Abstract
An accurate representation of ion solvation in aqueous solution is critical for meaningful computer simulations of a broad range of physical and biological processes. Polarizable models based on classical Drude oscillators are introduced and parametrized for a large set of monoatomic ions including cations of the alkali metals (Li+, Na+, K+, Rb+ and Cs+) and alkaline earth elements (Mg2+, Ca2+, Sr2+ and Ba2+) along with Zn2+ and halide anions (F−, Cl−, Br− and I−). The models are parameterized, in conjunction with the polarizable SWM4-NDP water model [Lamoureux et al., Chem. Phys. Lett. 418, 245 (2006)], to be consistent with a wide assortment of experimentally measured aqueous bulk thermodynamic properties and the energetics of small ion-water clusters. Structural and dynamic properties of the resulting ion models in aqueous solutions at infinite dilution are presented.
1 Introduction
Ions are fundamental to the structure and function of biological systems, where their local environment can be as diverse as are the roles that they play. Ions are involved in the folding of proteins and nucleic acids, enzyme catalysis, and in numerous cellular signaling processes. Monovalent ions such as Na+, K+, and Cl− play an important role in the homeostasis and electric activity of living cells and in modulating biomolecular stability through both specific and non-specific interactions.1,2 Divalent cations, such as Zn2+, Mg2+, and Ca2+, are often associated with catalytic or regulatory activities of proteins and enzymes,3,4 including the activation by Ca2+ of K+ channels,5 the stabilization by Zn2+ of zinc-finger proteins,6 and the condensation and folding of RNA and DNA by Mg2+.7-12
In order to have meaningful computational models to probe and explore the diverse and important roles of ions in biological phenomena, accurate and physically realistic descriptions of their microscopic interactions is crucial. This is not trivial, because a balanced description of the interactions between ion and ion, ion and water, and ion and biomolecules is required. A number of non-polarizable models for ions have been developed.13-18 The influence of induced electronic polarization, in particular, has been shown to be critical to the study of ion channels19-21 and it is expected to be generally important for the study of aqueous ionic systems. Striking a balance between accuracy and computational expense is an important consideration in designing useful models. Because proper statistical averaging in biological systems requires long simulations to sample over many configurations,22 achieving a sufficient sampling of the relevant configurations can become computationally prohibitive if the microscopic interactions are generated via sophisticated ab initio quantum mechanical (QM) approaches. In that regards, treatments of many-body polarization effects based on simple potential functions have important advantages over QM methods.
An alternative approach would be to adopt a polarizable force field.23-26 Generally speacking, there are three different methods to account for explicit electronic polarization into classical force fields, i.e., induced dipole model, fluctuation charge model and classical Drude Oscillator model. Correspondingly, a number of polarizable models for ions have been developed based on these three different approaches to study a variety of phenomena involving ions.27-30 Our own efforts have been focused on developing a complete all-atomic polarizable force field for proteins, nucleic acids and membranes based on the concept of the classical Drude oscillator.31 This model approximates the quantum mechanical electronic responses by using auxiliary massless charged particles that are harmonically bound to the polarizable nuclei.24,26,29,32-35 This approach is also referred to as the “shell model”36-38 or “charge-on-spring”.24,39 The polarizable Drude force field has been shown in previous studies to be accurate for the simulation of liquid water,33-35 aqueous ionic systems,29 condensed phases of small organic molecules,40-44 and lipid membranes.45 In these efforts, all new chemical entities must be compatible with the polarizable SWM4-NDP water model,35 which serves as the central reference for the polarizable Drude force field.
In the present paper, models for an extended set of ions are constructed and parameterized to be compatible with the SWM4-NDP water model.35 The set comprises the most abundant and biologically relevant monoatomic ions, including cations of the alkali metals (Li+, Na+, K+, Rb+ and Cs+) and alkaline earth elements (Mg2+, Ca2+, Sr2+ and Ba2+) along with Zn2+ and halide anions (F−, Cl−, Br− and I−). The present models of alkali and halide ions are similar to those developed previously,29 which were parameterized in conjunction with the SWM4-DP water model.34 Because the SWM4-DP and the SWM4-NDP models differ in the sign of the auxiliary Drude particle attached to the oxygen atom, a re-parametrization is necessary to have models compatible with the SWM4-NDP water model.35 In addition, models for divalent cations are introduced and parameterized. The latter pose new challenges for a classical polarizable force field, particularly regarding the treatment of overpolarization and Coulombic singularities. Approaches to overcome these problems are presented.
