Abstract
The development of fibrosis promotes the differentiation of myofibroblasts, pro-fibrotic cells, which contribute to tissue dysfunction. Myofibroblast differentiation is dependent on actin assembly, which in response to force, is mediated by various actin-binding proteins including the mammalian Diaphanous-related formins (mDia). We examined the role of mDia in the mechano-sensing pathway that leads to force-induced expression of α-smooth muscle actin (SMA), a marker and critical determinant of myofibroblast differentiation. In cells treated with siRNA to knockdown mDia and then subjected to tensile force using collagen-coated magnetite beads attached to β1 integrins, actin assembly was inhibited at bead contact sites. Force-induced nuclear translocation of MRTF-A, a transcriptional co-activator of SMA, was reduced 50% by mDia knockdown. The expression of the transcriptional co-activator of SMA, serum response factor, was reduced by 50% after siRNA knockdown of mDia or by 100% in cells transfected with catalytically inactive mDia. Force-induced activation of the SMA promoter and SMA expression were blocked by knockdown of siRNA of mDia. In anchored collagen gel assays to measure myofibroblast-mediated contraction, knockdown of mDia reduced contraction by 50%. We conclude that mDia plays an important role in the development of force-induced transcriptional activation of SMA and myofibroblast differentiation.
Keywords: Cell/Differentiation, Actin, Collagen, Fibroblast, Integrin, Force, Myocardin-related Transcription Factor, Myofibroblast
Introduction
During the development of fibrosis in heart, skin, liver, and kidney, fibroblasts are activated to become myofibroblasts (1). These cells strongly express α-smooth muscle actin (SMA)3 and secrete abundant, disorganized collagen (2, 3), and other extracellular matrix molecules (4). Fibroblasts adhere to extracellular matrices through integrins (5, 6), cell surface receptors that provide sites for force transfer to the actin cytoskeleton and that are required for force-induced stimulation of SMA expression and myofibroblast differentiation (7). At sites of force transfer, focal adhesion proteins act as proximal sensors (8, 9) for translating mechanical forces into the signals that activate the Rho pathway (10). As a result, actin assembly is triggered (8), and there is enhanced nuclear translocation of the SMA transcriptional co-activator, myocardin-related transcription factor-A (MRTF-A) (10). Previous studies have shown that Rho signaling regulates the nuclear translocation of MRTF-A (11). Currently, the mechanisms by which force application regulates actin assembly that leads to myofibroblast differentiation are not well-defined.
Actin assembly is regulated by formin proteins, which are involved in many cytoskeletal processes such as cytokinesis, actin stress fiber formation, neurite outgrowth, and intracellular trafficking (12). Members of a subfamily of formins, the mammalian Diaphanous-related formins (mDia), interact with activated, GTP-bound Rho family small GTPases. Interactions between Rho and mDia disrupt the binding of the Rho-binding domain to the diaphanous autoregulatory domain in formins, thereby exposing the formin homology (FH) domains FH1 and FH2 and inducing actin assembly (13). The assembly of actin filaments and the formation of actin stress fibers require a balance between the activity of mDia and the ROCK family of Rho effector kinases (14). Although the activation of ROCK has been implicated in the regulation of mechanical force-induced actin assembly (10), the role of mDia in this process has not been determined. External force-induced formation of focal contacts can be mediated by active mDia1, which can bypass the requirement for ROCK-mediated myosin II contractility in focal contact assembly (9). Further, it has been suggested that formins such as mDia may mediate force-induced actin assembly at barbed ends of actin filaments by “leaky capping” as a result of the elasticity of formins (15).
A separate actin-dependent function of mDia involves the activation of serum response factor (SRF) (16), a transcription factor that regulates many growth factor-inducible and muscle-specific genes, including SMA (17). Activated mDia up-regulates SRF transcription, a process that is directly related to its capacity to induce actin assembly and diminish the pool of actin monomers (18). We tested the hypothesis that in response to force, mDia promotes actin assembly and enhances SMA promoter activity as a result of the release of MRTF-A from actin monomers. Our results demonstrate that mDia is required for actin assembly at sites of mechanical force and that mDia induces the nuclear translocation of MRTF-A, the expression of SRF and the enhancement of SMA promoter activity. These processes lead ultimately to increased SMA expression and cell contractility, hallmarks of myofibroblast differentiation (1).
MATERIALS AND METHODS
Reagents
Latex (2-μm diameter) beads were purchased from Polysciences (Warrington, PA). Antibodies to β-actin (clone AC-15), human gelsolin (clone GS-2C4), and rabbit polyclonal IgG conjugated to horseradish peroxidase, as well as fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody, tetramethylrhodamine B isothiocyanate-phalloidin, and Y-27632 were from Sigma-Aldrich. FITC-goat anti-rabbit, anti-mouse β1 integrin, and anti-GAPDH antibodies were purchased from Cedarlane (Hornby, ON). A polyclonal antibody that recognizes MRTF-A was generously donated by H. Nakano (Juntendo University School of Medicine, Tokyo, Japan). Anti-ROCK-1 antibody was obtained from Millipore (Billerica, MA). A Myc-tagged, dominant-negative mDia construct in which the catalytically active F1F2 domains (pEFN.F1F2d1) were deleted and a constitutively active mDia construct in which the autoinhibitory domains (pEFN.F1F2+C) of mDia were deleted were produced by one of us (J. W. C.). Antibody to human mDia1 (clone C-20) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). The BCA protein assay kit was purchased from Pierce.
