SUMMARY
Antibiotics inhibiting translation can increase bacterial growth rate in the presence of DNA synthesis inhibitors. Here, we show that this extreme type of drug antagonism, termed suppression, results from non-optimal regulation of ribosomal genes, leading to sub-maximal growth in the presence of DNA stress. Using GFP-tagged transcription reporters in Escherichia coli, we find that ribosomal genes are not directly regulated by DNA stress, leading to an imbalance between cellular DNA and protein content. Sequential deletion of up to 6 of the 7 ribosomal RNA operons corrects this imbalance and leads to improved survival and growth under DNA synthesis inhibition. Further, this genetic manipulation completely removes the suppressive drug interaction. Mathematical modeling shows that non-optimal regulation of ribosome synthesis under DNA stress can be explained as a side-effect of optimal growth-rate-dependent regulation in different nutrient environments. Together, these results reveal the genetic mechanism underlying an important class of suppressive drug interactions.
INTRODUCTION
Drug combinations can be an important tool for studying biological systems and revealing relationships between different cellular processes (Keith et al., 2005; Lehar et al., 2008; Lehar et al., 2007; Tsui et al., 2004). The interaction between two drugs can be classified as additive, synergistic, or antagonistic according to their combined effect being equal, greater, or less than that expected based on their individual effects, Figure 1A (Bliss, 1939; Loewe, 1928; Loewe, 1953; Pillai et al., 2005). Much attention has been given to synergistic drug combinations due to their increased potency. Antagonism, however, may have an advantage in slowing down and even reversing the evolution of resistance (Chait et al., 2007; Hegreness et al., 2008; Michel et al., 2008; Yeh et al., 2006).
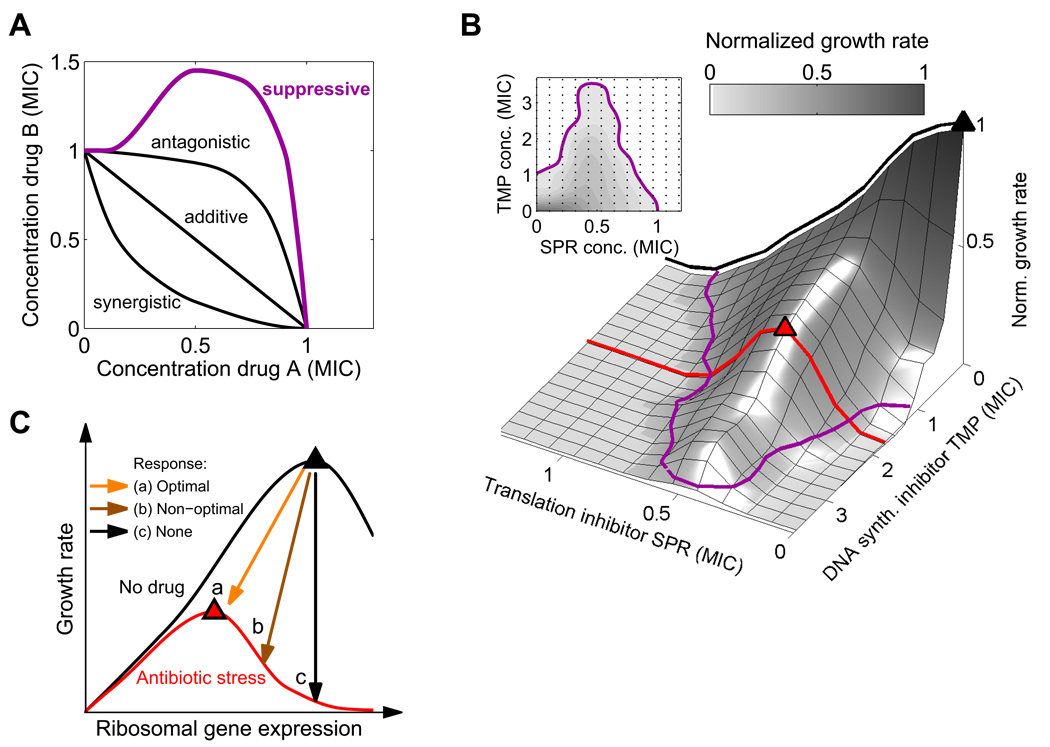
Figure 1. Suppression of DNA synthesis inhibitors by translation inhibitors suggests the hypothesis that ribosomal genes are not optimally regulated under DNA stress.
(A) MIC lines (isoboles) in the two-dimensional concentration space of two drugs. Two drugs are defined to interact additively if their combined effect is constant along linear lines of fixed total dosage (Loewe, 1928). Synergy and antagonism are defined as negative or positive deviations from this null line. A particularly strong type of antagonism – ‘suppression’ – characterizes drug pairs whose combined effect is weaker than that of one of the drugs alone (magenta line). (B) Suppressive interaction is seen in measurements of growth rates (gray levels) and MIC line (magenta) in a two-dimensional gradient of the translation inhibitor spiramycin (SPR) and the DNA synthesis inhibitor trimethoprim (TMP, inhibitor of DNA synthesis through folic acid deficiency). In the absence of DNA synthesis inhibitor (black line), growth rate is maximal without translation inhibition (black triangle) and reduces monotonically with the level of translation inhibitor. In contrast, at fixed finite concentration of DNA synthesis inhibitor (red line), growth rate increases initially as the translation inhibitor concentration increases, reaching an optimal value (red triangle) at intermediate translation inhibition level. (C) Schematic expectation for growth rate as a function of ribosomal gene expression in absence (black line) or presence (red line) of an antibiotic. Arrows show possible ribosomal gene expression regulation in response to antibiotic addition. The comparison of panels B and C suggests the hypothesis that a non-optimal, too high ribosome level in response to DNA synthesis inhibitors may cause this suppressive drug interaction. MICs for antibiotics are summarized in Table 1.
A particularly strong kind of antagonism, termed suppression, occurs when the combined inhibitory effect of two drugs is not only weaker than the expected additive sum, but also weaker than the effect of one of the drugs alone (Figure 1A; Pillai et al., 2005). We have previously reported that in the presence of a DNA synthesis-inhibiting antibiotic, the addition of a protein synthesis inhibitor increases the steady state growth rate of Escherichia coli (Figure 1B) and Staphylococcus aureus (Chait et al., 2007; Yeh et al., 2006). Many different pairings of DNA synthesis and translation inhibitors show this suppressive drug interaction (Yeh et al., 2006; see examples in Figure 1B and Figure S1), indicating that these interactions result from the effect of the drugs on bacterial physiology rather than from direct chemical interaction between the drugs. Considerable recent work has advanced our understanding of the effects of individual antibiotics on gene expression and cellular physiology (Brazas and Hancock, 2005; Davies et al., 2006; Drlica et al., 2008; Fajardo and Martinez, 2008; Goh et al., 2002; Hoffman et al., 2005; Kohanski et al., 2007; Kohanski et al., 2008; Kolodkin-Gal et al., 2008; Linares et al., 2006; Mason et al., 1995; Mesak et al., 2008; Piddock et al., 1990; Shaw et al., 2003; Yim et al., 2006; Yim et al., 2007), but the effects of drug combinations are less well understood and the mechanism that underlies suppressive drug interactions remains unknown.
It has been argued that many aspects of bacterial physiology have evolved to be ‘optimal’ - namely to maximize growth rate in a given condition (Dekel and Alon, 2005; Ibarra et al., 2002; Liebermeister et al., 2004). When protein synthesis inhibitors are added to DNA synthesis inhibitors, however, the cells actually grow faster. Thus, the overall rate of protein synthesis under DNA stress appears to be above the optimal value for maximum growth. This overall rate of protein synthesis is primarily determined by the number of ribosomes per cell, which is known to be tightly controlled (Gralla, 2005; Keener and Nomura, 1996; Moss, 2004; Paul et al., 2004). Precise regulation of ribosome synthesis is crucial for maximizing growth: under-production of ribosomes causes ineffective use of cellular resources, while over-production leads to an excess use of resources for protein synthesis at the expense of other cellular processes (Gralla, 2005; Keener and Nomura, 1996; Levy et al., 2007; Paul et al., 2004). As a result, in any particular environment, there exists an optimal level of ribosomes that maximizes the bacterial growth rate (Figure 1C). The observation that, under DNA stress, reduction in protein synthesis allows faster growth suggests that ribosome level is not optimally regulated in these conditions.
The rate of ribosome synthesis in E. coli is determined by the transcription rate of the ribosomal RNA operons (rrn operons; Keener and Nomura, 1996; Paul et al., 2004), which code for the three different ribosomal RNAs. Feedback mechanisms at the level of translation adjust ribosomal protein synthesis to stoichiometrically match rRNA production (Keener and Nomura, 1996). The standard E. coli lab strain K12 MG1655 has seven almost identical copies of the rrn operons, which are among the most highly transcribed loci in the genome. Multiple copies are needed because the maximal transcription rate from a single rrn operon is insufficient for the ribosome synthesis required at high growth rates (Condon et al., 1995; Stevenson and Schmidt, 2004).
