Abstract
Transcription factor Kruppel-like factor 4 (Klf4) is essential for somatic cell reprogramming. In addition, Klf4 seems to play a redundant role along with other Klf family proteins in embryonic stem (ES) cell self-renewal. However, how Klf4 regulates ES cell self-renewal and somatic cell reprogramming is still poorly understood. Here we report that Klf4 is required for both ES cell self-renewal and maintenance of pluripotency and that the expression of Klf4 prevents ES cell differentiation in response to withdrawal of leukemia inhibitory factor (LIF) or bone morphogenetic protein 4 (BMP4). In addition, Klf4 directly binds to the promoter region of Nanog and regulates its expression. Expression of Nanog prevents ES cell differentiation even when Klf4 gene expression is knocked down. On the other hand, knockdown of Nanog expression induces differentiation of ES cells that overexpress Klf4. Taken together, these results demonstrate that Klf4 functions upstream of Nanog in ES cell self-renewal and in preventing ES cell differentiation.
Keywords: Cell Differentiation, Chromatin Immunoprecipitation (ChiP), Embryonic Stem Cell, Gene Transcription, shRNA, Kruppel-like Factor 4, Nanog, Differentiation, Pluripotency, Self-renewal
Introduction
Self-renewal and pluripotency are two defining elements of embryonic stem (ES)3 cells. Self-renewal is the capability of ES cells to be maintained in a proliferative state for prolonged periods of time, whereas pluripotency is the ability of ES cells to differentiate into any cell type (1, 2). Understanding the mechanisms that maintain the self-renewal and pluripotency of ES cells is critical to the field of regenerative medicine.
Many transcription factors are involved in ES cell self-renewal and pluripotency regulation. Among them, Oct4, Sox2, and Nanog are thought to be the master regulators of ES cell pluripotency. All three factors have been well studied. Oct4 is a POU homeodomain transcription factor that has been established to be essential in vivo and in vitro for regulation of early embryonic differentiation and maintenance of pluripotency (3, 4). A certain amount of Oct4 is crucial for preventing differentiation and for sustaining ES cell self-renewal (4). Sox2 is a member of the Sox (SRY-related HMG box) gene family and is recognized as a transcriptional partner that works in collaboration with Oct4 to regulate gene expression (5–7). Nanog has been identified as a key factor in maintaining ES cell pluripotency (8, 9). Overexpression of Nanog in ES cells promotes self-renewal in the absence of leukemia inhibitory factor (LIF) (8, 9), a key cytokine for maintenance of ES cell pluripotency. If the Nanog gene is deleted, ES cells are prone to undergo differentiation (9). Nanog is thought to function in concert with other factors such as Oct4 and Sox2 in establishing ES cell identity (10).
Kruppel-like factor 4 (Klf4) belongs to the Krüppel-like factor (Klf) family of conserved zinc finger transcription factors (11). Klf4 is expressed in a variety of tissues and plays an important role in many physiological processes, including proliferation, terminal differentiation, and apoptosis (12, 13). Depending on the gene targeted, Klf4 can either activate or repress transcription and, in certain cellular contexts, it can function as either an oncogene or a tumor suppressor (14–16). The function of Klf4 has been thoroughly investigated in normal homeostasis, cell differentiation, and cancer formation. However, the role of Klf4 in ES cell self-renewal and pluripotency had been neglected until recent studies investigating reprogramming of somatic cells into pluripotent cells highlighted the critical role of Klf4 in remodeling cell fate (17–20). Controlled expression of Klf4, together with Oct4, Sox2, and c-Myc, is able to reprogram adult human fibroblasts to become induced pluripotent stem (iPS) cells that are similar to ES cells. Of the four factors, c-Myc function has been reported as an enhancer of reprogramming and can be removed (21, 22). The importance of Oct4 and Sox2 in ES cell self-renewal is well known but the function of Klf4 has not been as well investigated in previous studies. In fact, Klf4 has been found to be highly expressed in undifferentiated ES cells, with its expression decreasing dramatically during differentiation (23). Re-expression of Klf4 revert EpiSCs to pluripotent ES cell ground state (24). Klf4 and Klf2 have been found to co-localize with Oct4 in a subnuclear compartment of ES cells (23). These results suggest that Klf4 is very likely an important regulator of ES cell self-renewal and pluripotency.
Through both loss-of-function and gain-of-function assays, we report here that Klf4 is a critical factor for ES cell self-renewal and pluripotency. In response to extracellular signaling of LIF, Klf4 expression rapidly changes prior to a change in Nanog expression. At the same time, expression of Oct4 and Sox2 changes very little, apparently having no direct correlation with the change in Nanog expression. Knockdown of Klf4 expression in ES cells induces ES cell differentiation whereas overexpression of Klf4 maintains ES cells in a self-renewal state and negates the requirement for LIF. This indicates that Klf4 may be a direct downstream target of LIF and that Klf4 expression is sufficient for ES cell maintenance in the absence of LIF. The effect of Klf4 on ES cell self-renewal can be interrupted by Nanog shRNA, whereas overexpression of Nanog can rescue ES cell differentiation induced by Klf4 knockdown. This suggests that Klf4 is an upstream regulator of Nanog. Indeed, we show that Klf4 is able to bind to the Nanog promoter in the distal and proximal elements to regulate Nanog expression. Our results support a mechanism in which Klf4 acts as a mediator connecting LIF-Stat3 with Nanog to regulate ES cell self-renewal and pluripotency.
EXPERIMENTAL PROCEDURES
Cell Culture of ES Cells and 293T Cells
ES cell line E14 was cultured on plates coated with 0.1% gelatin and in Glasgow Minimum Essential Medium (GMEM, Sigma) supplemented with 15% fetal bovine serum (FBS), 100 nm nonessential amino acids, 1% sodium pyruvate, 200 mm glutamate, 1% penicillin streptomycin, 50 μm β-mercaptoethanol plus 10 ng/ml LIF (Chemicon). Medium was changed everyday and 10 ng/ml LIF (Chemicon) was added. 293T cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin streptomycin.