Cells
Human embryonic kidney (HEK-293) cells or human gingival fibroblasts (HGF; passages 5–12) were cultured at 37 °C in complete DME medium containing 10% fetal bovine serum and a 1:10 dilution of an antibiotic solution (0.17% w/v penicillin V, 0.1% gentamycin sulfate, and 0.01% μg/ml amphotericin). Cells were maintained in a humidified incubator gassed with 95% air and 5% CO2, and were passaged with 0.01% trypsin (Invitrogen, Burlington, ON). Prior to force experiments, cells were incubated in DME medium containing 0.5% serum, incubated with beads, and force was applied for various time periods.
Force Application
A force generation model was used as described previously (19, 20). Magnetite 2-μm diameter beads (Spherotech, Lake Forest, IL) were coated with collagen as described (20), rinsed in phosphate-buffered saline, and attached to the dorsal surface of cultured cells. A ceramic permanent magnet (Jobmaster, Mississauga, ON) was used to generate tensile forces (0.6 pN/μm2) (21) perpendicular to the dorsal surface of the cell. To restrict examination of mechanosensory events to periods of time prior to bead internalization (determined by trypan blue quenching of FITC-collagen-coated beads), forces were applied to cells only up to 60 min.
Collagen Bead-associated Proteins
Collagen or BSA-coated magnetite 2-μm diameter beads (Spherotech) were attached to 7 × 106 cells at a 10:1 bead/cell ratio for 30 min. Cells and collagen-coated magnetic beads were collected by scraping into ice-cold cytoskeletal extraction buffer (0.5% Triton X-100, 50 mm NaCl, 300 mm sucrose, 3 mm MgCl2, pH 6.8) containing protease inhibitors. BSA-coated beads attach to cells by nonspecific interactions and allow comparisons of proteins associated with a broad array of membrane proteins to be contrasted with those associating with β1 integrins (22–24). Beads were separated from cell lysates using a side-pull magnet. Bead-associated proteins were removed from beads with sample buffer, quantified (by BCA assay), separated on SDS-PAGE gels, and immunoblotted for specific proteins as indicated.
Immunofluorescence and Confocal Microscopy
Cells plated on beads were allowed to spread and bind to collagen beads for 30 min. Cells were fixed with 3% formaldehyde in PBS, permeabilized with 0.2% Triton X-100, and stained with the appropriate primary antibody followed by FITC or rhodamine-tagged secondary antibody. Spatial distributions of protein staining around beads were determined by confocal microscopy (Leica, Heidelberg, Germany; 40×, 1.4 numerical aperture oil immersion lens). Transverse optical sections were obtained at 1-μm nominal thickness.
siRNA Knockdown
Specific inhibition of human mDia and gelsolin were conducted with siRNAs (Dharmacon) with the following sequences: mDia (25)- 5′-AAAGGCAGAGCCACACUUCCU-3′, 5′-AGGAAGUGUGGCUCUGCCUUUUU-3′; gelsolin (26)- 5′-GCAAUCGGUAUGAAAGACUUU, 3′-AGUCUUUCAUACCGAUUGCUU-3′. Inhibition of Human ROCK1 was obtained from Silencer® Select Validated siRNA (Ambion) with the following sequences, 5′-CGGUUAGAACAAGAGGUAAUU-3′, 5′-UUACCUCUUGUUCUAACCGUU-3′. Cells were transfected with gene-specific siRNA (200 nm) or a negative control siRNA targeting GFP (Dharmacon) using DharmaFECT transfection reagent 2 for 48 h. If transfections of cDNA constructs were also required in experiments, the siRNA transfections were conducted for a total of 72 h. Cells were washed in phosphate-buffered saline, lysed with Laemmli buffer, and immunoblotted to assess the efficacy of the siRNA-dependent knockdown. After transfection and before force experiments, cells were incubated in DME medium containing 0.5% serum.
Transfection and Luciferase Promoter Studies
Cells were transfected with a 765-base pair rat SMA luciferase construct (Raphael Nemenoff, University of Colorado, Denver, CO) or an SRF reporter (p3D.ALuc) luciferase construct (27) and were co-transfected with a β-galactosidase construct as a control to normalize for variations of transfection efficiency. Transfections were done with FuGENE 6 (Roche Applied Science) according to the manufacturer's instructions. Cells were incubated with normal growth medium (5% serum in DME medium) within the first 36 h, and then cells were cultured in serum-reduced conditions (0.5% serum in DME medium) overnight. After transfections for 24 h, cells were loaded with collagen-coated beads, and magnetic force was applied for specific time periods. Cells were harvested, and luciferase and β-galactosidase activities were determined as described (28). Transfection data were computed as the fold change compared with basal promoter activity normalized to β-galactosidase activity.
Statistical Analyses
All experiments were repeated at least three times on separate days and with separate groups of cells. For continuous variables, means and standard errors of means were computed. Differences between groups were evaluated by Student's unpaired t test or analysis of variance for multiple comparisons. Statistical significance was set at p < 0.05. Post hoc comparisons were performed with Tukey's test. For all experiments, at least three independent experiments were evaluated, each performed in triplicate.