The paper is organized as follows. Section 2 recapitulates the relevant details of the Drude model for ion solvation and articulates the questions that arise when extending it to treat highly polarizable anions and divalent ions, along with steps that have been taken to solve the problem. Also included in Section 2 are the parameterization strategy and target data. In Section 3, results of the fitting are presented for our set of ions, along with an analysis of the structural properties and polarization effects that are predicted by the model.
2 Theory and Methods
2.1 Classical Drude polarizable model
The model ions are consistent with the SWM4-NDP polarizable water model with a negatively charged Drude oscillator bound to its oxygen site.35 The SWM4-NDP potential reproduces most properties of bulk water under ambient conditions (density, vaporization enthalpy, radial distribution function, dielectric constant, self-diffusion constant, shear viscosity, and free energy of hydration). In particular, the SWM4-NDP model yields a correct static dielectric constant, which makes it appropriate to study systems dominated by water-mediated electrostatic interactions. The SWM4-NDP water model comprises five interaction sites: (1) the oxygen atom “O” carrying a charge of , (2) the Drude particle “D” harmonically attached to the oxygen atom carrying a (negative) charge of , (3) the massless auxiliary site “M” carrying a charge , and (4-5) the hydrogen atoms“H1” and “H2”, each carrying a charge of . The interactions within and between water molecules are calculated according to the formulation of the polarizable Drude model described in Ref.33,34 The ion models comprise 2 sites: (1) the ion core atom “A” carrying a charge of , and (2) a Drude particle “D” attached to the ion atom carries a (negative) charge of . To be consistent with the SWM4-NDP model, polarization of an ion is represented by a negatively charged particle, representing electron density, bound to its nucleus (core). All atomic dispersion and electronic overlap effects are represented in a pairwise additive way using the Lennard-Jones (LJ) potential. The core repulsion and the van der Waals dispersive interactions is modeled by a LJ interaction between the water oxygen and the ion core atom via the Lorentz-Berthelot combination rule46. The potential energy of one ion and one water molecule can be written as,
| (1) |
where is the LJ interaction between the ion and the water-oxygen atom, represents the harmonic restoring springs associated with the Drude oscillator of the water molecule and of the ion, and includes all the Coulombic electrostatic interaction between the fixed and mobile charges carried by the ion (two sites) and the water molecule (fives sites). The spring constant KD is set to 1000 kcal/mol/Å2 for all Drude oscillators in the system. This value dictates the magnitude of the charge that the Drude particle must carry to produce the correct polarizability α, i.e., .35 For example, the charge on the Drude particle of I−, the most polarizable ion in the current study, is −4.733e with this restoring spring constant.
Simulations of the models are performed by considering the dynamics of an extended Lagrangian in which a small mass mD and kinetic energy are attributed to the Drude particles. The amplitude of the Drude oscillators is controlled with a low-temperature thermostat acting in the local center-of-mass reference frame of each atom-Drude pair.33 The mass of the Drude particles is set to 0.4 amu, which is subtracted from the mass of the physical polarizable nucleus such that the total mass of the pair remains constant. To ensure that the time course of the induced dipoles stays close to the self-consistent field (SCF) solution, a Nosé-Hoover thermostat at a temperature T* = 1 K is applied to the relative motion of each atom-Drude pair (in their local center-of-mass reference frame). It was shown that the trajectories generated according to this procedure are very close to those generated by the SCF regime of induced polarization.33,34 To control the global thermalization of the system, a second thermostat is applied to the center of mass of the atom-Drude pairs as well as the hydrogens atoms.
