Abstract
RPK1 (receptor-like protein kinase 1) localizes to the plasma membrane and functions as a regulator of abscisic acid (ABA) signaling in Arabidopsis. In our current study, we investigated the effect of RPK1 disruption and overproduction upon plant responses to drought stress. Transgenic Arabidopsis overexpressing the RPK1 protein showed increased ABA sensitivity in their root growth and stomatal closure and also displayed less transpirational water loss. In contrast, a mutant lacking RPK1 function, rpk1-1, was found to be resistant to ABA during these processes and showed increased water loss. RPK1 overproduction in these transgenic plants thus increased their tolerance to drought stress. We performed microarray analysis of RPK1 transgenic plants and observed enhanced expression of several stress-responsive genes, such as Cor15a, Cor15b, and rd29A, in addition to H2O2-responsive genes. Consistently, the expression levels of ABA/stress-responsive genes in rpk1-1 had decreased compared with wild type. The results suggest that the overproduction of RPK1 enhances both the ABA and drought stress signaling pathways. Furthermore, the leaves of the rpk1-1 plants exhibit higher sensitivity to oxidative stress upon ABA-pretreatment, whereas transgenic plants overproducing RPK1 manifest increased tolerance to this stress. Our current data suggest therefore that RPK1 overproduction controls reactive oxygen species homeostasis and enhances both water and oxidative stress tolerance in Arabidopsis.
Keywords: Arabidopsis, Membrane/Proteins, Oxygen/Reactive, Phosphorylation/Kinases/Serine-Threonine, Receptors/Threonine-Serine Kinases, Signal Transduction/Protein Kinases/Serine/Threonine, Abscisic Acid (ABA), Reactive Oxygen Species (ROS), Receptor-like Kinase, Drought Stress
Introduction
The acclimation of plants to ever-changing environmental conditions is mediated by an orchestrated but complex series of signaling networks that regulate cellular and molecular events. Water deficit conditions affect plant growth and development via various biological and physiological processes and thereby limit plant productivity. Improving the water deficit stress tolerance of plants is thus important for increasing crop productivity and yields. An increased understanding of the regulatory mechanisms underlying the plant responses to water deficit stress will provide key insights for future molecular breeding of such stress-tolerant plants. Under stress conditions, the expression of a variety of genes that function not only in stress tolerance but also in stress responses is up-regulated. The plant hormone abscisic acid (ABA)2 regulates a range of physiological events, including seed maturation and dormancy, the control of vegetative growth, and the tolerance of plants to various abiotic stresses, such as drought, salinity, and cold (1). ABA is dramatically produced under water deficit conditions and regulates the expression of various genes that also respond to water deficit stress. It has been shown that the bZIP-type transcription factors regulate the activation of the ABA-responsive gene expression in Arabidopsis (2). Other families of transcription factors, such as MYC, MYB, homeodomain protein, NAC, and AP2/EREBP, have been shown to play roles in ABA responses in plants (2).
To further elucidate the ABA signaling mechanisms, genetic and physiological analyses have been performed previously, and both negative and positive regulators of ABA signal transduction pathways have been identified (3). A number of recessive mutations resulting in insensitivity and hypersensitivity to ABA have been isolated, and the factors involved have been shown play important roles in ABA signal transduction as both positive and negative regulators (1, 4). Recently, also novel ABA receptor proteins, the PYR/PYL/RCARs (5, 6) and GTG1/2 (7), have been reported. Interactions between the PYR/PYL/RCARs and the protein phosphatase 2Cs are found to be stimulated by ABA and to control its signaling by inhibiting protein phosphatase 2C activity (5, 6). The PYR/PYL/RCARs encode the START proteins and are functionally redundant in terms of ABA perception (5, 6). Previous studies have also indicated that the relatively low number of recessive ABA-insensitive mutants is due to such genetic redundancy. Furthermore, interactions among signaling cascades involved in abiotic stress responses, such as those for drought, salinity, and cold, and between abiotic and the other pathways, such as biotic, phytohormone, and sugar responses, affect the complexity of the signaling networks and may cause functional redundancy among the components involved. Reactive oxygen species (ROS) are partially reduced or activated forms of oxygen (O2) and include superoxide free radicals, such as H2O2, and hydroxyl radicals, all of which are toxic and can cause the oxidative destruction of cells. It has also been suggested, however, that ROS also function as key regulators in plant growth and development, cell cycle, hormone signaling, biotic and abiotic stress responses, and programmed cell death (8–10). ROS production is induced by both abiotic and biotic stress insults, such as high light, osmotic stress, and pathogen attack. ROS have now been implicated as important second messengers of ABA signaling in guard cells (11–13). The guard cells of an Arabidopsis double mutant for the membrane-bound NADPH oxidase catalytic subunits, atbohD/atbohF, were shown to be impaired in ABA-induced H2O2 production and show a partially repressed ABA-induced stomatal closure (12). SRK2E/OST1 and ABI1, which catalyze phosphorylation and dephosphorylation during ABA signaling, control ABA-stimulated stomatal closure via ROS production (11, 14). Furthermore, recent studies have indicated that the activation of an antioxidant system for detoxification of ROS increases tolerance to drought stress (15, 16).
Receptor-like kinases (RLKs) belong to a large plant gene family (17) and contain an extracellular domain, a single transmembrane domain, and a cytosolic Ser/Thr protein kinase domain. The RLKs are classified on the basis of their putative extracellular ligand-binding domains, and the leucine-rich repeat receptor-like kinases are the largest group of RLKs within the Arabidopsis genome, with more than 200 members (18). Current studies indicate that they function in diverse signaling events in Arabidopsis, such as brassinosteroid perception by BRI1 and BAK1 (19); perception of bacterial flagellin fragments by FLS2 (20), which mediates a pathogen response; peptide plant hormone phytosulfokine perception by PSKR (21); and recognition of the CLE family peptide hormone, CLV3, by CLV1 (22). RPK1 is a leucine-rich repeat receptor-like kinase isolated from Arabidopsis, and its gene expression is induced by ABA, dehydration, high salt, and low temperatures (23, 24). To determine the function of RPK1 in ABA signaling in plants, we have previously isolated RPK1-knock-out mutants and constructed antisense-RPK1 transgenic lines. The loss of function of RPK1 shows ABA insensitivity in seed germination, plant growth, stomatal closure, and gene expression in Arabidopsis (24). In our present study, we generated transgenic Arabidopsis overproducing the RPK1 protein. We then compared the effects of RPK1 disruption and overproduction in Arabidopsis plants upon their sensitivity to water-deficit conditions and investigated whether RPK1 overproduction enhances the drought stress response pathways in these transgenic plants. We also show that transgenic plants overproducing RPK1 have an increased tolerance to drought stress as well as oxidative stress.
EXPERIMENTAL PROCEDURES
Plant Materials and Growth Conditions
Seeds of wild type Arabidopsis thaliana (Columbia; Co ecotype and Wassilewskija; WS ecotype), rpk1-1 (24), srk2e (25), and nced3-2 (26) mutants and transgenic plants were surface-sterilized, sown on germination medium (half-strength Murashige and Skoog salt and vitamin medium supplemented with 0.8% (w/v) agar and 1% (w/v) sucrose), and grown in a growth chamber at 2500 lux at 22 °C under a 16-h light/8-h dark photoperiod. Transgenic and control plants were grown on GM-agar plates containing kanamycin (20 mg/liter).
RNA Analyses
Total RNAs were extracted from the leaves or whole plants of 3-week-old Arabidopsis grown in soil pots or on GM-agar plates treated with or without ABA or H2O2 with TRIzol reagent (Invitrogen). RNA gel blot analysis was performed as described previously (27). For real-time quantitative RT-PCR, reverse transcription of the RNA samples was carried out with SuperScript III RNase H-reverse transcriptase (Invitrogen) with random hexamer primers. Real-time quantitative PCR was performed using real-time PCR system 7300 (Applied Biosystems, Foster City, CA) and SYBR Premix Ex Taq (Takara, Shiga, Japan). The primer pairs used for real-time PCR are listed in supplemental Table 1. Melting curve analysis confirmed that only one product was amplified. Specific cDNA was quantified with a standard curve based on known amounts of each amplified cDNA fragment. 18S rRNA was also amplified for calibration purposes, and three determinations were performed for each sample. We performed three biological replicates for real-time quantitative RT-PCR. Microarray analysis was performed using the Agilent Technologies (Palo Alto, CA) Arabidopsis 22K oligonucleotide array. Total RNA was used for the preparation of Cy5- and Cy3-labeled cDNA probes. The Arabidopsis 2 Oligo Microarray kit (Agilent Technologies) was used to compare transcript profiles as described previously (28).