RESULTS
mDia Mediates Force-induced Actin Assembly
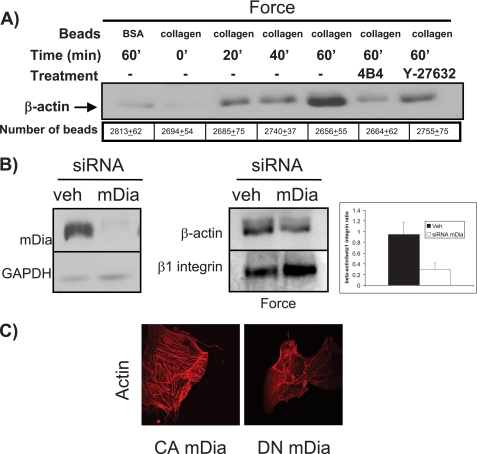
We examined the recruitment of β-actin to sites of mechanical force application by immunoblotting collagen-bead associated-proteins. There was a time-dependent increase of β-actin, which peaked at 60 min (Fig. 1A), indicating the formation of actin filaments at force-application sites (29). In cells incubated with BSA-coated beads or in cells preincubated with a β1 integrin blocking antibody (4B4) or in cells pretreated with the Rho-associated kinase inhibitor Y-27632, there was no increase of β-actin recruitment to beads after 60 min of force application. The numbers of beads isolated from each sample were counted to ensure that equivalent amounts (p > 0.2) of bead-associated proteins were isolated from cells in each experiment.
FIGURE 1.
Force-induced actin assembly is dependent on mDia. A, immunoblot analysis of collagen-coated bead-associated proteins over a 60-min time course after force application. The numbers of beads isolated in each sample were counted and used to estimate equivalence of protein loading. BSA-coated beads were used as nonspecific adhesion controls. A β1 integrin blocking antibody, 4B4, and the ROCK inhibitor, Y-27632, were used as negative controls. B, cells with mDia knockdown by siRNA were incubated with collagen-coated magnetite beads and subjected to force. Immunoblots of bead-associated proteins show β-actin recruitment. β1-Integrin immunoblots were used as loading controls. Left panel shows efficacy of siRNA knockdown of mDia. The ratios of β-actin to β1-integrin were computed from densitometry measurements of immunoblots (mean ± S.E., right panel). C, confocal images show actin-filament staining with rhodamine-phalloidin in cells transfected with constitutively active (CA)- or dominant negative (DN)-mDia.
We determined the effect of mDia on force-induced β-actin accumulation around collagen beads by knocking down mDia with siRNA. Compared with cells treated with an irrelevant siRNA, cells treated with mDia siRNA exhibited a 2-fold decrease in force-induced β-actin recruitment to beads (Fig. 1B). β1-Integrin immunoblots of bead-associated proteins from all samples showed similar amounts of protein, indicating that equivalent numbers of beads and bead-associated proteins were analyzed for each lane. We compared the organization of actin filaments in cells transfected with a constitutively active (CA) or a dominant-negative (DN) mDia construct. After staining with rhodamine-phalloidin and examination by confocal microscopy, we found very well-developed stress fibers in cells treated with CA mDia (Fig. 1C), whereas in cells treated with DN mDia stress fibers were sparse and poorly organized.
Transcriptional Activation of α-SMA
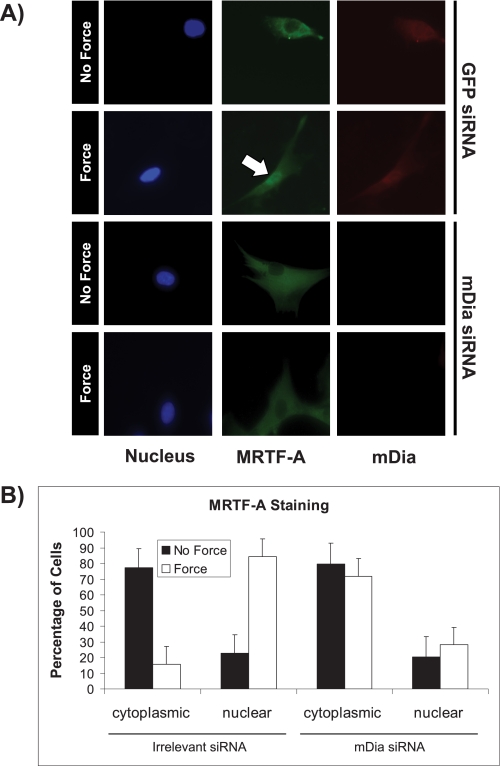
Actin polymerization requires the assembly of actin monomers into filaments, a process which releases myocardin-related transcription factor-A (MRTF-A) from actin monomers and enables the migration of MRTF to the nucleus where it can regulate transcription (18). Application of tensile force to fibroblasts triggers actin assembly and the nuclear translocation of cytoplasmic MRTF-A (10), which can then mediate serum response element (SRE)-regulated gene expression (28). We examined whether mDia is required for force-induced translocation of MRTF-A. In human gingival fibroblasts immunostained for endogenous MRTF-A, immunofluorescence images showed nuclear translocation of MRTF-A following force application (Fig. 2A). Quantification of the percentage of cells with nuclear or cytoplasmic localization of MRTF-A showed that in cells transfected with an irrelevant (green fluorescent protein) siRNA as a negative control, there was a 3.5-fold increase of the percentage of cells with MRTF-A nuclear translocation after 60 min of force application (p < 0.01). In contrast, cells treated with siRNA to knockdown mDia showed no change in the percentage of cells with force-induced MRTF-A nuclear translocation (Fig. 2B, p > 0.2).
FIGURE 2.
Force-induced MRTF-A nuclear translocation is dependent on mDia. A, immunofluorescence images showed the state of nuclear translocation of MRTF-A in cells treated with irrelevant or mDia siRNAs, incubated with collagen-coated magnetite beads and then subjected to force or no force. The white arrow shows the presence of MRTF-A in the nucleus. Cells were stained with DAPI to demonstrate nuclei (blue), and immunostained for MRTF-A (green) and mDia (red). B, immunofluorescence images were quantified after 60 min of force. Data are mean ± S.E. of the percentage of cells exhibiting MRTF-A nuclear translocation. In each experimental group 25 cells were quantified.
