Abstract
Wnt/β-catenin signaling is initiated at the cell surface by association of secreted Wnt with its receptors Frizzled (Fz) and low density lipoprotein receptor-related protein 5/6 (LRP5/6). The study of these molecular interactions has been a significant technical challenge because the proteins have been inaccessible in sufficient purity and quantity. In this report we describe insect cell expression and purification of soluble mouse Fz8 cysteine-rich domain and human LRP6 extracellular domain and show that they inhibit Wnt/β-catenin signaling in cellular assays. We determine the binding affinities of Wnts and Dickkopf 1 (Dkk1) to the relevant co-receptors and reconstitute in vitro the Fz8 CRD·Wnt3a·LRP6 signaling complex. Using purified fragments of LRP6, we further show that Wnt3a binds to a region including only the third and fourth β-propeller domains of LRP6 (E3E4). Surprisingly, we find that Wnt9b binds to a different part of the LRP6 extracellular domain, E1E2, and we demonstrate that Wnt3a and Wnt9b can bind to LRP6 simultaneously. Dkk1 binds to both E1E2 and E3E4 fragments and competes with both Wnt3a and Wnt9b for binding to LRP6. The existence of multiple, independent Wnt binding sites on the LRP6 co-receptor suggests new possibilities for the architecture of Wnt signaling complexes and a model for broad-spectrum inhibition of Wnt/β-catenin signaling by Dkk1.
Keywords: Biophysics, Cell Surface Receptor, Lipoprotein-like Receptor (LRP), Protein-Protein Interactions, Signal Transduction, Wnt Pathway, Biochemistry, Dickkopf (Dkk1)
Introduction
Since its discovery more than 30 years ago, the Wnt signaling pathway has captured the interest of researchers due to its role in development, progression of cancer, and self-renewal and differentiation of stem cells (1, 2). Wnt signaling is initiated at the cell surface where secreted Wnt glycoprotein forms a complex with the receptors Frizzled (Fz)2 (3) and low density lipoprotein receptor-related proteins 5 or 6 (LRP5/6) (4, 5). This results in stabilization of intracellular β-catenin (“Wnt/β-catenin pathway”) and its translocation to the nucleus. Nuclear β-catenin then initiates T-cell factor-dependent transcription of downstream Wnt target genes (6, 7). The interactions of LRP5/6 and Fz with Wnt are critical for mediating Wnt/β-catenin signaling (5, 8, 9). Underscoring the central role that these receptors play in Wnt signaling, suppression of either Fz or LRPs has been shown to generate phenotypes associated with Wnt mutations (10). Furthermore, mechanisms have evolved to regulate the Wnt·Fz·LRP complex through secreted proteins that interfere with these extracellular interactions. One such example is Dickkopf-1 (Dkk1), which inhibits Wnt signaling by binding to LRP6 (9, 11, 12). Dkk1 has been shown to play a crucial role in bone formation, vertebrate embryogenesis, and suppression of cancer cell proliferation (13–15). Therefore, Wnt ligands, receptors Fz and LRP, and their natural inhibitors are all of great therapeutic interest. Indeed, recent findings show that the soluble extracellular domain of Fz8 interferes with Wnt-driven tumor growth in vivo (16).
The detailed molecular arrangement of the Wnt·Fz·LRP ternary complex at the cell surface is still unknown, although it has been established that Fz binds to LRP6 in a Wnt-mediated fashion (5) and that this interaction is inhibited by conditioned media containing Dkk1 (12). Importantly, there is no evidence to date that supports formation of the ternary complex from purified extracellular domains, leaving open the possibility that other factors are required. Given that there are 19 Wnts, 10 Fzs, and 2 LRPs in mammals, it is likely that many different ternary complexes form. The combinatorial nature of this system makes it suitable for spatiotemporal control of development in an organism, a well established function of the Wnt pathway. However, there is still ambiguity about whether all of the Wnt·Fz·LRP combinations are biologically relevant (17–20). Moreover, the specificity of physical interactions among different Wnts and their corresponding Fz and LRP5/6 receptors remains largely unaddressed.
Several domains can be predicted with high confidence for the protein components that make up the Wnt signaling complex. Wnt molecules are lipid-modified proteins, possibly associated with lipoproteins and/or lipid vesicles in the extracellular environment (21, 22). Fz proteins are hepta-span transmembrane receptors with an additional conserved N-terminal cysteine-rich domain (CRD) responsible for Wnt binding (3). LRP5/6 receptors are single-span transmembrane proteins composed of a large extracellular domain of four predicted β-propellers, each followed by an epidermal growth factor-like domain. After this segment is an LDL type A repeat (LDLa) that is in close proximity to the transmembrane region. Finally, there is a short intracellular sequence that potentiates the Wnt signal (4, 5, 23).
The extracellular domains of Fz and LRP5/6 receptors are difficult to produce in good yields by utilizing common prokaryotic expression systems. In part, this is because they contain a large number of cysteines that form disulfide bonds in the folded protein. Mammalian expression systems have been used to obtain Fz8 CRD; in the case of LRP6, it is necessary to co-express with the chaperone mesoderm development (MESD) protein to ensure appropriate protein folding (24, 25). Hence, methods for producing high yields of soluble Wnt receptor proteins are needed to enable biochemical investigation of Wnt·Fz·LRP interactions and to facilitate future structural characterization. Because of the current difficulty in obtaining purified protein, researchers have used indirect methods to examine the role of LRP6 domains, such as assessing the effects of overexpression of truncated LRP6 forms on cellular Wnt signaling (9, 12). These studies provide important information but stop short of determining which interactions are direct and what the relevant binding affinities for LRP6 with Wnt or Dkk1 might be, both precursors to a full understanding of the signaling mechanism.