2.2 Overpolarization of Drude oscillators
The simple sum over Coulomb interactions of Uelec in Eq. (1) does not exclude singular r−1 attractive interactions between the Drude particles and other interaction sites carrying a net charge. Such singularities are generally not problematic in fixed-charge force fields, where the charges are buried within r−12 LJ core repulsive interactions. In the polarizable model based upon the Drude oscillator, however, the charge on the Drude particles is not as effectively shielded from other charges by such non-electrostatic core repulsive interactions. To clarify the situation leading to singularities, consider the interaction between a Drude oscillator bound to a heavy atom fixed at the origin and a point charge qi placed at some distance X. Along the one-dimensional axis, the potential energy is,
| (2) |
where x is the displacement of the oscillator, qD is the magnitude of the charge on each end of the Drude oscillator, and KD is the harmonic restoring force constant. The self-consistent field condition is:
| (3) |
The point of inflection for this solution becomes unstable when,
| (4) |
at which point x = X/3. For a fixed value of KD, instabilities can occur when the polarizability α is large, as in the case of some anions, or when the charge qi is large, as in the case of the small monoatomic divalent cations.
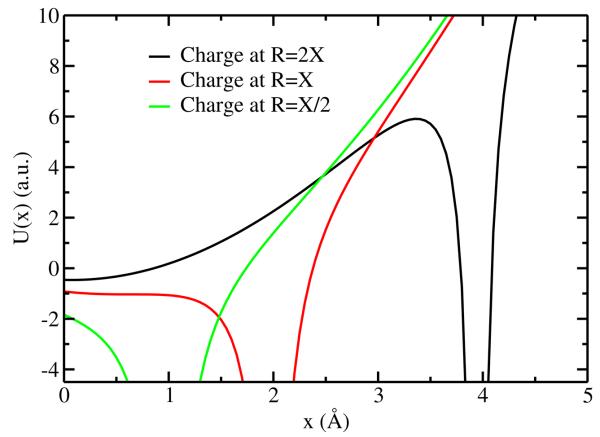
To illustrate the instabilities encountered with a divalent cation (qi = 2.0e), we substitute the charge and force constant parameters from the SWM4-NDP water model. The results are illustrated in Figure 1. Substituting parameters for the SWM4-NDP water model into Eq. (4), one finds that X = 1.974 Å. For example, when the divalent cation is placed at a distance of 2X from the origin, the Drude particle can reside in a stable minimum located at x ≈ 0.14 Å (Figure 1). But as shown in Figure 1, placing the divalent cation X < 1.974 Å will cause the SWM4-NDP oscillator to fall into the singular Coulomb well at X. In contrast, the system is stable with a monovalent cation (qi = 1.0e): according to Eq. (4), the instability appears only at X = 1.567 Å.29 This distance is considerably shorter than that observed for ion-water close contacts in simulation of ions in solution. Monovalent cations, unlike those for the divalent cations with the oxygen Drude sites on SWM4-NDP water molecules, therefore need no special treatment in order to avoid the “polarization catastrophe”.29
Figure 1.
SWM4-NDP water molecule with a point charge of +2e placed 2X, X and X/2 defined by Eq. 4 away from the oxygen atom.