Alkaline Phosphatase (AP) Staining and Immunostaining
Alkaline phosphatase staining was performed using an alkaline phosphatase detection kit (Sigma/Chemicon) in accordance with the manufacturer's instructions.
For SSEA1 immunostaining, cultures were fixed in 4% paraformaldehyde (PFA) then washed with phosphate-buffered saline (PBS). After blocking nonspecific binding sites with 10% normal goat serum, ES cells were stained with anti-SSEA-1 mouse monoclonal IgM antibodies (mc-480, University of Iowa, Developmental Studies Hybridoma Bank, 1:500) at room temperature for 1 h. After a PBS wash, the cells were incubated with Cy3-conjugated goat anti-mouse IgM (Jackson Laboratory) or Alexa Fluor 488 (Molecular Probes, Eugene, OR; 1:250) for 45 min. Cells were incubated in DAPI solution (Sigma, 0.05 μg/ml) for 20 min at room temperature for nuclear marking. A Zeiss Axiovert 200 microscope with a DVC-1310C digital camera (DVC, Austin, TX) was used, and images were processed with Photoshop and analyzed with Image-ProPlus software (Media Cybernetics, Silver Spring, MD) to measure Integrated Optical Density (IOD) of color-specific pixels. Statistical analyses were performed by using a two-tailed Student's t test and ANOVA with Instat software (GraphPad Software Inc., San Diego, CA).
Lentivirus Production and Transduction into ES Cells
Lentivirus was produced by using the polyethylenimine (PEI) transfection protocol. Briefly, 5 × 106 293T cells were seeded into a 10-cm plate and left overnight. Cells reached about 80% confluency and were ready to be transfected. Medium was changed 30 min prior to transfection. For a 10-cm plate, 1000 μl of DMEM, 10 mg of DNA (5:3:2, psPAX:vector:pMD2G), and 30 μl of PEI solution (1 mg/ml) were used. Virus was harvested over 48–72 h and concentrated by ultracentrifugation. Virus titer was tested before transduction into ES cells.
For virus transduction, ES cells were trypsinized, counted, and 2 × 106 cells were resuspended into 200 μl of ES culture medium in an Eppendorf tube and concentrated virus particles (1–10 μl) were added to the ES cells. After 1 h of incubation at 37 °C, the remaining medium was added, and the ES cells were plated into 0.1% gelatin-coated 6-well plates. Drug selection with puromycin (1 μg/ml) or neomycin (300 μg/ml) was started 48 h after the ES cells were transduced by viruses, and the culture was then maintained under drug selection conditions.
RT-PCR or qPCR
Total RNA was isolated using an RNeasy kit (Qiagen) according to the manufacturer's instructions and reverse-transcribed to generate cDNA with a SuperScript III First-Strand synthesis system (Invitrogen) using Random Hexamers. The PCRs were performed with Super Therm Taq polymerase. The cDNA pool was subjected to real-time PCR (qPCR) by using iQ SYBR Green Supermix (BioRad) on an AB 7300 Real-time PCR System (Applied Biosystems). The following conditions were used in qRT-PCR: 5 min at 95 °C, and 50 cycles of 15 s at 95 °C and 30 s at 60 °C. β-Actin or GAPDH was tested as an endogenous control to calculate the relative expression levels of target genes according to the manufacturer's instructions. Reactions were carried out in triplicate with the delta Ct method. Primer sequences are given in supplemental Table S1.
Immunoprecipitation and Immunoblotting
Cultures were washed three times with PBS then lysed with KLB buffer (1% Triton X-100, 0.05% SDS, 100 mm Na2HPO4, and 150 mm NaCl) and a protease inhibitor mixture for 30 min at 4 °C. Cells were then scraped and transferred to Eppendorf tubes and centrifuged for 15 min at 14,000 rpm at 4 °C. Protein lysate concentrations were determined by using a Spectramax M5, molecular devices, microplate readers, and bovine serum albumin protein quantification assay. A protein standard curve was made using a bovine serum albumin protein standard from Pierce.
Immunoprecipitation was performed on protein lysates from overexpression studies and endogenous level studies. For endogenous protein interactions, 1000–1500 mg of protein lysates were incubated with primary antibody at 4 °C on a rotator for 4 h, then 10 ml of Pierce immobilized protein A/G beads were added and incubated at 4 °C on a rotator overnight. For overexpression studies some constructs cloned into vectors were attached to an experimentally introduced tag, which further facilitated protein isolation via commercially available tag-conjugated beads. For FLAG-tagged constructs, mouse derived a FLAG M2-agarose beads (Sigma) were used. Once these beads were activated via washing several times with KLB, 10 ml of protein lysate containing 500 mg of protein was added to each Eppendorf tube and put on a rotator overnight at 4 °C. The next day, the beads were washed eight times with KLB, then resuspended with 1× SDS and put in a 95 °C heat block for 5 min. The supernatant was promptly used for Western blotting.
For immunoblotting, 10% acrylamide gels were used for Western blots, including analysis of immunoprecipitates of protein lysates. Separated proteins were transferred to Immobilon-P transfer membrane at 4 °C for 3 h at 250 A, and the membranes were blocked for 1 h in 5% milk/TBST, incubated with primary antibody overnight, washed three times for 10 min, incubated in secondary antibody for 1 h, washed five times for 10 min, incubated in Santa Cruz Biotechnology Western blot Luminescent Reagent for 2 min, exposed to film, and developed by a Kodak X-Omat 2000A Processor.
After primary and secondary antibody incubation followed by film development to view preliminary results, blots were rehydrated in PBS and washed in TBST. Restore Western blot Stripping Solution from Pierce was applied to the blot for 20 min in a 37 °C incubator and then left for an additional 10 min on a shaker at 20 °C. Stripped blots were then re-assayed to confirm that the beads used were indeed pulling down the proper protein. Appropriate antibodies were used to examine the blot for the presence of the corresponding proteins.