To dissect Wnt signaling at the cell surface, we examined the physical interactions of LRP6, Wnt3a, Wnt9b, Fz8, and Dkk1. Human Dkk1, mouse Fz8 CRD, and various truncated forms of the LRP6 extracellular domain were purified in high yield from insect cells. We established that the proteins are properly folded by showing that, as expected, they can inhibit Wnt/β-catenin signaling. We then measured binding constants for a number of binary complexes, providing clear evidence that the interactions are direct. Using these purified proteins, we reconstituted a Fz8·Wnt3a·LRP6 ternary complex, reflective of a complex that may indeed form at the cell surface. Formation of this complex is inhibited by binding of Dkk1 to LRP6, which blocks the Wnt3a-LRP6 interaction. Remarkably, Wnt3a and Wnt9b can bind to LRP6 simultaneously at distinct sites located within the E3E4 and E1E2 regions, respectively. The relatively straightforward process we describe here could be used, in principle, to characterize every possible Wnt·Fz, Wnt-LRP, and Fz-Wnt-LRP combination and to identify those likely to be biologically most relevant. Furthermore, our assays could be used as a screening platform for identifying ligands that inhibit the formation of Wnt signaling complexes.
EXPERIMENTAL PROCEDURES
Materials
Highly pure human Wnt3a and mouse Wnt5a, Wnt5b, and Wnt9b were obtained as carrier-free proteins (R&D Systems) for use in the binding assays. Mouse Wnt3a was purified from L-cells as described previously (22) for use in cell-based assays, unless otherwise indicated. For cloning and construct boundaries, see the supplemental data.
Protein Purification
Trichoplusia ni cells were maintained in suspension in ESF921 (Expression Systems LLC) at 27 °C as recommended by the vendor. Cells were grown to a density of 2 × 106 cells/ml in ESF921 and infected at an estimated multiplicity of infection of 1. Scale-up production of 5 liters or more was performed using a Wave Bioreactor (GE Bioscience). LRP6, Fz8 CRD, or Dkk1 proteins were expressed as secreted extracellular proteins. At 48 h post-infection, the medium was adjusted with 50 ml of 1 m Tris-HCl, pH 8, 5 ml of 0.25 m CaCl2, and 1 ml of 1 m NiCl2 per liter of medium. After an incubation period of 30 min, cells and debris were spun down at 3000 rpm for 20 min. The supernatant containing the proteins was subsequently filtered (0.22 μm). Nickel-nitrilotriacetic acid resin (1 ml, Qiagen) equilibrated in 50 mm Tris, pH 8, 300 mm NaCl was used for processing of 1 liter of insect cell media containing the His6-tagged proteins. The medium was passed twice through the column to increase the yield of purified protein. The column was washed twice with 20 ml of Tris, pH 8, 300 mm NaCl. Proteins were eluted from the resin by the addition of 4 × 1 ml of the same buffer containing 300 mm imidazole. The collected fractions were concentrated and resolved on a Superdex S200 gel-filtration column (GE Healthcare). Fractions were collected and concentrated to 10 μm stocks. Protein concentration was determined by absorbance at 280 nm.
Cellular β-Catenin Assay
Experiments were conducted with mouse fibroblast L-cells as previously described (26) and detailed in the supplemental data.
Binding Assays
Binding kinetics were measured by biolayer interferometry on an Octet Red instrument (ForteBio). Streptavidin high binding FA or anti-hIgG-Fc capture (AHC) biosensors were loaded with biotinylated hLRP6 or mFz8 CRD-Fc (16) in Tris, pH 8, 300 mm NaCl, 5% (v/v) glycerol, and 0.05% (w/v) Triton X-100. The loaded biosensors were washed in the same buffer before carrying out association and dissociation measurements for the indicated times. Kinetic parameters (kon and koff) and affinities (KD) were calculated from a nonlinear fit of the data using the Octet software Version 6.1; data were then plotted in Kaleidagraph (Synergy Software). Each reported value represents an average of three or more experiments at different concentrations, with a fitted experimental curve for which the correlation coefficient (R2) is above 0.96.
RESULTS
Expression of LRP6 Extracellular Domain and Fz8 CRD in Insect Cells
To study the biochemical interactions of Fz8, LRP6, Wnt3a, and Dkk1, we generated the extracellular domains of human LRP6 and mouse Fz8 as well as full-length human Dkk1. Rather than using transfected mammalian cells that produce low amounts of LRP6, we chose insect cells infected with AcNPV baculovirus to produce the 156-kDa, 47 cysteine-containing extracellular domain of LRP6 (FL ECD) (27, 28). We designed five constructs, choosing domain boundaries based on sequence alignment of the protein from different species, primary sequence analysis using programs such as SMART (29), and secondary structural prediction (Quick2D, MPI toolkit (30)) (supplemental Fig. S1A). To test the roles of the various LRP6 domains in Wnt signaling, we expressed the full-length extracellular domain of LRP6 (LRP6 FL ECD) as well as subfragments of this domain, which include E1E4 (the first four β-propellers with the epidermal growth factor-like domains that follow each of them), E1E2 (the first two β-propellers), and E3E4 (the third and fourth β-propellers) (supplemental Fig. S1A). For Fz8, we cloned and expressed the CRD (31); this domain has been shown both to be sufficient to bind Wnt3a and to be necessary for signaling (3, 16).
After testing expression in two insect cell lines and comparing the LRP6 native secretion signal to the secretion signal of the baculovirus coat protein GP67 (supplemental Fig. S1B), we selected Tni Pro cells and the GP67 secretion signal for expressing LRP6 and Fz8 proteins. Purification was on a nickel-nitrilotriacetic acid affinity column followed by size-exclusion chromatography. As a representative example, the purity of LRP6 E3E4 is shown at every step in the protocol (supplemental Fig. S1C). Proteins were purified to a high degree of homogeneity as determined by SDS-PAGE analysis (supplemental Fig. S1D), and their identities and glycan content were determined by mass spectrometry (supplemental Fig. S1, A and E). Overall yields ranged from 2 mg (LRP6 FL ECD) to 10 mg (Fz8 CRD) of purified protein per liter of expression medium. Thus, we have developed a method allowing access to significant quantities of LRP6 FL ECD and fragments without co-expression of the mesoderm development (MESD) chaperone.
