Abstract
Binding of the 4-1BB ligand (4-1BBL) to its receptor, 4-1BB, provides the T lymphocyte with co-stimulatory signals for survival, proliferation, and differentiation. Importantly, the 4-1BB-4-1BBL pathway is a well known target for anti-cancer immunotherapy. Here we present the 2.3-Å crystal structure of the extracellular domain of human 4-1BBL. The ectodomain forms a homotrimer with an extended, three-bladed propeller structure that differs from trimers formed by other members of the tumor necrosis factor (TNF) superfamily. Based on the 4-1BBL structure, we modeled its complex with 4-1BB, which was consistent with images obtained by electron microscopy, and verified the binding site by site-directed mutagenesis. This structural information will facilitate the development of immunotherapeutics targeting 4-1BB.
Keywords: Crystal Structure, Cytokine, Immunology, Receptors, Tumor Necrosis Factor (TNF), 4-1BB, 4-1BB Ligand, TNF Superfamily
Introduction
Members of the TNF6 superfamily are involved in the regulation of diverse immune functions. Some family members produce signals leading to apoptosis, whereas others are involved in lymphocyte activation and differentiation (1). Except for lymphotoxin α, members of the TNF superfamily are type II membrane proteins (2) and show relatively low sequence conservation (15–34%) within their C-terminal extracellular domains (3). Members of the TNF superfamily interact with members of the TNF receptor (TNFR) superfamily, which exist as type I membrane proteins or soluble decoy receptors. The extracellular domains of members of the TNFR superfamily are characterized by multiple cysteine-rich pseudo-repeats (4).
The T cell co-stimulatory receptor 4-1BB (CD137) is induced when T cells receive antigen-specific signals (5, 6). Its ligand, 4-1BBL (CD137L), is also induced on antigen-presenting cells, such as dendritic cells, macrophages, and B cells (7, 8). The 4-1BBL-4-1BB pathway co-stimulates T cells to carry out effector functions such as eradication of established tumors (9, 10) and the broadening of primary and memory CD8+ T cell responses (11, 12). 4-1BB-mediated signals have been shown to induce a novel subpopulation of CD11c+CD8+ T cells that have strong anti-cancer and anti-autoimmune effects (13).
Crystal structures of several TNF-TNFR complexes have already been solved, including those of the TNFβ-TNFR1 (14), TRAIL-DR5 (15), TALL1-BAFF-R (16), and OX40-OX40L (17) pairs. In general, trimeric TNF ligands bind to three receptor molecules, resulting in a 3-fold symmetric complex. Despite low sequence homology among members of the TNF family, they typically form a trimer with a bell-shaped structure. Therefore, other TNF family members were anticipated to form the same canonical bell-shaped trimer (18) (or the subsequently identified blooming flower-like structure; see below). TNF family members can be classified into three groups according to their sequence and structural characteristics (17). Group 1 (conventional group) is characterized by the distinctive bell shape of the trimer and the relatively long loops connecting the CD, DF, and DE strands. The members of group 2 (EF-disulfide group) possess an EF-disulfide bond and shorter CD and EF loops, resulting in a more globular shape (17). Based on sequence homology, 4-1BBL belongs to the third group, which also includes CD27, CD30L, GITRL, and OX40L. The members of this group are characterized by divergent sequences (17). They differ from each other and have relatively low sequence homology (15∼20%) with other TNF family members (17).
The trimers formed by OX40L and GITRL differ from those of other TNF members because they have a blooming flower-like structure in which the monomers are splayed out and form an angle of ∼45° with respect to the trimer axis (17, 19). In all other known structures of TNF members, this angle is between 20 and 30°. Furthermore, OX40L and GITRL have considerably shorter TNF homology domains (THDs) (∼120 residues) compared with conventional THDs (∼150 residues) (19). Although 4-1BBL belongs to the third group of TNF members, its THD has a typical length of ∼162 residues. It was therefore unclear whether the 4-1BBL trimer would have the traditional bell-shaped structure, the blooming flower-like structure seen with OX40L and GITRL, or even yet another different structure. To answer this question, we determined the structure of 4-1BBL, which indeed showed a novel trimer organization. To gain insight into the interaction of 4-1BBL with 4-1BB, we modeled the structure of the 4-1BBL-4-1BB complex, which was corroborated by single particle EM. The interaction of 4-1BBL with 4-1BB was further dissected by receptor binding studies with mutant 4-1BBL proteins. The results enable us to discuss the characteristics of human 4-1BBL and to compare them with those of other members of the TNF family.
EXPERIMENTAL PROCEDURES
Cloning, Protein Expression, and Purification
The biologically active ectodomain of 4-1BBL was expressed in Escherichia coli as a glutathione S-transferase (GST) fusion protein. The cDNA encoding the ectodomain of human 4-1BBL residues 58–254 was cloned into the pGEX6P-1 vector (Amersham Biosciences) downstream of GST using the restriction endonucleases BamHI and XhoI. Because this human 4-1BBL construct contains only one methionine residue, residues Leu147 and Gln168 were mutated to methionines to obtain enough phase information to be able to solve the structure by single-wavelength anomalous diffraction. The correct folding of the selenomethionine-substituted protein was verified by gel filtration chromatography, and its activity was verified by a receptor binding assay using FACScan on Jurkat 8-1 cells (data not shown).