A number of empirical remedies to the overpolarization problems are possible. The simplest treatment is to introduce an additional anharmonic restoring force to prevent excessively large excursions of the Drude particle away from the atom.39 The value of such anharmonic term can be adjusted to reduce the polarizability of atoms at high field. This approach was used to prevent any instabilities with the highly polarizable anions such as Br− and I−. The anharmonic restoring force was chosen to be active only beyond a certain stretching distance ΔRcut,
| (5) |
thus, preserving the linear polarization response with small displacements. The cutoff was chosen to be based on the maximum displacement of the Drude particles with respect to their nuclei in SWM4-NDP water models to ensure that its properties will not be affected. The force constant was chosen to be 40,000 kcal/mol/Å4 to reproduce the maximum induced dipole moment of halide anions estimated by MP2 calculations (See Supporting Information Figure S1). If the stiff anharmonic restoring force causes numerical instability with the finite time-step integrator, the problem could be treated using a multiple time-step algorithm, where the rapidly varying restoring forces are integrated with a smaller time-step than the remaining slowly varying forces.47
A second possibility is to associate a small repulsive core to the Drude particle implemented with the NBFIX option in the program CHARMM.48 Finally, another possibility is to introduce electrostatic screening functions that alter the charge-charge 1/r Coulombic interactions at short distances.30,49-52 The latter approach was used to construct stable and accurate models of the divalent cations. A screening function was introduced to modulate the electrostatic interactions between the divalent ion and the induced dipole component of the oxygen of the water molecules
| (6) |
where rij are the distances between the pairs of charges taken from and . The screening function Siw(r) is given by,
| (7) |
where aiw = (αiαw)(1/6)/tiw; αi and αw are the polarizabilities of the ion and the oxygen of the water molecule, respectively. This functional form of electrostatic screening was originally introduced for point dipoles by Thole.49 The dimensionless Thole parameter tiw modulates the strength of the screening for the i j pair. In the polarizable Drude model, this form has previously been utilized to modulate the intramolecular nearest bonded-neighbor, 1-2 interactions, and next-nearest-bonded-neighbor 1-3 interactions.43 For the divalent cations, the four terms representing the electrostatic interactions between the ion-Drude pair and the water oxygen-Drude pair were treated using Eq. (7).
2.3 Free energy calculations
The absolute hydration free energy of the ions was calculated and decomposed into three contributions following a free energy perturbation simulation protocol established previously,29,53
| (8) |
where and are the repulsive and attractive (dispersive) components, respectively, of the LJ interaction in Eq. (1). The electrostatic component of the hydration free energy is . Each component of the total hydration free energy was computed from independent simulations in which an ion was placed in a periodic box containing 216 explicit SWM4-NDP water molecules. Long range electrostatic interactions were computed using particle mesh Ewald summation.54 A smooth real space cutoff is applied between 10 and 12 Å with a Ewald splitting parameter of 0.34 Å−1, a grid spacing of ~ 1.0 Å and a sixth-order interpolation of the charge to the grid. The isothermal-isobaric ensemble was simulated using a Nosé-Hoover thermostat55,56 and the modified Andersen-Hoover barostat of Martyna et al.57 along with a 1 fs time-step. The internal geometry of the SWM4-NDP water molecule was kept rigidly fixed using the SHAKE/Roll and RATTLE/Roll algorithm.58,59 For each value of the thermodynamic coupling parameter, λ, after an initial equilibration of 200 ps, equilibrium properties were averaged over a 400 ps molecular dynamics simulation. For the dispersive and electrostatic components, λ took on values between λ = 0 and λ = 1 that were evenly spaced in increments of 0.1. For the repulsive term, λ took on the following values (0,0.05,0.1,0.15,0,2,0.25,0,3,0.35,0.4,0.5,0.6,0.7,0.8,0.9,1). The repulsive contribution, , was computed using a soft-core scheme as described elsewhere53 and was unbiased using the weighted histogram analysis method (WHAM),60 while and were computed using thermodynamic integration (TI). Based on multiple runs, the overall precision of the calculated absolute hydration free energies with the current protocol is on the order of 0.2 kcal/mol for monovalent ions and 0.5 kcal/mol for divalent ions.
In discussions of the hydration free energy of ionic species, one may consider the real physical value, which includes the contribution of the phase potential arising from crossing the physical air/water interface, or the intrinsic bulk phase value, which is independent of the interfacial potential.29,61,62 The relationship between real and intrinsic hydration free energies is defined as , where F is the Faraday constant (23.06 kcal/mol/V) and Φ is the electrostatic Galvani potential at the liquid-vacum interface, or the phase potential of the liquid relative to vacuum. It may be tempting to consider the intrinsic free energy as somehow reflecting more faithfully the true hydration free energy of the ion within the bulk phase because it is “disentangled” from the bias arising from the liquid-vacum interfacial potential. However, it should be noted that the actual value of Φ depends upon the convention to define the Galvani potential. One may construct an “internal” Galvani potential via a P-sum, where the potential at all points in space is the superposition of the total charge density from all the particles.63,64 Alternatively, one may construct an “external” Galvani potential via a M-sum, where the potential from each water molecule contributes only to points in space that are outside their repulsive core.63,64 Although the internal or external Galvani potentials can be defined mathematically without ambiguity by specifying which P- or M-sum convention has been used, they cannot be measured experimentally by a physical process.