Soluble LRP6 ECD Directly Binds to Wnt3a and Inhibits Wnt3a-mediated β-Catenin Stabilization
To study the interaction between LRP6 and Wnt3a, we developed a binding assay based on biolayer interferometry (32). Site-specifically biotinylated LRP6 E1E4 was obtained by co-expressing a C-terminal His6-Avi-tagged (33) version of LRP6 E1E4 with biotin ligase in insect cells. This version of LRP6 expressed at similar levels as the non-biotinylated form and was easily purified from insect cells. We loaded biotinylated LRP6 E1E4 at 20 μg/ml onto streptavidin biosensors and transferred the sensor tip to Wnt3a solutions of various concentrations. We observe direct binding between LRP6 E1E4 and Wnt3a with a KD of 9 ± 4 nm (Fig. 1A, Table 1), suggestive of a tight complex between LRP6 and Wnt3a on the cell surface. To our knowledge, this is the first demonstration of direct binding between purified LRP6 and Wnt3a.
FIGURE 1.
Soluble LRP6 ECD inhibits Wnt3a-mediated β-catenin stabilization by direct binding to Wnt3a. A, shown is a binding assay between C-terminal biotinylated LRP6 E1E4 (coated on the streptavidin biosensor tip) and Wnt3a in solution (KD = 9 nm). B, shown is the effect of LRP6 fragments on Wnt3a-mediated stabilization of β-catenin. Increasing concentrations of the indicated molecules were added to mouse L-cells in the presence of a constant concentration of Wnt3a (0.6 nm). This concentration was predetermined to stabilize intracellular β-catenin and produce a good signal-to-noise ratio in the assay. FL ECD (IC50 = 480 nm) and E1E4 fragment (IC50 = 360 nm) inhibit β-catenin stabilization, which indicates that the LDLa region is not required. E1E2 and E3E4 have IC50 values exceeding 2 μm, which indicates that the E1E4 region is required for full inhibition.
TABLE 1.
Binding affinities and kinetic constants for various Wnts and Dkk1 binding to LRP6 E1E4, E1E2, E3E4, and Fz8 CRD, as measured by biolayer interferometry
| KD | Kon | Koff | |
|---|---|---|---|
| nm | m−1.s−1.104 | s−1.10−4 | |
| Binding to LRP6 E1E4 | |||
| Wnt3a | 9.1 ± 3.8 | 3.62 ± 0.01 | 3.31 ± 0.01 |
| Wnt5aa | |||
| Wnt5ba | |||
| Wnt9b | 10.5 ± 3.0 | 3.66 ± 0.01 | 3.82 ± 0.01 |
| Dkk1 | 3.0 ± 0.6 | 9.81 ± 0.03 | 2.90 ± 0.01 |
| Binding to LRP6 E1E2 | |||
| Wnt3aa | |||
| Wnt9b | 7.4 ± 1.3 | 2.52 ± 0.01 | 1.85 ± 0.01 |
| Dkk1 | 64.4 ± 16.9 | 18.5 ± 0.01 | 119 ± 0.4 |
| Binding to LRP6 E3E4 | |||
| Wnt3a | 174 ± 10 | 0.38 ± 0.06 | 6.7 ± 0.03 |
| Wnt9ba | |||
| Dkk1 | 20.9 ± 1.5 | 2.24 ± 0.04 | 4.68 ± 0.04 |
| Binding to Fz8 CRD | |||
| Wnt3a | 3.6 ± 1.2 | 2.66 ± 0.01 | 0.96 ± 0.01 |
| Wnt5a | 50.4 ± 12.8 | 1.01 ± 0.41 | 5.07 ± 0.15 |
| Wnt5b | 36.5 ± 7.9 | 1.26 ± 0.33 | 4.59 ± 0.16 |
| Wnt9b | 482 ± 16 | 0.23 ± 0.06 | 11.3 ± 38 |
a No binding.
Next, we tested the biological activity of soluble LRP6 ECD in mouse fibroblast L-cells. For this, we used a cell-based infrared imaging assay recently developed in our laboratory (26). We treated L-cells with Wnt3a in the presence or absence of the various LRP6 fragments and monitored quantitatively β-catenin stabilization. To account for any variations in cell number, the β-catenin signal was normalized to the DNA signal. Our results show that purified soluble LRP6 FL ECD and E1E4 inhibit Wnt signaling in L-cells with IC50 values of 480 ± 50 and 360 ± 90 nm, respectively (Fig. 1B), suggesting that the LDLa domain of LRP6 is not important for Wnt binding. E1E2 (first two β-propellers) and E3E4 fragments (last two β-propellers) did not inhibit Wnt3a signaling at low concentrations in the cell-based assay. However, at concentrations higher than 2 μm, they start to exhibit an inhibitory effect, indicating that their IC50 values may fall in the 10–20 μm range (Fig. 1B). Altogether, the biochemical and cellular results are consistent with a model in which soluble LRP6 directly competes with the cell surface LRP6 receptor for binding to Wnt3a, hence disrupting Fz/Wnt/LRP6 complex formation and resulting in inhibition of cellular β-catenin stabilization.