The rrn operons are regulated to achieve maximal growth in different nutrient environments. The levels of factors that reflect intracellular levels of resources such as amino acids and energy, including nucleoside triphosphates (NTPs), guanosine pentaphosphate and tetraphosphate (collectively referred to as ppGpp), affect rrn transcription (Cashel et al., 1996; Dennis et al., 2004; Gaal et al., 1997; Keener and Nomura, 1996; Paul et al., 2004; Schneider et al., 2002; Schneider and Gourse, 2004). Overproduction of protein depletes these resources and thus down-regulates ribosome synthesis (Gralla, 2005; Paul et al., 2004). In many environmental conditions, this negative feedback loop is able to maintain ribosome concentration near its optimal level (the level that maximizes growth rate); in particular, ribosome synthesis is kept high in nutrient-rich environments and is shut down as a consequence of nutrient starvation (Cashel et al., 1996). However, it is unclear if the regulation of ribosome synthesis leads to optimal expression levels (maximal growth) under all stress conditions, and in particular under DNA stress (Figure 1C).
The observation that, under DNA stress, inhibition of protein synthesis actually increases the rate of cellular growth suggests that the number of ribosomes per cell in these conditions is too high. Overexpression of ribosomes under these conditions would lead to an inefficient use of cellular resources and a growth rate that is lower than could be maximally achieved. Here, we test the hypothesis that the rate of ribosome synthesis in bacteria is non-optimal under DNA stress, and that this non-optimality causes the suppressive drug interactions in which translation inhibitors allow faster growth under DNA synthesis inhibition. We address this hypothesis by measuring cell composition, morphology and gene expression changes in response to antibiotics, by genetically manipulating ribosome synthesis, and by using a theoretical analysis of resource allocation in the cell.
RESULTS
Protein-DNA ratio is skewed under DNA stress
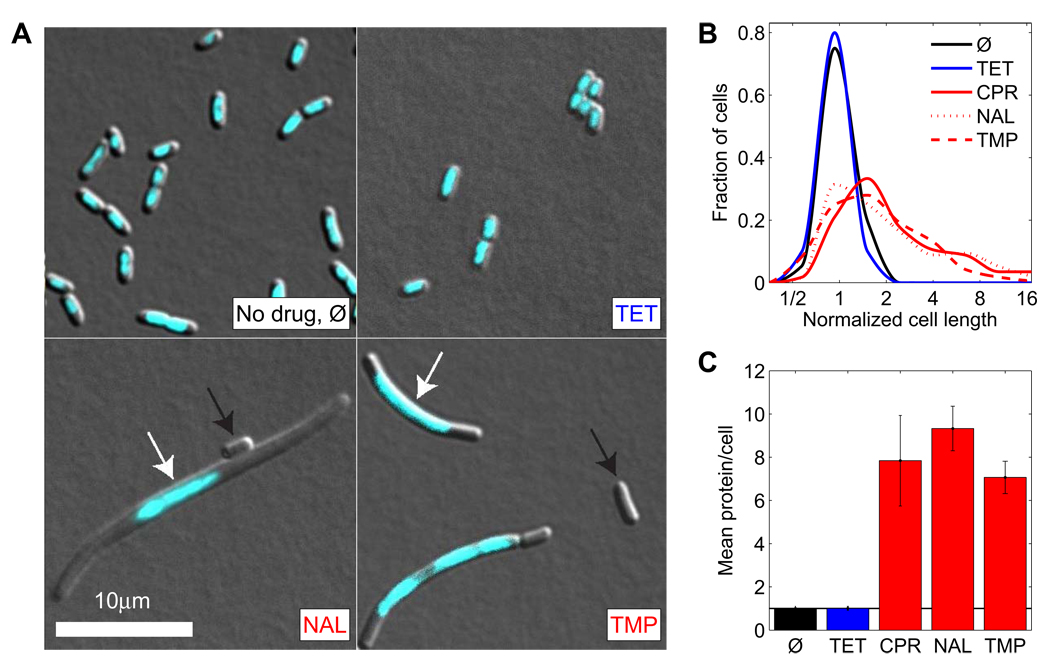
We first examined the changes in cell morphology and composition that result from treating cells with DNA synthesis inhibitors. These inhibitors cause DNA damage and trigger the SOS response, including expression of the cell division inhibitor sulA (Huisman and D'Ari, 1981; Mesak et al., 2008; Walker, 1996). This prevents cell division before chromosome replication is completed (Huisman and D'Ari, 1981), leading to an increased average cell size (Walker, 1996) and cell size variability, in particular at sub-inhibitory antibiotic concentrations where exponential growth occurs at a reduced rate (Figure 2A,B). The increased cell size under DNA stress correlates with an elevated average amount of protein per cell (Figure 2C; measured by a modified Lowry assay, Experimental Procedures). Cellular DNA, however, is typically still restricted to only one or two nucleoids (Figure 2A) and, with increasing concentration of DNA synthesis inhibitor, the mean DNA content decreases per volume and even per cell (Georgopapadakou and Bertasso, 1991). The ratio of protein to DNA therefore significantly increases in the presence of DNA synthesis inhibitors. We wondered whether this imbalance could be caused by excessive production of ribosomes in the cell, leading to overproduction of proteins and thus to reduced growth rates. We therefore examined whether and how ribosome synthesis is regulated in response to DNA synthesis inhibitors.
Figure 2. DNA synthesis inhibitors lead to increased cell size and protein-DNA ratio.
(A) Microscopy images of DAPI stained (cyan) E. coli cells growing in absence of antibiotics (Ø) and in presence of translation inhibitor TET and DNA synthesis inhibitors NAL and TMP. Scale bar, 10µm. DNA synthesis inhibitors lead to a mixed population of cells that are larger and contain only one or few nucleoids (white arrows). A small fraction of cells has no nucleoid (black arrows). (B) Histograms of cell lengths. Mean cell size and variability increases in presence of DNA synthesis inhibitors but not in presence of translation inhibitors. (C) Mean total protein per cell (measured by a variant of the Lowry assay) in presence of different antibiotics normalized to no drug control. All antibiotic concentrations are tuned to achieve the same normalized growth rate (~0.35). See Experimental Procedures.