Protein expression and purification of this 4-1BBL protein were performed by a method previously described (31), with slight modifications. The cells were harvested and resuspended in binding buffer (phosphate-buffered saline, 0.1% nonylphenylpolyethylene glycol (NonideAt® P40 substitute, Nonidet P-40), and 5% glycerol) and then sonicated on ice. After centrifugation of the cell extract, the supernatant containing 4-1BBL was incubated with glutathione excellose (Takara) resin that had been equilibrated with binding buffer. The GST fusion protein was released from the affinity matrix by competitive elution with glutathione. Separation of 4-1BBL from its GST fusion partner was accomplished by proteolytic cleavage for 4 h at 4 °C with PreScission protease (Amersham Biosciences Bioscience) in 20 mm reduced l-glutathione, 20 mm Tris-HCl (pH 7.8), 50 mm NaCl, and 0.1% dodecyl octaethylene glycol ether. The human 4-1BBL protein was separated from the GST protein using an ion exchange column (Resource Q; Amersham Biosciences) in 20 mm Tris-HCl (pH 7.8) and 50 mm NaCl and concentrated to ∼7 mg/ml for crystallization.
The ectodomain of human 4-1BB (residues 18–180) was also expressed as a GST fusion protein in E. coli and purified as described for 4-1BBL. The complex was formed by mixing 4-1BBL and 4-1BB at a molar ratio of 1:1 and purified by gel filtration chromatography using a Superdex 200 column (data not shown).
Crystallization
The initial crystallization of 4-1BBL was performed using commercially available screening solutions (Hampton Research) and the vapor diffusion method in microbatch at 290 K. After optimizing the initial crystallization conditions, crystals suitable for data collection were obtained in drops consisting of 1.5 μl of protein sample and 1.5 μl of reservoir solution (0.1 m NaCl, 0.1 m bis-Tris, pH 6.5, 1.5 m ammonium sulfate).
X-ray Diffraction and Data Collection
X-ray diffraction data of the crystals were collected on a Bruker proteum 300 CCD detector at the 4A (HFMX) beamline of the Pohang Light Source. Before mounting, the crystals were soaked in cryo-protective buffer containing 25% polyethylene glycol in the crystallization buffer. X-ray diffraction data were collected at a crystal-to-detector distance of 200 mm, with a 0.5-s exposure for each 1° oscillation frame. The collected data were processed using DENZO and SCALEPACK of the HKL package (32).
Structure Determination and Refinement
The 4-1BBL crystals diffracted to ∼2.3-Å resolution and belonged to space group P321 with unit cell dimensions a = b = 121.85 Å, c = 33.58 Å, α = β = 90°, γ = 120°. With one molecule/asymmetric unit, these unit cell dimensions correspond to a solvent content of 65.8%. The structure of 4-1BBL was solved by the single-wavelength anomalous diffraction method. Phasing and automatic tracing was performed using the SOLVE/RESOLVE package (33, 34). The expected selenium sites were identified using the SOLVE program, and density modification was carried out in RESOLVE. A tracing of most Cα atoms was automatically conducted by RESOLVE; the initial model was built with the program COOT until most of the residues were fit into the electron density map (35). The model was refined in CNS (36), and the quality of the structure was assessed using PROCHECK (37).
Site-directed Mutagenesis
Site-directed mutagenesis was carried out using a QuikChange site-directed mutagenesis kit (Stratagene). Constructs, confirmed by DNA sequencing, were expressed in the E. coli BL21 (DE3) strain. Mutant proteins were purified as described for wild-type e4-1BBL, and their sizes were analyzed by gel filtration chromatography and native gel electrophoresis. The following mutations were introduced: Q89A, L115G, V140A, Y142A, F199A, H205A, V240A, P242K, P242E, and P245D to test the interaction between 4-1BBL subunits and K127A, R171G, S172G, H205A, Q227A, and Q230A to test the interaction of 4-1BBL with 4-1BB.
Fluorescence-activated Cell Sorter Binding Experiments
To measure binding of 4-1BBL to 4-1BB, single cell suspensions of Jurkat 8-1 cells (T cell line expressing 4-1BB) were preincubated with 1 μg of Fc blocking antibody/106 cells in 100 μl of fluorescence-activated cell sorter staining buffer (0.1% fetal bovine serum in phosphate-buffered saline). Jurkat 8-1 cells were then incubated with 0.5 μg/ml (25 nm) of various 4-1BBL mutant proteins for 40 min at 20 °C. The cells were stained in the dark with 1 μg of phycoerythrin-conjugated anti-4-1BBL monoclonal antibody (C65–485) or 1 μg of isotype-control antibody. All of the samples were washed three times with staining buffer and immediately analyzed by flow cytometry. Prior to the experiments, 4-1BB expression was verified using a phycoerythrin-conjugated anti-4-1BB monoclonal antibody (4B4–1). All of the samples were analyzed by FACScan (BD Bioscience).
Electron Microscopy and Image Processing
The samples were prepared by conventional negative staining with 0.75% (w/v) uranyl formate as described previously (38). The images were collected with a Tecnai T12 electron microscope (FEI, Hillsboro, OR) equipped with an LaB6 filament and operated at an acceleration voltage of 120 kV. The images were recorded on imaging plates at a magnification of 67,000× and a defocus value of −1.5 μm using low dose procedures. The imaging plates were read out with a Ditabis micron imaging plate scanner (Ditabis Digital Biomedical Imaging System AG, Pforzheim, Germany) using a step size of 15 μm, a gain setting of 20,000, and a laser power setting of 30%, and 2 × 2 pixels were averaged to yield a pixel size of 4.5 Å on the specimen level.
Using the display program BOXER associated with the EMAN software package (39), 10,754 particles were interactively selected from 28 images of 4-1BBL, and 11,546 particles were selected from 56 images of the 4-1BBL-4-1BB complex. The particles were windowed into 40 × 40-pixel images. Using the SPIDER software package (40), the particles were rotationally and translationally aligned and subjected to 10 cycles of multi-reference alignment. Each round of alignment was followed by K-means classification specifying 100 output classes. The references used for the first multi-reference alignment were randomly chosen from the raw images.