Dkk1 Competes with Wnt3a for Binding to LRP6
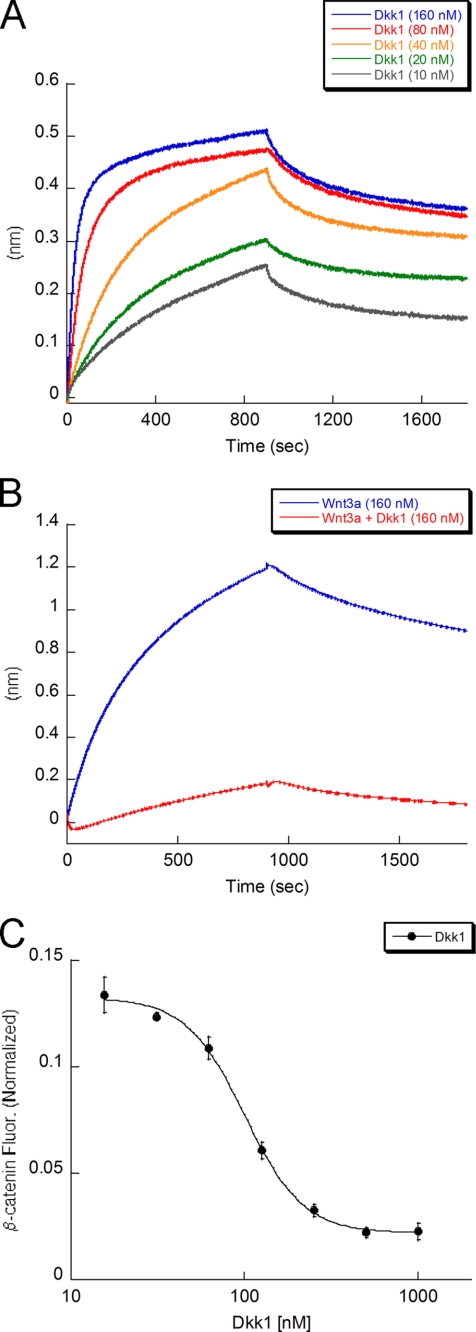
Although Dkk1 is established as an inhibitor of Wnt signaling (9, 12), a direct interaction with LRP6 has not been demonstrated with purified proteins. Instead, earlier reports utilized Scatchard analysis, co-immunoprecipitation, and overexpression of LRP6 deletion mutants to demonstrate that Dkk1 interacts with LRP6 on the cell surface (9, 11, 12). By using biolayer interferometry, we show that full-length soluble Dkk1 does indeed bind directly to LRP6 with a KD of 3 ± 1 nm (Fig. 2A and Table 1). We further demonstrate that Dkk1 inhibits Wnt3a binding to LRP6 (Fig. 2B and supplemental Fig. S2). These data are in agreement with a previous study utilizing Dkk1-conditioned media to study Fz8-Wnt1-LRP6 interactions (12). Taken together, these findings support a model in which Dkk1 antagonism of Wnt3a signaling is due to direct competition with Wnt3a for binding to the LRP6 receptor.
FIGURE 2.
Dkk1 inhibits Wnt3a-mediated β-catenin stabilization by competing with Wnt3a for binding to LRP6. A, shown is a direct binding assay between C-terminal-biotinylated LRP6 E1E4 coated on the streptavidin biosensor and Dkk1 in solution (KD = 3 nm). B, binding of Wnt3a to LRP6 is inhibited in the presence of Dkk1. Dkk1 pre-bound to LRP6 (raw data in supplemental Fig. S2) inhibits the binding of Wnt3a to LRP6. LRP6 is not saturated by Dkk1, resulting in residual Wnt3a binding. C, shown is Dkk1 inhibition of Wnt3a-mediated β-catenin stabilization (IC50 = 100 nm). Wnt3a concentration is 0.6 nm.
Using the β-catenin imaging assay, we show that Dkk1 inhibits Wnt3a-mediated stabilization of cellular β-catenin with an IC50 of 100 ± 5 nm (Fig. 2C). The difference observed between the cellular IC50 and the biochemical KD for Dkk1 and LRP6 may reflect differences in assay sensitivity and readout. Additionally, differences in LRP6 receptor occupancy or organization at the cell surface may decrease sensitivity to these inhibitors.
Soluble Fz8 CRD Binds Directly to Wnt3a and Inhibits Wnt3a-mediated β-Catenin Stabilization
To quantitatively measure the Fz8-Wnt3a interaction, we used a Fz8 CRD-Fc fusion protein (16) and immobilized it onto anti-human Fc biosensors. In this format we observed tight binding between Wnt3a and Fz8 CRD-Fc with a KD of 4 ± 1 nm (supplemental Fig. S3). Using the β-catenin imaging assay, we found that Fz8 CRD (monomer, see “Experimental Procedures” and supplemental Fig. S4) potently inhibits Wnt3a/β-catenin signaling with an IC50 of 0.8 ± 0.2 nm (supplemental Fig. S3B), in agreement with a previously reported IC50 value (16). Therefore, consistent with earlier reports on Fz8 CRD interactions with Wnt ligands (16, 19), inhibition of Wnt/β-catenin signaling by Fz8 CRD is due to direct competition with the endogenous Fz receptors for Wnt3a binding.
Reconstitution of a Fz8 CRD·Wnt3a·LRP6 Triple Complex in Vitro
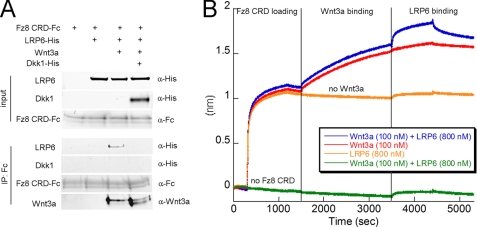
It is now commonly accepted that Wnt3a, Fz8 CRD, and LRP6 form a ternary signaling complex at the cell surface (10, 34). However, it has not been demonstrated that the purified proteins can form this complex. First we sought to reconstitute the Fz8 CRD·Wnt3a·LRP6 complex in solution by mixing Fz8 CRD-Fc, Wnt3a, and LRP6 E1E4. Fz8 CRD-Fc was precipitated with protein G beads, and the precipitate was analyzed by Western blot (Fig. 3A). In the absence of Wnt3a, LRP6 does not coprecipitate with Fz8 CRD. However, in the presence of Wnt3a, we observe coprecipitation of LRP6, suggesting formation of a Fz8 CRD·Wnt3a·LRP6 ternary complex. In agreement with the result above (Fig. 2B), LRP6 E1E4 coprecipitation does not occur in the presence of added Dkk1. These findings are consistent with a model in which Fz8 CRD forms a Wnt3a-mediated complex with LRP6, and Dkk1 prevents LRP6 binding to Wnt3a.