Ribosomal gene expression is not specifically regulated by DNA stress
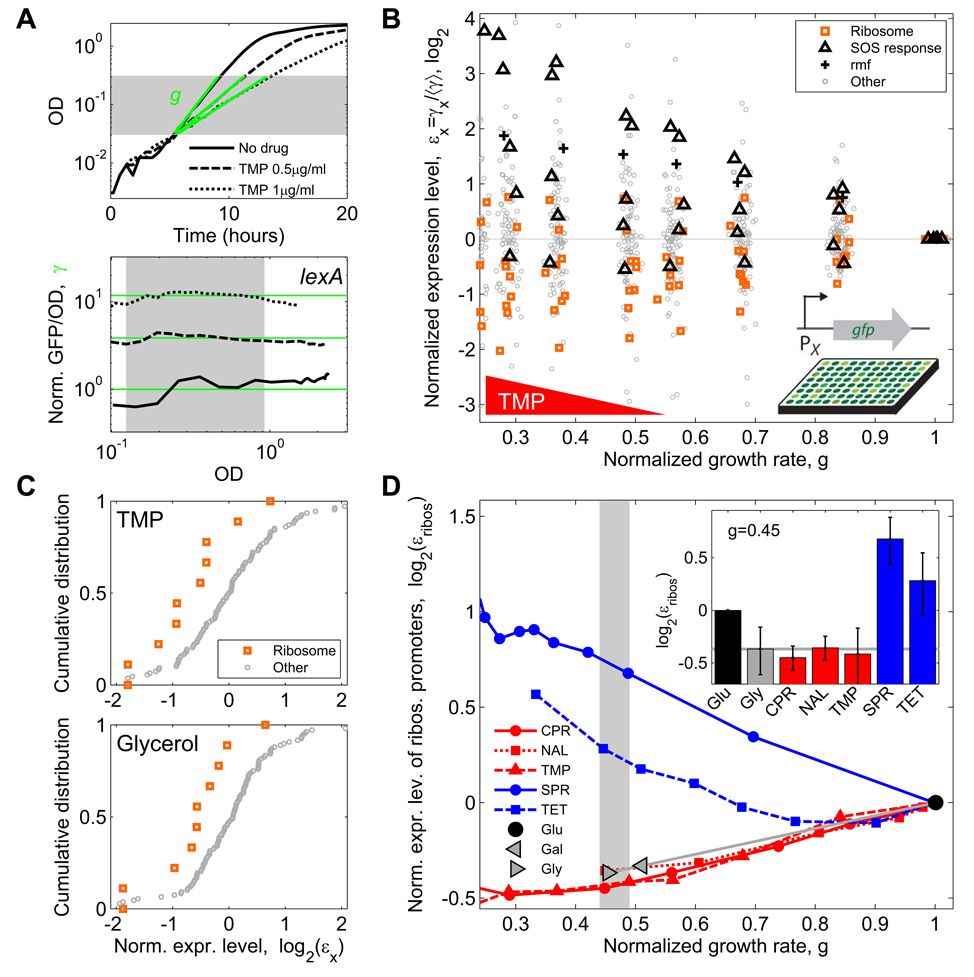
We used strains from a genome-wide GFP transcription reporter library (Zaslaver et al., 2006; Zaslaver et al., 2004) to measure changes in the expression level of promoters from almost 200 E. coli genes, representing key cellular functions including DNA stress response, metabolism and ribosome regulation and synthesis (Table S1). We obtained high time resolution measurements of optical density and GFP fluorescence of cultures growing in the presence of different antibiotics at a range of concentrations. We focused on measurements during exponential growth phase since our main interest in this work is to understand the combined effects of drugs on steady state growth. Growth rates (g) were determined from the increase in optical density over time (OD, Figure 3A). Changes in gene expression level (γ) were defined as the effect of the drug on the average GFP signal per OD during exponential phase (γ = [GFP/OD] / [GFP/OD]no drug; Figure 3A and Experimental Procedures). By repeating the measurement at a range of drug concentrations, we determined expression level changes in response to antibiotics as a function of growth inhibition (see example for trimethoprim (TMP) in Figure 3B; antibiotics used in this study are summarized in Table 1). As expected, most SOS response genes were up-regulated in response to DNA stress caused by any of three different DNA synthesis inhibitors (TMP, Figure 3B; ciprofloxacin (CPR) and nalidixic acid (NAL), Figure S2). On the other hand, most ribosomal genes were down-regulated in response to these DNA synthesis inhibitors (Figure 3B,C; Figure S2). Consistent with the down-regulation of ribosomal genes, the ppGpp-regulated ribosome inactivator gene rmf (Izutsu et al., 2001) was up-regulated in response to TMP (Figure 3B).
Figure 3. Ribosomal gene expression is down-regulated under DNA stress only as much as in the normal physiological response to slow growth.
(A) Example data demonstrating measurement of drug effect on growth rate and transcription reporters. Optical Density (OD) and GFP expression from various promoters (shown, as an example, is the promoter of lexA – the master regulator of the SOS response) are measured as a function of time for various drug concentrations (shown, 0, 0.5 and 1 µg/ml TMP). Top: growth rates are defined by linear regression (green lines) to the OD curves (black). Bottom: Expression level γ (green lines) is defined as GFP fluorescence intensity per OD, averaged over an OD range of exponential growth (shaded region) and normalized to no drug control. (B) Normalized expression levels εx of 110 promoters in E. coli as a function of growth rate in various concentrations of TMP. For each promoter x, εx is defined as expression level γx, normalized to the median expression level of all promoters 〈γ〉 (Experimental Procedures). SOS response promoters are up-regulated (black triangles). Most ribosomal promoters are down-regulated (orange squares) consistent with the up-regulation of the ribosome inactivator rmf (black crosses). Random scatter added to growth rate to enhance visibility. (C) Top: Cumulative distributions of normalized expression levels εx showing down-regulation of ribosomal genes (orange) relative to all other promoters (gray) at a fixed concentration of TMP (normalized growth rate ~0.49). Bottom: a similar regulation is seen with no drug when the same change in growth rate is achieved by changing the carbon source from glucose to glycerol. (D) Mean normalized expression level of ribosomal promoters εribos as a function of normalized growth rate for different DNA synthesis inhibitors (CPR, NAL, TMP; red), translation inhibitors (SPR, TET; blue), and in growth media with different carbon sources (glucose, galactose, glycerol; gray). Inset: εribos values at normalized growth rate of ~0.45; error bars show SEM. Ribosomal promoters are up-regulated in response to translation inhibitors and down-regulated in presence of DNA synthesis inhibitors. This down-regulation, however, is similar to the growth-rate dependent down-regulation that results from a change of carbon source (gray horizontal line).
Table 1.
Antibiotics used in this study, abbreviation, MIC in the wild type MG1655 strain, and main mode of action
| Antibiotic | Abbreviation | MIC in LB (µg/ml) |
MIC in M9 (µg/ml) |
Mode of action |
|---|---|---|---|---|
| Ciprofloxacin | CPR | 0.012 | 0.012 | DNA gyrase |
| Nalidixic acid | NAL | 6 | 6 | DNA gyrase |
| Trimethoprim | TMP | 0.42 | 1.5 | Folic acid synthesis |
| Spiramycin | SPR | 192 | 120 | Protein synthesis, 50S |
| Tetracycline | TET | 1.5 | 1.5 | Protein synthesis, 30S |
| Nitrofurantoin | NIT | 5 | 5 | Multiple mechanisms |
How much are ribosomal genes down-regulated under DNA synthesis stress? Ribosome production is normally reduced when the growth rate of the cell is reduced (Bremer and Dennis, 1996), but it is possible that the inhibition of DNA synthesis may also have a more specific effect on ribosome synthesis. To distinguish between non-specific (growth-mediated) and specific effects of DNA synthesis inhibition on growth, we compared the change in expression of ribosomal genes in the presence of antibiotics that inhibit DNA synthesis to the change seen when growth rate is reduced using a poor growth medium. We found that, for equivalent reductions in cellular growth rate caused by these two mechanisms, the degree to which ribosomal gene expression is down-regulated is essentially identical (Figure 3C,D). Thus, we see no evidence for specific regulation of ribosomal expression by DNA stress.
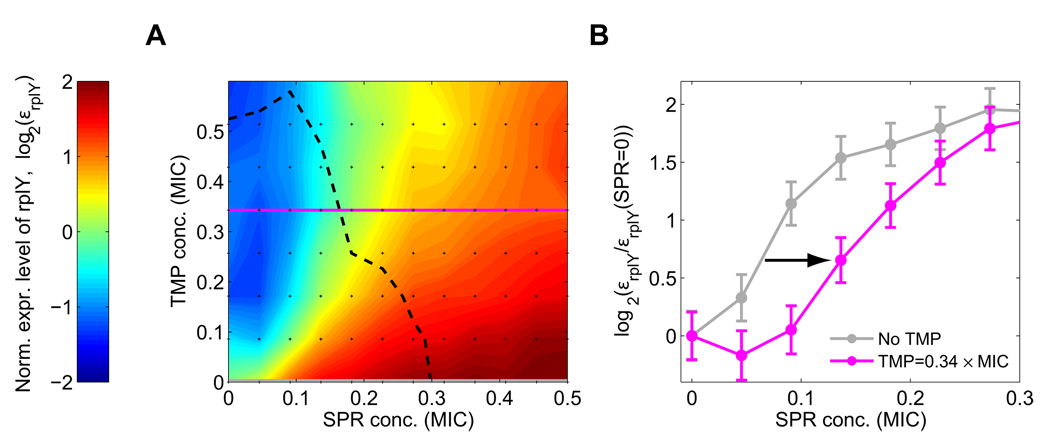
While DNA synthesis inhibitors did not specifically regulate ribosome production, the translation inhibitors spiramycin (SPR) and tetracycline (TET) elicit an up-regulation of ribosomal gene expression (Figure 3D) which counters the effect of these drugs, consistent with previous studies (Fraenkel and Neidhardt, 1961; Kurland and Maaloe, 1962; Schneider et al., 2002). We next asked how this up-regulation of ribosomal genes in response to protein synthesis inhibitors is affected by the presence of DNA synthesis inhibitors. We measured the regulation of 80 promoters including nine that control ribosomal genes (Table S1) in a two-dimensional concentration matrix of TMP and SPR (Experimental Procedures). For each promoter, we obtained its fold change expression level as a function of the two drug dosages (Figure 4A, Figure S3; Experimental Procedures; Kaplan et al., 2008; Tsui et al., 2004). We found that ribosomal gene expression levels in the presence of TMP are lower than in its absence for any SPR level (Figure 4A) and the up-regulation of ribosomal genes by SPR is significantly delayed (occurs at higher SPR concentrations) under TMP stress (arrow in Figure 4B). Consequently, the increase in growth resulting from the addition of a translation inhibitor in the presence of a DNA synthesis inhibitor occurs without substantial increase in ribosome production.
