription of nuclear receptors is mediated through functional recruitment of coactivators, which primarily use canonical LXXLL motifs to interact with the receptor LBD. To study hormone-bound AR/coactivator interactions, we used AlphaScreen assays to characterize interactions of the AR LBD with a panel of coactivator/corepressor motifs in the presence of DHT. In this assay, the interaction between a biotin-tagged SRC2-3 peptide with the DHT-bound AR LBD produced a binding signal of 74,000 photons (Fig. 1A). Incubation of an excess of unlabeled 50 μm SRC2-3 inhibited the binding signal by more than 70% (Fig. 1A).
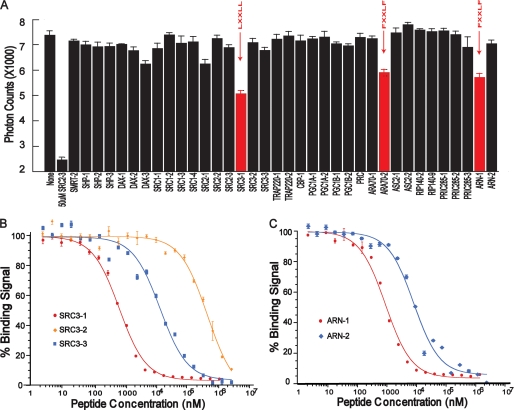
FIGURE 1.
Binding profile of peptide motifs to the AR LBD/DHT complex in the AlphaScreen assay. A, various unlabeled peptides at identical concentrations of 500 nm were used to compete off the binding of biotinylated SRC2-3 LXXLL motif to AR. The result shown is the average of triplicate experiments, with error bars showing S.D. The SRC3-1 LXXLL motif has the highest affinity for the AR LBD/DHT complex as determined by the peptide competitions. B, competition curves of the three LXXLL motifs from SRC3 to the AR LBD/DHT complex. C, competition curves of the FXXLF (ARN1) and WXXLF (ARN2) motifs to the AR LBD/DHT complex. Error bars represent S.D. from triplicate experiments.
To determine which coactivators are preferentially recruited, we performed a peptide profiling experiment using a panel of 36 unlabeled peptides to compete with the binding of biotinylated SRC2-3 to the DHT-bound AR (Fig. 1A). The sequences of these peptides, as reported previously (33, 45), were selected from endogenous nuclear receptor coregulators, including the p160 family of coactivators (SRC1, SRC2, and SRC3), PGC1, SHP, DAX1, and AR coactivators (ARA70). All coactivator peptides were designed to have an identical length of 15 residues with a central position of the LXXLL motif flanked by 6 and 4 residues at the N and C terminus (Table 1) because all nuclear receptor (NR)/coactivator structures determined to date reveal no structural information more than 2–3 residues outside of the core LXXLL motif. In this experiment, the unlabeled peptides were applied at a uniform concentration of 500 nm under identical experimental conditions to compete with the binding of biotinylated motif to AR. Most unlabeled peptide motifs showed little competition; only three peptide motifs, SRC3-1, ARA70-2, and ARN1, yielded significant competition, indicating that they contain motifs that bind strongly to AR. ARA70-2 is the second FXXLF motif from AR coactivator-70; ARN1 is the FXXLF motif from the AR N terminus (Table 1). Thus, inhibition by these two FXXLF motifs is expected. In contrast, strong binding of SRC3-1 to AR was surprising given that it does not contain a canonical FXXLF motif but rather an LXXLL motif. The SRC3-1 motif also shows strong binding to estrogen receptor (ER) but not to other steroid receptors such as GR and mineralocorticoid receptor (MR) (33, 38) (supplemental Fig. S1). These data thus provide the first hint that SRC3 may be a preferred coactivator of AR among the three steroid receptor coactivators, and these data are consistent with the fact that SRC3 (but not SRC1 and SRC2) is selectively phosphorylated in response to androgen and estrogen but not to glucocorticoid or progesterone (26).
TABLE 1.
IC50 of AR/coactivator peptides as determined by AlphaScreen
The substrate concentration used in the competition is 10 nm for the His6-GST-AR LBD and 20 nm for the biotinylated SRC2-3 LXXLL motif. All peptides have 15 residues except for SRC1-4, where its natural protein ends, and for ARN-1 and ARN-2, where 13-residue peptides were shown to have maximal activity in mammalian two-hybrid assays with the AR LBD (46). Residues of each peptide are numbered from the core LXXLL motif as 1, 2, 3, … and upstream of the core motif as −1, −2, −3 and so on.
| Coactivator motif | Sequence |
IC50 | ||
|---|---|---|---|---|
| −654321 | 12345 | 6789 | ||
| μm | ||||
| SRC1-1 | SQTSHK | LVQLL | TTTA | 61.3 ± 4.0 |
| SRC1-2 | TERHKI | LHRLL | QESS | 64.5 ± 2.6 |
| SRC1-3 | SKDHQL | LRYLL | DKDE | 9.4 ± 0.37 |
| SRC1-4 | AQQKSL | LQQLL | TE | 32.0 ± 1.8 |
| SRC2-1 | SKGQTK | LLQLL | TTKS | 15.6 ± 0.1 |
| SRC2-2 | KEKHKI | LHRLL | QDSS | 370 ± 94 |
| SRC2-3 | KKENAL | LRYLL | DKDD | 3.1 ± 0.4 |
| SRC3-1 | SKGHKK | LLQLL | TCSS | 0.71 ± 0.1 |
| SRC3-2 | QEKHRI | LHKLL | QNGN | 432 ± 67 |
| SRC3-3 | KENNAL | LRYLL | DRDD | 13.6 ± 2.2 |
| ARN-1 | YRGA | FQNLF | QSVR | 0.95 ± 0.15 |
| ARN-2 | ASSS | WHTLF | TAEE | 8.9 ± 0.13 |
| ARA70-2 | RETSEK | FKLLF | QSYN | 1.27 ± 0.35 |
To further explore the AR binding affinity, we measured the IC50 values under conditions in which the IC50 closely approximates the binding constant KD (see under “Experimental Procedures”). Consistent with the competition profile at a single concentration, the full-dose competition curve gave an IC50 of 0.71 μm for SRC3-1, which binds to AR slightly better than the two FXXLF motifs from ARN1 and ARA70-2, which have IC50 values of 0.95 and 1.27 μm, respectively (Fig. 1 and Table 1). Our affinity value (IC50) for the ARN1 FXXLF is in excellent agreement with that previously determined by isothermal titration calorimetry (46) or by surface plasma resonance (9), which gave a KD of 0.9–1.2 μm. Importantly, the affinity of SRC3-1 is 10–100-fold stronger than that of other LXXLL motifs in the SRC family (Table 1), suggesting that among the three SRC coactivators, AR may preferentially recruit SRC3.