Chromatin Immunoprecipitation
Mouse ES cells were chemically crosslinked by the addition of one-tenth volume of fresh 11% formaldehyde solution for 15 min at room temperature. Cells were rinsed twice with 1 × PBS and harvested using a silicone scraper and then flash-frozen in liquid nitrogen and stored at −80 °C. Upon thawing, the cells were lysed by using lysis buffer and the cell contents sonicated to solubilize and shear the cross-linked DNA by using a Sonicator 3000 (Misonix, Farmingdale, NY) in cold room. The resulting whole-cell extract was incubated at room temperature for 1 h with 100 μl of protein G magnetic beads that had been preincubated with the appropriate antibodies against Klf4 or IgG at 4 °C overnight. Beads were washed five times with KLB buffer and one time with TE buffer containing 50 mm sodium chloride. Bound complexes were eluted from the beads by heating at 65 °C with occasional vortexing, and cross-linking was reversed by overnight incubation at 65 °C. Whole-cell extract DNA (reserved from the sonication step) was also treated for cross-link reversal. Immunoprecipitated DNA and whole-cell extract DNA were then purified by treatment with RNase A, proteinase K, and multiple extractions with phenol:chloroform:isoamyl alcohol. Purified DNA was used as a template for PCR to amplify the proximal promoter and distal enhancer regions of Nanog. The primer sequences and locations are listed in supplemental Table S1.
Luciferase Reporter Assay
293T cells were transfected by using CaCl2, along with the reporter plasmid and various expression vectors. Plasmid DNA pcDNA3 was added to the transfections to make the total concentration of DNA the same for each reaction. Six hours after transfection, the cells were fed with fresh medium (DMEM with 10% fetal bovine serum) and incubated overnight. The cells were then serum-starved for 24 h in DMEM containing 0.5% FBS. Cell extracts were then prepared and luciferase assays were done by using a Luciferase Assay System (Promega) according to the manufacturer's instructions. The Spectra Max M5, Molecular Devices, Microplate Reader was used for sequential assay of firefly and Renilla luciferases. Duplicate wells were analyzed.
RESULTS
Klf4 Is Required for ES Cell Self-renewal and Pluripotency
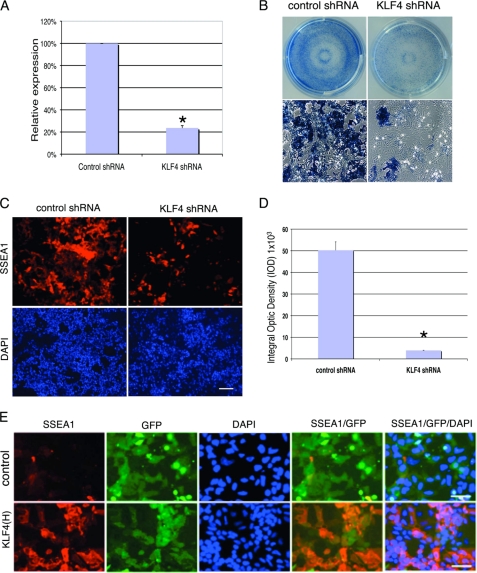
To understand the function of Klf4 in ES cell self-renewal and pluripotency, we knocked down the expression of Klf4 in ES cells. Lentivirus-expressing Klf4 shRNA was introduced into ES cells. Puromycin selection was implemented 2 days after viral infection, and the drug was kept in the culture. After 3–7 days of drug selection, the cells were passaged and cultured for an additional 4 days. Real-time qPCR results revealed that shKlf4 had knocked down the expression of Klf4 by 77.8% (Fig. 1A). Klf4 knockdown led to ES cell differentiation, as indicated by a dramatic reduction in both alkaline phosphatase (AP) (Fig. 1B) and SSEA1 expression (Fig. 1C). Quantitative analysis of SSEA1 staining by measuring the Integral Optic Density (IOD) showed that, compared with the level induced by control shRNA, more than 90% of ES cells were differentiated and became SSEA1-negative (Fig. 1D). Morphologically, Klf4 knockdown ES cells became flat and much larger than undifferentiated cells (data not shown).
FIGURE 1.
Knockdown of Klf4 expression induces differentiation of ES cells. A, quantitative RT-PCR results indicate Klf4 expression was inhibited by Klf4 shRNA by 77.8%. Data are presented as the mean ± S.D. of triplicates. B, AP staining was significantly reduced in Klf4 knockdown cells. Upper panel, whole-well view. Lower panel, higher magnification. C, SSEA1 staining of ES cells was strongly reduced by shKlf4 knockdown. The nuclear signal is shown by DAPI staining. Scale bar: 100 μm. D, quantitative analysis of SSEA1 staining in shKlf4 cells shown in C. SSEA1 signal was quantified by Integral Optic Density (IOD). Data are presented as the mean ± S.D. of triplicates. E, expression of human Klf4 can rescue the knockdown effect of mouse shKlf4 in ES cells. Upper panel, SSEA1 staining was significantly reduced in ES cells infected with shKlf4 and transfected with a control vector plus a GFP expression vector. GFP-positive cells are SSEA1-negative. Lower panel, compared with upper panel, SSEA1 staining increased significantly in ES cells infected with mouse shKlf4 and transfected with human Klf4 plus a GFP expression vector. GFP-positive cells, which expressed human Klf4, are also SSEA1-positive. Scale bar: 50 μm. F, (a) With Klf4 knockdown, expression levels of pluripotency-related genes Nanog, Oct4, and Sox2 decreased. b and c, genes associated with the undifferentiated state and stemness were down-regulated. d, genes associated with the differentiated state were up-regulated. Data are presented as the mean ± S.D. of triplicates.