Protein Extraction and Protein Gel Blot Analysis
To prepare total protein extracts, 3-week-old transgenic plants were ground in liquid nitrogen and thawed in extraction buffer (25 mm Tris-HCl, pH 7.0, 5 mm MgCl2, 1 mm CaCl2, 0.25 m sucrose, and a protease inhibitor mixture (Roche Applied Science)) using a mortar and pestle. The extracts were then centrifuged at 10,000 × g for 10 min at 4 °C, and the microsomal fraction was isolated by centrifugation of the supernatant at 100,000 × g for 60 min at 4 °C. The resulting microsomal membrane fraction was suspended in extraction buffer containing 0.5% Triton X-100. The concentration of extracted proteins was determined using the Bio-Rad protein assay. The microsomal fractions were then separated by SDS-PAGE and blotted onto a phenylmethylsulfonyl fluoride membrane. The RPK1 protein was detected with anti-RPK1 peptide antibody (24), anti- phosphothreonine antibody from Zymed Laboratories (Invitrogen), or monoclonal anti-hemagglutinin (HA) antibody (Sigma). Signals were developed using Pierce SuperSignal West Dura extended duration substrate (Thermo Fisher Scientific).
Construction of Transgenic Plants
The full-length cDNA fragment of RPK1 was cloned into the pGreenII vector (29), which contains the modified cauliflower mosaic virus 35S promoter and the Ω-sequence of the tobacco mosaic virus (30). A double HA-His tag sequence was obtained by annealing the primers 5′-ggccgcttacccatacgacgttccagactacgctggttacccatacgacgttccagactacgctagatccggtcaccaccaccaccaccactaagagct-3′ and 5′-cttagtggtggtggtggtggtgaccggatctagcgtagtctggaacgtcgtatgggtaaccagcgtagtctggaacgtcgtatgggtaagc-3′ and ligating into NotI- and SacI-digested pGreenII vector to produce HA/His fusion. The RPK1 sequence was amplified by PCR using the primers 5′-tctttctcttgtgaagcggccgctgaaaaatgaaact-3′ and 5′-ttcttctagcggccgcacacaatctagaaggctggat-3′, digested with NotI, and then cloned into the corresponding sites in pGreenII to obtain the expression plasmid 35S:RPK1 for plant transformation. This vector was introduced into Agrobacterium tumefaciens C58 cells, and plants were transformed as described previously (24). A single mutation was introduced into the RPK1 kinase domain by substituting lysine 289 with glutamic acid (K289E) to generate the mutant kinase protein (mRPK1). The mRPK1 fragment was cloned into the pGreenII vector to express this protein in Arabidopsis. To generate transgenic plants containing RPK1 promoter constructs, we cloned the RPK1 promoter region (24) also into pGreenII containing RPK1-HA (RPK1pro:RPK1) or mRPK1-HA (RPK1pro:mRPK1) inserts. We subsequently obtained 30 transgenic T2 lines overexpressing the RPK1 gene using each fusion vector. The lines showing the highest exogenous RPK1 expression were then selected for further experiments.
Preparation of Microsomal Membranes from Arabidopsis and Immunoprecipitation
Three-week-old transgenic plants were ground in liquid nitrogen and homogenized in an extraction buffer containing 50 mm Tris-HCl (pH 7.5), 5 mm MgCl2, 0.25 m sucrose, 1 mm EDTA with proteinase inhibitor mixture (Roche Applied Science). The homogenate was filtered through Miracloth and centrifuged at 10,000 × g for 20 min, and the supernatant was then ultracentrifuged at 100,000 × g for 30 min to pellet the microsomal membrane. This pellet was then solubilized with PreserveX™-QML (QBI Life Sciences, Madison, WI). RPK1 proteins were immunoprecipitated from the solubilized microsomal membranes with HA-agarose (Sigma).
ABA Sensitivity of Arabidopsis Root Growth
Arabidopsis seeds of wild type, rpk1-1, and 35S:RPK1 plants were sterilized and germinated on GM. Five-day-old young seedlings were then transferred to fresh MS medium containing 1% sucrose and various concentrations of ABA. The root length was measured after 10 days of culture.
Stomatal Movement Assay and Measurement of ABA Content
Stomatal movement assays were performed essentially as described previously (8). Briefly, rosette leaves of 4-week-old wild type, rpk1-1, 35S:RPK1 and control plants grown on soil at 22 °C under a 16-h light/8-h dark photoperiod in 50% RH were harvested and incubated for 2 h in a solution containing 10 mm KCl, 0.2 mm CaCl2, and 10 mm MES-KOH (pH 6.15) under white light (8). These leaves were subsequently transferred to a solution containing the same buffer and ABA and incubated for a further 2 h. Guard cells were photographed under a color laser three-dimensional profile microscope (Keyence, Tokyo, Japan). Twenty stomatal apertures were measured for each leaf experiment. The endogenous ABA contents of rosette leaves of 4-week-old wild type, rpk1-1, 35S:RPK1, and control plants were measured as previously described (31).
Resistance to Drought Stress
Survival rates associated with drought tolerance levels were measured as described previously (32) with minor modifications. Briefly, 2–3-week-old plants were grown on GM-agar plates at 22 °C under a 16-h light/8-h dark photoperiod and then transferred to filter paper on dishes. These plants were then left unwatered for 8 h in 30% RH under continuous white light and then rewatered for 2 days. The plants were then photographed, and the numbers of surviving green plants were calculated. In addition, 4-week-old plants grown on soil pots at 22 °C under a 16-h light/8-h dark photoperiod in 30–40% RH were subjected to dehydration stress by lack of watering. After 2 weeks of withholding water, the plants were rewatered for 1 week and then photographed. The surviving green plants were then counted. Thirty plants were used in each experiment, and all experiments were repeated three times.
Analyses of fresh weight loss were performed using detached rosette leaves at the same developmental stage and size as 3-week-old plants. Three leaves per plant were excised and maintained in a growth chamber in 30% RH. Fresh weights were measured at the indicated periods of time, and five plants of each genotype and transgenic line were analyzed in independent experiments repeated five times.
Detection of Reactive Oxygen Species
H2O2 was detected in leaves using 3,3′-diaminobenzidine staining as described previously (33). Briefly, the first fully expanded leaf from both a 5-week-old wild type and a 35S:RPK1 plant were vacuum-infiltrated with 3,3′-diaminobenzidine solution (1 mg/ml, pH 3.8; Sigma). The leaves were then placed in a plastic box under high humidity until a reddish brown precipitate was observed and then fixed with a solution of 3:1:1 ethanol/lactic acid/glycerol and photographed.
H2O2 Sensitivity and Superoxide Dismutase Activity Assay
Detached fully expanded leaves from the rosettes of 4-week-old wild type, rpk1-1, 35S:RPK1, and control plants grown on soil at 22 °C under a 16-h light/8-h dark photoperiod in 50% RH were harvested and incubated for 3 days in solution with or without H2O2 under white light. These leaves were also pretreated with 50 μm ABA for 3 h and then transferred to H2O2 solution. Photochemical efficiency (Fv/Fm ratio) measurements were taken after 3 days of treatment with H2O2 using a PAM chlorophyll fluorometer (Walz, Effeltrich, Germany), after the leaves had been allowed to dark-adapt for 60 min. Chlorophyll content measurements in the leaves were performed as described by Arnon (34) by measuring the absorbance at 664 and 647 nm of 80% acetone extracts. Twenty leaves of each genotype were treated and measured for each condition. All experiments were repeated three times. Superoxide dismutase activity was determined as described (35).
Stastical Analysis
Statistical analyses were performed using either Student's t test or one-way analysis of variance (ANOVA), followed by Turkey's honestly significant difference (HSD) test using Statistical Package, version 7.0.1 (Japanese edition; JMP Software).
Accession Numbers
Arabidopsis Genome Initiative locus identifiers for the genes described in this article are as follows: RPK1 (At1g69270), FRK1 (At2g19190), AtGLR2.7 (At2g29120), Cor15a (At2g42540), NCED3 (At3g14440), SRK2E (At4g33950), Kin2 (At5g15970), RD29A (At5g52310), and AtrbohD (At5g47910).