SRC3 Is a Potent AR Coactivator
The strong binding affinity between AR and the SRC3-1 motif is unexpected and suggests that AR may preferentially recruit SRC3 over SRC1 or SRC2. To validate the functional significance of this observation, we used cell-based assays to test the ability of the full-length SRCs to enhance activation by the AR LBD or the full-length receptor. In COS-7 cells, SRC3 can potently activate AR in the presence of DHT even at the level of 20 ng of SRC3 cotransfection plasmids (Fig. 2A). In contrast, SRC2 only weakly activates AR, and SRC1 did not activate AR. In contrast, SRC1 is capable of activating GR, ER, and progesterone receptor (PR) (supplemental Fig. S2), suggesting that its inability to activate AR is the result of its poor binding to AR. Overall, these results correlate well with the relative binding affinity of LXXLL motifs in these three coactivators (Table 1). Only one out of four LXXLL motifs (SRC1-3, 9.4 μm) in SRC1 has moderate affinity, and the remaining three SRC1 motifs have a very weak affinity for AR. The first and third motifs of SRC2 have moderate affinity (15.6 and 3.1 μm, respectively). In contrast, SRC3 has two AR-binding LXXLL motifs: SRC3-1 with a strong affinity (0.71 μm) and SRC3-3 with a moderate affinity (13.6 μm) (Table 1). The rank order of affinity of LXXLL motifs in the three SRC coactivators matched nicely with their ability to activate AR.
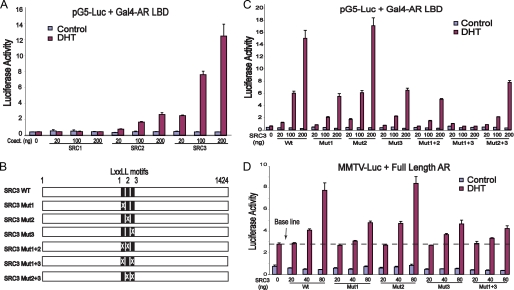
FIGURE 2.
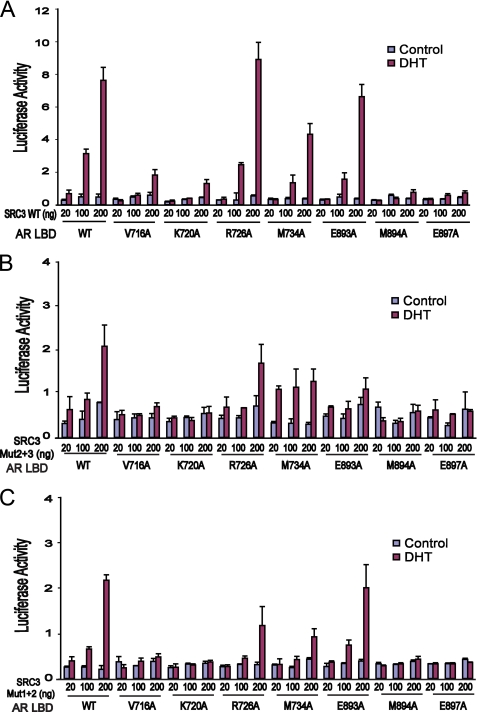
Activation of AR by SRC coactivators. A, SRC3 is the most potent among the three SRC coactivators of AR. The Gal4-AR LBD (50 ng) was transfected with 150 ng of pG5Luc and an increasing amount of three SRC plasmids into COS-7 cells. Activation of pG5Luc by the Gal4-AR LBD was determined in the presence or absence of 10 nm DHT. B, a schematic diagram of wild-type (WT) and mutated SRC3 showing the locations of its three LXXLL motifs. Mutation of individual LXXLL motifs to LXXAA is indicated by X. C, both the SRC3-1 and the SRC3-3 motifs are important for coactivation of the AR LBD. COS-7 cells were cotransfected with 50 ng Gal4-AR LBD, 150 ng of pG5Luc, and three different amounts of wild-type or mutated SRC3, and the luciferase activity was determined by triplicate experiments in the presence or absence of 10 nm DHT. Mutations in either the first or the third LXXLL motif of SRC3 reduced AR activation by nearly 50%, and simultaneous mutations in both motifs abolished coactivation of AR by SRC3. D, both the SRC3-1 and the SRC3-3 motifs are important for coactivation of the full-length AR. COS-7 cells were cotransfected with 50 ng of pCMV-AR plasmid containing the full-length AR, 150 ng of MMTV-Luc, and three different amounts of wild-type or mutated SRC3, and the luciferase activity was determined in the presence or absence of 10 nm DHT. Error bars represent S.D. from triplicate experiments.
To further study SRC3/AR interactions, we mutated the three LXXLL motifs to LXXAA in the context of the full-length SRC3 (Fig. 2, B and C). Mutations in the first or the third motif significantly decreased SRC3 coactivation, but mutation of the second motif had little effect. A combined mutation in both the first and the third motifs completely abolished the ability of SRC3 to activate AR (Fig. 2C, Mut1+3), whereas the effect of mutations in the second motif with either the first or the third motif was similar to that observed with a single mutation. Although SRC3 with the single intact motif (Mut1+2 or Mut2+3) can activate AR moderately, optimal activation of AR required the presence of both the first and the third motifs, implying that SRC3 uses those two motifs cooperatively to interact with the receptor dimer. Similar results were obtained with the MMTV promoter and full-length AR (Fig. 2D), suggesting the important roles of SRC3-1 and SRC3-1 motifs in AR activation.