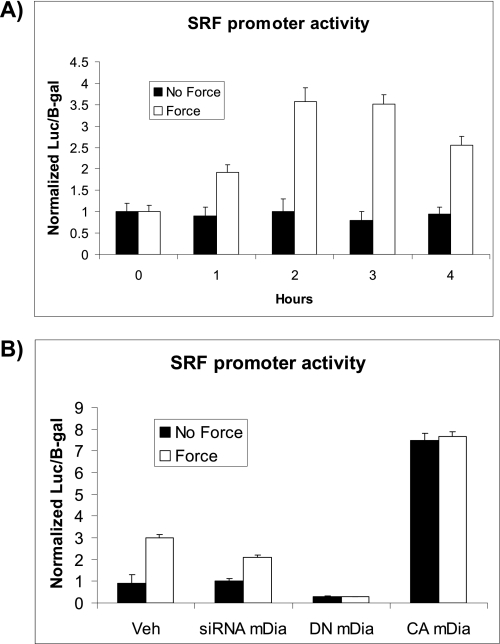
Mechanical force-induced nuclear translocation of MRTF-A alone is not sufficient to mediate SRE-regulated gene expression (30). MRTF-A also requires association with other transcriptional co-activators, most notably the serum response factor (SRF) (18). To determine if force altered the expression of SRF in this mechanotransduction model, we conducted time course experiments in which cells were co-transfected with a SRF reporter (3D.A; luciferase read-out) and lacZ constructs and then subjected to force application. This SRF reporter consists of three copies of the “D” binding site from the actin promoter and responds exclusively to the actin/MRTF pathway (27). The application of force to collagen-coated beads for 2 h caused a 3.5-fold increase of SRF-promoter activity (p < 0.02) in NIH 3T3 cells. In cells that were pretreated with siRNA to knockdown mDia, force-induced SRF-reporter activity after 2 h of force application was reduced by 33% (p < 0.02) compared with force-loaded cells with normal mDia levels (Fig. 3B). The potential regulation of SRF expression by mDia was investigated using a mutant mDia (13). Cells transfected with a DN-mDia construct exhibited no increase in force-induced SRF reporter activity, and the baseline expression was below that of untreated cells. However, transfection with a constitutively active (CA)-mDia construct enhanced SRF reporter activity equivalently in cells treated with or without force by 2.7-fold compared with force application in cells that were not transfected.
FIGURE 3.
mDia is required for force-induced SRF activation. A, force-induced SRF-luciferase promoter activity normalized to β-galactosidase was measured in NIH 3T3 cells. Cells were stimulated with force over a time course (1 to 4 h). Promoter activities are shown as fold-change compared with basal promoter activity. B, SRF promoter activity was measured after 2 h of force application in cells transfected with mDia siRNA, DN-mDia, CA-mDia, or vehicle controls.
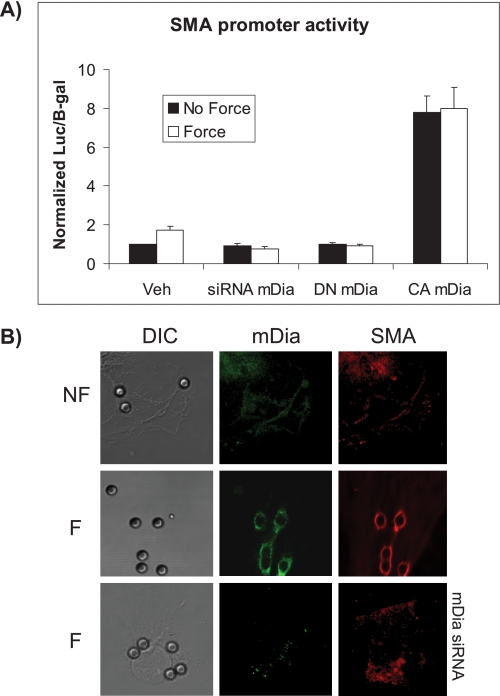
SRF and MRTF-A can up-regulate transcriptional activity of SRE-regulated genes, including SMA (10, 31). As the data above showed that mDia can affect force-induced nuclear translocation of MRTF-A, we examined the effect of mDia in force-induced SMA promoter activity. Cells that were transfected with a SMA-luciferase promoter construct and treated with siRNA mDia showed no significant enhancement (p > 0.2) of force-induced SMA promoter activity, but there was a 1.75-fold increase in cells treated with irrelevant siRNA (p < 0.02, Fig. 4A). Similarly, there was no increase in force-induced SMA promoter activity in cells transfected with DN-mDia. Compared with baseline levels, the transfection of CA mDia caused a 9-fold increase in SMA promoter activity that was indistinguishable in cells treated with or without force.
FIGURE 4.
Force-induced SMA activation is dependent on mDia. A, force-induced SMA-luciferase promoter activity normalized to β-galactosidase was measured after 4 h of force application in NIH 3T3 cells. Cells were also treated with mDia siRNA, DN mDia, or CA mDia. B, confocal images of cells with (F) or without (NF) 4 h of force application show staining of mDia with FITC-conjugated antibody and SMA with TRITC-conjugated antibody around single collagen-coated beads. DIC images of the same cells show the location of beads. Note that cells treated with siRNA for mDia show no detectable mDia.
By immunostaining and confocal microscopy we evaluated the spatial localization of mDia and SMA at collagen beads following force application. There was enhanced localization of both mDia and SMA at collagen-coated beads after 4 h of force application, consistent with the notion that formins such as mDia may act as proximal mechano-sensors in focal adhesions (9, 15) In cells without force treatment, there was little change of fluorescence intensity attributable to mDia (Fig. 4B), and in cells treated with mDia siRNA, there was very little accumulation of SMA around beads.