RESULTS
RPK1 Overproduction Increases ABA Sensitivity in Arabidopsis
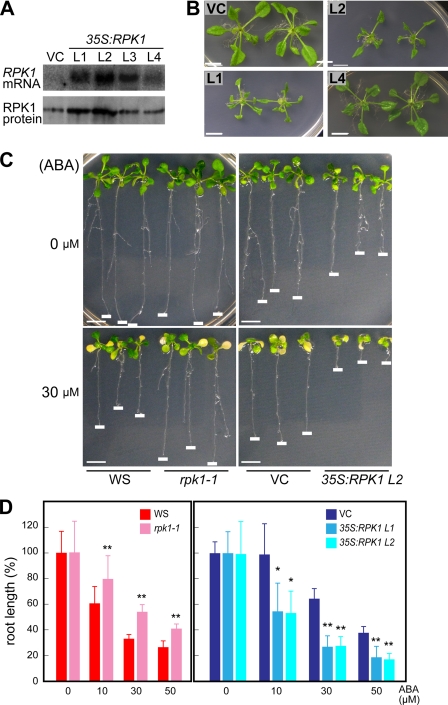
To further understand the molecular mechanisms controlling the RPK1-mediated ABA signaling pathway, we generated transgenic Arabidopsis plants (Co ecotype), which overexpress the RPK1 gene fused to an HA tag in the expression vector under the control of the cauliflower mosaic virus 35S promoter. Thirty transgenic T2 plants were subjected to RPK1 expression analysis by Northern blotting. Transgenic lines overexpressing RPK1 (35S:RPK1) were selected, and their homozygous T3 plants were used for further analysis. By both Northern and protein blot analyses, we detected the highest levels of RPK1 expression in transgenic lines L1 and L2, as compared with wild type control plants transformed with the empty vector (Fig. 1A). We also generated transgenic lines overexpressing non-tagged RPK1 under the control of the 35S promoter in Arabidopsis but could not obtain lines overproducing the RPK1 protein (data not shown). As shown in Fig. 1B, the 35S:RPK1 plants showed weak growth retardation (supplemental Fig. 1). Previously, we reported that rpk1 mutants manifest impaired root growth inhibition when exposed to ABA. We thus analyzed root growth inhibition by ABA in our current experiments using the 35S:RPK1 lines L1 and L2 and compared these results with those obtained for the rpk1 mutant. The negative effects of ABA on root growth were found to be reduced in the rpk1 mutants, whereas the 35S:RPK1 plants showed increased sensitivity to ABA (Fig. 1, C and D). The roots of the rpk1-1 plants grew faster than those of the wild type plants on ABA-containing media, whereas the root growth of 35S:RPK1 plants was more severely inhibited by ABA compared with the control plants.
FIGURE 1.
ABA sensitivity of transgenic Arabidopsis overexpressing RPK1. A, expression levels of RPK1 in transgenic plants. Total RNAs were extracted from transgenic plants carrying the 35S vector (vector control (VC)) and from four transgenic lines of 35S:RPK1 (L1–L4; Columbia ecotype) and were blotted and hybridized with a cDNA probe for RPK1. Protein blot analysis was performed with microsomal fractions of 35S:RPK1 lines (L1–L4) using an anti-RPK1 antibody (23). B, 35S:RPK1 (lines L1–L3) and vector control plants grown on GM-agar plates for 2 weeks. C and D, assay for root growth inhibition by ABA. Young seedlings of 35S:RPK1 (lines L1 and L2), vector control, wild type Arabidopsis (WS), and rpk1-1 were transferred to ABA-containing medium and incubated for 10 days. The relative root lengths were measured and are shown as a percentage of the root lengths grown in the absence of ABA. Values are the means ± S.D. (n = 50). The asterisks indicate statistically significant differences in comparisons between WS and rpk1-1, determined by Student's t test, or in comparisons between vector control and 35S:RPK1, evaluated by one-way ANOVA with Tukey's HSD (*, p < 0.0001; **, p < 0.005). Bars, 1 cm.
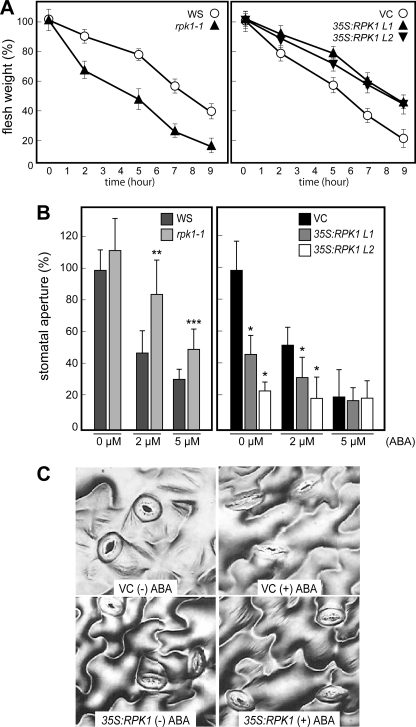
ABA signaling plays a crucial role in reducing water loss by regulating stomatal closing under conditions of drought stress. To investigate the role of RPK1 in this process, we measured the fresh weight loss in detached leaves from rpk1-1 and 35S:RPK1 plants (Fig. 2A). The disruption of RPK1 resulted in an increased water loss rate when compared with the wild type WS ecotype. In contrast, the overproduction of RPK1 in transgenic Arabidopsis led to a decrease in water loss. To further examine the nature of the ABA sensitivity that controls water loss during drought stress, we analyzed stomatal movement responses to ABA in the 35S:RPK1 plants (Fig. 2, B and C). Compared with the wild type controls, the guard cells of 35S:RPK1 plants exhibited smaller stomatal apertures without ABA treatment, whereas rpk1-1 mutant plants showed insensitivity in ABA-induced stomatal closure, similar to the results of our previous study (24). A slightly increased ABA content in rpk1-1 and decreased ABA content in 35S:RPK1 was observed (supplemental Fig. 2), suggesting that the endogenous ABA levels in the knock-out and transgenic plants did not affect their phenotypes. These data thus suggest that RPK1 overproduction enhances ABA signaling during root growth and also the stomatal closing of guard cells in the transgenic plants.
FIGURE 2.
Response of the rpk1-1 mutant and RPK1 overexpression lines to the regulation of transpiration under water stress conditions. A, transpirational water loss in wild type (WS), rpk1-1, RPK1 overexpression (35S:RPK1 lines L1 and L2), and control plants (VC) at the indicated time points after leaf detachment. Water loss is expressed as a percentage of the initial fresh weight. Values are the means ± S.D. of five samples of three leaves each. B, effects of ABA on stomatal closure in wild type, rpk1-1, 35S:RPK1 L1 and L2, and control plants. Leaves were treated with or without ABA for 2 h after stomatal preopening under light for 2 h, and the stomatal aperture was measured. Values are the means ± S.D. of 20 measurements. The asterisks indicate statistically significant differences in comparisons between wild type and rpk1-1, determined by Student's t test, or in comparisons between control and 35S:RPK1, evaluated by one-way ANOVA with Tukey's HSD (*, p < 0.0001; **, p < 0.0005; ***, p < 0.005). C, guard cells of 35S:RPK1 L1 and control plants treated with or without 5 μm ABA for 2 h. Bars, 30 μm.
RPK1 Overproduction Increases the Tolerance to Drought Stress in Arabidopsis
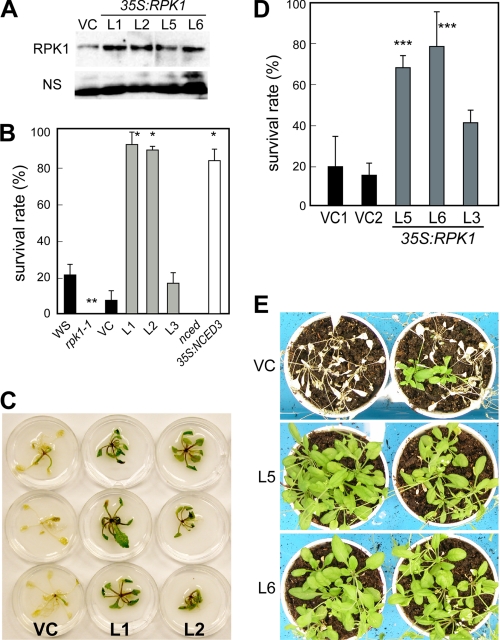
To investigate the physiological function of RPK1 during drought stress, we examined whether the disruption or overproduction of RPK1 would affect the tolerance to this condition in plants (Fig. 3). Wild type, rpk1-1, 35S:RPK1 (L1, L2, and L3 lines), nced3-2 (a mutant of a key enzyme in ABA biosynthesis during drought stress; 9-cis-epoxicarotenoid dioxygenase 3), and 35S:NCED3 (31) plants grown for 2 weeks on GM-agar plates under normal conditions were transferred onto filter paper and then exposed to drought stress by ceasing the water supply in 40% RH for 8 h. After this drought stress treatment, the plants were watered again, incubated for a further 2 days, and then analyzed for surviving green plants (Fig. 3, B and C). The survival rates of rpk1-1 and nced3-2 were lower than those of the control plants after drought stress. However, the survival rates of the 35S:RPK1 plants were significantly higher (>80%) than those of the control plants and were similar to those of the 35S:NCED3 plants (Fig. 3B). To further examine drought stress tolerance, we selected two additional 35S:RPK1 lines (L5 and L6) that showed characteristics similar to those of the L1 and L2 lines (Fig. 3B). Fig. 3, D and E, shows the survival rates of 35S:RPK1 lines L5, L6, and L3 grown on soil pots under long term drought stress. The transgenic plants grown on pots were exposed to drought stress by stopping watering in a 30–40% RH environment for 2 weeks. After this drought stress treatment, the plants were watered and cultured for a further 1 week and then analyzed for surviving green plants (Fig. 3D). The 35S:RPK1 plants showed improved stress tolerance to drought compared with the wild type plants. These results indicate important roles for the RPK1-mediated signaling pathway in both short term and long term drought stress tolerances in Arabidopsis.