FIGURE 3.
Fz8 CRD and LRP6 form a triple complex mediated by Wnt3a. A, shown is pulldown of purified LRP6 E1E4 by purified Fz8 CRD in the presence of purified Wnt3a. Purified full-length Dkk1 prevents ternary complex formation (fourth lane) by competing with Wnt3a for LRP6 binding. IP, immunoprecipitation. B, shown is direct binding of C-terminal Fc fusion of Fz8 CRD (coated on the anti-human Fc biosensor) to Wnt3a and LRP6. The first step corresponds to Fz8 CRD loading and shows the interaction between the Fc fusion and the anti-human Fc bio-layer. After a washing step, Wnt3a is loaded on Fz8 CRD for an extended period of time to establish formation of the binary complex. The third step corresponds to the binding of LRP6 E1E4 to the preformed Fz8·Wnt3a binary complex (blue trace) in the presence of constant concentration of Wnt3a. LRP6 does not interact with Fz8 CRD in the absence of Wnt3a (orange trace). The green trace serves as a negative control for which there is no loading of Fz8 CRD.
Having demonstrated that Wnt3a binds to both LRP6 E1E4 and Fz8 CRD by using biolayer interferometry (Figs. 1A and supplemental S3A), we investigated the formation of the ternary complex in the same manner. Fz8 CRD-Fc was loaded onto anti-human Fc biosensors. We then established the formation of a Fz8 CRD·Wnt3a binary complex (Fig. 3B) in the presence of Wnt3a (100 nm). Next, the binary complex was exposed to LRP6 E1E4 (800 nm) while maintaining a constant concentration of Wnt3a (100 nm). The observation of a second binding event demonstrates that LRP6 forms a ternary complex with Wnt3a that is already bound to Fz8 CRD (Figs. 3B and supplemental Fig. S5). As a control, we conducted the above steps without adding Wnt3a and detect no interaction between LRP6 and Fz8 CRD (Fig. 3B). These results indicate that LRP6 and Fz8 CRD must bind to different locations on Wnt3a and that LRP6 does not bind to Fz8 CRD in the absence of Wnt3a, in agreement with the immunoprecipitation data (Fig. 3A). It is worth noting that we could not perform this experiment in the reverse format, i.e. by coating LRP6 first on the biosensor tip and measuring Fz8 CRD binding as a function of prebound Wnt3a. This could be due to the display of LRP6 on the biosensor tip hindering binding of Fz8 CRD-Fc. Nevertheless, our studies establish formation of a Fz8 CRD·Wnt3a·LRP6 ternary complex with purified components, both in solution and on a biosensor tip.
Wnt3a and Wnt9b Bind Simultaneously to Distinct Sites on LRP6
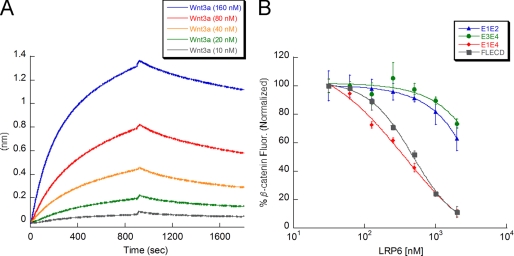
Having established a biophysical assay for probing the interactions of Wnt3a with its receptors, we investigated the binding of other representative Wnts to LRP6 and to Fz8 CRD. Our results demonstrate that Wnt5a and Wnt5b bind with lower affinity than Wnt3a to Fz8 CRD (Wnt5a, KD = 50 nm; Wnt5b, KD = 36 nm; Table 1 and supplemental Fig. S6), whereas Wnt9b only binds very weakly. On the other hand, Wnt5a and Wnt5b do not interact with LRP6, whereas Wnt9b binds tightly to LRP6 with a KD of 11 ± 3 nm (Table 1 and supplemental Fig. S7A). Furthermore, we found that Dkk1 inhibits Wnt9b binding to LRP6 (supplemental Fig. S7, B and C).