Figure 4. Up-regulation of ribosomal promoters by protein synthesis inhibitors is delayed under DNA stress.
(A) Color map of normalized expression level εrplY of ribosomal promoter rplY in a two-dimensional concentration matrix (black dots) of DNA synthesis inhibitor (TMP) and translation inhibitor (SPR). Other ribosomal promoters behave similarly, Figure S3. Dashed line, line of constant growth rate (isobole; g=0.38). (B) Relative change in expression level εrplY(SPR) / εrplY(SPR=0) as a function of SPR concentration, at no TMP (TMP=0, grey), and at a fixed TMP concentration (TMP=0.34 MIC, magenta). Up-regulation requires higher SPR concentration in the presence of TMP (arrow). Error-bars in B were estimated from the standard deviation of replicate measurements done on different days (see Figure S17).
This delayed response allows a translation inhibitor to substantially reduce overall protein synthesis and restore the protein-DNA ratio to near its normal value (Piddock et al., 1990), plausibly explaining the ability of translation inhibitors to increase the survival and growth of cells suffering DNA synthesis inhibition. Hence, the results discussed so far are consistent with the hypothesis that the ribosome synthesis rate is not optimally controlled under prolonged DNA stress; that is, it is not sufficiently down-regulated to maximize cellular growth rate. But how can we test this hypothesis more directly? The hallmark of non-optimality is the possibility for improvement: if cellular production of ribosomes is indeed non-optimal under DNA stress, we should be able to manipulate it to increase cellular survival and growth.
Manipulating ribosome synthesis increases growth rate and survival in the presence of DNA synthesis inhibitors
To test whether direct manipulation of ribosome levels affects growth and survival in the presence of DNA synthesis inhibitors, we measured responses to antibiotics in strains that are engineered to decrease or increase ribosome synthesis. Following previous work (Asai et al., 1999; Condon et al., 1993), we constructed strains in which up to six of the seven rrn operons were incrementally deleted (designated Δ1, Δ2, Δ3, Δ4, Δ5 and Δ6; Experimental Procedures; Table S2; no plasmid-borne rrn operons were added to these strains). Our construction method removes the selection marker linked to each of the rrn operon deletions, allowing a direct comparison of the physiology of these strains with wild-type (Experimental Procedures). The relationship between the number of rrn operons and ribosome levels is not necessarily linear, since feedback regulation of ribosome synthesis partially compensates for deletions by increasing the expression of the remaining rrn operons (Condon et al., 1993). Nevertheless, ribosome levels and the rRNA concentration – an upper bound for ribosome level – is reduced by deleting rrn operons, particularly in rich growth media where rrn operon transcription rates are close to saturation and cannot be increased much further (Asai et al., 1999; Condon et al., 1993). We also examined strains deleted for the genes relA and spoT; these double-deletion mutants are devoid of ppGpp, a key negative regulator of ribosome synthesis (Xiao et al., 1991), and thus show increased rrn expression (Barker et al., 2001; Bartlett and Gourse, 1994). The relA spoT deletion strain has a longer lag time for the transition from stationary phase to exponential growth and a slightly reduced steady state growth rate (Figure S16; Gaal and Gourse, 1990).
We measured growth rates of these modified strains in normal conditions as well as under conditions where different antibiotics were added to the cultures. In the absence of antibiotics, the wild type strain grows faster than all mutants with altered ribosome synthesis; genetically increasing or decreasing ribosome synthesis leads to reduced growth rates (Figure 5A). This observation confirms previous results (Asai et al., 1999) and is in agreement with the idea that ribosomal synthesis is optimally regulated to maximize growth in the absence of stress (Gralla, 2005; Paul et al., 2004).
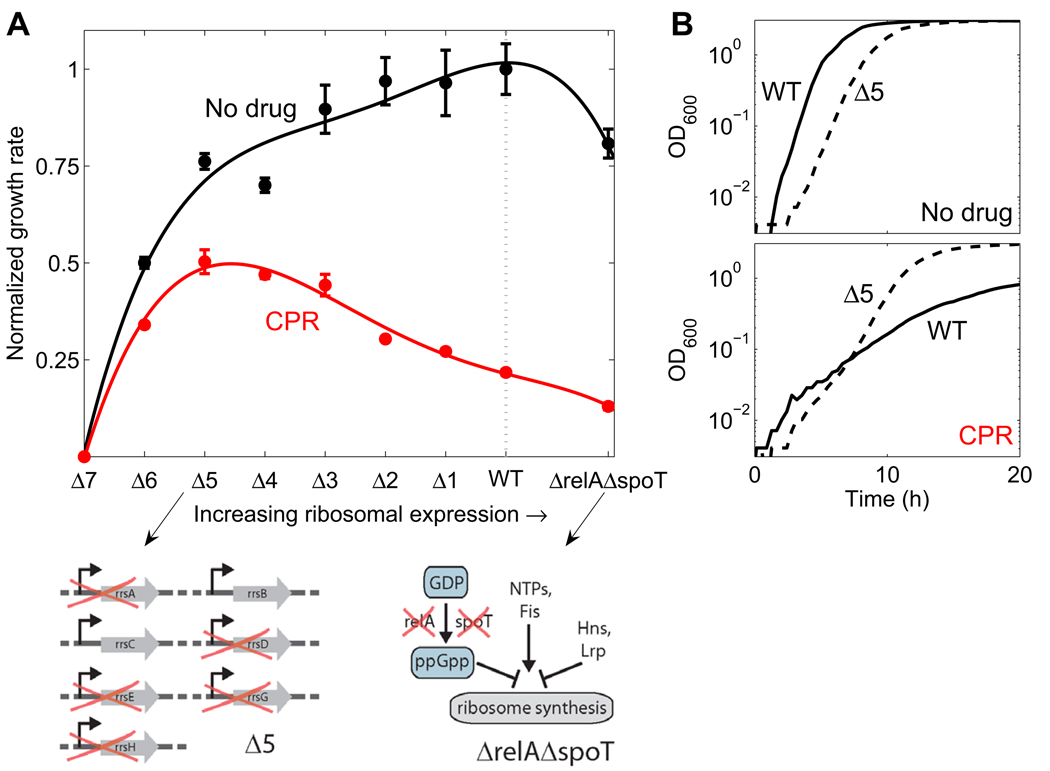
Figure 5. Wild-type regulation of ribosomal expression level under DNA stress is non-optimal: genetically manipulating ribosome synthesis can increase survival and growth.
(A) Normalized growth rates of wild-type (WT) and strains with incremental deletions of one to six of the seven rrn operons as well as for ΔrelA ΔspoT strain, in rich medium (LB) in the absence (black) and presence of the DNA synthesis inhibitor CPR (red). Lines, 4th order polynomial fit to guide the eye. Schematic on left: strain Δ5 in which 5 of 7 rrn operons are deleted. Schematic on right: ΔrelA ΔspoT strain which is devoid of ppGpp, a key negative regulator of ribosome synthesis, while other factors regulating ribosome synthesis remain. While wild-type expression level is optimized for maximal growth under no drug conditions, it is not optimized for maximal growth under DNA synthesis inhibition: Reduced ribosome synthesis in rrn deletion strains increases growth. (B) Sample data showing the growth curves (OD versus time) for WT (solid line) and Δ5 strain (dashed line) in no drug or under CPR at the concentration of A.
In DNA stress conditions, however, the picture is profoundly different (Figure 5A and Figure S4A,B). As expected, all strains grow more slowly under DNA stress than in a stress-free environment (Figure 5A, CPR curve lower than no-drug curve). But, in the presence of the DNA synthesis inhibitors CPR and NAL, the strain with maximal growth is not the wild type, but rather a strain with reduced ribosome synthesis (Δ5 at the drug concentration shown in Figure 5, Figure S4A,B). Complementing the deletion strains with a plasmid expressing one of the rrn operons (rrnB), partially revokes the increase in growth of these deletion mutants under DNA stress, confirming that this phenotype is directly related to the reduction in rrn operons (Figure S13). Coincident with their increased growth rate compared to wild type, the deletion stains also have a closer to normal cell size under DNA synthesis inhibition (Figure S5). Thus, in the presence of DNA synthesis inhibitors the expression of rrn genes in the wild type appears to be higher than optimal.
We next tested if optimizing ribosome synthesis also allows cells to tolerate higher concentrations of DNA synthesis inhibitors. We determined changes of the minimal inhibitory concentration (MIC) in the rrn deletion mutants for a range of antibiotics (Experimental Procedures). Indeed, as we delete rrn operons we see an incremental increase in MIC for the DNA synthesis inhibitors CPR and NAL (TMP behaves differently in this assay; see below), compared to no change or even a decrease for antibiotics with other modes of action (Figure S4F).