Relative Contribution of mDia in SMA Expression
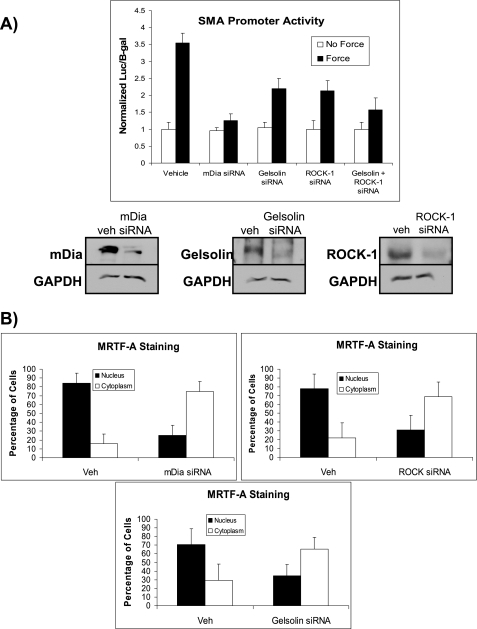
In addition to the effect of mDia on force-induced SMA transcription shown above, previous studies have also established roles for gelsolin and ROCK in regulating SMA expression (8, 10). To determine the relative contribution of each of these pathways on force-induced expression of SMA, we compared the effects of reducing the expression of gelsolin, ROCK and mDia on force-induced SMA promoter activity. In cells pretreated with siRNA to knockdown gelsolin, ROCK, or mDia, forced-induced SMA-promoter activity was reduced by 60, 68, and 80%, respectively compared with force-loaded cells with irrelevant siRNAs (Fig. 5A). The combined siRNA knockdown of gelsolin and Rho kinase produced an additional (75%) reduction of SMA promoter activity after double siRNA knockdown.
FIGURE 5.
Relative contribution of gelsolin, ROCK, and mDia pathways in SMA expression. A, NIH 3T3 cells were pretreated with siRNA to knockdown gelsolin, ROCK, or mDia. Force-induced SMA-luciferase promoter activity normalized to β-galactosidase was measured after 4 h of force application. Immunoblots show efficacy of knockdown of gelsolin, ROCK, and mDia with GAPDH as loading controls. B, measurements of nuclear translocation of MRTF-A in cells. Immunofluorescence images were quantified after 60 min of force in cells with or without siRNA knockdown of gelsolin, ROCK, and mDia. Cells were stained for MRTF-A (green) and with DAPI to identify location of nuclei (blue). Data are mean ± S.E. percentage of cells with MRTF-A nuclear translocation, from 25 cells for each experimental group.
Because tensile force-mediated nuclear translocation of MRTF-A is required for activation of SMA promoter, we compared the effect of gelsolin, ROCK, and mDia on force-induced MRTF-A nuclear translocation. Quantification of fixed cells immunostained for endogenous MRTF-A showed that force-induced MRTF-A nuclear translocation was inhibited by gelsolin, ROCK and mDia siRNA knockdown (Fig. 5B). However, there was no significant difference in the amount of reduction of MRTF-A nuclear translocation between the three treatment groups (p > 0.2).
mDia Is Required for Myofibroblast Differentiation
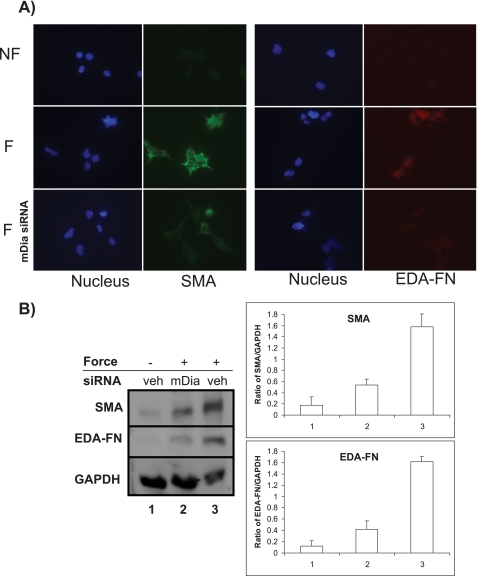
MRTF-A nuclear translocation contributes to the up-regulation of SMA, which, along with ED-A fibronectin, is a marker for myofibroblast differentiation (1). To determine the impact of mDia on force-induced myofibroblast differentiation, we examined the expression of SMA and ED-A fibronectin in NIH 3T3 cells subjected to tensile force. Cells incubated with collagen-coated magnetite beads in low serum (0.2%) were exposed to tensile forces (0.6 pN/μm2) by magnetic forces perpendicular to the dorsal surface of the cells for 3 days and were evaluated by immunofluorescence. Immunostaining for SMA and ED-A fibronectin was very low in cells not exposed to force (Fig. 6A) but there was abundant SMA and ED-A staining in force-loaded cells. In contrast, force-loaded cells in the presence of mDia siRNA showed no increase of SMA and ED-A fibronectin staining. Immunoblots of SMA and ED-A fibronectin showed increased SMA and ED-A expression in force-treated cells that was reduced in force-treated cells in which mDia siRNA was knocked down (Fig. 6B). Immunoblots for GAPDH were used as loading controls.
FIGURE 6.