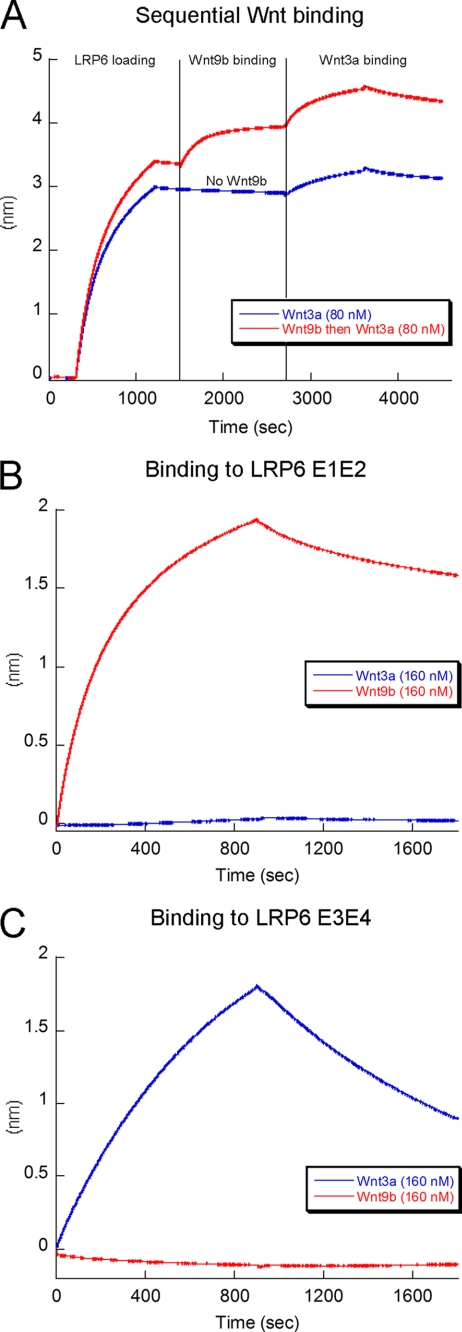
Given that Wnt3a and Wnt9b both bind to LRP6 and that both interactions are inhibited by Dkk1 (Fig. 2B and supplemental Fig. S7, B and C), we hypothesized that Wnt3a and Wnt9b might bind to the same site on LRP6. To test this, we measured whether pre-binding of Wnt9b to immobilized LRP6 blocks Wnt3a binding. Strikingly, our data show instead that Wnt3a binds to LRP6 that is already bound to Wnt9b (Fig. 4A). This finding is remarkable, as it indicates that different binding sites on LRP6 may recruit different Wnts and, provocatively, that simultaneous binding of more than one Wnt may initiate different types of intracellular signaling responses.
FIGURE 4.
Multiple Wnts bind at distinct sites on LRP6. A, shown is prebinding of Wnt9b (red trace) to LRP6 E1E4 does not prevent Wnt3a binding (compare with the blue trace), suggesting two independent Wnt binding sites on LRP6. B, Wnt9b binds tightly to LRP6 E1E2, whereas Wnt3a shows no binding. C, Wnt3a binds to LRP6 E3E4, whereas Wnt9b does not.
To further explore this unexpected result, we investigated binding of Wnt3a and Wnt9b to biotinylated LRP6 E1E2 and E3E4 subdomains. We demonstrate that Wnt9b binds exclusively to E1E2, whereas Wnt3a binds instead to E3E4 (Fig. 4, B and C). This suggests that E3E4 may be the primary region on LRP6 that binds Wnt3a on the cell surface. It is worth noting that Wnt9b affinity for E1E2 (KD = 7 nm, Table 1, and supplemental Fig. S8A) is similar to its affinity for E1E4 (KD = 11 nm). On the contrary, Wnt3a affinity for E3E4 (KD = 174 nm, Table 1 and supplemental Fig. S8B) is significantly decreased compared with its affinity for E1E4 (KD = 9 nm). This observation might be explained by E1E2 being necessary for E3E4 stabilization and, therefore, for full affinity for Wnt3a. The LRP6 E3 region has been reported to be less stable than the other β-propellers (35), which would support this hypothesis.
The existence of independent Wnt3a and Wnt9b sites on LRP6 raises the question of how the relatively small Dkk1 protein can block binding of both Wnts to LRP6. Two possibilities exist; 1) Dkk1 may bind to more than one site on LRP6, or 2) Dkk1 may bind at a single site that is in close proximity to both the Wnt3a and Wnt9b binding sites, such that Dkk1 either sterically interferes with Wnt binding or induces a conformational change on LRP6 that disrupts both Wnt binding sites. To discriminate between these two models, we assessed binding of Dkk1 to LRP6 E1E2 and E3E4 fragments. We found that Dkk1 binds to both fragments with KD values of 64 and 21 nm for E1E2 and E3E4, respectively (Table 1, supplemental Fig. S8, C and D). This supports the conclusion that Dkk1 has multiple binding sites on the LRP6 extracellular domain.
DISCUSSION
The Wnt co-receptors LRP6 and Fz8 are critical for mediating Wnt/β-catenin signaling, but study of their precise physical interactions with Wnt3a has been hampered due to technical challenges with their production. In this report we describe a simple protocol for generating biologically active extracellular domains of these receptors from insect cells. It is likely that a similar protocol could be used to generate other proteins involved in Wnt/β-catenin signaling such as LRP5 and other Fz homologues. Using a cell-based β-catenin imaging assay, we show that the extracellular domains of LRP6 and Fz8 are effective at inhibiting Wnt/β-catenin signaling. Our studies support previous findings about the Wnt inhibitory activity of Fz8-Fc in cells and in vivo models (16). In addition, the LRP6 fragments reported herein add to the toolbox of reagents available to investigate Wnt-receptor interactions.
To corroborate models of Wnt/β-catenin signaling derived from cellular data, we investigated the physical interactions among the Wnt receptors, their ligands, and the Wnt inhibitor Dkk1. The notion that Wnt forms a complex with Fz and LRP5/6 has been established in studies utilizing conditioned media and receptor overexpression (5, 12). However, direct binding with purified components had not been demonstrated. In this study we detect high affinity (nm) interactions between 1) Wnt3a and LRP6 E1E4 and 2) Dkk1 and LRP6 E1E4. Armed with these findings and knowledge of the previously established high affinity interactions between Fz proteins and Wnt ligands (19), we asked whether the Fz/Wnt3a/LRP6 biochemical interaction could be reconstituted from purified components. Our data show the formation of a ternary complex on a biosensor tip and in solution, demonstrating clearly that Wnt3a and the extracellular regions of co-receptors Fz8 and LRP6 are sufficient to mediate complex formation.