To confirm the effects of the Klf4-specific shRNA, we performed a rescue experiment using human Klf4. ES cells were transduced by mouse Klf4 shRNA lentivirus and then cultured for 2 days. Then, human Klf4 expression constructs or a control vector were co-transfected with GFP expression vector by electroporation into the Klf4 knockdown ES cells that already contained mouse Klf4 shRNA. The culture was allowed to grow for 5–7 days with puromycin addition before being fixed for staining. In contrast to Klf4 knockdown ES cells transfected with control vector, SSEA1 staining of the cells transfected with human Klf4 was increased significantly (Fig. 1E). This result demonstrated that the sequence of mouse shKlf4 specifically knocked down the mKlf4 gene in ES cells and could be rescued by overexpression of human Klf4.
Gene expression profiling by TLDA card array analysis showed that when Klf4 expression was knocked down most genes associated with pluripotency such as Sox2, Oct4, and Nanog (Fig. 1F, a), genes related to an undifferentiated state, such as TDGF1, GDF3 (Fig. 1F, b), and other stemness genes such as FGF4, FoxD3, LIFR (Fig. 1F, c), were all down-regulated. However, genes related to differentiation such as T, FLT1, CDX2, COL1A1 were up-regulated (Fig. 1F, d). These results suggest that Klf4 prevents ES cell differentiation and is required for ES cell self-renewal and pluripotency.
Klf4 Functions to Prevent ES Cell Differentiation
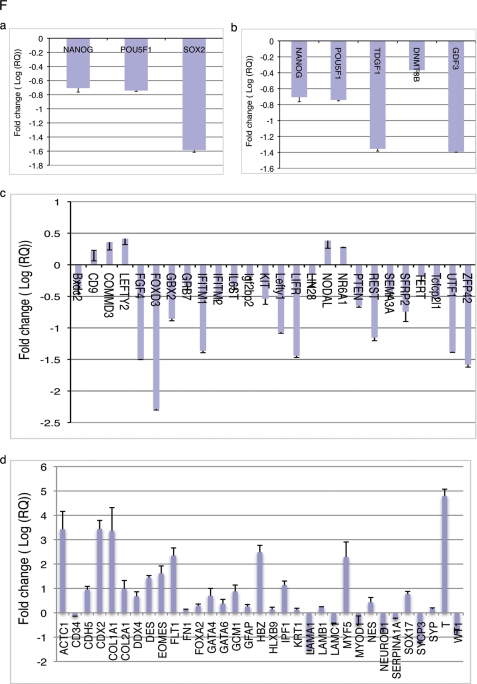
Mouse ES cells are generally cultured in serum supplemented with LIF. LIF is an IL6 family cytokine that activates Stat3 to maintain stem cells in an undifferentiated state (25, 26). Mouse ES cells can also be cultured under serum-free conditions with LIF and BMP4 (27). In serum-free cultures, LIF alone is unable to sustain ES cell self-renewal whereas BMP4 has been shown to aid in maintaining ES cell self-renewal (27). We hypothesized that Klf4 is also downstream of LIF and is able to prevent ES cell differentiation and maintain self-renewal and pluripotency of ES cells. To test this hypothesis, we generated mouse ES cell lines stably overexpressing human Klf4 and cultured the cells in the presence or absence of serum. The overexpression of Klf4 mRNA and protein was confirmed by RT-PCR and Western blot (Fig. 2A), and then the cell line was cultured in medium containing serum with or without LIF. As shown in Fig. 2, B–D, both AP and SSEA1 staining decreased significantly in control ES cells upon LIF withdrawal, and statistical analysis showed that the reduction of SSEA1 staining was about 80%. AP and SSEA1 staining were normal in the Klf4-overexpressing ES cells cultured without LIF (Fig. 2, B and C). These results support that that overexpressed Klf4 was able to maintain mouse ES cells in the pluripotent undifferentiated stage in the absence of LIF.
FIGURE 2.
Overexpression of Klf4 prevents differentiation of ES cells in serum and serum-free culture conditions. ES cells were infected with lentivirus expressing human Klf4 or control vector, and then cultured in differentiation conditions for 4 days. A, Human Klf4 (hKlf4) was stably expressed in ES cells. The mouse Klf4 gene (upper panel) and human Klf4 gene (middle panel) were detected by RT-PCR. The human Klf4 protein was detected by immunoblot (IB) with anti-FLAG tag of Klf4 extracts from ES cells infected with hKlf4 lentivirus (lower panel). B, AP staining of ES cells and ES cells expressing human Klf4 cultured in medium containing serum with or without LIF. C, SSEA1 staining of ES cells overexpressing Klf4 or control ES cells under serum-containing conditions with and without LIF addition. Without LIF, SSEA1 staining in control ES cells strongly decreased. However, SSEA1 staining in Klf4-expressing ES cells did not change despite LIF removal. The scale bar represents a distance of 100 μm. D, quantification of IOD of SSEA1staining. Data are presented as the mean ± S.D. of triplications. E, AP staining of ES cells and ES cells expressing human Klf4 cultured in serum-free (N2/B27) medium with either BMP4 or LIF addition. F, pluripotency gene expression analysis in Klf4-expressing ES cells and control ES cells described in B and E. These cells were cultured in the serum-containing condition with or without LIF supplement, or in serum-free conditions with or without BMP4 or LIF or both. The expression of Nanog, Oct4, Sox2, Klf4, and Nat1 was determined by RT-PCR.
Next we cultured the Klf4-overexpressing ES cells and control ES cells for 5 days in serum-free medium containing N2/B27 supplemented with LIF or BMP4 or both. As expected, AP staining was reduced in the control ES cell culture under either LIF-alone or BMP4-alone conditions (Fig. 2E), indicating that ES cells differentiated only when supplemented with BMP4 or LIF alone. However, AP staining was still normal in ES cells overexpressing Klf4 when cultured under similar conditions, supporting a role for Klf4 in preventing ES cell differentiation. These results were further confirmed by RT-PCR of pluripotency genes in serum or serum-free ES cell cultures with or without addition of LIF or BMP (Fig. 2F). Under serum-free N2/B27 culture conditions, mouse endogenous Klf4 was strongly expressed in ES cells transduced by Klf4, regardless of whether LIF or BMP was added. In addition to Klf4, other genes associated with undifferentiated states of ES cells, specifically Oct4, Sox2, and Nanog, were also strongly expressed. However, in control ES cells, Klf4 was greatly reduced, and Oct4, Sox2, and Nanog were very low in LIF-only and BMP4-only cultures (Fig. 2F). Taken together, these results indicate that Klf4 functions downstream of LIF or BMP4 in regulating self-renewal and pluripotency of ES cells. Overexpression of Klf4 enhances self-renewal and maintenance of the undifferentiated state of ES cells.