RESULTS
Overall Structure of the 4-1BBL Ectodomain
To determine the crystal structure of human 4-1BBL, which has only one methionine residue, we introduced methionine substitutions for residues Leu147 and Gln168. The structure of this construct was determined at 2.3-Å resolution using the single-wavelength anomalous diffraction method. The crystallographic data are summarized in supplemental Table S1. Human 4-1BBL is composed of a cytoplasmic domain (residues 1–25), a transmembrane domain (residues 26–48), and an extracellular domain (residues 49–254). The extracellular domain can be further divided into the tail region (residues 49–92) and the THD (residues 93–254) (20). Although most crystal structures of TNF family members cover only the THD, our 4-1BBL construct, which we designated e4-1BBL, also includes part of the tail region (residues 58–92). Although present in our construct, residues Ala58–Asp79 (N terminus), Leu170–Gly175 (DE loop), Ala188–Ala192 (EF loop), and Gly247–Glu254 (C terminus) were not well enough defined in the electron density map to be modeled, presumably because of disorder caused by a high flexibility in these regions.
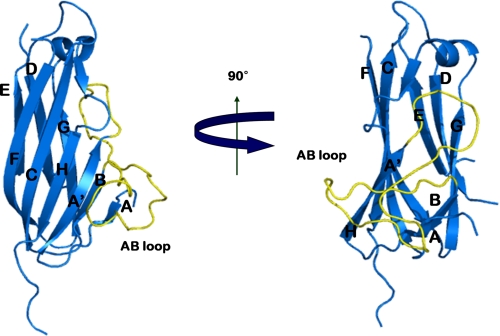
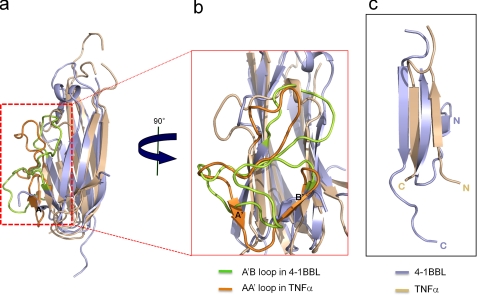
Sequence alignment of e4-1BBL with THDs of other TNF family members shows a sequence homology of only 17–23% (supplemental Fig. S1). Still, e4-1BBL adopts a similar β-sandwich jelly roll structure that has already been seen in the THDs of other TNF ligand members. THDs typically have two antiparallel β-sheets formed by strands A′AHCF (inner sheet) and B′BGDE (outer sheet) (18). In contrast, the β-sheets in e4-1BBL consist of strands A′HCF and ABGDE (Fig. 1). The lack of the two strands corresponding to the A′ and B′ strands in TNFα (supplemental Fig. S1) results in a longer A′B loop in e4-1BBL, making its structure different from those of other TNF ligands, including OX40L and GITRL.
FIGURE 1.
Overall structure of the 4-1BBL monomer. The structure shows a two-layered jellyroll β-sandwich topology similar to the canonical structure of other TNF family members. The inner and outer sheets of the jellyroll β-sandwich are composed of A′HCF and ABGDE, respectively. The long A′B loop is colored in yellow.
The height of the 4-1BBL THD is ∼60 Å, which is similar to those of other TNF members, such as TNFα and TRAIL, but longer than the THDs of GITRL or OX40L, which are only ∼40 Å (19). Thus, although 4-1BBL belongs to the same group as GITRL and OX40L, in terms of the THD domain it is more similar to canonical TNF members.
Structure of the 4-1BBL Trimer
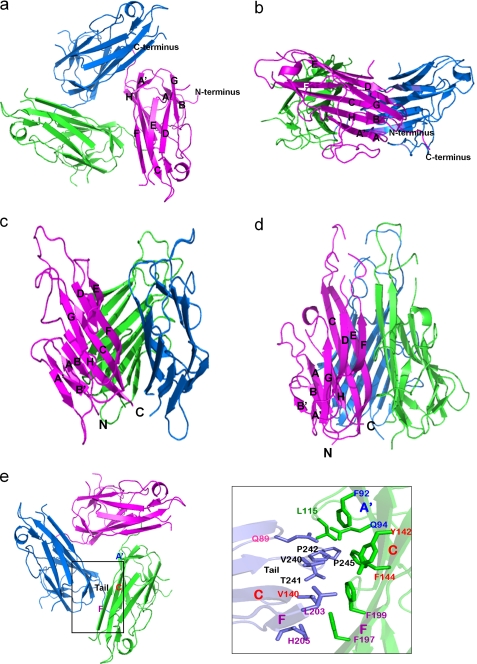
All of the previously determined structures of TNF members showed a trimer with a canonical bell shape or a blooming flower-like structure (18, 21). In contrast, the structure of the e4-1BBL trimer has the appearance of a 3-fold symmetric, three-bladed propeller (Fig. 2). The difference in structure is unlikely to be caused by the two introduced methionine substitutions (L147M in the C strand and Q168M in the DE loop), because these residues are located far from the monomer-monomer interface in the conventional bell-shaped trimer. In addition, gel filtration chromatography showed that the trimer formed by the e4-1BBL double mutant has the same size as the wild-type protein (data not shown), and the e4-1BBL mutant binds its receptors on Jurkat 8-1 cells with similar affinity as the wild-type protein (see Fig. 4). These results indicate that the two methionine substitutions do not affect trimerization of e4-1BBL nor its binding to 4-1BB.
FIGURE 2.
4-1BBL forms a trimer that is atypical for members of the TNF family. a, top view of the 4-1BBL trimer shown in ribbon representation. 4-1BBL forms a 3-fold symmetric assembly resembling a three-bladed propeller. b, side view of the 4-1BBL trimer. c, crystal structure of GITRL in ribbon representation (Protein Data Bank code 2Q1M). The shape of the GITRL trimer resembles a blooming flower. d, crystal structure of TNFα (Protein Data Bank code 1TNF), which shows the canonical bell-shaped trimer. e, the interactions between 4-1BBL subunits in the trimer are primarily mediated by hydrophobic interactions between the C-terminal tail of one subunit and the A′B loop and the A′, C, and F strands of the adjacent subunit. The residues involved in the hydrophobic interactions are: Gln89 (AA′ loop), Val140 (C strand), Leu203 (F strand), His205 (F strand), Val240 (C-terminal tail), Thr241 (C-terminal tail), Pro242 (C-terminal tail), and Pro245 (C-terminal tail) in one protomer (blue) and Gln94 (A′ strand), Phe92 (A′ strand), Leu115 (A′B loop), Tyr142 (C strand), Phe144 (C strand), Phe197 (F strand), and Phe199 (F strand) in the adjacent protomer (green).