As described previously (9, 11, 12, 36), we find that Dkk1 inhibits Wnt/β-catenin signaling (IC50 100 nm). The mechanism of Dkk1 inhibition has been controversial. One idea is that Dkk1 promotes LRP6 internalization that is mediated by Kremen receptors present in certain cell types (37, 38). This was recently challenged by a study showing that Dkk1 antagonizes Wnt signaling without LRP6 internalization (36). However, this latter study did not provide evidence that Dkk1 competes with Wnt3a for binding to LRP6. Our sequential binding data show that neither Wnt3a nor Wnt9b can bind to LRP6 in the presence of saturating Dkk1 and, therefore, suggest strongly that Dkk1 competes directly with Wnt ligands for binding to LRP6 on the cell surface. This is consistent with a model of Dkk1 inhibition that does not require LRP6 internalization. Instead, as concluded previously (12, 36), Dkk1 prevents formation of the Fz8·Wnt3a·LRP6 extracellular signaling complex (Fig. 5A). Dkk1 inhibits binding of LRP6 to Wnt3a but does not disrupt the Wnt3a-Fz8 CRD interaction.
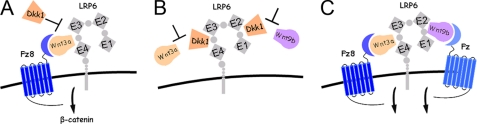
FIGURE 5.
Proposed model for Fz8·Wnt3a·LRP6 complex formation and its regulation by Dkk1. A, Fz8 CRD binding to LRP6 is mediated by Wnt3a binding to E3E4. Dkk1 competes with Wnt3a for binding to LRP6. B, two Dkk1 molecules bind at distinct sites on LRP6 and inhibit binding of both Wnt3a and Wnt9b. C, Wnt3a and Wnt9b simultaneously bind to the LRP6 receptor at distinct sites. One LRP6 molecule may recruit two Wnt·Fz complexes at the cell surface, thereby promoting clustering of Fz receptors and initiating different downstream Wnt responses.
Unexpectedly, we find that LRP6 has at least two independent binding sites for Wnt ligands and for Dkk1. Data supporting this include our observation of simultaneous binding of Wnt3a and Wnt9b to LRP6 E1E4, binding of Wnt3a and Wnt9b to LRP6 E3E4 and E1E2, respectively, and binding of Dkk1 to both E1E2 and E3E4. In addition, we show that Dkk1 that is prebound to E3E4 is no longer able to bind to E1E2 (supplemental Fig. S9), suggesting that a common binding surface on Dkk1 recognizes both sites. We, therefore, propose a more detailed model for Dkk1 inhibition (Fig. 5B) in which two (or more) Dkk1 molecules bind to multiple Wnt binding sites on LRP6. Furthermore, our data suggest that E1E2 and E3E4 may be the primary regions on LRP6 that interact with Wnt9b and Wnt3a on the cell surface, respectively. Our results challenge the prevailing view that Wnt3a interacts with the E1E2 region of LRP6 and that Dkk1 interacts only with LRP6 E3E4 (9). Although Wnt3a has been shown to bind to E1E2 in an enzyme-linked immunosorbent assay (35), this experiment was performed with conditioned media containing E1E2, raising questions about whether other proteins are involved in mediating this binding event. The differences seen between the direct binding studies reported here and interaction analyses based either on enzyme-linked immunosorbent assay (35) or indirect transcriptional readouts (9) stress the importance of well defined biochemical assays using purified proteins.
It is remarkable that Wnt3a and Wnt9b can bind at the same time to two exclusive binding sites on LRP6. It is, therefore, conceivable that one LRP6 receptor recruits two Wnt·Fz complexes (Fig. 5C). Because Wnt3a has a much higher affinity for Fz8 CRD than does Wnt9b, Wnt9b may interact with another Fz protein instead. This would promote clustering of different Fz receptors within a single signaling complex as a way to propagate and enhance the Wnt signal or to initiate different types of downstream responses (17).
In conclusion, we have reconstituted a Fz8·Wnt3a·LRP6 ternary complex and revealed new insight about its molecular arrangement and regulation by Dkk1. It is now clear that Dkk1 inhibits assembly of Wnt3a or Wnt9b signaling complexes by competing with Wnt ligand binding to LRP6. Understanding the molecular details of Dkk1 binding to LRP6 through high resolution structural studies could help in the design of potent Wnt signaling inhibitors that mimic the ability of Dkk1 to bind to multiple biologically relevant sites. These may have therapeutic applications in treating Wnt-related diseases. Finally, our study demonstrates Wnt-receptor interactions with purified proteins allowing further dissection of the biochemical interactions among other Wnts, Fzs, and LRPs.
Supplementary Material
Acknowledgments
We thank Paul Polakis for helpful discussions, Bonnee Rubinfeld for the cDNA of LRP6 and Fz8 CRD, Natalia Arenas Ramirez for help with the cell-based assays, Alberto Estevez, Kyle Mortara, and Krista Bowman for protein expression in insect cells, and Qui Phung and Tommy Cheung for help with the mass spectrometry analysis.

The on-line version of this article (available at http://www.jbc.org) contains supplemental data and Figs. S1–S9.
- Fz
- Frizzled
- LRP
- low density lipoprotein receptor-related protein
- Dkk1
- Dickkopf-1
- CRD
- cysteine-rich domain
- LDLa
- LDL type A repeat
- FL ECD
- full-length extracellular domain.