Klf4 Expression Dramatically Changes Prior to Nanog Expression in Response to LIF Signaling in ES Cells
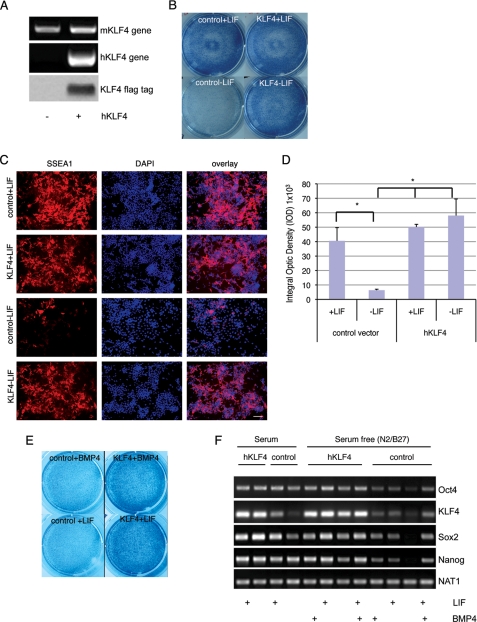
Among many essential pluripotency factors like Oct4 and Sox2, Nanog is another protein that can prevent ES cell differentiation. To address whether Klf4 may function through Nanog to maintain ES cell pluripotency and prevent ES cell differentiation, we sought to determine the expression pattern of Klf4 and Nanog in response to LIF stimulation and withdrawal. ES cells were starved of LIF for 16 h, and then gene expression of pluripotency factors were harvested 0.5, 1, 2, 4, 6, 12, 24, 48, and 72 h after LIF addition. qPCR results showed that expression of Klf4 was quickly induced, producing a small peak at 1 h and maximal expression beginning around 24 h (Fig. 3A). Following Klf4 induction, the expression of Nanog gradually increased to a small peak at around 2 h, subsiding by 6 h, and then increasing again between 12 and 72 h during the first 72 h of LIF induction (Fig. 3A). This experiment suggests that expression of Nanog follows expression of Klf4 in response to LIF stimulation.
FIGURE 3.
Changes in Klf4 expression precede the change in Nanog expression during ES cell differentiation. The expression of Klf4 and Nanog in ES cells and embryoid bodies was determined by quantitative RT-PCR. A, ES cells were cultured under starvation conditions without LIF for overnight. Then LIF was added, followed by a time course analysis of Klf4 and Nanog expression. B, time course analysis of Klf4 and Nanog expression upon LIF withdrawal. C, expression of Sox2 and Oct4 levels did not change upon LIF withdrawal.
When LIF was withdrawn from the medium, the expression of Klf4 declined to its lowest point 2 h after LIF withdrawal, whereas the decline in Nanog expression followed that of Klf4 and reached its lowest level 6-h post-LIF withdrawal (Fig. 3B). It is of interest to note that the expression of Oct4 and Sox2 exhibited no significant decrease during our observation period after LIF was removed from the medium (Fig. 3C). The down-regulation of Nanog expression after LIF removal seemed to have no direct correlation with Oct4 and Sox2 expression.
Taken together, these results show that the expression of Klf4 quick and dramatically changed upon the LIF removal or addition assay. The onset and peak increase of Klf4 expression preceded that of Nanog. The rapid response (shift in expression) of Klf4, the delayed response of Nanog, and the lack of response of Oct4 and Sox2 suggest that Klf4 might be a direct target of LIF signaling and that Nanog might be regulated by Klf4. Klf4 may function as a bridge, connecting LIF signaling with Nanog in regulating ES cell self-renewal and pluripotency.
Klf4 Is Upstream of Nanog in Regulating Self-renewal and Pluripotency and Preventing ES Cell Differentiation
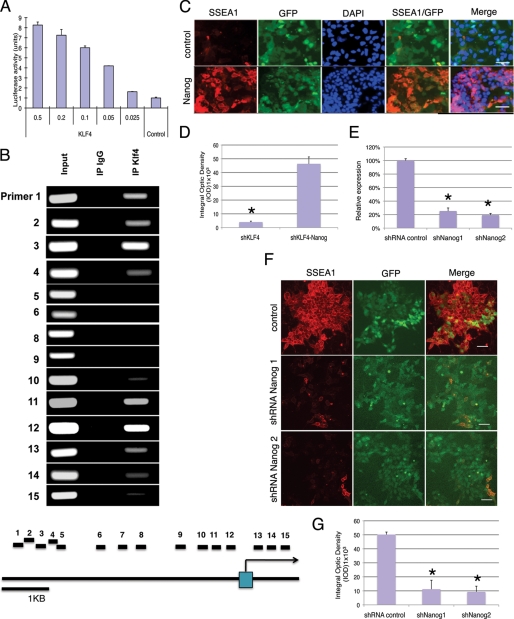
To determine if Klf4 can regulate Nanog gene expression, a luciferase reporter assay was performed on a luciferase gene driven by the Nanog promoter. When this luciferase reporter construct was co-transfected into 293T cells with a construct expressing Klf4, the Nanog promoter was activated by Klf4 in a dosage-dependent manner (Fig. 4A). Furthermore, chromatin immunoprecipitation using Klf4 antibody demonstrated that Klf4 binds to two different sites on the Nanog promoter in ES cells (Fig. 4B). These results support the notion that Nanog is a direct downstream target gene of Klf4 in ES cells.