To avoid any ambiguity, only real hydration free energies are considered throughout the present study. corresponds to the reversible thermodynamic work to move a single ion from vacuum to the interior of a pure water phase (i.e., across the physical liquid-vacuum interface). Our free energy calculations with periodic boundary conditions (PBC) and particle mesh Ewald are carried out within the P-sum convention, and the implicit reference phase potential of the liquid is the “internal” Galvani potential. Such free energy calculations yield , which then needs to be shifted by zΦ (calculated within the same P-sum convention) to yield the physically meaningful . For our polarizable SWM4-NDP water model, the P-sum internal Galvani potential Φ is equal to −545 mV35 (negative in the liquid phase relative to vacuum), giving rise to an energy shift of ∓12.6 kcal/mol for the monovalent cations/anions and twice that amount for the divalent species. Finally, for a direct comparison with published experimental tables, it is necessary to convert the results into the free energy of an ion going from an ideal gas at 1 atmosphere to an idealized bulk solution at 1 M concentration. To account for the compression from a volume of 24.465 liter per mole in the ideal gas to the 1 M solution, a small entropic contribution of −kBT ln(1/24.465) = 1.9 kcal/mol must be added per ion.29
2.4 Target Data and Parametrization Strategy
2.4.1 Ionic charges and polarizabilities
For the alkali cations and halide anions, the values of the polarizabilities from the earlier parameterization compatible with the SWM-DP water model were used here without changes.29 In Table 1 included among these polarizabilities are gas-phase values, reported by Mahan,65 for the alkali cations and a set of “solvent-renormalized” values for the anion halides, with the bare gas-phase polarizability66 scaled down by a factor of 0.724. Although the actual values vary, such reduced anion polarizabilities have also been noted in previous quantum mechanical studies,67-70 see Ref.29 for a more complete discussion. For the divalent cations, gas-phase values were again taken from Mahan:65 0.075 Å3 for Mg2+, 0.49 Å3 for Ca2+, 0.87 Å3 for Sr2+, 1.56 Å3 for Ba2+ and 0.42 Å3 for Zn2+.
Table 1.
Parameters for alkali cations, halide anions and divalent cations with negatively charged Drude oscillators.
| q(|e|) | α(Å3)a | −δq(|e|) | Emin (kcal/mol) | Rmin/2 (Å) | Thole tiw | |
|---|---|---|---|---|---|---|
| Li+ | +1 | 0.032 (0.032) | 0.310427 | 0.0300000 | 1.1000000 | - |
| Na+ | +1 | 0.157 (0.157) | 0.687597 | 0.0315100 | 1.4616800 | - |
| K+ | +1 | 0.830 (0.830) | 1.580968 | 0.1419265 | 1.6866521 | - |
| Rb+ | +1 | 1.370 (1.370) | 2.031161 | 0.2730669 | 1.7855083 | - |
| Cs+ | +1 | 2.360 (2.360) | 2.665877 | 0.2766036 | 2.0238218 | - |
| F− | −1 | 1.786 (2.467) | 2.319199 | 0.0026181 | 2.4622406 | - |
| Cl− | −1 | 3.969 (5.482) | 3.457187 | 0.0719737 | 2.4811139 | - |
| Br− | −1 | 5.262 (7.268) | 3.980713 | 0.0823440 | 2.6262883 | - |
| I− | −1 | 7.439 (10.275) | 4.733085 | 0.2084343 | 2.7579694 | - |
| Zn2+ | +2 | 0.420 (0.420) | 1.124637 | 0.2500000 | 0.9349678 | 2.14773 |
| Mg2+ | +2 | 0.075 (0.075) | 0.475246 | 0.0500000 | 1.1264156 | 1.51567 |
| Ca2+ | +2 | 0.490 (0.490) | 1.214747 | 0.2100000 | 1.2708552 | 1.50877 |
| Sr2+ | +2 | 0.870 (0.870) | 1.618629 | 0.3400000 | 1.3059400 | 1.23792 |
| Ba2+ | +2 | 1.560 (1.560) | 2.167454 | 0.6000000 | 1.5717385 | 1.45869 |
2.4.2 Ionic monohydrates
For the monovalent ions, the target monohydrate properties are the same as those used in a previous effort29 and are summarized in Table 2. Namely, the target binding enthalpies, ΔH, for the alkali cations and halide anions are the gas-phase binding enthalpies measured by Džidić and Kebarle71 and Tissandier et al.72 In addition, the binding energies are taken from the ab initio computations.73-76 A set of target ion-oxygen distances was established for the monovalent ions as described in detail by Lamoureux and Roux.29 For the divalent cations, ab initio quantum chemical computations were carried out at the MP2 level using Gaussian03.77 The details of the basis sets used and the resulting target data are presented in Table 2. In these computations, the water geometry was fixed to that of the SWM4-NDP model while the ion-oxygen separation was varied.