FIGURE 3.
Drought tolerance of the 35S:RPK1 plants. A, protein blot analysis was performed with microsomal fractions of the 35S:RPK1 transgenic plants (lines L1, L2, L5, and L6) using an anti-RPK1 antibody. NS, nonspecific proteins. B and C, enhanced tolerance to rapid drought stress in the 35S:RPK1 (lines L1 and L2) plants. Transgenic plants, including vector control (VC), grown on GM-agar plates under normal conditions were transferred onto filter paper, and water was withheld for 8 h under 40% RH conditions. After drought stress treatment, the plants were watered and incubated for a further 2 days and then photographed (B). Survival rates were determined as the number of visible green plants after rehydration (C), and 20 plants were used in each experiment. Values are the means ± S.D. calculated from the results of three independent experiments. Bars, 1.0 cm. D and E, enhanced tolerance to long term drought stress in the 35S:RPK1 (lines L5 and L6) plants. The transgenic plants were grown on pots under normal conditions, and water was withheld for 2 weeks in 30–40% RH conditions. After this drought stress treatment, the plants were watered and incubated for 1 week and then photographed (D). Survival rates were determined as the number of visible green plants after rehydration (E), and 20 plants were used in each experiment. Values are the means ± S.D. calculated from the results of three independent experiments. The asterisks indicate statistically significant differences in comparisons between WS and rpk1-1, determined by Student's t test, or in comparisons between VC, 35S:RPK1, and 35S:NCED3, evaluated by one-way ANOVA with Tukey's HSD (*, p < 0.0001; **, p < 0.005; ***, p < 0.05).
RPK1 Overproduction Enhances the Transcription of Stress-responsive Genes
To further elucidate the effects of RPK1 overproduction on ABA and stress signaling pathways in Arabidopsis during drought stress, we performed comparative microarray analysis of wild type and 35S:RPK1 plants grown under normal conditions using an Agilent Arabidopsis 2 Oligo Microarray (Agilent Technologies), covering >21,000 genes. Two experiments were performed using RNA samples from two independent lines overexpressing the RPK1 protein (L1 and L2). Each sample was labeled differently (Cy3 or Cy5). Genes exhibiting an expression ratio (35S:RPK1/wild type) higher than 3 are listed in Table 1 and supplemental Table 2. Genes for which the expression ratio (wild type/35S:RPK1) was lower than 3 are shown in supplemental Table 3. In comparison with the vector control plants, 27 genes were found to be up-regulated with an average expression ratio greater than 3 (Table 1). Among these, 12 genes were identified as water stress/ABA-responsive genes induced by drought, high salinity, cold, osmotic stress, and ABA treatment. These included genes such as Cor15a, Cor15b, RD29A, and GolS3 (2), which were observed to be up-regulated in the RPK1 transgenic plants (Table 1 and supplemental Table 2). Other genes were also found to be up-regulated not only by water stress and ABA treatment but also by H2O2 treatment and heat stress, including ANAC036, heat shock protein (HSP) (36), and calcium-binding proteins (Table 1). Genes for stress-responsive transcription factors, such as TEM1, RAV1, and RAV2/TEM2 (37), and a senescence-related transcription factor, WARKY53 (38), were also found to be up-regulated in the RPK1 transgenic plants.
TABLE 1.
The up-regulated genes in the microarray data of untreated 35S:RPK1 (35S:RPK1/wild type)
| Locus | Gene | -Fold change |
||||||
|---|---|---|---|---|---|---|---|---|
| 35S:RPK1/wild typea | Dryb,c | NaClb,c | Coldb,c | ABAc,d | Heatb,c | H2O2c | ||
| -fold | ||||||||
| Stress-responsive genes | ||||||||
| At2g42540.1 | Cold-regulated protein, Cor15a | 5.4 | 39.8 | 38.3, 7.8 | 67.4, 30.9 | 8.2, 40.4 | ||
| At5g52310.1 | Low temperature-responsive protein 78 (LTI78)/rd29A | 3.5 | 13.4 | 16.7, 20.1 | 17.9, 44.4 | 30.7, 25.20 | ||
| At4g30650.1 | Hydrophobic protein/low temperature and salt responsive protein | 3.4 | 41.3, 6.2 | 3.7 | ||||
| At2g42530.1 | Cold-regulated protein, Cor15b | 3.2 | 20.3, 2.5 | 36.9, 18.1 | 6.3, 6.5 | |||
| Metabolism | ||||||||
| At1g09350.1 | Galactinol synthase (GolS3) | 4.0 | 53.9, 103.4 | |||||
| At1g75900.1 | Family II extracellular lipase 3 EXL3 (EXL3) | 3.8 | 2.3 | 3.0, 3.0 | 2.2 | |||
| At1g64400.1 | Long-chain acyl-CoA synthetase, putative | 3.6 | ||||||
| At3g44870.1 | S-Adenosyl-l-methionine:carboxyl methyltransferase family protein | 3.2 | 4.0 | 11.1 | 63.3 | |||
| Disease | ||||||||
| At2g43620.1 | Chitinase | 5.1 | 2.5, 2.2 | 9.0 | 5.5, 11.9 | 3.0 | ||
| At3g52430.1 | Phytoalexin-deficient 4 protein (PAD4) | 3.4 | 3.8 | 2.3 | ||||
| TF | ||||||||
| At1g25560.1 | AP2 domain-containing transcription factor | 4.4 | 3.2 | 2.3 | 4.5 | |||
| At1g13260.1 | DNA-binding protein RAV1 (RAV1) | 3.9 | 2.0 | 4.5, 6.6 | 2.0 | |||
| At1g68840.1 | DNA-binding protein RAV2 (RAV2) | 3.9 | 3.8 | 3.0 | 2.0 | |||
| At4g23810.1 | WRKY family transcription factor AR411 (WRKY53) | 3.9 | 4.1, 12.5 | 3.1 | 2.2 | |||
| At2g17040.1 | ANAC036 no apical meristem (NAM) family protein | 3.6 | 2.4, 8.4 | 3.2 | 51.9 | |||
| Hsp | ||||||||
| At3g09440.1 | Heat shock cognate 70 kDa protein 3 (HSC70-3) | 3.7 | 5.9 | 2.5 | 8.6, 2.0 | 2.3 | ||
| At5g52640.1 | Heat shock protein 81-1 (HSP81-1)/heat shock protein 83 (HSP83) | 3.4 | 4.3 | 2.0 | 32.7, 8.9 | 11.9 | ||
| At5g56010.1 | Heat shock protein (HSP81-3) | 3.3 | 4.0 | 6.9, 4.5 | ||||
| At3g12580.1 | Heat shock protein 70, putative/HSP70 | 3.0 | 31.0 | 5.2, 3.7 | 147.7, 4.1 | 47.6, 5.6 | 14.3 | |
| At5g23240.1 | DNAJ heat shock N-terminal domain-containing protein | 3.0 | 3.7 | |||||
| Calcium | ||||||||
| At5g39670.1 | Calcium-binding EF-hand family protein | 4.2 | 6.1 | 4.5 | 8.9 | 39.4 | 6.7 | |
| At5g62070.1 | Calmodulin-binding family protein | 3.5 | 2.1 | |||||
| At2g41090.1 | Calmodulin-like calcium-binding protein, 22 kDa (CaBP-22) | 3.0 | 2.9 | 2.5 | 2.3 | |||
| Transporter | ||||||||
| At5g50200.1 | Nitrate transporter, NRT3.1 | 5.2 | 4.3 | 2.4 | ||||
| Other | ||||||||
| At2g24600.1 | Ankyrin repeat family protein | 3.4 | 4.0, 6.2 | 6.5 | 6.8 | |||
| Unknown | ||||||||
| At2g20142.1 | Expressed protein | 4.2 | 2.8 | 3.3 | 61.8 | 2.4 | ||
| At4g38560.1 | Expressed protein | 3.1 | 3.0 | 3.2 | ||||
a Values are the means and S.D. of two different 35S:RPK1 lines (ratio >3.0).
b The microarray data (boldface characters) from Maruyama et al. (58) (ratio >2.0).
c The microarray data are available in Genevestigator (available on the World Wide Web) (ratio >2.0).
d Microarray data (boldface characters) from Y. Fujita and K. Yamaguchi-Shinozaki (unpublished data) (ratio >2.0).