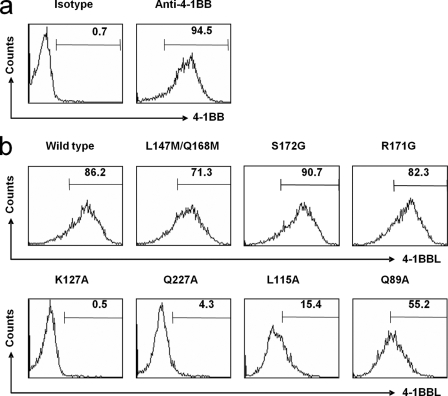
FIGURE 4.
Binding affinity of the 4-1BBL-4-1BB interaction measured by FACScan. a, control experiments using phycoerythrin-conjugated anti-4-1BBL monoclonal antibody as positive control and isotype-control antibody as negative control. b, measurements of binding affinities of wild-type and mutant 4-1BBL for 4-1BB. Mutations L115G, K127A, and Q227A decreased the binding affinity, whereas mutations R171G and S172G showed no effect. The Q89A mutation resulted in a 40% decrease in binding affinity.
The crystal structure of the e4-1BBL trimer revealed unique features that distinguish it from trimers formed by other TNF ligands. In the OX40L and GITRL trimers, the subunits are more splayed out with respect to the trimer axis than in trimers formed by most other canonical TNF ligands (17, 19). Remarkably, the e4-1BBL trimer is much more planar than those formed by other TNF family members, giving it the appearance of a three-bladed propeller (Fig. 2).
The trimeric organization of TNF members such as TNFα, TNFβ, and TRAIL is well conserved (18, 22). The typical trimer is assembled such that one edge of each subunit (strands E and F) is packed against the inner sheet of its adjacent subunit, forming large and mostly hydrophobic interfaces, resulting in a very stable interaction. The subunit interactions are usually mediated by hydrophobic and aromatic residues (including tyrosine and phenylalanine) in the inner β-strand of each subunit, called “tiles.” In the case of 4-1BBL, however, instead of the two long strands E and F, the hydrophobic C terminus of one subunit interacts with the A′B loop and strands A′, C, and F of its neighboring subunit. The contact surface between e4-1BBL subunits is much smaller compared with those of other trimers of TNF members. Only 14 residues of a 4-1BBL subunit participate in interactions with the adjacent subunit (Fig. 2e). In canonical TNF ligands many more residues are involved in trimer interactions (e.g. nearly 40 residues in TNFα) (22).
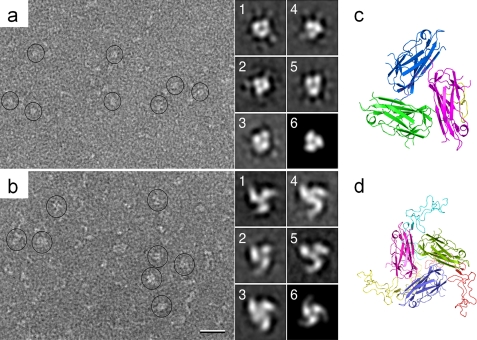
To further study the oligomeric state of 4-1BBL in solution, negatively stained samples were imaged by EM. The EM images revealed individual particles (marked by circles in the left panel of Fig. 6a). With a molecular mass of only 20 kDa, monomeric 4-1BBL would be too small to be seen by negative stain EM, indicating that the protein forms oligomers in solution. 10,754 particles were selected from 28 images and classified into 100 classes (supplemental Fig. S2). Some of the averages showed clear indications of a trimeric organization of the particles (see Fig. 6a, panels 1–5) and were similar to a projection calculated from the crystal structure filtered to a resolution of 25 Å (see Fig. 6a, panel 6). The single particle EM averages thus confirmed the trimeric state of 4-1BBL in solution.
FIGURE 6.
Electron microscopy of 4-1BBL and its complex with 4-1BB. a, raw image of negatively stained 4-1BBL and representative class averages (panels 1–5), which indicate a trimeric organization of 4-1BBL in solution. b, raw image of the negatively stained 4-1BBL-4-1BB complex and representative class averages of these complexes (panels 1–5). The scale bar represents 25 nm, and the side length of the individual panels is 18 nm. c, the 4-1BBL trimer in the orientation used to calculate the projection shown in panel 6 of A. d, model of the 4-1BBL-4-1BB complex in the orientation used to calculate the projection shown in panel 6 of B. The 4-1BBL subunits are colored in blue, pink, and green, and the 4-1BB subunits are yellow, red, and cyan. The 4-1BB structure was modeled based on the structure of DR5 (Protein Data Bank code 1DU3). The 4-1BBL-4-1BB complex has a windmill-like appearance, which is consistent with the EM averages of the complex.
To confirm that the three-bladed propeller structure of e4-1BBL is mediated by hydrophobic residues, we performed site-directed mutagenesis experiments. Based on the crystal structure, residues Gln89, Leu115, Val140, Tyr142, Phe199, His205, Val240, Pro242, and Pro245 are involved in the hydrophobic interactions that stabilize the trimer. The single mutations Q89A, L115A, H205A, V240A, P242K, P242E, and P245D had, however, no effect on trimer formation when analyzed by native gel electrophoresis and gel filtration chromatography (supplemental Table S2). By contrast, the V140A mutant exists as a dimer, and the F199A mutant exists as a mixture of dimers and trimers. The Y142A and the Δ240–254 truncation mutants were insoluble and could not be studied. Considering that the interaction sites form a long, widely distributed hydrophobic patch, it is not too surprising that most of the single point mutations had no effect on trimerization, but the results suggest that residues Val140 and Phe199 may play more important roles in stabilizing the 4-1BBL trimer.