Structural Basis of AR/SRC3 Interactions
To investigate the molecular basis for the selective recruitment of SRC3 coactivator by AR, we determined the crystal structure of the DHT-bound AR LBD bound to SRC3-1 or SRC3-3 motifs at a resolution of 1.55 and 2.00 Å, respectively (PDB codes: 3L3X and 3L3Z). The statistics for the data and the refined structures are summarized in supplemental Table S1. The overall structural features of the complexes are shown in Fig. 3, A and B. Each motif is clearly defined in the AR coactivator-binding site, which is bordered by the “charge-clamp” residues, which are Lys-720 from the end of H3 and Glu-897 from the center of the AF2 helix. The core LXXLL motif of SRC3-1 and SRC3-3 adopts a two-turn α-helix, which fits between the 2 charge-clamp residues (Fig. 3, C and D). The C-terminal carbonyls of the LXXLL helix are capped by hydrogen bonds with Lys-720, a key interaction conserved in all NR/coactivator complexes. However, the AF2 residue Glu-897 does not cap the N-terminal amide of the LXXLL helical motif, an interaction that is normally observed in many other NR/coactivator complexes. This is because the LXXLL helix is shifted half a helical turn (about 2.5 Å) from the AF2 helix toward Lys-720 of H3 (Fig. 3E, purple helix). The lack of the helix-capping interactions by the AF2 charge-clamp residue was also observed in previous structures of AR bound to other LXXLL motifs (9, 47, 48), thus helping to explain why most LXXLL coactivator motifs do not interact well with AR.
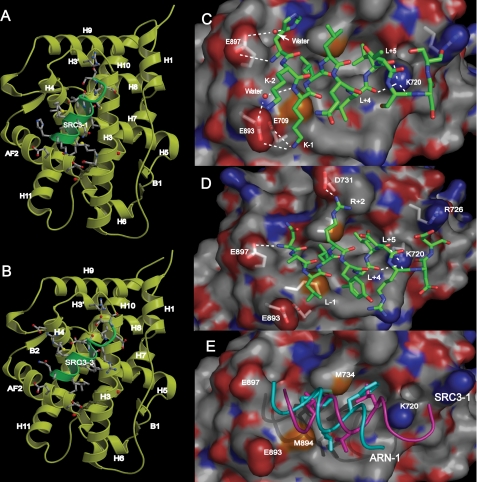
FIGURE 3.
Structure of the AR LBD/SRC3 interface. A and B, overall structures of the AR LBD/DHT complex bound to the SRC3-1 LXXLL motif (A) or the SRC3-3 LXXLL motif (B). The AR LBD is shown as yellow ribbons, and SRC motifs are green. C and D, the interface between the AR LBD and the LXXLL motifs of SRC3-1 (C) and SRC3-3 (D). Hydrogen bonds or charged interactions are indicated by dashed lines; key residues labeled. The AR LBD is in surface representation with carbon atoms in gray, oxygen atoms in red, and nitrogen atoms in blue. The SRC3-1 and SRC3-3 motifs are in stick representation with carbon atoms in green, oxygen atoms in red, and nitrogen atoms in blue. E, the different binding modes between the SRC3-1 LXXLL motif (magenta) and the ARN1 FXXLF motif (cyan, PDB code: 1XOW) in the AR coactivator-binding pocket.
However, our structure reveals that the SRC3-1 LXXLL motif makes new interactions with AR to compensate for its lack of interaction with Glu-897, the AR AF2 charge-clamp residue. The new interactions are made by the first and second lysine residues (K-1 and K-2) upstream of the core LXXLL motif of SRC3-1, both of which form a charged interaction with the negative residues Glu-709 and Glu-893 of AR (Fig. 3C). In addition, Glu-893 forms a water-mediated hydrogen bond to the backbone amide of K-1. Glu-897 of the AF2 helix also has a polar interaction and a water-mediated interaction with the third upstream residue (H-3) of SRC3-1 (Fig. 3C). Mutagenesis and binding studies (below) confirmed that both Glu-893 and Glu-897 are important for AR binding to the SRC3-1 motif, and the specific interaction between Glu-893 of AR and K-1 and K-2 of SRC3-1 is a key for AR selectivity toward SRC3.
The structure of the AR LBD bound to the SRC3-3 motif reveals the structural basis for the moderate affinity of this motif for AR. In the center of the motif, Arg+2 forms a charge interaction with Asp-731 of AR (Fig. 3D), analogous to the GR LBD structure bound to the SRC2-3 motif (49). In the C terminus, the stretch of negatively charged residues (Asp+8 and Asp+9) is close enough to form charged interactions with Lys-720 and Arg-726. In the N terminus, the backbone amide of H-3 of the LXXLL motif forms a long distance hydrogen bond with the AF2 charge-clamp residue Glu-897 (Fig. 3D). Thus, in both structures, the AR AF2 residue Glu-897 makes important interactions with the flanking residues at the N terminus of the LXXLL motif, compensating for the lack of classical charge-clamp interactions.
Biochemical Basis of AR Selectivity toward the SRC3-1 Motif
To further determine the molecular basis for AR selectivity toward the SRC3-1 motif, we generated a series of swap mutations between SRC3-1 and SRC3-2 because these two motifs show more than a 600-fold difference in AR binding affinity (Fig. 1B and supplemental Table S2). We divided the coactivator motifs into three segments: the N-terminal flanking segment, the central core LXXLL motif, and the C-terminal flanking segment. Swapping the core LXXLL motif or the C-terminal flanking segment between SRC3-1 and SRC3-2 motif caused a moderate 2.9- and 9.7-fold decrease in binding to AR (supplemental Table S2). In contrast, swapping the N-terminal flanking sequence resulted in a nearly 500-fold decrease in AR binding affinity, suggesting that the primary SRC3-1 selectivity for AR is determined by the N-terminal flanking sequence. We also performed swap mutations at individual residues between SRC3-1 and SRC3-2. Most of these single swap mutations had a mild effect on AR binding except for the residue at the −1 position; a change of Lys to Ile caused a 42-fold decrease in AR binding. The decrease from this mutation is similar to that of alanine mutations in the core leucine residues of the LXXLL motif (supplemental Table S2). Together, these data suggest that AR selectivity for the SRC3-1 motif is primarily determined by the N-terminal flanking residues, particularly by K-1 and K-2 residues. This result is consistent with the structure of the AR/SRC3-1 complex, showing that the side chains of K-1 and K-2 form electrostatic interactions with the oxygen atoms of the carboxylic side chains Glu-893 and Glu-709 of AR (Fig. 3C).