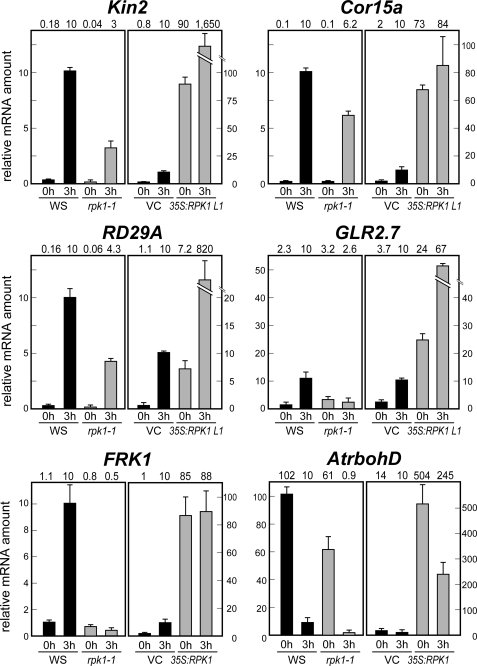
Fig. 4 illustrates the relative transcript levels of several stress-responsive genes, standardized using absolute 18 S rRNA transcript levels, during drought stress in Arabidopsis. The expression levels of the ABA-responsive genes Kin2, Cor15a, and RD29A (2) were significantly reduced in the rpk1-1 mutant after drought stress. In contrast, the expression levels of these same genes were strongly increased in 35S:RPK1 plants, indicating that RPK1 overproduction enhances their transcription. The expression levels of the putative glutamate receptor, AtGLR2.7 (AT2g29120) were also found to be affected by drought stress (i.e. decreased in rpk1-1 and increased in 35S:RPK1) (Fig. 4 and supplemental Table 2), suggesting that RPK1 also regulates the expression of drought stress-responsive GLR. The expression levels of a receptor-like kinase FRK1, which is induced by drought stress in wild type plants, were found to be down-regulated in rpk1-1 and up-regulated in 35S:RPK1 (Fig. 4 and supplemental Table 2). The expression levels of the NADPH oxidase gene, which has roles in ROS production, were also up-regulated by RPK1 overproduction (Fig. 4 and supplemental Table 2). In contrast, several stress-responsive genes, such as RD20, DREB, and NAC (2), were shown to be down-regulated in the 35S:RPK1 plants (supplemental Table 3). This down-regulation is somewhat unexpected but may be caused by a feedback control mechanism whereby the activation of the RPK1-responsive stress-related pathway represses the expression of stress-responsive genes regulated by other such pathways in the transgenic plants. Further experiments will be needed to clarify this phenomenon. Expression of the cytokinin-responsive ARR genes (27) and GATA type transcription factor genes (39) was also found to be decreased in 35S:RPK1.
FIGURE 4.
Expression levels of RPK1 downstream genes in rpk1-1 and 35S:RPK1. Relative mRNA levels were determined in rpk1-1 and 35S:RPK1 plants under dehydration conditions by quantitative RT-PCR. Similar results were obtained from three independent experiments, and a typical result is shown with S.D. values calculated from three independent PCRs from the same biological sample. 0h, plants grown under normal conditions; 3h, plants subjected to dehydration stress for 3 h. VC, vector control; WS, wild type.
RPK1 Overproduction Enhanced ROS Scavenging Activity
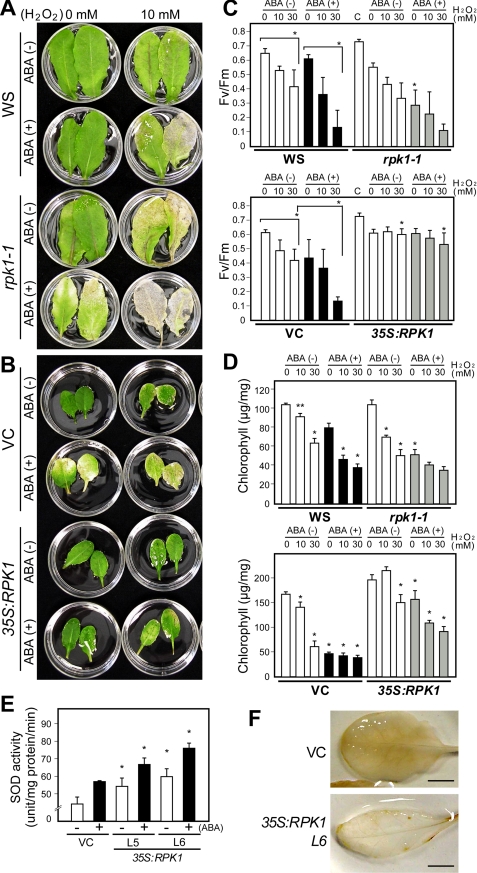
ROS is implicated in ABA signaling as an essential second messenger, and indeed ABA-induced stomatal closure involves the ROS production that activates Ca2+ channels (12). In our present microarray analysis, ROS-responsive genes were found to be up-regulated in the RPK1-overproducing plants (Table 1 and supplemental Table 2), suggesting that ROS homeostasis might be affected. To further evaluate the effects of RPK1 overproduction in the transgenic plants during oxidative stress, we investigated the H2O2 sensitivity of rpk1-1 and 35S:RPK1 leaves by detecting their ability to remove exogenous H2O2. In wild type leaves, pretreatment of ABA caused more severe bleaching when they were floated on 10 mm H2O2 under light (Fig. 5A). Leaves of the rpk1-1 mutant showed higher sensitivity to exogenous H2O2 than those of wild type, suggesting that the RPK1 mutation impairs the ability to scavenge exogenous H2O2. In contrast, the 35S:RPK1 leaves remained green during incubation with H2O2 both with and without pretreatment of ABA, indicating that RPK1 overproduction enhances the resistance to oxidative stress (Fig. 5B).
FIGURE 5.
The rpk1-1 mutant shows increased sensitivity to H2O2, whereas 35S:RPK1 plants show enhanced tolerance to H2O2. A and B, response of detached leaves from wild type, rpk1-1, 35S:RPK1, and vector control plants (VC) to treatment with 10 mm H2O2. The plants were grown on soil, and the leaves were harvested and incubated under white light for 3 days in H2O2 solution. Chlorotic lesions were formed in response to H2O2 treatment and were increased by pretreatment with 50 μm ABA for 3 h. Bars, 1.0 cm. C and D, photochemical efficiency (C) and chlorophyll contents (D) are shown in leaves treated with various concentrations of H2O2 for 3 days with or without pretreatment of 50 μm ABA for 3 h. Values are the means ± S.D. of three replicates (n = 10). The asterisks for wild type (WS) and vector control plant data indicate significant differences versus those obtained under control conditions, respectively. The asterisks for rpk1-1 and 35S:RPK1 values indicate significant differences versus those obtained for the wild type and control plants under equivalent experimental conditions, respectively, evaluated by one-way ANOVA with Turkey's HSD (*, p < 0.0001; **, p < 0.005). C, values of non-treated leaves. E, superoxide dimutase activities in the leaves of 35S:RPK1 lines L5 and L6 and control plants (vector control) grown on soil pots for 5 weeks. Enzyme activities were assayed in three independent replicates, and the values shown are the means ± S.D. The asterisks indicate statistically significant differences in a comparison of vector control and 35S:RPK1 under the same conditions, determined by one-way ANOVA with Tukey's HSD (*, p < 0.0001). F, ROS production in leaves of 35S:RPK1 and control plants. The detached leaves from these plants were stained with 3,3′-diaminobenzidine (DAB). The photographs shown are of representative leaves from three independent experiments. Bar, 0.25 cm.