Comparison of 4-1BBL with Other TNF Family Members
Although the overall structure of the 4-1BBL monomer appears similar to that of other TNF members, superposition of the e4-1BBL monomer with other TNF ligand monomers results in large root mean square deviations (rmsd) between the Cα atoms (e.g. an rmsd of ∼3 Å between e4-1BBL and GITRL and an rmsd of ∼7 Å between e4-1BBL and TNFα). Other TNF family members are much more similar to each other as indicated by rmsd values ranging from 2 to 3 Å (e.g. the rmsd between TNFα and OX40L is 1.9 Å). Although in the same TNF group, the molecular dimensions of the THDs of GITRL and OX40L are very different from those of 4-1BBL. For structural comparisons we therefore chose the THD of TNFα, which is, despite TNFα being a member of conventional group 1, more similar to that of 4-1BBL (Fig. 3a). The major differences between 4-1BBL and other TNF family members lie in the relative orientations of the N and C termini and the positions of the flexible loops. The N and C termini of 4-1BBL extend from the inner and outer sheets on opposite sides of the molecule, whereas those of other TNF family members are located near to each other on the same side of the molecule (Fig. 3c). Furthermore, the A′ and B′ strands of TNFα are missing in 4-1BBL, and the A strand of 4-1BBL occupies the position of and corresponds to the B′ strand of TNFα (although with opposite direction). As a result, the A′B loop in 4-1BBL is much longer than the corresponding AA′ loop in TNFα (Fig. 3, a and b). Binding studies of 4-1BBL to Jurkat 8-1 cells indicate that the long A′B loop of e4-1BBL is important for receptor binding. When residue Leu115 in the A′B loop was mutated to alanine, the binding affinity decreased dramatically (Fig. 4). The A′B loop is also involved in the interaction with the C-terminal tail of an adjacent subunit in the trimer. Taken together, these results demonstrate that the unique orientation of the N terminus and the longer A′B loop change the characteristics of 4-1BBL, resulting in a new trimer organization resembling a three-bladed propeller.
FIGURE 3.
Comparison of the structures of 4-1BBL and TNFα. a and b, superimposition of 4-1BBL (light blue) and TNFα (wheat). The A′B loop in 4-1BBL is colored in light green, and the AA′ loop in TNFα is colored in orange. The 4-1BBL structure has a much longer A′B loop because of its lack of strands corresponding to the A′ and B′ strands in the TNFα structure. c, positions of the N and C termini in 4-1BBL and TNFα.
The EF and DE loops are disordered in the e4-1BBL structure, whereas they are resolved in the TNFα structure. The conserved hydrophobic residues (especially tyrosine residues) in the DE loop of TNFα, TNFβ, and FASL have been reported to be energetically important for receptor binding (23–25). The hydrophobic residues in the DE loop of 4-1BBL are, however, not conserved, and the loop is expected to be structurally different from those in other canonical TNF members including TNFα, TNFβ, and TRAIL. The 4-1BBL-4-1BB complex may thus not feature the typical hydrophobic interactions. We mutated residues Arg171 and Ser172 in e4-1BBL. If the 4-1BBL-4-1BB interaction is similar to the TNFβ-TNFR1 binding mode, these two 4-1BBL residues could potentially be involved in forming salt bridges or hydrogen bonds with 4-1BB. The mutations had, however, no effect on receptor binding (Fig. 4b). Taken together, the DE loop of e4-1BBL does not appear to play a major role in receptor binding.
Interaction between 4-1BBL and 4-1BB
All of the known structures of TNF-TNFR complexes show that trimeric TNF ligands bind to three receptor molecules with a 3:3 stoichiometry (1, 14, 18), but the contact regions between ligands and receptors are very diverse. For example, the OX40 receptor uses its CRD1, CRD2, and CRD3 regions to bind OX40L, whereas in the TNFβ-TNFR1 and TRAIL-DR5 complexes only the CRD2 and CRD3 regions of the receptors contribute to ligand binding (17).
To gain further insight into the interaction of 4-1BBL with 4-1BB, we tried to determine the structure of 4-1BBL bound to 4-1BB, but we were not successful in producing crystals. We therefore decided to model the structure of the complex. We first generated a homology model of 4-1BB using the SWISS-MODEL server (26). Because their sequences align well, we used the TNFR1 structure (Protein Data Bank code 1TNR) as template for the 4-1BB structure. We then used the structure of the TNFβ-TNFR1 complex (14) to define the positions of the 4-1BB molecules. The 4-1BBL trimer was superimposed with the TNFβ trimer, and the positions of the TNFR1 molecules were used to define the positions of the 4-1BB molecules in the 4-1BBL-4-1BB complex. In conventional TNF-TNFR complexes such as TNFβ-TNFR1 and TRAIL-DR5, one receptor molecule binds almost equally to two subunits of the ligand trimers (17). In our model of the 4-1BBL-4-1BB complex, however, one receptor interacts predominantly with just one ligand and only slightly with the C-terminal tail of the adjacent subunit (Fig. 6d).