FIGURE 4.
Klf4 functions upstream of Nanog to prevent ES cell differentiation. A, expression of Klf4 activates the PNanog-luciferase reporter in a dose-dependent manner. B, ChIP analysis indicates that Klf4 binds to both the proximal and distal regions of the Nanog promoter. The location of primers (C) Nanog expression inhibited ES cell differentiation induced by Klf4 shRNA expression. Upper panel, SSEA1 staining was strongly reduced in ES cells infected with shKlf4 and then transfected with control vector. GFP (control vector)-positive cells are not SSEA1-positive. Lower panel, SSEA1 staining increased significantly in Klf4 shRNA-expressing ES cells transfected with a Nanog expression construct. GFP-positive (Nanog-expressing) cells are also SSEA1-positive. Scale bar: 50 μm. D, quantitative analysis of IOD of SSEA1 staining shown in C. Data are presented as the mean ± S.D. of triplicates. E, Nanog shRNA1 and shRNA2 inhibit Nanog expression by 74 and 80%, respectively. Expression of Nanog was determined by real-time RT-PCR. Data are presented as the mean ± S.D. of triplicates. F, Nanog shRNA expression induces differentiation in Klf4-expressing ES cells. A Nanog shRNA expression construct or a control vector was co-transfected with a GFP construct into Klf4-expressing ES cells. SSEA1 staining was performed 4 days after transfection. shNanog-positive cells (middle and lower panels) were SSEA1-negative, whereas control cells (upper panel) were SSEA1-positive. Scale bar: 50 μm. G, quantitation of SSEA1 staining shown in F. Data are presented as the mean ± S.D. of triplicates.
To further validate that Klf4 is upstream of Nanog and functions through Nanog to prevent ES cell differentiation, we performed experiments whether Nanog can rescue the differentiation induced by Klf4 knockdown, and whether Nanog knockdown can induce differentiation in Klf4-overexpressing ES cells. Two days after infection of the Klf4 shRNA lentivirus, a Nanog expression vector was co-transfected with a GFP expression construct into the ES cells. The same as in Fig. 1, SSEA1 expression was dramatically reduced by Klf4 knockdown (Fig. 4C, upper panels). However, SSEA1 expression was strongly increased in the Klf4 knockdown cells by transfection of Nanog expression vectors, and most of the GFP-positive transfected cells were also SSEA1-positive (Fig. 4C, lower panels). SSEA1 intensities in these cells were quantified (Fig. 4D). These results suggest that Nanog can rescue ES cell differentiation induced by Klf4 knockdown and that Nanog functions downstream of Klf4.
To further confirm the correlation of Klf4 with Nanog in ES cells, we performed a parallel experiment in which Nanog expression was knocked down in ES cells overexpressing human Klf4. Two Nanog shRNAs that target two different sequence of Nanog gene were used. Real-time RT-PCR analysis suggested that both shRNAs inhibited Nanog gene expression by 75–80% (Fig. 4E). Each of these Nanog shRNAs were then co-transfected with a GFP expression construct into Klf4-expressing ES cells. Five days after transfection, the ES cells were fixed, and SSEA1 expression was examined. Both Nanog shRNAs significantly reduced the SSEA1 signal in the Klf4-expressing ES cells compared with the control (Fig. 4, F and G). Because both shRNA gave the same phenotype, the phenotype could not be due to the off-target effect of shRNA. This result suggests that reduction of Nanog blocks the effect of Klf4 overexpression on preventing ES cell differentiation. Together, these data further suggest that Klf4 functions upstream of Nanog in preventing ES cell differentiation and in maintaining ES cell pluripotency.
DISCUSSION
In this study, we have demonstrated the important roles of Klf4 in preventing ES cell differentiation and in maintaining ES cell self-renewal and pluripotency. We also demonstrated that Klf4 functions through regulating Nanog gene expression in preventing ES cell differentiation.
Self-renewal and maintenance of pluripotency in mouse ES cells requires LIF (25, 26). LIF is a member of the IL6 cytokine family and is used to maintain ES cell cultures in an undifferentiated state through activation of the Stat3 gene (28, 29). Oct4 and Sox2 have been established as two essential transcription factors that form a heterodimer that binds to the Nanog promoter to regulate the expression of downstream genes that contribute to the maintenance of self-renewal (30). Oct4 and Sox2, however, are not direct targets of Stat3 (31), and experiments showed that Oct4 expression cannot prevent ES cell differentiation in the absence of LIF (4).
Our experiments indicate that Klf4 might be a direct downstream target of LIF-Stat3 signaling, as we observed the rapid response of extracellular LIF signaling in Klf4 expression. This observation is also confirmed by recent publications that Klf4 is a direct downstream target of LIF/Stat3 (32–34). When activated by Stat3 signaling, Klf4 leads to activation of Sox2 expression for the maintenance of ES cells (33).
Klf4 is upstream of Nanog because Klf4 expression alteration was followed by a corresponding change in Nanog expression. Klf4 is also able to bind to the Nanog promoter in the proximal and distal regions thereby regulating the activity of the Nanog promoter. The differentiation of the ES cell induced by Klf4 knockdown is able to rescue by Nanog overexpression, whereas knockdown of Nanog induces differentiation even in Klf4-expressing cells. Taken together, Klf4 play an important role in linking extracellular LIF signaling to the transcription of Nanog to regulate ES cell pluripotency.
We recently reported that Klf4 forms a protein complex with Oct4 and Sox2 by direct protein binding (35). In that report, our sequential ChIP results show that Klf4 may co-localize or may form a complex with Oct4 and Sox2 to regulate Nanog gene expression. Klf4 may help regulate Nanog expression through action on Oct4 and Sox2. Because LIF activates Klf4 expression rapidly, Oct4 and Sox2 may be recruited or assembled to form a complex with Klf4 that co-localizes on the Nanog promoter to regulate downstream stemness gene expression in the presence of LIF. When LIF is removed, Klf4 levels rapidly decrease, the Oct4-Sox2 heterodimer may not bind to the Nanog promoter without assembly factor Klf4, and it may cause Nanog down-regulation. In an alternate explanation, Klf4 may act as an enhancer of the effect of Oct4 and Sox2 on the Nanog promoter. In the absence of Klf4, Oct4 and Sox 2 can still bind to the Nanog promoter to regulate Nanog expression, though the effect is insufficient for maintaining pluripotency.