To determine the role of AR residues in coactivator selectivity, we mutated several key AR residues that contact the SRC3-1 motif in the crystal structure and measured their binding to SRC3-1 and ARN1 motif (supplemental Table S3). The K720A mutation abolished AR binding to the SRC3-1 LXXLL motif and to the ARN1 FXXLF motif, indicating the critical role of Lys-720 in the binding of these motifs. Interestingly, the E897A mutation of the AF2 charge-clamp residue selectively affected the binding to the ARN1 FXXLF motif; it had little effect on binding to the SRC3-1 LXXLL motif. In contrast, the E893A mutation selectively decreased AR binding to the SRC3-1 motif but only mildly affected the binding to ARN1 motif. This result complements the mutation data of K-1 in the SRC3-1 motif (supplemental Table S2) and is in excellent agreement with the interactions between K-1 of SRC3-1 and Glu-893 of AR observed in the structure. Mutations in several other AR residues showed modest effects on binding to the SRC3-1 motif. However, the M894A mutation enhanced the AR N/C interaction (where N/C indicates the interaction between the N terminus and the C terminus of AR) by 50% but decreased AR binding to SRC3-1 by 22.5-fold (supplemental Table S3).
In cell-based reporter assays, SRC3 failed to activate AR with mutations of K720A and M894A (Fig. 4A). The E893A mutation, which reduced AR/SRC3-1 binding, only affected AR activation by the SRC3 mutant Mut2+3, which has only the SRC3-1 motif (Fig. 4B), but did not affect activation by wild-type SRC3 (Fig. 4A) or by the SRC3 mutant Mut1+2 (Fig. 4C). This indicates that Glu-893 is mainly responsible for AR/coactivator binding selectivity toward the SRC3-1 motif but not the overall binding affinity of SRC3. Together, these data further support the role of Glu-893/K-1 interaction in determining the AR coactivator specificity toward the SRC3-1 motif.
FIGURE 4.
Mutational studies of the AR coactivator-binding site. A, the ability of wild-type (WT) SRC3 to activate AR having a wild-type or mutated coactivator-binding pocket. COS-7 cells were cotransfected with 150 ng of pG5Luc, 100 ng of wild-type or mutated Gal4-AR LBD, and three different amounts of wild-type or mutated SRC3. The luciferase activity was determined by triplicate experiments in the presence or absence of 10 nm DHT. Mutation in either charge-clamp residue (Glu-897 or Lys-720) disrupts AR activation by SRC3, whereas mutation in Glu-893 mildly reduced AR activation. Mutations in hydrophobic residues (V716A, M734A, and M894A) that contact the LXXLL motif also reduce AR activation by SRC3. B and C, the same experiments as in A, except that mutated SRC3 was used. When SRC3 containing only the first LXXLL motif (Mut2+3) was used, the mutation E893A of AR significantly reduced AR activation by SRC3 (B). In contrast, when SRC3 containing only the third motif (Mut1+2) was used, the E893A mutation did not affect AR activation by SRC3 (C). Error bars represent S.D. from triplicate experiments.
We further performed “gain-of-function” mutations to validate the role of SRC3-1 LXXLL motif in the AR coactivator selectivity. In COS-7 cells, SRC2 weakly activates AR (Fig. 2A and supplemental Fig. S3C). Replacing the SRC2-1 motif with the SRC3-1 motif in the full-length SRC2 greatly increased its ability to activate AR (supplemental Fig. S3C). To corroborate this, replacing the two N-terminal residues of SRC2-1 with the SRC3-1 residues (SRC2-1KK) increased its binding affinity to AR by 3–4-fold (supplemental Fig. S3, A and B). These data further confirm that AR selectivity toward SRC3 is primarily determined by the SRC3-1 LXXLL motif via the charge interaction of its N-terminal flanking lysine residues (K-1 and K-2) with the negative residues of AR.
Cooperative Binding of SRC3-1 and SRC3-3 Motifs to AR
It has been a challenge to determine how AR recruits coactivators via their LXXLL motifs given that the AR LBD forms intramolecular interactions with its own N-terminal FXXLF motif. Our results indicate that SRC3 contains two AR-binding LXXLL motifs, one motif (SRC3-1) having approximately the same AR binding affinity as that of AR binding to its own N-terminal FXXLF motif and the other motif (SRC3-3) having modest affinity for AR (Table 1).
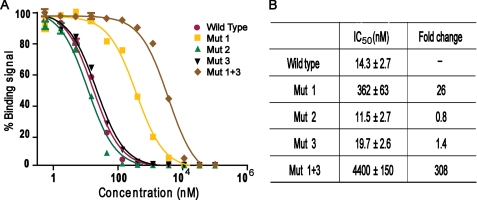
As a hormone-bound receptor, AR binds to DNA and activates transcription as a homodimer. The presence of two AR-binding LXXLL motifs in SRC3 suggests that they may be used to interact with the AR dimer synergistically. To test this idea, we expressed and purified an SRC3 fragment (residues 615–714) that encompasses all three LXXLL motifs. This fragment binds to AR with an IC50 of about 14 nm, an affinity that is 50- and 1000-fold better, respectively, than the isolated SRC3-1 and SRC3-3 motifs (compare Fig. 5 and Table 1), suggesting a cooperative binding of the LXXLL motifs of SRC3 to AR. We also expressed and purified the corresponding fragments of SRC1 and SRC2 containing three LXXLL motifs. The IC50 value of the SRC1 and SRC2 fragments to AR is 647 and 562 nm, respectively (supplemental Fig. S4). Thus, the binding of SRC1 and SRC2 to AR is ∼50-fold weaker than the binding of SRC3 to AR.