Force-induced myofibroblasts differentiation is dependent on mDia. A, fluorescence images of NIH3T3 cells incubated with collagen-coated magnetite beads, and with (F) or without (NF) exposure to tensile force applied by a ceramic permanent magnet. Cells were immunostained for SMA (green) and ED-A fibronectin (red). Nuclei were stained with DAPI (blue). The presence of SMA and ED-A fibronectin indicates myofibroblastic phenotype. Middle panels show force-loaded cells after treatment with mDia siRNA. B, immunoblots show protein expression of SMA and ED-A fibronectin from experiments conducted in the same conditions as A. GAPDH is shown as loading controls.
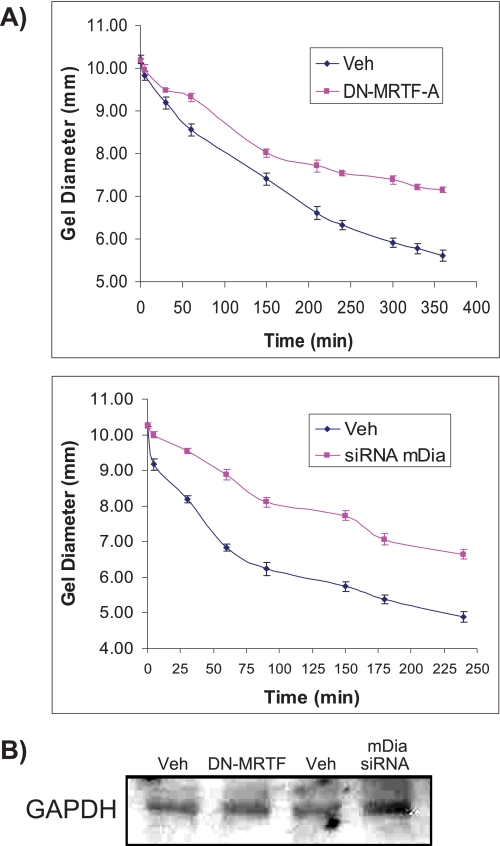
In addition to the expression of ED-A fibronectin and SMA, increased tractional remodelling of the extracellular matrix is a hallmark of myofibroblast differentiation (1). Accordingly we examined the role of mDia in the contraction of stress-relaxed collagen gels. Cells treated with siRNA for mDia or irrelevant GFP siRNA were incubated in collagen gels plated on rigid tissue culture plastic for 3 days. On day 3, the collagen gels were released from the dish (stress-relaxed gels), and gel diameter was measured over time. Knockdown of mDia compared with an irrelevant siRNA control, reduced by 2-fold the contraction of stress-relaxed collagen gels (Fig. 7A; p < 0.001 at all sampling times). Similarly, transfection of DN-MRTF-A inhibited the contraction of stress-relaxed collagen gels. Immunoblots of GAPDH from cells extracted from the collagen gels showed that equivalent numbers of cells were present in the gels (Fig. 7B).
FIGURE 7.
Gel contraction by fibroblasts requires MRTF-A and mDia. A, contraction of stress-relaxed collagen gels in cells treated with DN-MRTF-A or siRNA for mDia or vehicle controls. The data are means + S.E. for gel diameter over time after release of the gel from the dish. B, immunoblots of GAPDH from cell lysates isolated from collagen gels indicate equivalent number of cells were analyzed in each collagen gel sample.
DISCUSSION
Cultured fibroblasts respond to applied mechanical forces by undergoing alterations of shape and structure that include actin cytoskeletal remodelling and the formation of stress fibers, which lead to the development of the myofibroblast phenotype (8, 10, 28). Our principal finding is that mDia promotes actin assembly, which facilitates transfer of mechanical signals into downstream processes that promote SMA expression and myofibroblast differentiation. Overexpression or depletion of the actin-nucleating properties of mDia can disrupt this process by interfering with actin assembly. We have identified mDia as a crucial molecule in regulating the force-induced function of the transcriptional co-activators, MRTF-A and SRF. Further, we have shown that the impact of mDia on force-induced activation of SMA is comparable in magnitude to the actin assembly pathways regulated by gelsolin and ROCK (10). These data provide evidence for a mechanotransduction system involving mDia in the regulation of actin assembly and SMA expression in myofibroblast differentiation (Fig. 8) and are consistent with earlier suggestions that formins such as mDia may be important in force-driven actin assembly and could be a key element cellular mechanosensation (15).
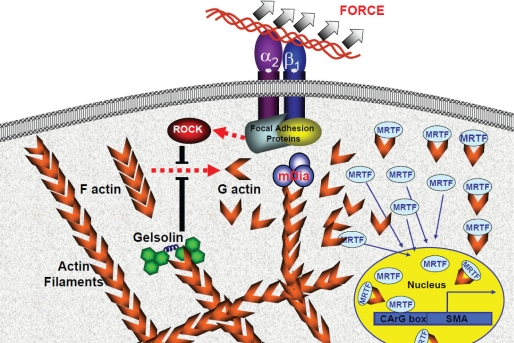
FIGURE 8.
Proposed model of mechanotransduction. Application of tensile forces to collagen beads triggers the recruitment and activation of focal adhesion proteins. In turn, focal adhesion proteins promote actin assembly by activating gelsolin and ROCK, which prevent actin filament disassembly. The nucleation of actin barbed ends by mDia enables actin filament growth and elongation. The dissociation of MRTF-A from actin monomers during actin assembly initiates the nuclear translocation of MRTF-A. In the nucleus, MRTF-A acts as transcriptional co-activator for the induction of SMA.