REFERENCES
- 1.Nusse R. (2008) Cell Res. 18, 523–527 [DOI] [PubMed] [Google Scholar]
- 2.Polakis P. (2007) Curr. Opin. Genet. Dev. 17, 45–51 [DOI] [PubMed] [Google Scholar]
- 3.Bhanot P., Brink M., Samos C. H., Hsieh J. C., Wang Y., Macke J. P., Andrew D., Nathans J., Nusse R. (1996) Nature 382, 225–230 [DOI] [PubMed] [Google Scholar]
- 4.Pinson K. I., Brennan J., Monkley S., Avery B. J., Skarnes W. C. (2000) Nature 407, 535–538 [DOI] [PubMed] [Google Scholar]
- 5.Tamai K., Semenov M., Kato Y., Spokony R., Liu C., Katsuyama Y., Hess F., Saint-Jeannet J. P., He X. (2000) Nature 407, 530–535 [DOI] [PubMed] [Google Scholar]
- 6.Clevers H. (2006) Cell 127, 469–480 [DOI] [PubMed] [Google Scholar]
- 7.van Amerongen R., Mikels A., Nusse R. (2008) Sci. Signal 1, re9. [DOI] [PubMed] [Google Scholar]
- 8.Bilic J., Huang Y. L., Davidson G., Zimmermann T., Cruciat C. M., Bienz M., Niehrs C. (2007) Science 316, 1619–1622 [DOI] [PubMed] [Google Scholar]
- 9.Mao B., Wu W., Li Y., Hoppe D., Stannek P., Glinka A., Niehrs C. (2001) Nature 411, 321–325 [DOI] [PubMed] [Google Scholar]
- 10.He X., Semenov M., Tamai K., Zeng X. (2004) Development 131, 1663–1677 [DOI] [PubMed] [Google Scholar]
- 11.Bafico A., Liu G., Yaniv A., Gazit A., Aaronson S. A. (2001) Nat. Cell Biol. 3, 683–686 [DOI] [PubMed] [Google Scholar]
- 12.Semënov M. V., Tamai K., Brott B. K., Kühl M., Sokol S., He X. (2001) Curr. Biol. 11, 951–961 [DOI] [PubMed] [Google Scholar]
- 13.Niehrs C. (2006) Oncogene 25, 7469–7481 [DOI] [PubMed] [Google Scholar]
- 14.Aguilera O., Fraga M. F., Ballestar E., Paz M. F., Herranz M., Espada J., García J. M., Muñoz A., Esteller M., González-Sancho J. M. (2006) Oncogene 25, 4116–4121 [DOI] [PubMed] [Google Scholar]
- 15.MacDonald B. T., Joiner D. M., Oyserman S. M., Sharma P., Goldstein S. A., He X., Hauschka P. V. (2007) Bone 41, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeAlmeida V. I., Miao L., Ernst J. A., Koeppen H., Polakis P., Rubinfeld B. (2007) Cancer Res. 67, 5371–5379 [DOI] [PubMed] [Google Scholar]
- 17.van Amerongen R., Nusse R. (2009) Development 136, 3205–3214 [DOI] [PubMed] [Google Scholar]
- 18.Wu C. H., Nusse R. (2002) J. Biol. Chem. 277, 41762–41769 [DOI] [PubMed] [Google Scholar]
- 19.Hsieh J. C., Rattner A., Smallwood P. M., Nathans J. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3546–3551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rulifson E. J., Wu C. H., Nusse R. (2000) Mol. Cell 6, 117–126 [PubMed] [Google Scholar]
- 21.Morrell N. T., Leucht P., Zhao L., Kim J. B., ten Berge D., Ponnusamy K., Carre A. L., Dudek H., Zachlederova M., McElhaney M., Brunton S., Gunzner J., Callow M., Polakis P., Costa M., Zhang X. M., Helms J. A., Nusse R. (2008) PLoS ONE 3, e2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willert K., Brown J. D., Danenberg E., Duncan A. W., Weissman I. L., Reya T., Yates J. R., 3rd, Nusse R. (2003) Nature 423, 448–452 [DOI] [PubMed] [Google Scholar]
- 23.Wehrli M., Dougan S. T., Caldwell K., O'Keefe L., Schwartz S., Vaizel-Ohayon D., Schejter E., Tomlinson A., DiNardo S. (2000) Nature 407, 527–530 [DOI] [PubMed] [Google Scholar]
- 24.Hsieh J. C., Lee L., Zhang L., Wefer S., Brown K., DeRossi C., Wines M. E., Rosenquist T., Holdener B. C. (2003) Cell 112, 355–367 [DOI] [PubMed] [Google Scholar]
- 25.Koduri V., Blacklow S. C. (2007) Biochemistry 46, 6570–6577 [DOI] [PubMed] [Google Scholar]
- 26.Hannoush R. N. (2008) PLoS ONE 3, e3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Nocera P. P., Dawid I. B. (1983) Proc. Natl. Acad. Sci. U.S.A. 80, 7095–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith G. E., Summers M. D., Fraser M. J. (1983) Mol. Cell. Biol. 3, 2156–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Biegert A., Mayer C., Remmert M., Söding J., Lupas A. N. (2006) Nucleic Acids Res. 34, W335–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dann C. E., Hsieh J. C., Rattner A., Sharma D., Nathans J., Leahy D. J. (2001) Nature 412, 86–90 [DOI] [PubMed] [Google Scholar]
- 32.Abdiche Y., Malashock D., Pinkerton A., Pons J. (2008) Anal. Biochem. 377, 209–217 [DOI] [PubMed] [Google Scholar]
- 33.Smith P. A., Tripp B. C., DiBlasio-Smith E. A., Lu Z., LaVallie E. R., McCoy J. M. (1998) Nucleic Acids Res. 26, 1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angers S., Moon R. T. (2009) Nat. Rev. Mol. Cell Biol. 10, 468–477 [DOI] [PubMed] [Google Scholar]
- 35.Liu C. C., Pearson C., Bu G. (2009) J. Biol. Chem. 284, 15299–15307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semënov M. V., Zhang X., He X. (2008) J. Biol. Chem. 283, 21427–21432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mao B., Wu W., Davidson G., Marhold J., Li M., Mechler B. M., Delius H., Hoppe D., Stannek P., Walter C., Glinka A., Niehrs C. (2002) Nature 417, 664–667 [DOI] [PubMed] [Google Scholar]
- 38.Yamamoto H., Sakane H., Yamamoto H., Michiue T., Kikuchi A. (2008) Dev. Cell 15, 37–48 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.