FIGURE 5.
Synergistic binding of the two LXXLL motifs of SRC3 toward AR. A, inhibition of biotinylated SRC2-3 binding to AR by unlabeled wild-type or mutated SRC3 fragments. Mutated SRC3 fragment labels correspond to the labels in Fig. 2B. Error bars show S.D. B, IC50 values of the wild-type and mutated SRC3 fragments derived from the competition curves in panels A and B. The -fold decrease in affinity is relative to the wild-type fragment, which is set to 1.
We also expressed and purified the same SRC3 fragment with mutations in the LXXLL motifs. Mutations in the second motif had little effect. Mutations in the third motif caused a slight decrease of binding affinity to AR, and mutations in the first motif resulted in a 26-fold loss of AR binding affinity (Fig. 5B). These results again corroborate the binding data of individual motifs (Table 1) and the mutational data from cell-based assays (Fig. 2). Furthermore, mutations in both the first and the third motifs result in greater than a 300-fold decrease in AR binding, indicating a cooperative binding of these two SRC3 motifs. The strong affinity (IC50 of 10–14 nm) resulting from the synergistic binding of the two LXXLL motifs in SRC3 thus provide a biochemical basis for AR recruitment of the SRC3 coactivator.
AR Mutations in Prostate Cancer Increase Binding to SRC3
Both AR and SRC3 have been implicated in the proliferation and survival of prostate cancer cells (23, 25). Identification of SRC3 as an AR-selective coactivator suggests that one effect of AR mutations in prostate cancer may be increased interaction with SRC3. To test this idea, we made 14 individual mutations in the AR LBD corresponding to AR mutations found in prostate cancer (via the Androgen Receptor Gene Mutations Database) (50). Eight of these mutated AR proteins can be expressed and purified to a level comparable with the wild-type protein (Fig. 6A). Quantitative binding assays showed that most AR mutations increased binding with SRC3 by 2–3-fold (V715M, R726L, V757A, H874Y, T877A, and M886I in Fig. 6B). The binding affinity of these AR mutations for SRC3 is also increased by 2–3-fold as indicated by competition curves (Fig. 6, C and D). Based on structural analysis (supplemental material), the AR mutations increase SRC3 binding by increasing the stability of the AR protein (as also revealed by the protein gel in Fig. 6A for the V715M, R726L, V757A, H874Y, T877A, and M886I mutations). In contrast, the mutations that decrease SRC3 binding either reduce AR protein stability (A748T) or directly affect AR coactivator binding (V730M) (Fig. 6A). Interestingly, V715M and M886I were found previously as gain-of-function mutations in binding of coactivators or transcription activity (51, 52). R726L, V757A, H874Y, and T877A are found frequently as somatic mutations that are associated with prostate cancer or resistance to antiandrogen therapy (53–56). In contrast, the two mutations (V730M and A748T) that showed decreased binding to SRC3 were from patients having lower Gleason scores than patients with wild-type AR (the Gleason score is used to grade prostate cancer, with advanced disease having a higher score) (57). The correlation of these AR mutations with SRC3 binding further supports the importance of the functional interactions between AR and SRC3 in the development of prostate cancer.
DISCUSSION
We have identified SRC3 as an AR-preferential coactivator that binds to AR synergistically through its two LXXLL motifs (SRC3-1 and SRC3-3). Mutagenesis studies indicate that both motifs are required for SRC3 to fully activate AR. In addition, the crystal structure of the AR LBD bound to these two motifs provides key insights into how the selectivity of AR for SRC3 is achieved. The discovery of SRC3 as an AR preferred coactivator has important implications in AR regulation and coactivator complex assembly. As both AR and SRC3 serve as key regulators in the development and progression of prostate cancer, our results elucidate an important mechanism in the functional relationship between these two proteins and their roles in prostate cancer.
Basis of AR/SRC3 Binding Specificity
As a ligand-dependent nuclear receptor, AR activates transcription through selective recruitment of coactivators, but how AR achieves ligand-dependent recruitment of a coactivator has been an enigmatic problem. Unlike other nuclear receptors, AR is thought to interact strongly with FXXLF motifs presented in its own N terminus (ARN1) but bind weakly to canonical LXXLL motifs found in many nuclear receptor coactivators. Thus, the identification of the SRC3-1 LXXLL motif as a potent motif that binds to AR is somehow surprising, yet it makes sense in terms of their functional relationship in prostate cancer. AR is the key player in prostate cancer, its transcriptional activity being essential for prostate cancer initiation and development (2). SRC3, also emerging as a key regulator, is found overexpressed in many prostate cancers (23, 25). The activity of SRC3 is required for prostate cancer cell survival and proliferation; deletion of SRC3 in mice inhibits spontaneous prostate cancer progression (17). Establishing the SRC3 as the AR-preferential coactivator thus provides a molecular mechanism for the functional relationship of these two proteins in prostate cancer.
The molecular basis for AR selectivity toward SRC3 is uncovered by our structural and mutagenesis studies, which reveal that AR uses two negatively charged residues (Glu-893 and Glu-709) to interact with the lysine at the −1 position of the SRC3-1 motif. This selective mechanism is not found in other steroid hormone receptors such as the GR and the MR. The GR and MR do not contain residues analogous to Glu-893 and Glu-709, and they bind poorly to the SRC3-1 motif (33, 38) The binding affinity of MR to SRC3-1 is 16.8 μm, which is nearly 24-fold weaker than that of AR (38).
The only steroid receptor other than AR that binds SRC3-1 with high affinity is ERα, which contains negatively charged residues (Asp-538 and Asp-351) analogous to those in AR for interacting with K-1 of the SRC3-1 motif. ERα also interacts with the second LXXLL motif from all three SRC coactivators (supplemental Fig. S1), and the basis of such binding has been well illustrated by the structure of ERα bound to the second LXXLL motif of GRIP1 (also called TIF2 and SRC-2) (58). Thus, SRC3 also contains two LXXLL motifs (SRC3-1 and SRC3-2) with high binding affinity to ERα, which may serve as the physical basis for functional coupling of these two proteins.