Actin cytoskeletal remodelling has been suggested as a mechanosensory process that can translate molecular signals in response to applied mechanical force (32, 33). mDia, which is an important actin-nucleating protein (34) and a critical mediator of actin nucleation, has been extensively studied in the maintenance of cell polarity, vesicular trafficking, signaling to the nucleus, and embryonic development (12). Recent studies have indicated that mDia is required in stress fiber formation in response to mechanical stretch (35) and is involved in sphingosine-1-phosphate signaling to stress fiber formation in fibroblasts (36). Mechanical tension triggers the localization of mDia to focal adhesions (9) and may mediate force-induced actin assembly involving formins such as mDia (15). When we examined collagen bead-associated proteins of cells that had been subjected to tensile force, we found an enrichment of β-actin filaments that was dependent on the expression of mDia. Previous data showed that shear stress-induced actin reorganization is dependent on Rho (37) and Rho activation has been invoked in tensile-force-induced SMA expression in myofibroblast differentiation (10). Because Rho binding to mDia has been reported to increase actin remodeling (38) and the activation of Rho causes MRTF-A nuclear translocation, which correlates with SMA expression (10), we examined how mDia is involved in SMA regulation. We found that increased nuclear translocation of MRTF-A by force was dependent on mDia. MRTF-A normally binds by its RPEL motif to actin monomers but dissociation of MRTF-A from actin monomers occurs when the cytoplasmic pool of actin monomers is diminished because of increased actin filament formation (39). Subsequently, MRTF-A is enriched in nuclei where it becomes available as a transcriptional co-activator for binding of transcription factors to the promoter region of genes with a force-responsive region, such as the CArG boxes of the core sequences in the serum response element of the SMA promoter (28).
The SRE is a binding site for many transcription factors and the serum response factor (SRF) is a primary transcriptional co-activator of the SRE that is responsible for controlling a number of genes that encode actin cytoskeletal and contractile proteins (40). Previous studies showed that serum- and LIMK-induced SRF expression is linked to the activity of mDia (16). We found that maximal transcriptional expression of SRF occurs 2 h after force application to cells. This response was dependent on the relative abundance of mDia because down-regulation of mDia by siRNA or transfection with a dominant negative mDia construct inhibited force-induced up-regulation of SRF. However, any observed force-induced up-regulation of SRF was likely not detectable in cells transfected with constitutively active mDia because of the very high level of expression of SRF.
Because mDia regulates the transcriptional modifying activities of SRF (16) and MRTF-A (18), which in turn control the expression of mechano-sensitive genes like SMA (10, 28), we investigated the requirement of mDia in the pathway leading to force-induced up-regulation of the SMA promoter. In cells transfected with a SMA-luciferase promoter construct, we found that mDia was required for force-induced up-regulation of the SMA promoter. The co-localization of mDia and SMA was also found at magnetite bead sites, which indicate that force-induced signaling to mDia may be through a specialized cell membrane domain containing the lipid raft marker, ganglioside GM1 (41). Further, we assessed the relative importance of mDia with other known actin assembly pathways (gelsolin (8) and ROCK (10)) that regulate force-induced SMA expression. The incomplete suppression of force-induced SMA-promoter activity after siRNA knockdown of gelsolin, ROCK or mDia may indicate that each of these actin assembly pathways contributes separately but incompletely to SMA activation. However, in our current study, since we did not achieve 100% knockdown of any of these proteins, we cannot estimate precisely their relative importance to regulation of force-induced SMA expression.
We studied the effect of mDia on myofibroblast differentiation by exposing fibroblasts for 3 days to chronic tensile forces and then examined SMA and ED-A fibronectin, markers of myofibroblast differentiation. Whereas it is likely that the attached collagen beads were internalized within 24 h of incubation and therefore were not applying tensile forces to cell-surface integrins after 24 h, nonetheless we found that 3-day force exposure enhanced the expression of SMA and ED-A fibronectin, which was suppressed by siRNA knockdown of mDia. Further, knockdown of mDia or transfection of cells with dominant negative MRTF-A, markedly reduced the ability of cells to contract collagen gels. Taken together, these data suggest a mechanism for force-induced myofibroblast differentiation by which force induces the actin nucleating activity of mDia (9, 15) that leads subsequently to MRTF-A translocation and SMA expression.
This work was supported by CIHR Operating Grant MGP-37783, a CIHR resource grant, and Heart and Stroke Foundation Grant T-6022 (to C. A. M.).