Oct4, Sox2, and Nanog have been identified as central regulators in the transcriptional hierarchy specifying ES cell identity (10, 36). These factors act in ES cell regulatory circuitry and display a high degree of autoregulation, feed-forward regulation, and overall interconnectivity (10, 37, 38). Whereas previous studies showed that Klf4 also displays autoregulatory loops, Klf4 appears to be exempt from the circuitry dictating the behavior of other central transcription factors (39). Klf4 serves as a master regulator for ES cell core transcription factors by occupying numerous promoters regulating expansive feed-forward loops that include Oct4, Sox2, Nanog, and other common downstream targets. Our results in this and previous reports strongly support this reasoning based on the following three observations. First, Klf4 is much more sensitive to extracellular signaling than are Oct4, Sox2, and Nanog. Second, Klf4 is required for maintaining pluripotency, and overexpression of Klf4 maintains an undifferentiated state independent of LIF. Third, Klf4 interacts with Oct4 and Sox2 to form a complex that binds to the Nanog promoter and positively regulates Nanog expression. Furthermore, no binding sites for Oct4, Sox2, and Nanog are found in the Klf4 promoter region. From this, Klf4 seems exempt from ES cell regulatory circuitry but also plays an important role in ES cell self-renewal and pluripotency through recruiting ES cell regulatory factors responding to extracellular signals.
A previous study by Jiang et al. (40) shows that members of the Klf family (Klf2, Klf4, and Klf5) exhibit a great degree of redundancy and that individual Klfs are therefore not necessary for ES self-renewal. Our results indicate that knockdown of Klf4 alone is enough to induce ES cell differentiation. Discrepancy in RNAi may explain the difference. Through lentiviral infection methods, Klf4 shRNA is stably expressed, resulting in a lasting knockdown of Klf4 function. Insufficient or short term window of Klf4 knockdown may be compensated by Klf2 and Klf5. If Klf4 is sufficiently knocked down, the redundancy effects of Klf2 and Klf5 may be insufficient to maintain ES cell identity. In fact, although Klf5 has been reported to be required for ES cell growth in the inner cell mass of early embryos, Klf5 cannot prevent ES cell differentiation with extended culture, and ES cells still maintain a typical morphology and Nanog expression when Klf5 is disrupted (41).
Despite the fact that Klf2 and Klf5 share 46 and 24% similarity with Klf4, respectively, and although all of them are expressed in ES cells and are down-regulated during ES cell differentiation (42), their regulation behavior on the ES cell is different. For example, Klf4 is a direct LIF/Stat3 downstream target but Klf5 responds to LIF in a Stat3-independent pattern and Klf2 does not respond to LIF (33). Unlike Klf4 regulating cell self-renewal and pluripotency by acting on Nanog, Klf5 functions for ES cell maintenance through suppression of expression of genes related to differentiation. Klf2 and Klf4 can activate the Lefty1 promoter to regulate ES cell identity but Klf5 has no such activity (43). This dissimilarity helps to illustrate the varying roles of independent Klfs and the unique characteristics within the family. Different Klfs may exhibit some level of redundancy and cooperation, but regarding specific cell-regulatory functions, each Klf member may have a discrete role. Put simply, the role of Klf4 on ES cell self-renewal and pluripotency is essential and may not be replaced by another member of the Klf family under natural conditions.
In summary, this study has elucidated the role of Klf4 in ES cell identity, reinforcing and aiding in the confirmation of its importance in the maintenance of pluripotency as well as self-renewal. Klf4 may carry out this function by acting as a fast responding mediator to LIF-Stat3 signal changes, eventually affecting the expression of Nanog, which is well established as a key factor in maintenance of ES cell stemness.
Supplementary Material
Acknowledgments
We thank Kouichi Hasagawa and Jason Tso for critically reading the manuscript.
This work was supported in part by a grant from the California Institute of Regenerative Medicine (CIRM).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- ES
- embryonic stem
- Klf4
- Kruppel-like factor 4
- LIF
- leukemia inhibitory factor
- DMEM
- Dulbecco's modified Eagle's medium
- PBS
- phosphate-buffered saline
- DAPI
- 4′,6-diamidino-2-phenylindole
- PEI
- polyethylenimine
- IL
- interleukin
- GFP
- green fluorescent protein
- BMP
- bone morphogenetic protein
- Stat
- signal transducer and activator of transcription
- ChIP
- chromatin immunoprecipitation assay
- AP
- alkaline phosphatase.