It is worth noting that SRC3 was originally cloned as amplified-in-breast cancer 1 (AIB1), whose activity has since been shown to be essential for breast cancer development and progression (20). Interestingly, SRC3 phosphorylation is required for its coactivation function, and SRC3 is phosphorylated upon treatment with androgen or estrogen but not with glucocorticoid or progesterone (26), which further supports that SRC3 is the preferential coactivator for ER and AR. The emerging roles of SRC3 in the oncogenic process of prostate and breast cancers suggests that the SRC3 interaction with ER and AR could serve as a target for therapeutical intervention in these diseases.
Assembly of the AR/SRC3 Complex in the Presence of the AR N/C Interaction
Our finding that SRC3 contains a high affinity LXXLL motif (SRC3-1) and a moderate affinity motif (SRC3-3) has a mechanistic implication for AR/coactivator assembly. Coactivator recruitment by the AR LBD is required for activation of target genes in chromatin as the AR N-terminal activation domain poorly activates chromatinized genes (59), although the AR N-terminal AF1 can recruit SRC coactivators and activate transient reporter genes in a hormone-independent manner (60–62). However, it has been puzzling how AR recruits coactivators via the LXXLL motifs because the C-terminal AR LBD is considered to have high affinity for its own N-terminal FXXLF motif (31). Such N/C interaction would exclude the binding of coactivators containing LXXLL motifs (30). Identification of SRC3-1 as a potent AR-binding motif thus provides a half-solution to this problem; SRC3 is capable of competing with the AR N-terminal FXXLF motif for the AF2-binding site. The other half of the solution is provided by the synergistic binding of the two LXXLL motifs (SRC3-1 and SRC3-3) to AR, as discussed below.
As a DNA-binding transcriptional activator, AR functions as a homodimer. The presence of the two LBDs in the homodimer provides two AF2-binding sites for cooperative binding of multiple LXXLL motifs in a single coactivator. We have shown that it is indeed the case that synergistic binding of the two LXXLL motifs in SRC3 dramatically increases SRC3 binding affinity to AR. This cooperative binding of SRC3 to AR is reminiscent of SRC2 (TIF2/GRIP1); a fragment containing two LXXLL motifs (the second and third of SRC2) binds to mineralocorticoid receptor with 25-fold higher affinity than either single motif (38). Thus, synergistic binding to a receptor dimer by multiple coactivator LXXLL motifs is a common theme of nuclear receptor/coactivator assembly. The high affinity resulting from synergistic binding should allow SRC3 to use these two motifs to wrap around the AR LBD dimer.
This model of SRC3 binding to the AR LBD dimer has implications of AR recruitment of coactivators. It has been shown that all three SRC members are recruited to the prostate-specific antigen (PSA) upstream distal enhancer with similar efficiency in response to androgens (63, 64). This seems to contradict the AR LBD selectivity for SRC3, but it can be explained by the fact that SRC3 can form heterodimers with SRC1 or SRC2 onto the classical hormone-responsive element found in the PSA enhancer (65). Thus, upon hormone binding, AR uses its C-terminal AF2 to bind with SRC3, which then recruits SRC1 or SRC2 via a mechanism of heterodimerization. This model is consistent with the results of RNA interference studies, which reveal that PSA activation is critically dependent on SRC3 but less so on SRC1 and SRC2 (65). RNA interference studies from two other groups also reveal that SRC3, but not SRC1 and SRC2, is required for prostate cancer cell growth and survival in both an AR-dependent and an AR-independent manner (23, 25). Furthermore, many AR mutations in prostate cancer increase the binding affinity to SRC3 (Fig. 6), and SRC3 is often expressed at higher levels that correlate with the prognosis for and progression of prostate cancer (23, 25). Either of these scenarios will drive the balance toward the assembly of an active AR/SRC3 coactivator complex in prostate cancer cells. The selective utilization of SRC3 by AR thus correlates the functional activity of AR with the emerging role of SRC3 as an oncogenic product in the development and growth of prostate cancer.
Acknowledgments
We thank D. Nadziejka for editing the manuscript, E. Wilson and M. Tsai for cDNAs of the androgen receptor and SRC coactivators, S. Triezenberg for discussion, and Z. Wawrzak and J. S. Brunzelle for assistance in data collection at the beamlines of Sector 5 (DuPont, Northwest and Dow Collaborative Access Team), and sector 21 (Life Sciences Collaborative Access Team), which is in part funded by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of the Advanced Photon Source was supported by the Office of Science of the United States Department of Energy.
This work was supported, in whole or in part, by National Institutes of Health Grants DK071662, DK066202, and HL089301 (to H. E. X.). This work was also supported in part by a grant from the Jay and Betty Van Andel Foundation and a Department of Defense Prostate Cancer Grant W81XWH0510043 (to H. E. X.).
This article was selected as a Paper of the Week.
The atomic coordinates and structure factors (codes 3L3X and 3L3Z) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental text, supplemental Figs. S1–S4, and supplemental Tables S1–S3.
- AR
- androgen receptor
- GR
- glucocorticoid receptor
- ER
- estrogen receptor
- MR
- mineralocorticoid receptor
- NR
- nuclear receptor
- SRC
- steroid receptor coactivator
- DHT
- 5-α-dihydrotestosterone
- LBD
- ligand-binding domain
- GST
- glutathione S-transferase
- SUMO
- small ubiquitin-like modifier
- CMV
- cytomegalovirus
- MMTV
- murine mammary tumor virus
- MOPS
- 4-morpholinepropanesulfonic acid
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid.