Table 2.
The properties of alkali cations, halide anions and divalent cations with Drude polarizable models.
| Ion | Umina | dminb | ΔHc | rmax | gmax | rmin | gmin | Nc | Dd | ΔGe | ΔΔGf |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Li+ | −35.9 (−35.2) | 1.91 (1.87) | −35.6 (−34.0,−34.0) | 2.02 | 12.50 | 2.56 | 0.00 | 4.0 | 1.30 (1.03) | −120.5 | −24.2 (−23.8~-26.2) |
| Na+ | −24.6 (−24.3) | 2.25 (2.26) | −24.4 (−24.0,−25.0) | 2.38 | 7.42 | 3.24 | 0.20 | 5.6 | 1.58 (1.33) | −96.3 | −17.3 (−16.7~−17.7) |
| K+ | −17.9 (−17.8) | 2.62 (2.64) | −17.6 (−17.9,−18.1) | 2.74 | 4.80 | 3.56 | 0.45 | 6.9 | 2.20 (1.98) | −78.6 | −5.2 (−4.9~−5.4) |
| Rb+ | −15.7 (−16.1) | 2.79 (2.79) | −15.2 (−15.9,−16.0) | 2.90 | 4.04 | 3.80 | 0.62 | 8.1 | 2.44 (2.07) | −73.7 | −7.1 (−5.5~−7.7) |
| Cs+ | −13.3 (−14.1) | 3.05 (2.99) | −12.5 (−13.7, —) | 3.16 | 3.25 | 4.10 | 0.75 | 9.7 | 2.56 (2.06) | −66.5 | |
| F− | −23.5 (−25.9) | 2.53 (2.44) | −23.2 (−23.3,−23.3) | 2.72 | 4.77 | 3.34 | 0.37 | 5.5 | 1.33 (1.48) | −108.0 | −30.0 (−13.4~−30.6) |
| Cl− | −14.0 (−14.4) | 3.09 (3.11) | −13.7 (−13.1,−14.4) | 3.16 | 3.15 | 3.78 | 0.72 | 6.5 | 1.82 (2.03) | −78.4 | −6.5 (−3.3~−6.9) |
| Br− | −12.4 (−12.7) | 3.26 (3.26) | −11.9 (−12.6,−13.0) | 3.28 | 2.70 | 3.96 | 0.75 | 6.8 | 1.85 (2.08) | −71.6 | −8.5 (−7.7~−11.1) |
| I− | −10.2 (−10.6) | 3.56 (3.50) | −9.7 (−10.2,−10.5) | 3.50 | 2.28 | 4.16 | 7.13 | 7.1 | 2.02 (2.05) | −63.1 | |
| Zn2+ | −100.0 (−96.3) | 1.82 (1.86) | −99.4 (−103.1) | 2.14 | 17.2 | 3.08 | 0.0 | 6.0 | 0.61 (0.70) | −460.2 | −90.3 (−107.6) |
| Mg2+ | −89.4 (−77.9) | 1.86 (1.93) | −89.0 (−81.8) | 2.06 | 19.0 | 2.72 | 0.0 | 6.0 | 0.82 (0.71) | −447.2 | −77.3 (−77.7~−80.3) |
| Ca2+ | −55.6 (−54.9) | 2.18 (2.25) | −55.1 (−56.5) | 2.28 | 16.9 | 2.76 | 0.0 | 6.0 | 0.96 (0.79) | −369.9 | −32.7 (−29.8~32.9) |
| Sr2+ | −45.2 (−40.6) | 2.30 (2.52) | −44.7 (−) | 2.42 | 11.9 | 3.20 | 0.0 | 7.2 | 0.96 (0.79) | −337.2 | −27.2 (−27.9~31.1) |
| Ba2+ | −37.7 (−34.0) | 2.56 (2.73) | −37.3 (−) | 2.68 | 10.4 | 3.30 | 0.1 | 8.2 | 0.97 (0.85) | −310.0 |
The reference binding energy in parentheses Umin of monovalent monohydrates are taken from various sources: 29 Li+; 73 Na+; 74 K+; 75 Rb+; 74 Cs+; 74 F−; 76 Cl−; 76 Br−; 76 I−; 76 The reference values for divalent were obtained with MP2 calculations with basis set 6-311++G(3df,3pd) for hydrogen, oxygen, Mg2+, Ca2+, and Zn2+ and LANL2DZ pseudopotential and associated basis set for Sr2+ and Zn2+. The energies are in kcal/mol.
The reference distance in parentheses Rmin of the alkali cations are taken from various sources 29 while those of the halide anions are taken from. 76 The reference values for divalent cations were obtained from MP2 calculations (See a for details). The distances are in Å.