REFERENCES
- 1.Evans M. J., Kaufman M. H. (1981) Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 2.Martin G. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nichols J., Zevnik B., Anastassiadis K., Niwa H., Klewe-Nebenius D., Chambers I., Schöler H., Smith A. (1998) Cell 95, 379–391 [DOI] [PubMed] [Google Scholar]
- 4.Niwa H., Miyazaki J., Smith A. G. (2000) Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 5.Ambrosetti D. C., Basilico C., Dailey L. (1997) Mol. Cell Biol. 17, 6321–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Avilion A. A., Nicolis S. K., Pevny L. H., Perez L., Vivian N., Lovell-Badge R. (2003) Genes Dev. 17, 126–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuan H., Corbi N., Basilico C., Dailey L. (1995) Genes Dev. 9, 2635–2645 [DOI] [PubMed] [Google Scholar]
- 8.Chambers I., Colby D., Robertson M., Nichols J., Lee S., Tweedie S., Smith A. (2003) Cell 113, 643–655 [DOI] [PubMed] [Google Scholar]
- 9.Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 10.Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuh R., Aicher W., Gaul U., Côté S., Preiss A., Maier D., Seifert E., Nauber U., Schröder C., Kemler R. (1986) Cell 47, 1025–1032 [DOI] [PubMed] [Google Scholar]
- 12.Rowland B. D., Bernards R., Peeper D. S. (2005) Nat. Cell Biol. 7, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 13.McConnell B. B., Ghaleb A. M., Nandan M. O., Yang V. W. (2007) Bioessays 29, 946–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowland B. D., Peeper D. S. (2006) Nat. Rev. Cancer 6, 11–23 [DOI] [PubMed] [Google Scholar]
- 15.Chen X., Johns D. C., Geiman D. E., Marban E., Dang D. T., Hamlin G., Sun R., Yang V. W. (2001) J. Biol. Chem. 276, 30423–30428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shields J. M., Christy R. J., Yang V. W. (1996) J. Biol. Chem. 271, 20009–20017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 18.Wernig M., Meissner A., Foreman R., Brambrink T., Ku M., Hochedlinger K., Bernstein B. E., Jaenisch R. (2007) Nature 448, 318–324 [DOI] [PubMed] [Google Scholar]
- 19.Yamanaka S. (2007) Cell Stem Cell 1, 39–49 [DOI] [PubMed] [Google Scholar]
- 20.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. (2007) Cell 131, 861–872 [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa M., Koyanagi M., Tanabe K., Takahashi K., Ichisaka T., Aoi T., Okita K., Mochiduki Y., Takizawa N., Yamanaka S. (2008) Nat. Biotechnol. 26, 101–106 [DOI] [PubMed] [Google Scholar]
- 22.Wernig M., Meissner A., Cassady J. P., Jaenisch R. (2008) Cell Stem Cell 2, 10–12 [DOI] [PubMed] [Google Scholar]
- 23.Bruce S. J., Gardiner B. B., Burke L. J., Gongora M. M., Grimmond S. M., Perkins A. C. (2007) BMC Genomics 8, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo G., Yang J., Nichols J., Hall J. S., Eyres I., Mansfield W., Smith A. (2009) Development 136, 1063–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith A. G., Heath J. K., Donaldson D. D., Wong G. G., Moreau J., Stahl M., Rogers D. (1988) Nature 336, 688–690 [DOI] [PubMed] [Google Scholar]
- 26.Williams R. L., Hilton D. J., Pease S., Willson T. A., Stewart C. L., Gearing D. P., Wagner E. F., Metcalf D., Nicola N. A., Gough N. M. (1988) Nature 336, 684–687 [DOI] [PubMed] [Google Scholar]
- 27.Ying Q. L., Nichols J., Chambers I., Smith A. (2003) Cell 115, 281–292 [DOI] [PubMed] [Google Scholar]
- 28.Matsuda T., Nakamura T., Nakao K., Arai T., Katsuki M., Heike T., Yokota T. (1999) EMBO J. 18, 4261–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa H., Burdon T., Chambers I., Smith A. (1998) Genes Dev. 12, 2048–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodda D. J., Chew J. L., Lim L. H., Loh Y. H., Wang B., Ng H. H., Robson P. (2005) J. Biol. Chem. 280, 24731–24737 [DOI] [PubMed] [Google Scholar]
- 31.Kidder B. L., Yang J., Palmer S. (2008) PLoS One 3, e3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bourillot P. Y., Aksoy I., Schreiber V., Wianny F., Schulz H., Hummel O., Hubner N., Savatier P. (2009) Stem Cells 27, 1760–1771 [DOI] [PubMed] [Google Scholar]
- 33.Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R., Tomlinson S., Smith A. (2009) Cell Stem Cell 5, 597–609 [DOI] [PubMed] [Google Scholar]
- 34.Niwa H., Ogawa K., Shimosato D., Adachi K. (2009) Nature 460, 118–122 [DOI] [PubMed] [Google Scholar]
- 35.Wei Z., Yang Y., Zhang P., Andrianakos R., Hasegawa K., Lyu J., Chen X., Bai G., Liu C., Pera M., Lu W. (2009) Stem Cells 27, 2969–2978 [DOI] [PubMed] [Google Scholar]
- 36.Orkin S. H. (2005) Cell 122, 828–830 [DOI] [PubMed] [Google Scholar]
- 37.Masui S., Nakatake Y., Toyooka Y., Shimosato D., Yagi R., Takahashi K., Okochi H., Okuda A., Matoba R., Sharov A. A., Ko M. S., Niwa H. (2007) Nat. Cell Biol. 9, 625–635 [DOI] [PubMed] [Google Scholar]
- 38.Odom D. T., Dowell R. D., Jacobsen E. S., Nekludova L., Rolfe P. A., Danford T. W., Gifford D. K., Fraenkel E., Bell G. I., Young R. A. (2006) Mol. Syst. Biol. 2, 2006.0017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang J., Chan Y. S., Loh Y. H., Cai J., Tong G. Q., Lim C. A., Robson P., Zhong S., Ng H. H. (2008) Nat. Cell Biol. 10, 353–360 [DOI] [PubMed] [Google Scholar]
- 41.Ema M., Mori D., Niwa H., Hasegawa Y., Yamanaka Y., Hitoshi S., Mimura J., Kawabe Y., Hosoya T., Morita M., Shimosato D., Uchida K., Suzuki N., Yanagisawa J., Sogawa K., Rossant J., Yamamoto M., Takahashi S., Fujii-Kuriyama Y. (2008) Cell Stem Cell 3, 555–567 [DOI] [PubMed] [Google Scholar]
- 42.Bruce S. J., Gardiner B. B., Burke L. J., Gongora M. M., Grimmond S. M., Perkins A. C. (2007) BMC Genomics 8, 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakatake Y., Fukui N., Iwamatsu Y., Masui S., Takahashi K., Yagi R., Yagi K., Miyazaki J., Matoba R., Ko M. S., Niwa H. (2006) Mol. Cell Biol. 26, 7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.