To obtain experimental evidence for this unexpected ligand-receptor interaction, we imaged the 4-1BBL-4-1BB complex by negative stain EM. Although the particle population appeared heterogeneous, the individual particles (marked by circles in the left panel of Fig. 6b) were larger than those seen in the images of 4-1BBL alone (Fig. 6a). Classification of 11,546 particles from 56 images into 100 classes confirmed the heterogeneity of the particles both in size and shape (supplemental Fig. S2), but some of the class averages (Fig. 6b, from 1 to 5) showed a clear indication of the trimeric organization of the complex. The heterogeneity in the particle population is most likely due to the presence of only partially assembled complexes, and the differences between the class averages may be related to variability in the binding of 4-1BB to the 4-1BBL trimer. Considering the small size of the complex (∼120 kDa), the variability may also be caused by slight differences in the stain embedding of the molecules. Nonetheless, the experimental class averages in Fig. 6b show strong similarities with a projection calculated from the modeled structure of the complex filtered to a resolution of 25 Å (Fig. 6b, panel 6). Projections calculated from other TNF-TNFR complexes were much less similar to the EM class averages (supplemental Fig. S3). The EM results thus support the asymmetric interaction of the receptors with the 4-1BBL trimer suggested by the model of the 4-1BBL-4-1BB complex (Fig. 6, c and d).
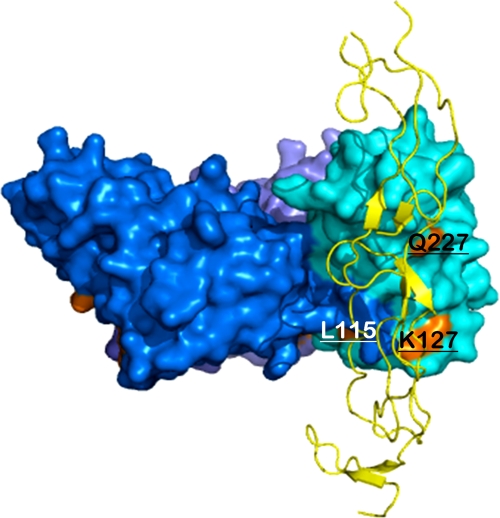
To map the receptor binding sites of the 4-1BBL trimer, we performed site-directed mutagenesis experiments guided by our modeled structure of the 4-1BBL-4-1BB complex and known structures of other TNF-TNFR complexes. According to our model, three residues of 4-1BBL (Lys127, Gln227, and Gln230) (Fig. 5) should be in close proximity to 4-1BB. We mutated these residues and measured the binding affinity of the mutants to 4-1BB expressed on Jurkat 8-1 cells. The Q230A mutant protein was not soluble and could not be used for binding studies, but mutation of residues Lys127 and Gln227 to alanine caused 170- and 80-fold decreases in binding affinity, respectively. Because Lys127 (A′B loop) and Gln227 (H strand) are located in an exposed surface away from the trimerization interface, the mutations are likely to have a direct effect on receptor binding rather than to interfere with trimerization. Interestingly, a L115G mutation (A′B loop) (Fig. 5) also caused a 6-fold decrease in affinity, whereas a Q89A mutation (AA′ loop) caused a 2-fold decrease (Fig. 4). Residues Leu115 and Gln89 are key residues in the assembly of the trimer because they interact with the C-terminal tail of the adjacent subunit. Thus, the decrease in affinity caused by these two mutations is most likely due to a destabilization of the trimer structure. Our mutagenesis data and known structures of other TNF-TNFR complexes suggest that the 4-1BBL-4-1BB interface is not concentrated at one site, but rather dispersed over the surface of 4-1BBL. However, from the structure of 4-1BBL the A′B loop is expected to be the major interaction site with 4-1BB, which correlates well with our model of the complex. The model also shows that the AB, BC, CD, and FG loops of the lower part in 4-1BBL are close to 4-1BB and are potentially involved in 4-1BB recognition. The AA′, CD, DE, and GH loops in other TNF family members have already been shown to be involved in receptor binding. Based on the overall structure of the complex visualized by EM and the fact that the DE loop in 4-1BBL does not seem to be involved in 4-1BB binding, it appears as if 4-1BBL binds 4-1BB in a somewhat different way from the interaction seen in the TNFβ-TNFR1 complex (14).
FIGURE 5.
Model of the 4-1BBL-4-1BB complex showing the interaction sites of 4-1BBL and 4-1BB. The structure of the 4-1BBL-4-1BB complex was modeled based on the known structure of the TNFβ-TNFR1 complex (Protein Data Bank code 1TNR). 4-1BBL residues involved in receptor binding, Leu115, Lys127, and Gln227, are labeled and colored in orange.
DISCUSSION
In this study, we present the crystal structure of human 4-1BBL, which shows structural features distinct from other TNF ligands. Rather than extending from the inner sheet of the molecule, the N terminus, which was well defined in the electron density map, extends from the outer sheet, on the opposite side from the C terminus. 4-1BBL also has the longest A′B loop of all known members of the TNF family because of the lack of strands corresponding to the A′ and B′ strands in other TNF molecules. In particular, the 4-1BBL trimer resembles a three-bladed propeller, which is different from other TNF trimers. Based on known structures of other TNF-TNFR complexes, receptor binding is unlikely to cause substantial conformational changes in 4-1BBL. For example, the rmsd between the structures of free hTRAIL (1D2Q) and DR5-bound trail (1DU3) is only 0.470 Å (121 atoms).
Compared with the canonical TNFα trimer, the trimer formed by e4-1BBL is stabilized by only a very small monomer-monomer interface, which is due to the more planar organization of the subunits in the trimer. The buried solvent-accessible surface area in 4-1BBL is 2,889 Å2, whereas it is 7,236 Å2 in TNFα. The buried solvent-accessible surface area of hGITRL (3,172 Å2) is comparable with that of 4-1BBL (19). However, whereas hGITRL exists in solution in a dynamic monomer-trimer equilibrium (19), gel filtration chromatography showed that the 4-1BBL trimer is as stable in solution as trimers formed by other members of the TNF family. The unique organization of the 4-1BBL trimer raised the question of whether the trimeric crystal structure represents the physiological form of 4-1BBL or whether it is a crystallization artifact. This question could be answered by negative stain EM because the class averages also revealed trimeric particles that resembled the three-bladed propeller, thus confirming that 4-1BBL also exists as a trimer in solution.