REFERENCES
- 1.Tindall D. J. (2000) Mayo Clin. Proc. 75, S26–S31 [PubMed] [Google Scholar]
- 2.Dehm S. M., Tindall D. J. (2005) Expert Rev. Anticancer Ther. 5, 63–74 [DOI] [PubMed] [Google Scholar]
- 3.Heinlein C. A., Chang C. (2004) Endocr. Rev. 25, 276–308 [DOI] [PubMed] [Google Scholar]
- 4.Culig Z., Bartsch G., Hobisch A. (2004) Curr. Cancer Drug Targets 4, 455–461 [DOI] [PubMed] [Google Scholar]
- 5.Grossmann M. E., Huang H., Tindall D. J. (2001) J. Natl. Cancer Inst. 93, 1687–1697 [DOI] [PubMed] [Google Scholar]
- 6.Miyamoto H., Messing E. M., Chang C. (2004) Prostate 61, 332–353 [DOI] [PubMed] [Google Scholar]
- 7.Chen C. D., Welsbie D. S., Tran C., Baek S. H., Chen R., Vessella R., Rosenfeld M. G., Sawyers C. L. (2004) Nat. Med. 10, 33–39 [DOI] [PubMed] [Google Scholar]
- 8.Chang C. Y., McDonnell D. P. (2005) Trends Pharmacol. Sci. 26, 225–228 [DOI] [PubMed] [Google Scholar]
- 9.Hur E., Pfaff S. J., Payne E. S., Grøn H., Buehrer B. M., Fletterick R. J. (2004) PLoS Biol. 2, E274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heemers H. V., Tindall D. J. (2007) Endocr. Rev. 28, 778–808 [DOI] [PubMed] [Google Scholar]
- 11.Rahman M., Miyamoto H., Chang C. (2004) Clin Cancer Res. 10, 2208–2219 [DOI] [PubMed] [Google Scholar]
- 12.Lonard D. M., O'Malley B, W. (2007) Mol. Cell 27, 691–700 [DOI] [PubMed] [Google Scholar]
- 13.Yan J., Tsai S. Y., Tsai M. J. (2006) Acta Pharmacol. Sin. 27, 387–394 [DOI] [PubMed] [Google Scholar]
- 14.Li L. B., Louie M. C., Chen H. W., Zou J. X. (2008) Cancer Lett. 261, 64–73 [DOI] [PubMed] [Google Scholar]
- 15.Louie M. C., Zou J. X., Rabinovich A., Chen H. W. (2004) Mol. Cell. Biol. 24, 5157–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fereshteh M. P., Tilli M. T., Kim S. E., Xu J., O'Malley B. W., Wellstein A., Furth P. A., Riegel A. T. (2008) Cancer Res. 68, 3697–3706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung A. C., Zhou S., Liao L., Tien J. C., Greenberg N. M., Xu J. (2007) Cancer Res. 67, 5965–5975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mussi P., Yu C., O'Malley B. W., Xu J. (2006) Mol. Endocrinol. 20, 3105–3119 [DOI] [PubMed] [Google Scholar]
- 19.Kuang S. Q., Liao L., Zhang H., Lee A. V., O'Malley B. W., Xu J. (2004) Cancer Res. 64, 1875–1885 [DOI] [PubMed] [Google Scholar]
- 20.Anzick S. L., Kononen J., Walker R. L., Azorsa D. O., Tanner M. M., Guan X. Y., Sauter G., Kallioniemi O. P., Trent J. M., Meltzer P. S. (1997) Science 277, 965–968 [DOI] [PubMed] [Google Scholar]
- 21.Su Q., Hu S., Gao H., Ma R., Yang Q., Pan Z., Wang T., Li F. (2008) Oncology 75, 159–168 [DOI] [PubMed] [Google Scholar]
- 22.Kirkegaard T., McGlynn L. M., Campbell F. M., Müller S., Tovey S. M., Dunne B., Nielsen K. V., Cooke T. G., Bartlett J. M. (2007) Clin. Cancer Res. 13, 1405–1411 [DOI] [PubMed] [Google Scholar]
- 23.Zhou H. J., Yan J., Luo W., Ayala G., Lin S. H., Erdem H., Ittmann M., Tsai S. Y., Tsai M. J. (2005) Cancer Res. 65, 7976–7983 [DOI] [PubMed] [Google Scholar]
- 24.Yan J., Erdem H., Li R., Cai Y., Ayala G., Ittmann M., Yu-Lee L. Y., Tsai S. Y., Tsai M. J. (2008) Cancer Res. 68, 5460–5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zou J. X., Zhong Z., Shi X. B., Tepper C. G., deVere White R. W., Kung H. J., Chen H. (2006) Prostate 66, 1474–1486 [DOI] [PubMed] [Google Scholar]
- 26.Zheng F. F., Wu R. C., Smith C. L., O'Malley B. W. (2005) Mol. Cell. Biol. 25, 8273–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lonard D. M., O'Malley B. W. (2006) Cell 125, 411–414 [DOI] [PubMed] [Google Scholar]
- 28.Glass C. K., Rosenfeld M. G. (2000) Genes Dev. 14, 121–141 [PubMed] [Google Scholar]
- 29.He B., Lee L. W., Minges J. T., Wilson E. M. (2002) J. Biol. Chem. 277, 25631–25639 [DOI] [PubMed] [Google Scholar]
- 30.He B., Bowen N. T., Minges J. T., Wilson E. M. (2001) J. Biol. Chem. 276, 42293–42301 [DOI] [PubMed] [Google Scholar]
- 31.He B., Kemppainen J. A., Wilson E. M. (2000) J. Biol. Chem. 275, 22986–22994 [DOI] [PubMed] [Google Scholar]
- 32.Dubbink H. J., Hersmus R., Pike A. C., Molier M., Brinkmann A. O., Jenster G., Trapman J. (2006) Mol. Endocrinol. 20, 1742–1755 [DOI] [PubMed] [Google Scholar]
- 33.Suino-Powell K., Xu Y., Zhang C., Tao Y. G., Tolbert W. D., Simons S. S., Jr., Xu H. E. (2008) Mol. Cell. Biol. 28, 1915–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suino K., Peng L., Reynolds R., Li Y., Cha J. Y., Repa J. J., Kliewer S. A., Xu H. E. (2004) Mol. Cell 16, 893–905 [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Choi M., Cavey G., Daugherty J., Suino K., Kovach A., Bingham N. C., Kliewer S. A., Xu H. E. (2005) Mol. Cell 17, 491–502 [DOI] [PubMed] [Google Scholar]