The reference enthalpies in parentheses ΔH for the monovalent monohydrates are taken from the experimental studies. 71,72 The reference enthalpies in parentheses ΔH for the divalent monohydrates are taken from the theoretical study. 87 The enthalpies are in kcal/mol.
The experimental self diffusion coefficients are taken from the Handbook of Chemistry and Physics. 95 The self diffusion coefficients are in 10−5cm2/s.
The real hydration free energies ΔGreal in kcal/mol are obtained from the intrinsic free energies from periodic boundary conditions (PBC) simulations corrected with the phase potential and entropy related contributions.
2.4.3 Absolute hydration free energies
The experimentally determined absolute hydration free energies of ions at infinite dilution is a central piece of information to optimize the model.72,78-83 However, as noted previously,29 there is a large spread in the reported values for the alkali and halide ions (see Fig. 2 of Ref.29). The same is true for the divalent cations. Considerations based on interfacial potentials may help explain the origin of some of those discrepancies, although it is important to realize that the overall offset of the absolute hydration free energy scale measured experimentally remains undetermined. The most reliable target data that can be extracted from the experimental measurements are the relative hydration free energies between different ions of identical charge, and the total solvation free energies of neutral salts (ΔGhydr(ABn) = ΔGhydr(A) + n × ΔGhydr(B)). Notwithstanding those issues, it should be noted there are also some quantitative differences among the various experimental values. For example, Tissandier et al.72 reports the total hydration free energy of KCl to be −156.8 kcal/mol, while Randles78 has reported −151.3 kcal/mol. During the parametrization monovalent ions, we principally aimed to reproduce the target data generated from the measurements of Tissandier.72 The values from Tissandier,72 Schmid,83 and Klots81 are quite close for the monovalent ions. For the divalent ions, the target data was taken from the measurements of Schmid,83 except in the case of Zn2+ where our model falls between the data from Schmid83 and Gomer and Tryson.80
Figure 2.
Binding energies of the divalent monohydrates as a function of the distance between the ion and the oxygen atoms: Solid line: ab initio results; Dashed line: Drude. See Table 2 caption for the theory and basis set used to obtain the reference data.