H2O2 sensitivity was further assessed by measuring of photochemical efficiency of photosystem II and also the chlorophyll contents in the H2O2-treated leaves of rpk1-1 and 35S:RPK1. A decrease in photosynthetic activities was detected during incubation of these plants in H2O2 solution, whereas it is accelerated after pretreatment with ABA in the wild type control (Fig. 5C). Severe decreases in photosynthetic activities were observed in rpk1-1 leaves after pretreatment with ABA, an approximate 50% decrease in the solution without H2O2 compared with wild type in the same solution. In contrast, 35S:RPK1 leaves showed similar photosynthetic activities even after treatment with H2O2. Similar results were obtained for the chlorophyll content. Thus, the rpk1-1 leaves were found to be more sensitive to H2O2, particularly after pretreatment with ABA, whereas 35S:RPK1 leaves exhibited a higher chlorophyll content compared with wild type controls (Fig. 5D). The regulation of antioxidant enzymes is crucial for the redox states of cells. The transcription levels of genes encoding ROS scavenging enzymes were not severely altered in 35S:RPK1 (Table 1 and supplemental Table 2), and we thus further investigated these enzyme activities in the transgenic plants. The activity of superoxide dismutase was determined in 35S:RPK1 and wild type leaves with or without ABA treatment (Fig. 5E). In the wild type control, superoxide dismutase activity was induced by ABA treatment, but its activities in the 35S:RPK1 leaves were significantly higher than wild type. We then evaluated ROS generation in 35S:RPK1 Arabidopsis leaves after treatment with ABA. H2O2 formation in the transgenic plants was visualized by polymerization with 3,3′-diaminobenzidine. H2O2 accumulation was observed as dark brown deposits in the leaves of wild type plants (Fig. 5F). The leaves of 35S:RPK1 plants, however, did not accumulate H2O2, indicating that RPK1 overproduction stimulates scavenging ROS. These data thus suggest that RPK1 plays an important role in the control of ABA-responsive H2O2 homeostasis.
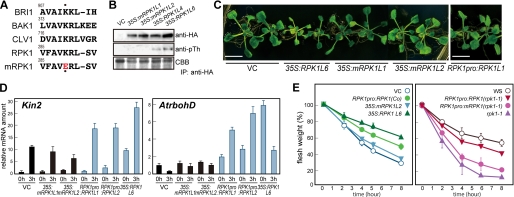
Mutation of the RPK1 Phosphorylation Site Suppresses RPK1 Function
To further elucidate the activation mechanisms for the RPK1 receptor-like kinase, we examined the effects of amino acid substitutions on its activity in vivo (Fig. 6). Previously, substitution of the conserved lysine within the kinase domains of the plant RLKs (to glutamic acid) has been shown to abolish their activity (40). We thus created an enzymatically inactive form of RPK1 in the same way (Fig. 6A; K289E mutant RPK1). We then produced transgenic Arabidopsis plants overexpressing this K289E mutant RPK1 (mRPK1) driven by the cauliflower mosaic virus 35S promoter. The proteins immunoprecipitated with anti-HA antibody from the microsomal fractions of 35S:RPK1 and 35S:mRPK1 transgenic plants were then subjected to Western blot analysis. Immunoprecipitated RPK1-HA, but not mRPK1-HA, cross-reacted with anti-phosphothreonine antibody (Fig. 6B). We then evaluated the effects of kinase inactivation on these plant phenotypes. Plants that had been transformed with mRPK1 did not show an altered size, whereas 35S:RPK1 plants showed growth retardation (Fig. 6C). We quantified both water stress- and ROS-related gene expression during drought stress. As shown in Fig. 6D, the expression levels of Kin2 and AtrbohD in the 35S:mRPK1 transgenic plants were low, even under drought conditions, compared with 35S:RPK1. The calculated water loss rates further indicated that mutation of the RPK1 kinase domain abolishes RPK1 overproduction phenotypes in the transgenic plants (Fig. 6E, left).
FIGURE 6.
Mutation of the RPK1 phosphorylation site suppresses the RPK1 function. A, alignment of the RPK1 kinase domain with other plant RLKs. A conserved lysine in the kinase domain is indicated by dots, and a single mutation of this residue was introduced by a substitution with glutamic acid (K289E; yielding mRPK1). B, RPK1 is phosphorylated on threonine residues in vivo. Proteins immunoprecipitated (IP) with anti-HA antibody from the microsomal fractions of 35S:RPK1 and 35S:mRPK1 transgenic plants were analyzed by Western blot. RPK1-HA and mRPK1-HA proteins were both detectable on immunoblots using anti-HA antibody; phosphorylated RPK1-HA proteins could be detected with a phosphothreonine antibody. C, phenotypes of the 35S:RPK1, 35S:mRPK1, and RPK1pro:RPK1 transgenic plants. Two-week-old plants are shown. Bar, 1 cm. D, expression levels of RPK1 downstream genes in 35S:RPK1, 35S:mRPK1, and RPK1pro:RPK1 transgenic plants. The relative mRNA levels were determined as shown in Fig. 4. 0h, plants grown under normal conditions; 3h, plants subjected to dehydration stress for 3 h. E, transpirational water loss in the 35S:RPK1, 35S:mRPK1, and RPK1pro:RPK1 plants of the Columbia ecotype (left). The introduction of the RPK1pro:mRPK1 construct into the rpk1-1 mutant plant did not rescue the mutant phenotype (right). The transpirational water loss rate was measured as shown in Fig. 2. VC, control plants.
Given the inherent nonspecific effects of ectopic gene expression under the control of the cauliflower mosaic virus 35S promoter, we additionally generated transgenic plants expressing RPK1 driven by its own promoter (RPK1pro:RPK1) (24). The RPK1pro:RPK1 plants did not show the growth retardation (Fig. 6C). We then estimated the expression levels of stress-responsive genes during drought stress in the RPK1pro:RPK1 plants. As shown in Fig. 6D, the expression levels of Kin2 and AtrbohD in the RPK1pro:RPK1 plants were higher than in the control plants. Furthermore, in the rpk1-1 mutant, the introduction of RPK1pro:RPK1 rescued the water loss phenotype. In contrast, RPK1pro:mRPK1 did not function in the rpk1-1 mutant (Fig. 6E, right). Taken together, our data indicate that RPK1 under the control of its native promoter shows enhanced function in transgenic Arabidopsis plants and that inactivation of the RPK1 kinase activity abolishes its function.
DISCUSSION
Plants have acquired the ability to survive in conditions of scarce water by developing highly systematic signaling networks, in which ABA is one of the important regulators of increased plant tolerance to water stress (1, 2). In our current study, we investigated the effects of overproduction of a membrane-localized receptor-like kinase, RPK1, in Arabidopsis and found that it induces various stress-responsive genes and enhances the physiological responses to drought stress and ABA sensitivity. Our data suggest that unregulated RPK1 enhances the downstream stress signal transduction pathways and increases the tolerance to drought stress in transgenic Arabidopsis.
In our previous study (24), we showed that rpk1 knock-out mutants have a decreased ABA sensitivity. We were thus interested in what the effects of overexpressed RPK1 protein would be on the plant responses to drought stress. In our current study, we now show that the rpk1-1 mutant has decreased ABA sensitivity with regard to root growth inhibition, transpirational water loss, and stomata closure, whereas RPK1 overproduction increases the sensitivity of these processes to ABA (Figs. 1 and 2). In our current transgenic plants overexpressing RPK1, the expression of many water stress-inducible genes was found to be up-regulated during drought stress, and these plants also showed strong tolerance to both short term and long term drought stress insults (Table 1 and Fig. 3). These results additionally suggest that the RPK1-signaling pathway positively controls the transcription of these genes and thereby enhances drought stress tolerance in plant cells. Signaling proteins that function as transducers or mediators need to be activated in response to cell signals. Receptor proteins perceive such extracellular signals and transduce them to the appropriate downstream pathway. Enhancement of the water stress and ABA signaling pathways by RPK1 overexpression may thus be caused by an increased frequency of extracellular signal perception by RPK1 or by amplification of the signaling cascade downstream of RPK1.