- 36.Li Y., Choi M., Suino K., Kovach A., Daugherty J., Kliewer S. A., Xu H. E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 9505–9510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou X. E., Suino-Powell K., Ludidi P. L., McDonnellb D. P., Xu H. E. (December6, 2009) Protein Expr. Purif. 10.1016/j.pep.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Suino K., Daugherty J., Xu H. E. (2005) Mol. Cell 19, 367–380 [DOI] [PubMed] [Google Scholar]
- 39.Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 40.Quarmby V. E., Kemppainen J. A., Sar M., Lubahn D. B., French F. S., Wilson E. M. (1990) Mol. Endocrinol. 4, 1399–1407 [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 42.Matias P. M., Donner P., Coelho R., Thomaz M., Peixoto C., Macedo S., Otto N., Joschko S., Scholz P., Wegg A., Bäsler S., Schäfer M., Egner U., Carrondo M. A. (2000) J. Biol. Chem. 275, 26164–26171 [DOI] [PubMed] [Google Scholar]
- 43.Navaza J., Gover S., Wolf W. (1992) in Molecular Replacement: Proceedings of the CCP4 Study Weekend, January 31–February 1, 1992 (Dodson E. J. ed) SERC Daresbury Laboratory, Warrington, UK [Google Scholar]
- 44.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 45.Li Y., Kovach A., Suino-Powell K., Martynowski D., Xu H. E. (2008) J. Biol. Chem. 283, 19132–19139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He B., Wilson E. M. (2003) Mol. Cell. Biol. 23, 2135–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He B., Gampe R. T., Jr., Kole A. J., Hnat A. T., Stanley T. B., An G., Stewart E. L., Kalman R. I., Minges J. T., Wilson E. M. (2004) Mol. Cell 16, 425–438 [DOI] [PubMed] [Google Scholar]
- 48.Estébanez-Perpiñá E., Moore J. M., Mar E., Delgado-Rodrigues E., Nguyen P., Baxter J. D., Buehrer B. M., Webb P., Fletterick R. J., Guy R. K. (2005) J. Biol. Chem. 280, 8060–8068 [DOI] [PubMed] [Google Scholar]
- 49.Bledsoe R. K., Montana V. G., Stanley T. B., Delves C. J., Apolito C. J., McKee D. D., Consler T. G., Parks D. J., Stewart E. L., Willson T. M., Lambert M. H., Moore J. T., Pearce K. H., Xu H. E. (2002) Cell 110, 93–105 [DOI] [PubMed] [Google Scholar]
- 50.Gottlieb B., Lehvaslaiho H., Beitel L. K., Lumbroso R., Pinsky L., Trifiro M. (1998) Nucleic Acids Res. 26, 234–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen G., Wang X., Zhang S., Lu Y., Sun Y., Zhang J., Li Z., Lu J. (2005) Prostate 63, 395–406 [DOI] [PubMed] [Google Scholar]
- 52.Culig Z., Hobisch A., Cronauer M. V., Cato A. C., Hittmair A., Radmayr C., Eberle J., Bartsch G., Klocker H. (1993) Mol. Endocrinol. 7, 1541–1550 [DOI] [PubMed] [Google Scholar]
- 53.Marcelli M., Ittmann M., Mariani S., Sutherland R., Nigam R., Murthy L., Zhao Y., DiConcini D., Puxeddu E., Esen A., Eastham J., Weigel N. L., Lamb D. J. (2000) Cancer Res. 60, 944–949 [PubMed] [Google Scholar]
- 54.Taplin M. E., Bubley G. J., Ko Y. J., Small E. J., Upton M., Rajeshkumar B., Balk S. P. (1999) Cancer Res. 59, 2511–2515 [PubMed] [Google Scholar]
- 55.Mononen N., Syrjäkoski K., Matikainen M., Tammela T. L., Schleutker J., Kallioniemi O. P., Trapman J., Koivisto P. A. (2000) Cancer Res. 60, 6479–6481 [PubMed] [Google Scholar]
- 56.Taplin M. E., Bubley G. J., Shuster T. D., Frantz M. E., Spooner A. E., Ogata G. K., Keer H. N., Balk S. P. (1995) N. Engl. J. Med. 332, 1393–1398 [DOI] [PubMed] [Google Scholar]
- 57.Sanchez D., Rosell D., Honorato B., Lopez J., Arocena J., Sanz G. (2006) BJU Int. 98, 1320–1325 [DOI] [PubMed] [Google Scholar]
- 58.Shiau A. K., Barstad D., Loria P. M., Cheng L., Kushner P. J., Agard D. A., Greene G. L. (1998) Cell 95, 927–937 [DOI] [PubMed] [Google Scholar]
- 59.Li J., Fu J., Toumazou C., Yoon H. G., Wong J. (2006) Mol. Endocrinol. 20, 776–785 [DOI] [PubMed] [Google Scholar]
- 60.Bevan C. L., Hoare S., Claessens F., Heery D. M., Parker M. G. (1999) Mol. Cell. Biol. 19, 8383–8392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma H., Hong H., Huang S. M., Irvine R. A., Webb P., Kushner P. J., Coetzee G. A., Stallcup M. R. (1999) Mol. Cell. Biol. 19, 6164–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Alen P., Claessens F., Verhoeven G., Rombauts W., Peeters B. (1999) Mol. Cell. Biol. 19, 6085–6097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Q., Carroll J. S., Brown M. (2005) Mol. Cell 19, 631–642 [DOI] [PubMed] [Google Scholar]
- 64.Louie M. C., Yang H. Q., Ma A. H., Xu W., Zou J. X., Kung H. J., Chen H. W. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2226–2230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang H., Yi X., Sun X., Yin N., Shi B., Wu H., Wang D., Wu G., Shang Y. (2004) Genes Dev. 18, 1753–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]