- SMA
- α-smooth muscle actin
- mDia
- mammalian diaphanous
- FITC
- fluorescein isothiocyanate
- BSA
- bovine serum albumin
- GFP
- green fluorescent protein
- SRE
- serum response element
- SRF
- serum-response factor
- DAPI
- 4′,6-diamidino-2-phenylindole
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1.Tomasek J. J., Gabbiani G., Hinz B., Chaponnier C., Brown R. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 349–363 [DOI] [PubMed] [Google Scholar]
- 2.Leslie K. O., Taatjes D. J., Schwarz J., vonTurkovich M., Low R. B. (1991) Am. J. Pathol. 139, 207–216 [PMC free article] [PubMed] [Google Scholar]
- 3.Sun Y., Weber K. T. (1996) Cardiovasc. Res. 31, 518–525 [PubMed] [Google Scholar]
- 4.Webber J., Meran S., Steadman R., Phillips A. (2009) J. Biol. Chem. 284, 9083–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacKenna D. A., Dolfi F., Vuori K., Ruoslahti E. (1998) J. Clin. Invest. 101, 301–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gullberg D., Gehlsen K. R., Turner D. C., Ahlén K., Zijenah L. S., Barnes M. J., Rubin K. (1992) EMBO J. 11, 3865–3873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hinz B., Gabbiani G. (2003) Curr. Opin. Biotechnol. 14, 538–546 [DOI] [PubMed] [Google Scholar]
- 8.Chan M. W., Arora P. D., Bozavikov P., McCulloch C. A. (2009) J. Cell Sci. 122, 2769–2781 [DOI] [PubMed] [Google Scholar]
- 9.Riveline D., Zamir E., Balaban N. Q., Schwarz U. S., Ishizaki T., Narumiya S., Kam Z., Geiger B., Bershadsky A. D. (2001) J. Cell Biol. 153, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X. H., Laschinger C., Arora P., Szászi K., Kapus A., McCulloch C. A. (2007) J. Cell Sci. 120, 1801–1809 [DOI] [PubMed] [Google Scholar]
- 11.Wang D., Chang P. S., Wang Z., Sutherland L., Richardson J. A., Small E., Krieg P. A., Olson E. N. (2001) Cell 105, 851–862 [DOI] [PubMed] [Google Scholar]
- 12.Wallar B. J., Alberts A. S. (2003) Trends Cell Biol. 13, 435–446 [DOI] [PubMed] [Google Scholar]
- 13.Copeland J. W., Copeland S. J., Treisman R. (2004) J. Biol. Chem. 279, 50250–50256 [DOI] [PubMed] [Google Scholar]
- 14.Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. (1999) Nat. Cell Biol. 1, 136–143 [DOI] [PubMed] [Google Scholar]
- 15.Kozlov M. M., Bershadsky A. D. (2004) J. Cell Biol. 167, 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Geneste O., Copeland J. W., Treisman R. (2002) J. Cell Biol. 157, 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Browning C. L., Culberson D. E., Aragon I. V., Fillmore R. A., Croissant J. D., Schwartz R. J., Zimmer W. E. (1998) Dev. Biol. 194, 18–37 [DOI] [PubMed] [Google Scholar]
- 18.Miralles F., Posern G., Zaromytidou A. I., Treisman R. (2003) Cell 113, 329–342 [DOI] [PubMed] [Google Scholar]
- 19.Glogauer M., Arora P., Chou D., Janmey P. A., Downey G. P., McCulloch C. A. (1998) J. Biol. Chem. 273, 1689–1698 [DOI] [PubMed] [Google Scholar]
- 20.Glogauer M., Ferrier J., McCulloch C. A. (1995) Am. J. Physiol. 269, C1093–C1104 [DOI] [PubMed] [Google Scholar]
- 21.Glogauer M., Ferrier J. (1998) Pflugers Arch. 435, 320–327 [DOI] [PubMed] [Google Scholar]
- 22.Arora P. D., Chan M. W., Anderson R. A., Janmey P. A., McCulloch C. A. (2005) Mol. Biol. Cell 16, 5175–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arora P. D., Glogauer M., Kapus A., Kwiatkowski D. J., McCulloch C. A. (2004) Mol. Biol. Cell 15, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arora P. D., Manolson M. F., Downey G. P., Sodek J., McCulloch C. A. (2000) J. Biol. Chem. 275, 35432–35441 [DOI] [PubMed] [Google Scholar]
- 25.Unsworth K. E., Way M., McNiven M., Machesky L., Holden D. W. (2004) Cell Microbiol. 6, 1041–1055 [DOI] [PubMed] [Google Scholar]
- 26.Walsh N., Dowling P., O'Donovan N., Henry M., Meleady P., Clynes M. (2008) J. Proteomics 71, 561–571 [DOI] [PubMed] [Google Scholar]
- 27.Sotiropoulos A., Gineitis D., Copeland J., Treisman R. (1999) Cell 98, 159–169 [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Su M., Fan J., Seth A., McCulloch C. A. (2002) J. Biol. Chem. 277, 22889–22895 [DOI] [PubMed] [Google Scholar]
- 29.Glogauer M., Arora P., Yao G., Sokholov I., Ferrier J., McCulloch C. A. (1997) J. Cell Sci. 110, 11–21 [DOI] [PubMed] [Google Scholar]
- 30.Cen B., Selvaraj A., Burgess R. C., Hitzler J. K., Ma Z., Morris S. W., Prywes R. (2003) Mol. Cell. Biol. 23, 6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanna M., Liu H., Amir J., Sun Y., Morris S. W., Siddiqui M. A., Lau L. F., Chaqour B. (2009) J. Biol. Chem. 284, 23125–23136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Botvinick E. L., Zhao Y., Berns M. W., Usami S., Tsien R. Y., Chien S. (2005) Nature 434, 1040–1045 [DOI] [PubMed] [Google Scholar]
- 33.Dickinson R. B., Caro L., Purich D. L. (2004) Biophys. J. 87, 2838–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Narumiya S., Tanji M., Ishizaki T. (2009) Cancer Metastasis Rev. 28, 65–76 [DOI] [PubMed] [Google Scholar]
- 35.Kaunas R., Nguyen P., Usami S., Chien S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15895–15900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Syuto T., Abe M., Yokoyama Y., Ishikawa O. (2009) Wound Repair Regen. 17, 589–597 [DOI] [PubMed] [Google Scholar]
- 37.Wojciak-Stothard B., Ridley A. J. (2003) J. Cell Biol. 161, 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li F., Higgs H. N. (2003) Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 39.Mouilleron S., Guettler S., Langer C. A., Treisman R., McDonald N. Q. (2008) EMBO J. 27, 3198–3208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miano J. M., Long X., Fujiwara K. (2007) Am. J. Physiol. Cell Physiol. 292, C70–C81 [DOI] [PubMed] [Google Scholar]
- 41.Palazzo A. F., Eng C. H., Schlaepfer D. D., Marcantonio E. E., Gundersen G. G. (2004) Science 303, 836–839 [DOI] [PubMed] [Google Scholar]