To understand the interaction of 4-1BBL with its receptor, we modeled the 4-1BBL-4-1BB complex based on the crystal structure of the TNFβ-TNFR1 complex. The resulting model of the complex had a windmill-like appearance. Projection averages of the 4-1BBL-4-1BB complex obtained by negative stain EM showed a similar shape, thus providing support for the modeled structure of the complex (Fig. 6,b and d). Because the three-bladed propeller structure of the 4-1BBL trimer is unique among TNF family members, we anticipated that the receptor-binding site might also differ from that of other TNF family members. According to our model of the complex and supported by our mutagenesis experiments, 4-1BBL may not use the DE loop, which has been reported to be important for receptor binding in other members of the TNF family. 4-1BBL may, however, still use the A′B loop for receptor binding like all of the other members of the TNF family.
In general, members of the TNF family form a symmetric 3:3 complex with their receptors, with each receptor binding to the monomer-monomer interface of the ligand trimer. Upon ligand binding, the cytoplasmic domains of the receptors become fixed in a specific conformation, resulting in the recruitment of adaptor and signaling molecules for downstream signaling. The organization of the TNF trimer will thus determine the structure of the assembled receptor trimer and in this way affect downstream signaling. The crystal structure presented here shows that the 4-1BBL trimer has a more extended organization, suggesting that the distance between the cytoplasmic tails of 4-1BB might be greater than the distance between those of other canonical TNFR family members. A crystal structure of the 4-1BBL-4-1BB complex will be needed, however, to obtain more detailed information on the ligand-receptor interface.
The effects of TNF receptor-ligand interactions are bidirectional, with both the T cell and the APC receiving activation signals. 4-1BBL has been implicated in reverse signaling to APCs (27, 28) and is thought to influence the maturation of monocytes and macrophages. Although 4-1BBL delivers co-stimulatory signals to T cells for proliferation and survival by binding its receptor 4-1BB, 4-1BBL also transmits positive or negative signals to APCs depending on the antigen-specific T cell response. These 4-1BBL-induced reverse signals involve several molecules known to play roles in receptor-mediated signal transduction (29). For example, protein-tyrosine kinases p38 MAPK, ERK1, ERK2, MAPK/ERK kinase (MEK), phosphatidylinositol 3-kinase, and cAMP-dependent protein kinase are all involved in 4-1BBL-mediated signaling (29). In general, the cytoplasmic domains of TNF family members do not have conserved motifs for signal transmission, and many of them function in a secreted form. However, TNFα and the CD27 ligand carry a casein kinase I motif in their cytoplasmic domains, and these phosphorylation sites indicate that they can instigate reverse signaling (30). Interestingly, 4-1BBL also has a casein kinase I motif that may participate in reverse signaling (30). The distances between the C termini of adjacent subunits in TNFα, hGITRL, OX40L, and 4-1BBL trimers are ∼8, ∼20, ∼35, and ∼40 Å, respectively. The distance between the C termini of adjacent 4-1BBL subunits in the trimer is comparable with that of hGITRL and OX40L, which mediate co-stimulation through reverse signaling. These observations suggest that 4-1BBL may also use reverse signaling for its immunomodulatory effects in the 4-1BBL-4-1BB pathway. The height of the 4-1BBL-4-1BB complex (∼60 Å) is less than that of other TNF-TNFR complexes (∼80 Å) and the major histocompatibility complex-T cell receptor complex (∼120 Å). This is due to the planar structure of 4-1BB, which results in a more extended arrangement of the 4-1BBL-4-1BB complex. From this structural point of view, we propose that the additional tight interactions resulting from the binding of 4-1BBL to 4-1BB may allow the T cell and the APC to come closer to each other.
The present study on the structure of 4-1BBL and its interaction with 4-1BB shows that the 4-1BBL trimer differs from trimers formed by other known TNF family members, including hGITRL and OX40L. In addition, our model of the 4-1BBL-4-1BB complex suggests that the structure of the complex also differs from structures of previously studied TNF-TNFR complexes. Our structural analysis thus provides new insight into the molecular mechanism of 4-1BBL-4-1BB signaling, whose modulation is closely linked to cancer and autoimmune diseases.
Supplementary Material
Acknowledgments
We thank Dr. Kyung-Jin Kim and Ghyung Hwa Kim for assistance at Beamlines 6C and 4MX of the Pohang Light Source.
This work was supported, in whole or in part, by National Institutes of Health Grant GM62580. This work was also supported by Korea Research Foundation Grants KRF-2006-005-J04502 and KRF-2008-313-C00745 (to H.-S. C.), KRF-C00040 (to K. C.), and KRF-2005-201-E00008 and KRF-2005-084-E00001 (to B. S. K.); by the SRC Fund from the Korea Science and Engineering Foundation; and by the Arthritis Foundation Innovative Research Award (to B. S. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 2WAK) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TNF
- tumor necrosis factor
- TNFR
- TNF receptor
- EM
- electron microscopy
- THD
- TNF homology domain
- GST
- glutathione S-transferase
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- rmsd
- root mean square deviation
- MAPK
- mitogen-activated protein kinase
- ERK
- extracellular signal-regulated kinase
- GITRL
- glucocorticoid-induced TNF receptor ligand.