Our microarray analysis indicates that various stress-responsive genes are controlled by RPK1 overproduction. The expression levels of water stress-responsive genes, such as Cor15a, Cor15b, RD29A, and GolS, which encode LEA (late embryogenesis-abundant) proteins and a key enzyme in biosynthesis of osmoprotectants (raffinose family oligosaccharides), are up-regulated in RPK1 transgenic Arabidopsis. These LEA proteins and osmoprotectants have been shown to function in the protection of cells during osmotic stress (41, 42). The expression level of a drought stress-responsive receptor-like kinase gene, FRK1 was also found to be up-regulated in the RPK1 transgenic plants. Furthermore, the expression of ROS-responsive genes, such as the HSP genes, is also up-regulated in RPK1 transgenic plants. These HSP proteins function as chaperones, which play important roles in the acquisition of not only thermotolerance but also the adaptation to various environmental stresses (36). Genes encoding enzymes involved in fatty acid metabolism, such as a long-chain acyl-coenzyme A synthetase, are also up-regulated in the RPK1 transgenic plants (Table 1 and supplemental Table 2). Long-chain acyl-coenzyme A synthetase catalyzes free fatty acids to acyl-coenzyme A thioesters, and this is a key step in fatty acid metabolism, including phospholipid, triacylglycerol, and jasmonate biosynthesis and fatty acid β-oxidation (43). β-Oxidation represents the degradation step for fatty acids in peroxisomes and plays important roles in ROS production and JA biosynthesis in response to environmental stresses (44–46). The expression level of the NADPH oxidase gene is also up-regulated by RPK1 overproduction during drought stress. NADPH oxidases have been suggested to function in ABA-induced ROS production in guard cells (8, 11, 12). In the RPK1pro:RPK1 plants, the transcription of water stress- and ROS-related genes was also found to have increased, although growth retardation, which can sometimes result from the strong overexpression of stress-responsive genes (e.g. see Ref. 28), did not occur in these plants. Our current results thus suggest that RPK1 overproduction causes the increased transcription of water stress- and ROS-responsive genes via the enhancement of these signaling pathways.
Different types of environmental stress cause an increase in oxidative damage to photosynthetic proteins and pigments. ROS signaling is controlled and modulated by its production and subsequent scavenging, which is mediated by scavenging enzymes and non-enzymatic antioxidants (14, 47). ROS homeostasis is maintained by a large gene network in Arabidopsis to control the steady-state level of these molecules (48). ABA-induced ROS production has been described in several plant species (12, 15), and ABA is also thought to regulate ROS scavenging enzymes, such as catalases, superoxide dismutases, glutathione peroxidases, and ascorbate peroxidases (13, 15, 48–50). To further elucidate the ROS scavenging properties of the rpk1-1 mutant and RPK1 overexpressor, we analyzed their phenotypes under oxidative stress conditions. Leaf longevity is regulated during developmental stages and by environmental stresses and phytohormones (44, 51, 52), and the photochemical efficiency of photosystem II and chlorophyll content have been used as physiological senescence markers. We then investigated the effect of oxidative stress on the levels of these senescence markers of the transgenic and mutant plants. The treatment with exogenous H2O2 caused severe damage in photochemical efficiency and chlorophyll contents of wild type plants (Fig. 5), and pretreatment with ABA caused further damage on the wild type plants. This enhanced damage on the leaves might be caused by the endogenous ROS production increased by the pretreatment with ABA. On the other hand, the rpk1-1 mutant showed increased sensitivity to H2O2, whereas the transgenic plants overproducing RPK1 enhanced tolerance to oxidative stress compared with the wild type plants (Fig. 5). We found that the activity of an antioxidant enzyme, superoxide dismutase, was increased in the transgenic plants overexpressing RPK1 protein, and the leaves of 35S:RPK1 did not accumulate H2O2 after treatment with ABA (Fig. 5). These results indicate that overproduction of RPK1 enhances the scavenging activity of oxidative stress and increases the levels of tolerance to this stress in plants. The higher tolerance of the RPK1 transgenic plants to drought stress may be due to the combined effects of a decrease in water loss, an up-regulation of stress-inducible gene expression, and an increase in the scavenging activity of oxidative stress. This in turn suggests the possibility that these transgenic plants will also show tolerance to the various different kinds of abiotic and biotic stresses. Further analyses are needed to elucidate the effects of RPK1 overproduction on plant tolerance to various stress conditions.
To further elucidate the RPK1-mediated signaling, we analyzed the kinase activity of RPK1 and found that this protein is phosphorylated on threonine residues in vivo, which is suggestive of kinase domain activation (Fig. 6). Activation of the kinase domain is a key signaling step for several plant RLKs. The mutation of conserved lysine residue abolishes RPK1 kinase activity, as seen for many other RLKs (40). Furthermore, the inactivation of the kinase function of RPK1 diminishes the enhanced transcription of stress-responsive genes and the decrease of water loss in the corresponding transgenic plants. These results suggest that RPK1 activity is essential for RPK1 function during stress signaling. Hence, phosphorylation of RPK1 downstream factor(s) may be a component of the signaling system in response to stress.
Recent studies have suggested multiple roles for the receptor-like kinases (53, 54). BAK1 is a signaling molecule for brassinosteroid perception and also acts in pathogen-associated molecular pattern signaling as a coreceptor of FLS2 and in brassinosteroid-independent cell death via ROS accumulation (53, 54). ROS production and mitogen-activated protein kinase cascades are involved downstream of the FLS2 signaling pathway, which regulates the transcription of target genes, such as FRK1 (55). RPK1 also controls the expression level of FRK1 (Fig. 4) (24), suggesting a cross-talk between the pathway controlled by RPK1 and FLS2 during drought stress. Recently, RPK1 and RPK2/TOAD2 (56) have been identified as important receptor proteins during embryonic pattern formation (57), suggesting the possibility that RPK1 may play dual roles in cell differentiation during early embryogenesis and stress signaling. There may also be control mechanisms for RPK1 and RPK2/TOAD2-mediated signaling that operate during embryogenesis via ROS signaling as well as stress signaling. Furthermore, the down-regulation of important cytokinin signaling genes was observed in our current microarray analysis of RPK1 overexpressors (supplemental Table 3). Hence, there is a possibility that the cytokinin signaling pathway is down-regulated by RPK1 during both stress and developmental processes. Taken together, our current results indicate that the stress-responsive RLKs function in multiple signaling systems and that they cross-talk in diverse plant responses during plant development and growth.
Supplementary Material
Acknowledgments
We acknowledge E. Ohgawara, K. Amano, K. Yoshiwara, and M. Toyoshima (JIRCAS) and K. Ishiyama (RIKEN) for technical assistance.
This work was supported by Japan Society for the Promotion of Science Grant-in aid for Scientific Research C 21580125 (to Y. O.) and by the Ministry of Agriculture, Forestry and Fisheries, Japan.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2 and Tables 1–3.
- ABA
- abscisic acid
- ROS
- reactive oxygen species
- RLK
- receptor-like kinase
- RT
- reverse transcription
- HA
- hemagglutinin
- ANOVA
- analysis of variance
- HSD
- honestly significant difference
- GM
- germination medium
- RH
- relative humidity.