In principle, the increased MIC for DNA synthesis inhibitors could be an indirect effect caused by the lower growth rate of the mutants with rrn operon deletions. To discriminate between such general growth rate effects and the specific effect of modified ribosomal expression in the rrn deletion strains, we reduced the growth rate of the wild type by changing the carbon source in the growth medium, and asked if this change has a similar effect on the MIC as the rrn operon deletions (Experimental Procedures). We found that changing the growth rate in this way does not lead to a detectable change in MIC for CPR and NAL, but TMP shows a two-fold lower MIC when growth rate is reduced to a level comparable to that of the Δ6 mutant (data not shown). This allows us to rationalize the observation that, unlike the case for CPR and NAL, the MIC for TMP is not increased in the Δ6 strain (Figure S4F): the increased MIC due to the reduction in ribosomal synthesis may be masked by the reduction in MIC caused by the decreased growth rate. Overall, our results show that strains with genetically reduced ribosome synthesis survive better in the presence of DNA synthesis inhibitors, and thus that the wild-type regulation of ribosome synthesis is non-optimal for growth in these conditions.
Reducing ribosome synthesis removes the suppressive drug interactions between DNA synthesis inhibitors and translation inhibitors
If non-optimality in the regulation of ribosome synthesis under DNA stress is the cause for the suppressive interactions between inhibitors of DNA synthesis and translation (Figure 1), then these suppressive drug interactions should disappear in the genetically altered strains. To test this prediction, we measured growth rates of the wild type and the strains with genetically altered ribosome expression levels in a two-dimensional drug matrix of a DNA synthesis and a translation inhibitor (Experimental Procedures). Strikingly, we find that genetically reducing ribosome synthesis reduces the magnitude of the suppressive drug interaction and can even remove it entirely. Indeed, in contrast to the wild type, the Δ6 strain shows an almost additive interaction (linear MIC line in Figure 6A compared to non-monotonic line in 6B; see also definition of drug interactions in Figure 1A). We observed this phenomenon for different antibiotic pairs that inhibit DNA synthesis and translation, for differently constructed rrn deletion strains (Figure S12), and for strains grown in both rich and minimal growth medium (Figure 6A,B and Figure S1). Further, complementing the deletion strains with a plasmid expressing one of the rrn operons (rrnB), partially restores the suppressive interaction between the drugs (Figure S14). Together, these results support the notion that the suppressive drug interaction is caused by non-optimal regulation of ribosome synthesis: the reduced ribosome synthesis rate in the Δ6 strain is closer to the optimal level for maximal growth rate under DNA stress and, consequently, the addition of an antibiotic that inhibits translation no longer has a beneficial effect.
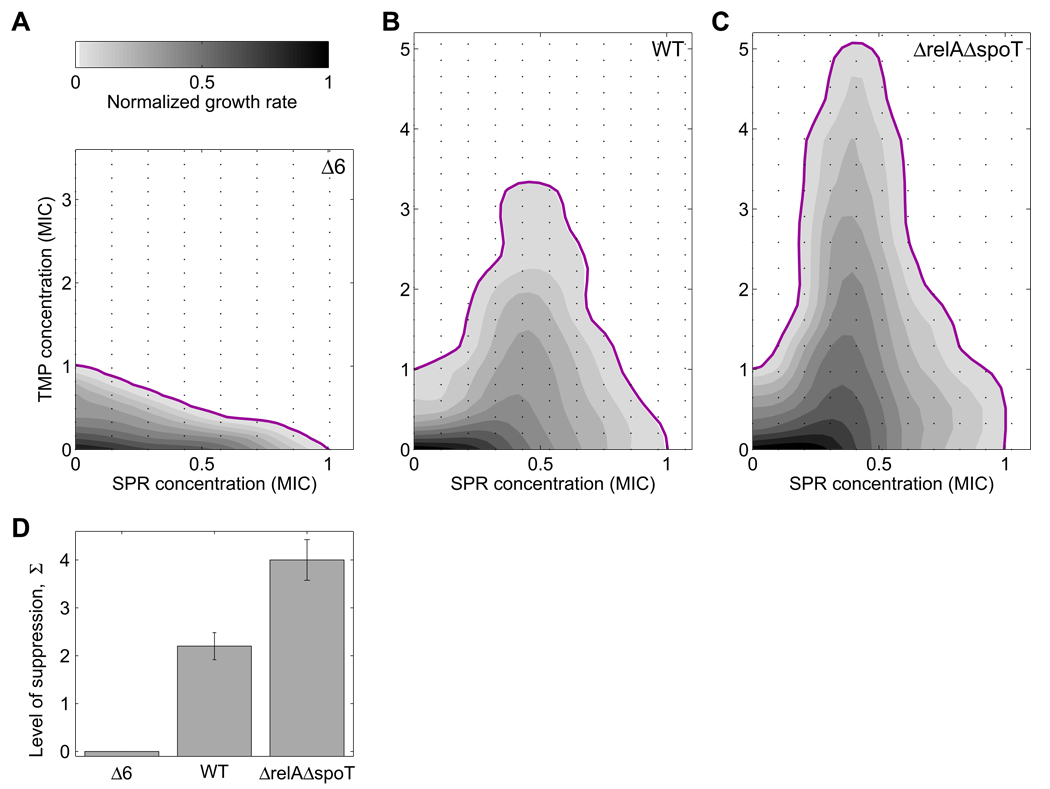
Figure 6. Genetically optimizing ribosome synthesis removes suppressive drug interaction between inhibitors of DNA synthesis and translation.
Growth rates (gray levels) and MIC line (magenta) of Δ6 (A), WT (B), and ΔrelA ΔspoT strain (C) in two-dimensional concentration matrices (black dots) of DNA synthesis inhibitor (TMP) and translation inhibitor (SPR). The suppressive drug interaction (B) disappears when ribosome synthesis is reduced (A) and is amplified when down-regulation of ribosome synthesis is impaired (C). The disappearance of suppression is incremental with number of rrn deletions and does not depend on the specific DNA synthesis or translation inhibitor used, Figure S1. (D) Quantified level of suppression in the three strains. The level of suppression Σ is defined as Σ=(MICmax − MIC0)/ MIC0, where MICmax is the maximal TMP MIC over all SPR concentrations and MIC0 the TMP MIC in absence of SPR. Cultures grown in rich medium (LB).
Conversely, we tested whether impairing the down-regulation of ribosome synthesis can amplify suppressive drug interactions. We can force ribosome synthesis in the presence of TMP even further above its optimal level by using a relA spoT deletion mutant. In this ppGpp-deficient mutant, the down-regulation of ribosome synthesis in response to TMP is impaired since it cannot elicit the wild type up-regulation of ppGpp in response to TMP (Khan and Yamazaki, 1972; Smith and Midgley, 1973). Indeed, we found that the impaired regulation of ribosome synthesis in a relA spoT deletion mutant amplifies the suppressive drug interaction between the DNA synthesis inhibitor TMP and the translation inhibitor SPR (compare Figure 6C to 6B). A very similar effect is observed in a relA deletion mutant (Figure S7). The observation that increasing ribosomal expression amplifies suppression while decreasing ribosomal expression reduces it (Figure 6D) provides persuasive evidence that this drug interaction is due to non-optimal regulation of ribosome expression.