REFERENCES
- 1.Locksley R. M., Killeen N., Lenardo M. J. (2001) Cell 104, 487–501 [DOI] [PubMed] [Google Scholar]
- 2.Armitage R. J. (1994) Curr. Opin. Immunol. 6, 407–413 [DOI] [PubMed] [Google Scholar]
- 3.Gruss H. J., Dower S. K. (1995) Blood 85, 3378–3404 [PubMed] [Google Scholar]
- 4.Smith C. S., Parker L., Shearer W. T. (1994) J. Immunol. 153, 3997–4005 [PubMed] [Google Scholar]
- 5.Kwon B. S., Weissman S. M. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1963–1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollok K. E., Kim Y. J., Zhou Z., Hurtado J., Kim K. K., Pickard R. T., Kwon B. S. (1993) J. Immunol. 150, 771–781 [PubMed] [Google Scholar]
- 7.Pollok K. E., Kim Y. J., Hurtado J., Zhou Z., Kim K. K., Kwon B. S. (1994) Eur. J. Immunol. 24, 367–374 [DOI] [PubMed] [Google Scholar]
- 8.Vinay D. S., Kwon B. S. (1998) Semin. Immunol. 10, 481–489 [DOI] [PubMed] [Google Scholar]
- 9.Melero I., Shuford W. W., Newby S. A., Aruffo A., Ledbetter J. A., Hellström K. E., Mittler R. S., Chen L. (1997) Nat. Med. 3, 682–685 [DOI] [PubMed] [Google Scholar]
- 10.Ye Z., Hellström I., Hayden-Ledbetter M., Dahlin A., Ledbetter J. A., Hellström K. E. (2002) Nat. Med. 8, 343–348 [DOI] [PubMed] [Google Scholar]
- 11.Halstead E. S., Mueller Y. M., Altman J. D., Katsikis P. D. (2002) Nat. Immunol. 3, 536–541 [DOI] [PubMed] [Google Scholar]
- 12.Bertram E. M., Lau P., Watts T. H. (2002) J. Immunol. 168, 3777–3785 [DOI] [PubMed] [Google Scholar]
- 13.Vinay D. S., Cha K., Kwon B. S. (2006) J. Mol. Med. 84, 726–736 [DOI] [PubMed] [Google Scholar]
- 14.Banner D. W., D'Arcy A., Janes W., Gentz R., Schoenfeld H. J., Broger C., Loetscher H., Lesslauer W. (1993) Cell 73, 431–445 [DOI] [PubMed] [Google Scholar]
- 15.Cha S. S., Sung B. J., Kim Y. A., Song Y. L., Kim H. J., Kim S., Lee M. S., Oh B. H. (2000) J. Biol. Chem. 275, 31171–31177 [DOI] [PubMed] [Google Scholar]
- 16.Liu Y., Xu L., Opalka N., Kappler J., Shu H. B., Zhang G. (2002) Cell 108, 383–394 [DOI] [PubMed] [Google Scholar]
- 17.Compaan D. M., Hymowitz S. G. (2006) Structure 14, 1321–1330 [DOI] [PubMed] [Google Scholar]
- 18.Bodmer J. L., Schneider P., Tschopp J. (2002) Trends Biochem. Sci. 27, 19–26 [DOI] [PubMed] [Google Scholar]
- 19.Chattopadhyay K., Ramagopal U. A., Mukhopadhaya A., Malashkevich V. N., Dilorenzo T. P., Brenowitz M., Nathenson S. G., Almo S. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 19452–19457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabu C., Quéméner A., Jacques Y., Echasserieau K., Vusio P., Lang F. (2005) J. Biol. Chem. 280, 41472–41481 [DOI] [PubMed] [Google Scholar]
- 21.Fesik S. W. (2000) Cell 103, 273–282 [DOI] [PubMed] [Google Scholar]
- 22.Eck M. J., Sprang S. R. (1989) J. Biol. Chem. 264, 17595–17605 [DOI] [PubMed] [Google Scholar]
- 23.Goh C. R., Loh C. S., Porter A. G. (1991) Protein Eng. 4, 785–791 [DOI] [PubMed] [Google Scholar]
- 24.Yamagishi J., Kawashima H., Matsuo N., Ohue M., Yamayoshi M., Fukui T., Kotani H., Furuta R., Nakano K., Yamada M. (1990) Protein Eng. 3, 713–719 [DOI] [PubMed] [Google Scholar]
- 25.Schneider P., Bodmer J. L., Holler N., Mattmann C., Scuderi P., Terskikh A., Peitsch M. C., Tschopp J. (1997) J. Biol. Chem. 272, 18827–18833 [DOI] [PubMed] [Google Scholar]
- 26.Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arens R., Nolte M. A., Tesselaar K., Heemskerk B., Reedquist K. A., van Lier R. A., van Oers M. H. (2004) J. Immunol. 173, 3901–3908 [DOI] [PubMed] [Google Scholar]
- 28.Eissner G., Kolch W., Scheurich P. (2004) Cytokine Growth Factor Rev. 15, 353–366 [DOI] [PubMed] [Google Scholar]
- 29.Söllner L., Shaqireen D. O., Kwajah M. M., Wu J. T., Schwarz H. (2007) Cell Signal 19, 1899–1908 [DOI] [PubMed] [Google Scholar]
- 30.Watts A. D., Hunt N. H., Wanigasekara Y., Bloomfield G., Wallach D., Roufogalis B. D., Chaudhri G. (1999) EMBO J. 18, 2119–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Byun J. S., Kim D. U., Ahn B., Kwon B. S., Cho H. S. (2006) Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 23–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 33.Terwilliger T. C., Berendzen J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 571–579 [DOI] [PubMed] [Google Scholar]
- 34.Terwilliger T. C. (2001) Acta Crystallogr. D Biol. Crystallogr. 57, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 36.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 37.Pontius J., Richelle J., Wodak S. J. (1996) J. Mol. Biol. 264, 121–136 [DOI] [PubMed] [Google Scholar]
- 38.Ohi M., Li Y., Cheng Y., Walz T. (2004) Biol. Proced. Online 6, 23–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ludtke S. J., Baldwin P. R., Chiu W. (1999) J. Struct. Biol. 128, 82–97 [DOI] [PubMed] [Google Scholar]
- 40.Frank J., Radermacher M., Penczek P., Zhu J., Li Y., Ladjadj M., Leith A. (1996) J. Struct. Biol. 116, 190–199 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.