REFERENCES
- 1.Finkelstein R. R., Gampala S. S., Rock C. D. (2002) Plant Cell 14, S15–S45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaguchi-Shinozaki K., Shinozaki K. (2006) Annu. Rev. Plant Biol. 57, 781–803 [DOI] [PubMed] [Google Scholar]
- 3.Li S., Assmann S. M., Albert R. (2006) PLoS Biol. 4, e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirayama T., Shinozaki K. (2007) Trends Plant Sci. 12, 343–351 [DOI] [PubMed] [Google Scholar]
- 5.Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. (2009) Science 324, 1064–1068 [DOI] [PubMed] [Google Scholar]
- 6.Park S. Y., Fung P., Nishimura N., Jensen D. R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T. F., Alfred S. E., Bonetta D., Finkelstein R., Provart N. J., Desveaux D., Rodriguez P. L., McCourt P., Zhu J. K., Schroeder J. I., Volkman B. F., Cutler S. R. (2009) Science 324, 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pandey S., Nelson D. C., Assmann S. M. (2009) Cell 136, 136–148 [DOI] [PubMed] [Google Scholar]
- 8.Pei Z. M., Murata Y., Benning G., Thomine S., Klüsener B., Allen G. J., Grill E., Schroeder J. I. (2000) Nature 406, 731–734 [DOI] [PubMed] [Google Scholar]
- 9.Foreman J., Demidchik V., Bothwell J. H., Mylona P., Miedema H., Torres M. A., Linstead P., Costa S., Brownlee C., Jones J. D., Davies J. M., Dolan L. (2003) Nature 422, 442–446 [DOI] [PubMed] [Google Scholar]
- 10.Apel K., Hirt H. (2004) Annu. Rev. Plant Biol. 55, 373–399 [DOI] [PubMed] [Google Scholar]
- 11.Murata Y., Pei Z. M., Mori I. C., Schroeder J. I. (2001) Plant Cell 13, 2513–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kwak J. M., Mori I. C., Pei Z. M., Leonhardt N., Torres M. A., Dangl J. L., Bloom R. E., Bodde S., Jones J. D., Schroeder J. I. (2003) EMBO J. 22, 2623–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neill S., Desikan R., Hancock J. (2002) Curr. Opin. Plant Biol. 5, 388–395 [DOI] [PubMed] [Google Scholar]
- 14.Mustilli A. C., Merlot S., Vavasseur A., Fenzi F., Giraudat J. (2002) Plant Cell 14, 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao Y., Lv D., Wang P., Wang X. C., Chen J., Miao C., Song C. P. (2006) Plant Cell 18, 2749–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu H., Chen X., Hong Y. Y., Wang Y., Xu P., Ke S. D., Liu H. Y., Zhu J. K., Oliver D. J., Xiang C. B. (2008) Plant Cell 20, 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shiu S. H., Bleecker A. B. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diévart A., Clark S. E. (2004) Development 131, 251–261 [DOI] [PubMed] [Google Scholar]
- 19.Kinoshita T., Caño-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. (2005) Nature 433, 167–171 [DOI] [PubMed] [Google Scholar]
- 20.Gómez-Gómez L., Bauer Z., Boller T. (2001) Plant Cell 13, 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- 21.Matsubayashi Y., Ogawa M., Morita A., Sakagami Y. (2002) Science 296, 1470–1472 [DOI] [PubMed] [Google Scholar]
- 22.Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006) Science 313, 845–848 [DOI] [PubMed] [Google Scholar]
- 23.Hong S. W., Jon J. H., Kwak J. M., Nam H. G. (1997) Plant Physiol. 113, 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osakabe Y., Maruyama K., Seki M., Satou M., Shinozaki K., Yamaguchi-Shinozaki K. (2005) Plant Cell 17, 1105–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshida R., Hobo T., Ichimura K., Mizoguchi T., Takahashi F., Aronso J., Ecker J. R., Shinozaki K. (2002) Plant Cell Physiol. 43, 1473–1483 [DOI] [PubMed] [Google Scholar]
- 26.Umezawa T., Okamoto M., Kushiro T., Nambara E., Oono Y., Seki M., Kobayashi M., Koshiba T., Kamiya Y., Shinozaki K. (2006) Plant J. 46, 171–182 [DOI] [PubMed] [Google Scholar]
- 27.Osakabe Y., Miyata S., Urao T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2002) Biochem. Biophys. Res. Commun. 293, 806–815 [DOI] [PubMed] [Google Scholar]
- 28.Sakuma Y., Maruyama K., Qin F., Osakabe Y., Shinozaki K., Yamaguchi-Shinozaki K. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 18822–18827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellens R. P., Edwards E. A., Leyland N. R., Bean S., Mullineaux P. M. (2000) Plant Mol. Biol. 42, 819–832 [DOI] [PubMed] [Google Scholar]
- 30.Qin F., Sakuma Y., Tran L. S., Maruyama K., Kidokoro S., Fujita Y., Fujita M., Umezawa T., Sawano Y., Miyazono K., Tanokura M., Shinozaki K., Yamaguchi-Shinozaki K. (2008) Plant Cell 20, 1693–1707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iuchi S., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2000) Plant Physiol. 123, 553–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakamoto H., Maruyama K., Sakuma Y., Meshi T., Iwabuchi M., Shinozaki K., Yamaguchi-Shinozaki K. (2004) Plant Physiol. 136, 2734–2746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thordal-Christensen H., Zhang Z., Wei Y., Collinge D. B. (1997) Plant J. 11, 1187–1194 [Google Scholar]
- 34.Arnon D. I. (1949) Plant Physiol. 24, 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z., Gallie D. R. (2004) Plant Cell 16, 1143–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang W., Vinocur B., Shoseyov O., Altman A. (2004) Trends Plant Sci. 9, 244–252 [DOI] [PubMed] [Google Scholar]
- 37.Hilson P., Allemeersch J., Altmann T., Aubourg S., Avon A., Beynon J., Bhalerao R. P., Bitton F., Caboche M., Cannoot B., Chardakov V., Cognet-Holliger C., Colot V., Crowe M., Darimont C., Durinck S., Eickhoff H., de Longevialle A. F., Farmer E. E., Grant M., Kuiper M. T., Lehrach H., Léon C., Leyva A., Lundeberg J., Lurin C., Moreau Y., Nietfeld W., Paz-Ares J., Reymond P., Rouzé P., Sandberg G., Segura M. D., Serizet C., Tabrett A., Taconnat L., Thareau V., Van Hummelen P., Vercruysse S., Vuylsteke M., Weingartner M., Weisbeek P. J., Wirta V., Wittink F. R., Zabeau M., Small I. (2004) Genome Res. 14, 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miao Y., Zentgraf U. (2007) Plant Cell 19, 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brenner W. G., Romanov G. A., Köllmer I., Bürkle L., Schmülling T. (2005) Plant J. 44, 314–333 [DOI] [PubMed] [Google Scholar]
- 40.Wang X., Kota U., He K., Blackburn K., Li J., Goshe M. B., Huber S. C., Clouse S. D. (2008) Dev. Cell 15, 220–235 [DOI] [PubMed] [Google Scholar]
- 41.Battaglia M., Olvera-Carrillo Y., Garciarrubio A., Campos F., Covarrubias A. A. (2008) Plant Physiol. 148, 6–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taji T., Ohsumi C., Iuchi S., Seki M., Kasuga M., Kobayashi M., Yamaguchi-Shinozaki K., Shinozaki K. (2002) Plant J. 29, 417–426 [DOI] [PubMed] [Google Scholar]
- 43.Shockey J. M., Fulda M. S., Browse J. A. (2002) Plant Physiol. 129, 1710–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franke R., Schreiber L. (2007) Curr. Opin. Plant Biol. 10, 252–259 [DOI] [PubMed] [Google Scholar]
- 45.Goepfert S., Poirier Y. (2007) Curr. Opin. Plant Biol. 10, 245–251 [DOI] [PubMed] [Google Scholar]
- 46.Weber H. (2002) Trends Plant Sci. 7, 217–224 [DOI] [PubMed] [Google Scholar]
- 47.Davletova S., Rizhsky L., Liang H., Shengqiang Z., Oliver D. J., Coutu J., Shulaev V., Schlauch K., Mittler R. (2005) Plant Cell 17, 268–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mittler R., Vanderauwera S., Gollery M., Van Breusegem F. (2004) Trends Plant Sci. 9, 490–498 [DOI] [PubMed] [Google Scholar]
- 49.Mittler R. (2002) Trends Plant Sci. 7, 405–410 [DOI] [PubMed] [Google Scholar]
- 50.Kwak J. M., Nguyen V., Schroeder J. I. (2006) Plant Physiol. 141, 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schippers J. H., Nunes-Nesi A., Apetrei R., Hille J., Fernie A. R., Dijkwel P. P. (2008) Plant Cell 20, 2909–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lim P. O., Kim Y., Breeze E., Koo J. C., Woo H. R., Ryu J. S., Park D. H., Beynon J., Tabrett A., Buchanan-Wollaston V., Nam H. G. (2007) Plant J. 52, 1140–1153 [DOI] [PubMed] [Google Scholar]
- 53.Gendron J. M., Wang Z. Y. (2007) Curr. Opin. Plant Biol. 10, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López M. A., Bannenberg G., Castresana C. (2008) Curr. Opin. Plant Biol. 11, 420–427 [DOI] [PubMed] [Google Scholar]
- 55.Asai T., Tena G., Plotnikova J., Willmann M. R., Chiu W. L., Gomez-Gomez L., Boller T., Ausubel F. M., Sheen J. (2002) Nature 415, 977–983 [DOI] [PubMed] [Google Scholar]
- 56.Mizuno S., Osakabe Y., Maruyama K., Ito T., Osakabe K., Sato T., Shinozaki K., Yamaguchi-Shinozaki K. (2007) Plant J. 50, 751–766 [DOI] [PubMed] [Google Scholar]
- 57.Nodine M. D., Yadegari R., Tax F. E. (2007) Dev. Cell 12, 943–956 [DOI] [PubMed] [Google Scholar]
- 58.Maruyama K., Takeda M., Kidokoro S., Yamada K., Sakuma Y., Urano K., Fujita M., Yoshiwara K., Matsukura S., Morishita Y., Sasaki R., Suzuki H., Saito K., Shibata D., Shinozaki K., Yamaguchi-Shinozaki K. (2009) Plant Physiol. 150, 1972–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.