A simple mathematical model of ribosome synthesis regulation captures non-optimal response to DNA stress and suppressive drug interactions
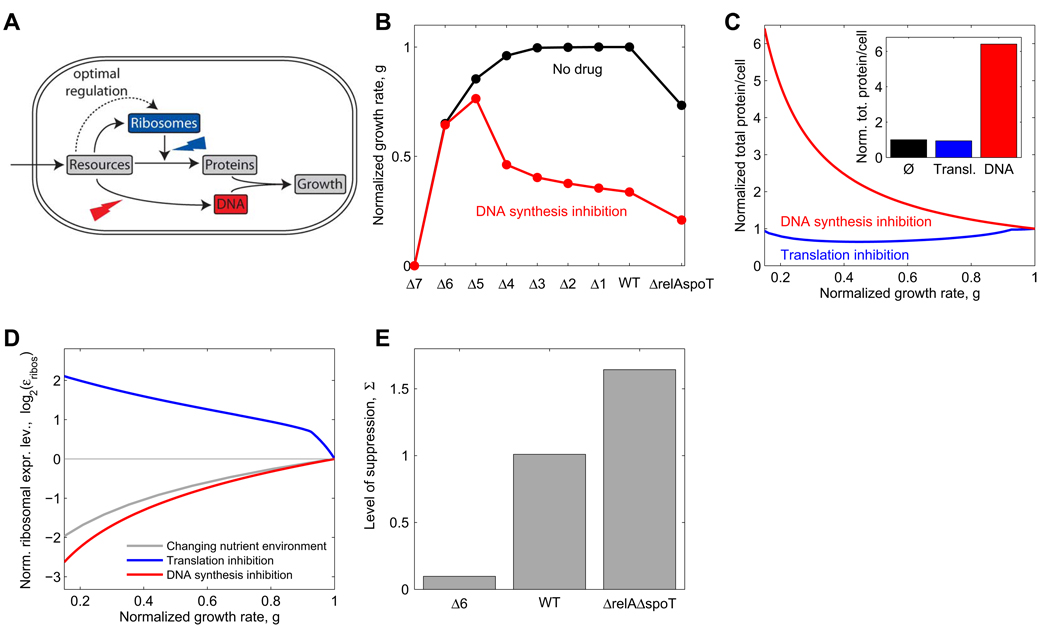
Why is ribosome synthesis so inappropriately regulated in response to DNA synthesis inhibitors? To explore this issue, we developed a coarse-grained mathematical model of ribosome synthesis regulation in bacterial growth (Figure 7A and Supplemental Data). This model describes the interdependencies of the cellular concentrations of DNA, proteins, ribosomes, resources, and cellular growth at steady state. Resources enter the cell at a fixed rate and are distributed between the production of proteins, ribosomes and DNA. In the model, ribosome synthesis is regulated based on the intracellular concentration of these resources. We optimize this regulation function to maximize the growth rate at different resource uptake rates, corresponding to different nutrient environments in the absence of antibiotics. We then assume that this same regulation function based on intracellular resource concentrations also applies when antibiotics are present. The effect of antibiotics is modeled as a reduction in the rate of translation or DNA synthesis. We further assume that a threshold amount of protein per replication origin must be produced to initiate DNA replication and cell division (Donachie, 1968; Donachie and Blakely, 2003). For simplicity, we assume in the model that the cellular protein concentration is constant so that the cell size is proportional to the total amount of protein per cell (this approximation may not be true in general). Importantly, most parameters that enter into the model are known or fully constrained by experimental data (Bremer and Dennis, 1996; Table S3). This simple model quantitatively reproduces the changes in cell composition and growth rate that have been observed in different nutrient environments (Bremer and Dennis, 1996; Figure S6).
Figure 7. Mathematical model with optimal growth rate-dependent regulation of ribosome synthesis yields non-optimal response to DNA synthesis and thereby suppression by translation inhibitors.
(A) Schematic depiction of a very simplified model of bacterial growth capturing resource allocation to DNA, ribosomes and proteins. Ribosome synthesis is assumed to be optimally regulated by resource concentration. See Supplemental Data for the complete mathematical model. (B) Growth rate obtained from the model for WT, rrn operon deletions and relA spoT deletions in the absence of antibiotics (black line) and in the presence of DNA synthesis inhibitor (red line), cf. Figure 5A. (C) Change of total protein per cell under translation inhibition (blue) or DNA synthesis inhibition (red), cf. Figure 2C. Inset shows total protein per cell at g=0.15. (D) Normalized ribosomal expression level εribos (corresponds to ribosomal protein fraction η in the model, see Supplemental Data) as a function of growth rate under reduced nutrient availability (gray), translation inhibition (blue), or DNA synthesis inhibition (red). Ribosome synthesis is similarly down-regulated in response to reduced nutrient availability or DNA synthesis inhibition and is up-regulated in response to translation inhibition, cf. Figure 3D. (E) Quantified level of suppression in the different strains, cf. Figure 6D. Parameters as in Table S3 with resource influx νa=15 h−1 which leads to a growth rate g=1.3 h−1 in absence of antibiotics, for details see Supplemental Data.
The model and its resource-based optimization of ribosome production faithfully describes our key experimental observations and in particular leads to non-optimal regulation under DNA stress. Specifically, it correctly captures the up-and down-regulation of ribosome synthesis in the presence of translation and DNA synthesis inhibitors, respectively (cf. Figure 3D and Figure 7D). As in our experimental results, this reduction in the level of ribosomal synthesis under DNA synthesis inhibition is similar to the reduction seen when growth is attenuated by nutrient deprivation (red versus grey lines in Figure 7D; compare to experiments in Figure 3D). This reduction in expression of ribosomal synthesis is insufficient, leading to a skewed protein-DNA ratio (cf. Figure 2 and Figure 7C) and to sub-maximal growth rate (cf. Figure 5A and Figure 7B). Importantly, this simple model also reproduces the suppressive drug interaction between DNA synthesis and translation inhibitors, its attenuation as a result of rrn operon deletions, and its amplification as a result of relA spoT deletions (cf. Figure 6D and Figure 7E).
DISCUSSION
We showed that ribosome synthesis is not specifically regulated by DNA synthesis inhibiting drugs, leading to a skewed DNA to protein ratio and sub-maximal growth rate. Genetically reducing ribosome synthesis allows cells to grow faster under DNA stress. Importantly, this genetic optimization of ribosome synthesis also eliminates the suppressive drug interactions between protein and DNA synthesis inhibitors. A simple mathematical model proposes that optimal regulation of ribosome production based on intracellular resource concentrations in normal conditions can lead to non-optimal resource allocation between DNA and protein synthesis under DNA synthesis inhibition, and thereby to decreased growth. This explanation, while fully consistent with our data, does not exclude the possibility that other mechanisms contribute to the suppressive interactions between DNA synthesis inhibitors and translation inhibitors. For example, reduced protein synthesis leads to reduced growth rate and thereby to a smaller number of replication forks, which ultimately may reduce the impact of DNA synthesis inhibitors, especially of gyrase inhibitors which cause double strand breaks and cell death through oxidative stress (Dwyer et al., 2007; Kolodkin-Gal et al., 2008). Up-regulation of drug efflux pumps may also play a role (Poole, 2005).
Our result that regulation of ribosomal gene expression in response to sustained DNA stress is not optimal for maximal growth in laboratory conditions raises the question of whether this response might be optimized for another goal or for other more natural conditions. The lack of specific regulation of ribosomal genes under DNA stress is particularly puzzling given E. coli’s ability to specifically regulate genes through the SOS response (Friedman et al., 2005; Michel, 2005; Radman, 1975; Tippin et al., 2004). There are several ways in which the observed lack of specific response to DNA stress could actually be beneficial in natural conditions. First, it is possible that in the natural environment in which the organism evolved, DNA synthesis inhibition is usually encountered at the same time as nutrient deprivation, removing the need for a specific mechanism to down-regulate protein synthesis (Cashel et al., 1996; Gralla, 2005; Paul et al., 2004). Indeed, gene regulation responses can exploit correlations between environmental changes, even if they do not occur simultaneously (Mitchell et al., 2009; Tagkopoulos et al., 2008). Second, it is possible that DNA stress is usually short-lived in the natural environment, and so the global response to DNA damage and the formation of larger, filamentous cells could be optimized to ensure a fast recovery when the stress is relieved (Guan and Burnham, 1992). Finally, it is possible that phenotypic variability between cells, which increases under DNA stress (Figure 2A,B), plays a role in the survival strategy under these conditions (Balaban et al., 2004; Guido et al., 2007; Kussell and Leibler, 2005; Pearl et al., 2008). In any case, while the lack of specific regulation of ribosomal genes under DNA stress could be optimal in some conditions, it is clearly non-optimal in the laboratory condition.
In summary, we showed that non-optimal regulation of ribosome synthesis is at the heart of the suppressive drug interactions between protein and DNA synthesis inhibitors. Understanding the underlying mechanism of the interaction allowed us to genetically manipulate whether and to what extent these two drug classes interact. More generally, these results show that cellular systems, even those critical for growth and survival, are not always optimally regulated, and that tight optimal control in some conditions can lead to non-optimal regulation in other conditions. Such non-optimal regulation may open possibilities for new ways to manipulate cellular growth in the lab and in the clinic.
EXPERIMENTAL PROCEDURES
Media, strains and drugs
Experiments were conducted as indicated in rich Luria-Bertani (LB) broth or M9 minimal medium with different carbon sources (glucose, galactose, glycerol) at 0.4%. Glucose M9 was supplemented with 0.2% amicase. Drug solutions were made from powder stocks, filter-sterilized, stored at −20°C in the dark and added as indicated. All strains used were derived from E. coli K-12 strain MG1655 (Supplemental Data and Table S2).
Growth rate and MIC assays
Overnight cultures were diluted ~2000-fold and grown on an automated robotic system (Caliper) at 30°C with rapid shaking in 96-well microtiter plates (Costar) containing 200 µl medium per well. Absorbance at 600nm (A600, proportional to optical density OD600, proportionality constant 3.1) and GFP fluorescence were recorded by a plate reader (Victor III or EnVision, Perkin-Elmer) at intervals of ~30min for at least 24h, and background subtracted. Growth rates were calculated using Matlab by linear regression of log(OD600) (Matlab function regress) during exponential growth (0.01<A600<0.1). The measurement error was evaluated as the 95% confidence interval of the linear regression (error bars in Figure 5A). Growth was annotated as no data if the regression error was greater than 20%. Also removed are some cases where resistant mutants occurred, in particular for CPR and NAL; these were identified by large variations between replicates and by no growth for 12h or longer followed by fast growth. Two-dimensional drug concentration matrices were set up on one 96-well plate (11×8 format) or on four plates (22×16 format) leaving one column per plate for controls. To reduce noise, a smoothed function was fitted to the measured growth rates by using a smoothing cubic spline, and linearly interpolated isoboles were plotted (Matlab functions csaps and contour).
MIC was defined as the lowest concentration at which background subtracted A600 did not exceed 0.02 after 24h. MICs were first determined crudely in logarithmic antibiotic concentration gradients with two-fold dilutions and then more accurately with linear gradients ranging from zero to about 2 times the MIC.
Gene expression assay
GFP reporter strains were grown in glucose M9 medium supplemented with 0.2% amicase. GFP background was subtracted as described (Zaslaver et al., 2006). We defined the expression level as the mean GFP/A600 in the interval 0.04<A600<0.3, Figure 3A. Only promoters with a clearly detectable GFP signal were used for analysis, reducing the total number to 110 promoters, Figure 3B. Expression level changes γ relative to the drug-free control were normalized to the median expression level change 〈γ〉 of all promoters in the same drug environment. Changes in the median expression level of all promoters reflect non-specific effects such as pH changes or changes of the reporter plasmid copy number. We verified that the effect of plasmid copy number on the measured expression level is independent of the GFP promoter, by comparing to strains in which the same GFP reporters were integrated into the chromosome (Supplemental Data).
DNA and protein assay
Cultures were grown to OD600~0.2 in glucose M9 medium, DAPI stained (5 µg/ml, 5 minutes), mounted on agar pads, and imaged (Figure 2A). Cell lengths were measured manually using ImageJ (http://rsbweb.nih.gov/ij/) for ~100 cells in each condition (Figure 2B). To calculate protein per cell, we combined 8 identical 200 µl cultures and determined the total protein concentration using the DC protein assay (Biorad). Cell concentration was estimated by colony plate count. We slightly over-estimate protein per cell because cells devoid of DNA (less than 10% of cells) do not form colonies. Error bars in Figure 2C represent the error N1/2 for the cell count N.
Supplementary Material
ACKNOWLEDGMENTS
We thank U. Alon, J. Collins, M. Elowitz, M. Ernebjerg, A. Gassett, A. Hilfinger, E. Kelsic, M. Kohanski, D. Landgraf, R. Milo, A. Palmer, D. Rudner, M. Springer, M. Staller, C. Squires, R. Ward, P. Yeh, and all members of the Kishony lab for discussions, comments, and technical help and M. Traxler for his gift of strains. This work was supported in part by National Institutes of Health Grant R01GM081617 (to RK). TB acknowledges funding from the Alexander von Humboldt foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires CL. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- Barker MM, Gaal T, Gourse RL. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- Bartlett MS, Gourse RL. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss CI. The toxicity of poisons applied jointly. Annals of Applied Biology. 1939;26:585–615. [Google Scholar]
- Brazas MD, Hancock RE. Using microarray gene signatures to elucidate mechanisms of antibiotic action and resistance. Drug Discov Today. 2005;10:1245–1252. doi: 10.1016/S1359-6446(05)03566-X. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of Chemical Composition and Other Parameters of the Cell by Growth Rate. In: Neidhardt FC, editor. Echerichia coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- Cashel M, Gentry DR, Hernandez VJ, Vinella D. The Stringent Response. In: Neidhardt FC, editor. Echerichia coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Chait R, Craney A, Kishony R. Antibiotic interactions that select against resistance. Nature. 2007;446:668–671. doi: 10.1038/nature05685. [DOI] [PubMed] [Google Scholar]
- Condon C, French S, Squires C, Squires CL. Depletion of functional ribosomal RNA operons in Escherichia coli causes increased expression of the remaining intact copies. Embo J. 1993;12:4305–4315. doi: 10.1002/j.1460-2075.1993.tb06115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C, Liveris D, Squires C, Schwartz I, Squires CL. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol. 1995;177:4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J, Spiegelman GB, Yim G. The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol. 2006;9:445–453. doi: 10.1016/j.mib.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Dekel E, Alon U. Optimality and evolutionary tuning of the expression level of a protein. Nature. 2005;436:588–592. doi: 10.1038/nature03842. [DOI] [PubMed] [Google Scholar]
- Dennis PP, Ehrenberg M, Bremer H. Control of rRNA synthesis in Escherichia coli: a systems biology approach. Microbiol Mol Biol Rev. 2004;68:639–668. doi: 10.1128/MMBR.68.4.639-668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donachie WD. Relationship between cell size and time of initiation of DNA replication. Nature. 1968;219:1077–1079. doi: 10.1038/2191077a0. [DOI] [PubMed] [Google Scholar]
- Donachie WD, Blakely GW. Coupling the initiation of chromosome replication to cell size in Escherichia coli. Curr Opin Microbiol. 2003;6:146–150. doi: 10.1016/s1369-5274(03)00026-2. [DOI] [PubMed] [Google Scholar]
- Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Kohanski MA, Hayete B, Collins JJ. Gyrase inhibitors induce an oxidative damage cellular death pathway in Escherichia coli. Mol Syst Biol. 2007;3:91. doi: 10.1038/msb4100135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo A, Martinez JL. Antibiotics as signals that trigger specific bacterial responses. Curr Opin Microbiol. 2008;11:161–167. doi: 10.1016/j.mib.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Fraenkel DG, Neidhardt FC. Use of chloramphenicol to study control of RNA synthesis in bacteria. Biochim Biophys Acta. 1961;53:96–110. doi: 10.1016/0006-3002(61)90797-1. [DOI] [PubMed] [Google Scholar]
- Friedman N, Vardi S, Ronen M, Alon U, Stavans J. Precise temporal modulation in the response of the SOS DNA repair network in individual bacteria. PLoS Biol. 2005;3:e238. doi: 10.1371/journal.pbio.0030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaal T, Bartlett MS, Ross W, Turnbough CL, Jr, Gourse RL. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- Gaal T, Gourse RL. Guanosine 3'-diphosphate 5'-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopapadakou NH, Bertasso A. Effects of quinolones on nucleoid segregation in Escherichia coli. Antimicrob Agents Chemother. 1991;35:2645–2648. doi: 10.1128/aac.35.12.2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh EB, Yim G, Tsui W, McClure J, Surette MG, Davies J. Transcriptional modulation of bacterial gene expression by subinhibitory concentrations of antibiotics. Proc Natl Acad Sci U S A. 2002;99:17025–17030. doi: 10.1073/pnas.252607699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla JD. Escherichia coli ribosomal RNA transcription: regulatory roles for ppGpp, NTPs, architectural proteins and a polymerase-binding protein. Mol Microbiol. 2005;55:973–977. doi: 10.1111/j.1365-2958.2004.04455.x. [DOI] [PubMed] [Google Scholar]
- Guan L, Burnham JC. Postantibiotic effect of CI-960, enoxacin and ciprofloxacin on Escherichia coli: effect on morphology and haemolysin activity. J Antimicrob Chemother. 1992;29:529–538. doi: 10.1093/jac/29.5.529. [DOI] [PubMed] [Google Scholar]
- Guido NJ, Lee P, Wang X, Elston TC, Collins JJ. A pathway and genetic factors contributing to elevated gene expression noise in stationary phase. Biophys J. 2007;93:L55–L57. doi: 10.1529/biophysj.107.118687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegreness M, Shoresh N, Damian D, Hartl D, Kishony R. Accelerated evolution of resistance in multidrug environments. Proc Natl Acad Sci U S A. 2008;105:13977–13981. doi: 10.1073/pnas.0805965105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman LR, D'Argenio DA, MacCoss MJ, Zhang Z, Jones RA, Miller SI. Aminoglycoside antibiotics induce bacterial biofilm formation. Nature. 2005;436:1171–1175. doi: 10.1038/nature03912. [DOI] [PubMed] [Google Scholar]
- Huisman O, D'Ari R. An inducible DNA replication-cell division coupling mechanism in E. coli. Nature. 1981;290:797–799. doi: 10.1038/290797a0. [DOI] [PubMed] [Google Scholar]
- Ibarra RU, Edwards JS, Palsson BO. Escherichia coli K-12 undergoes adaptive evolution to achieve in silico predicted optimal growth. Nature. 2002;420:186–189. doi: 10.1038/nature01149. [DOI] [PubMed] [Google Scholar]
- Izutsu K, Wada A, Wada C. Expression of ribosome modulation factor (RMF) in Escherichia coli requires ppGpp. Genes Cells. 2001;6:665–676. doi: 10.1046/j.1365-2443.2001.00457.x. [DOI] [PubMed] [Google Scholar]
- Kaplan S, Bren A, Zaslaver A, Dekel E, Alon U. Diverse two-dimensional input functions control bacterial sugar genes. Mol Cell. 2008;29:786–792. doi: 10.1016/j.molcel.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener J, Nomura M. Regulation of Ribosome Synthesis. In: Neidhardt FC, editor. Escherichia Coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 1417–1431. [Google Scholar]
- Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat Rev Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- Khan SR, Yamazaki H. Trimethoprim-induced accumulation of guanosine tetraphosphate (ppGpp) in Escherichia coli. Biochem Biophys Res Commun. 1972;48:169–174. doi: 10.1016/0006-291x(72)90358-0. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Wierzbowski J, Cottarel G, Collins JJ. Mistranslation of membrane proteins and two-component system activation trigger antibiotic-mediated cell death. Cell. 2008;135:679–690. doi: 10.1016/j.cell.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodkin-Gal I, Sat B, Keshet A, Engelberg-Kulka H. The communication factor EDF and the toxin-antitoxin module mazEF determine the mode of action of antibiotics. PLoS Biol. 2008;6:e319. doi: 10.1371/journal.pbio.0060319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurland CG, Maaloe O. Regulation of ribosomal and transfer RNA synthesis. J Mol Biol. 1962;4:193–210. doi: 10.1016/s0022-2836(62)80051-5. [DOI] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Lehar J, Krueger A, Zimmermann G, Borisy A. High-order combination effects and biological robustness. Mol Syst Biol. 2008;4:215. doi: 10.1038/msb.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar J, Zimmermann GR, Krueger AS, Molnar RA, Ledell JT, Heilbut AM, Short GF, 3rd, Giusti LC, Nolan GP, Magid OA, et al. Chemical combination effects predict connectivity in biological systems. Mol Syst Biol. 2007;3:80. doi: 10.1038/msb4100116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Ihmels J, Carmi M, Weinberger A, Friedlander G, Barkai N. Strategy of transcription regulation in the budding yeast. PLoS ONE. 2007;2:e250. doi: 10.1371/journal.pone.0000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebermeister W, Klipp E, Schuster S, Heinrich R. A theory of optimal differential gene expression. Biosystems. 2004;76:261–278. doi: 10.1016/j.biosystems.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Linares JF, Gustafsson I, Baquero F, Martinez JL. Antibiotics as intermicrobial signaling agents instead of weapons. Proc Natl Acad Sci U S A. 2006;103:19484–19489. doi: 10.1073/pnas.0608949103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewe S. Die quantitativen Probleme der Pharmakologie. Ergebnisse der Physiologie. 1928;27:47–187. [Google Scholar]
- Loewe S. The problem of synergism and antagonism of combined drugs. Arzneimittelforschung. 1953;3:285–290. [PubMed] [Google Scholar]
- Mason DJ, Power EG, Talsania H, Phillips I, Gant VA. Antibacterial action of ciprofloxacin. Antimicrob Agents Chemother. 1995;39:2752–2758. doi: 10.1128/aac.39.12.2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesak LR, Miao V, Davies J. Effects of subinhibitory concentrations of antibiotics on SOS and DNA repair gene expression in Staphylococcus aureus. Antimicrob Agents Chemother. 2008;52:3394–3397. doi: 10.1128/AAC.01599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B. After 30 years of study, the bacterial SOS response still surprises us. PLoS Biol. 2005;3:e255. doi: 10.1371/journal.pbio.0030255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel JB, Yeh PJ, Chait R, Moellering RC, Jr, Kishony R. Drug interactions modulate the potential for evolution of resistance. Proc Natl Acad Sci U S A. 2008;105:14918–14923. doi: 10.1073/pnas.0800944105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Romano GH, Groisman B, Yona A, Dekel E, Kupiec M, Dahan O, Pilpel Y. Adaptive prediction of environmental changes by microorganisms. Nature. 2009;460:220–224. doi: 10.1038/nature08112. [DOI] [PubMed] [Google Scholar]
- Moss T. At the crossroads of growth control; making ribosomal RNA. Curr Opin Genet Dev. 2004;14:210–217. doi: 10.1016/j.gde.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;38:749–770. doi: 10.1146/annurev.genet.38.072902.091347. [DOI] [PubMed] [Google Scholar]
- Pearl S, Gabay C, Kishony R, Oppenheim A, Balaban NQ. Nongenetic individuality in the host-phage interaction. PLoS Biol. 2008;6:e120. doi: 10.1371/journal.pbio.0060120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ, Walters RN, Diver JM. Correlation of quinolone MIC and inhibition of DNA, RNA, and protein synthesis and induction of the SOS response in Escherichia coli. Antimicrob Agents Chemother. 1990;34:2331–2336. doi: 10.1128/aac.34.12.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pillai SK, Moellering RC, Eliopoulos GM. Antimicrobial combinations. In: Lorian V, editor. Antibiotics in Laboratory Medicine. Philadelphia: Lippincott Williams and Wilkins; 2005. pp. 365–440. [Google Scholar]
- Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Radman M. SOS repair hypothesis: phenomenology of an inducible DNA repair which is accompanied by mutagenesis. Basic Life Sci. 1975;5A:355–367. doi: 10.1007/978-1-4684-2895-7_48. [DOI] [PubMed] [Google Scholar]
- Schneider DA, Gaal T, Gourse RL. NTP-sensing by rRNA promoters in Escherichia coli is direct. Proc Natl Acad Sci U S A. 2002;99:8602–8607. doi: 10.1073/pnas.132285199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider DA, Gourse RL. Relationship between growth rate and ATP concentration in Escherichia coli: a bioassay for available cellular ATP. J Biol Chem. 2004;279:8262–8268. doi: 10.1074/jbc.M311996200. [DOI] [PubMed] [Google Scholar]
- Shaw KJ, Miller N, Liu X, Lerner D, Wan J, Bittner A, Morrow BJ. Comparison of the changes in global gene expression of Escherichia coli induced by four bactericidal agents. J Mol Microbiol Biotechnol. 2003;5:105–122. doi: 10.1159/000069981. [DOI] [PubMed] [Google Scholar]
- Smith RJ, Midgley JE. The effect of trimethoprim on macromolecular synthesis in Escherichia coli. Regulation of ribonucleic acid synthesis by 'Magic Spot' nucleotides. Biochem J. 1973;136:249–257. doi: 10.1042/bj1360249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson BS, Schmidt TM. Life history implications of rRNA gene copy number in Escherichia coli. Appl Environ Microbiol. 2004;70:6670–6677. doi: 10.1128/AEM.70.11.6670-6677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagkopoulos I, Liu YC, Tavazoie S. Predictive behavior within microbial genetic networks. Science. 2008;320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tippin B, Pham P, Goodman MF. Error-prone replication for better or worse. Trends Microbiol. 2004;12:288–295. doi: 10.1016/j.tim.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Tsui WH, Yim G, Wang HH, McClure JE, Surette MG, Davies J. Dual effects of MLS antibiotics: transcriptional modulation and interactions on the ribosome. Chem Biol. 2004;11:1307–1316. doi: 10.1016/j.chembiol.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Walker GC. The SOS Response of Escherichia coli. In: Neidhardt FC, editor. Echerichia coli and Salmonella. Washington, D.C.: ASM Press; 1996. pp. 1400–1416. [Google Scholar]
- Xiao H, Kalman M, Ikehara K, Zemel S, Glaser G, Cashel M. Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J Biol Chem. 1991;266:5980–5990. [PubMed] [Google Scholar]
- Yeh P, Tschumi AI, Kishony R. Functional classification of drugs by properties of their pairwise interactions. Nat Genet. 2006;38:489–494. doi: 10.1038/ng1755. [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J. The truth about antibiotics. Int J Med Microbiol. 2006;296:163–170. doi: 10.1016/j.ijmm.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Yim G, Wang HH, Davies J. Antibiotics as signalling molecules. Philos Trans R Soc Lond B Biol Sci. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat Methods. 2006;3:623–628. doi: 10.1038/nmeth895. [DOI] [PubMed] [Google Scholar]
- Zaslaver A, Mayo AE, Rosenberg R, Bashkin P, Sberro H, Tsalyuk M, Surette MG, Alon U. Just-in-time transcription program in metabolic pathways. Nat Genet. 2004;36:486–491. doi: 10.1038/ng1348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.