2.4.4 Optimization procedure
The parameters for the ion-water oxygen interaction are constructed via the Lorentz-Berthelot combination rule,46 and . The adjustable parameters for the monatomic ions within the classical Drude oscillator scheme are the LJ parameters of the ion, and . Rather than exploring the LJ parameters of the ions directly, it is more convenient, to explore the space of monohydrate interaction energies and minimum-energy ion-oxygen distances {dmin, Umin},29 which can be compared to the available ab initio or experimental data. Furthermore, quadratic response functions are fitted to the data from explicit computations, defined by coordinates in {dmin, Umin}, to interpolate predicted properties (e.g. hydration free energy ) between simulated models:29,35
| (9) |
A set of polarizable models for the monovalent ions were thus constructed by determining the LJ parameters spanning a grid in the {Umin, dmin} coordinates and simultaneously fitting all ions using the interpolated polynomials. For divalent cations that use the Thole screening as described by Eq. (7), there are three parameters to fit per ion: the LJ radius and well depth of the ion, and the ion-water Thole screening factor. To simplify the parameterization process and ensure physically reasonable dispersive interactions, it proved useful to first assign the LJ well depth by making use of the familiar London dispersion formula for the leading-order dispersion coefficient,
| (10) |
where , , αi, αw are the ionization energy and polarizabilty of the ion and the water, respectively. Converting into the attractive coefficient of the LJ interaction, , yields an estimate of . The resulting values are 0.05 for Mg2+, 0.21 for Ca2+, 0.34 for Sr2+, 0.60 for Ba2+, and 0.25 for Zn2+ (all in kcal/mol). Once the LJ well depth is assigned, a grid in {dmin, Umin} was generated for each divalent ion by varying the LJ radius and Thole parameter for each ion. The parameterization of the complete set of ion models is fairly consistent and robust. As observed previously,29,84 alternative sets of parameters can be found with small variations in the ion monohydrate energies (±0.25 kcal/mol) or in the ion-water distances (±0.1 Å) resulting in small shifts in the absolute hydration free energies (±3 kcal/mol). At this stage, the parameterization procedure is aimed at designing accurate models for the infinite dilution limit. Solutions at high concentrations, where ion-ion interactions become important, will be examined in a later stage. The final fitted values are summarized in Table 1.
3 Results and Discussion
The previously developed K+ model was shown to capture the essential properties in both gas phase and aqueous solution.84 The parameters for the polarizable Drude ion models were optimized to be consistent with bulk hydration free energies of the neutral salts while yielding accurate energies and geometries for the ionic monohydrates. An important advantage of reproducing the bulk hydration free energies of neutral salts with our model ions is that these data contain no undetermined offset constant, as do the absolute hydration free energies of individual ions.29 Achieving an accurate description for the ionic monohydrates becomes critical, therefore, to “lock down” the absolute scale for the bulk hydration free energies. As shown in Table 2, this procedure defines a “solvation-consistent” scale for the absolute hydration free energies of ionic species despite the uncertainties in the experimental data. Overall, the properties of small gas phase clusters given in Table 4 and bulk solvation given in Table 3 are in excellent agreement with experiments.
Table 4.
The solvation enthalpies of ion M in water clusters (H2O)n. The enthalpies are in kcal/mol.
| Ions | Model/Exp | Solvation Enthalpy | |||||
|---|---|---|---|---|---|---|---|
| M | 1 | 2 | 3 | 4 | 5 | 6 | |
| Li+ | Drude | −35.6 | −65.3 (−29.7) | −87.0 (−21.7) | −103.0 (−16.0) | −114.2 (−11.2) | −125.3 (−11.1) |
| Exp.72 | −34.0 | −59.8 (−25.8) | −80.5 (−20.7) | −96.9 (−16.4) | −110.8 (−13.9) | −122.9 (−12.1) | |
| Exp.71 | −34.0 | −59.8 (−25.8) | −80.5 (−20.7) | −96.9 (−16.4) | −110.8 (−13.9) | −122.9 (−12.1) | |
|
| |||||||
| Na+ | Drude | −24.4 | −45.9 (−21.5) | −63.4 (−17.5) | −77.2 (−13.8) | −87.0 (−9.8) | −95.1 (−8.1) |
| Exp.72 | −25.0 | −44.8 (−19.8) | −60.2 (−15.4) | −73.4 (−13.2) | −84.9 (−11.5) | −95.6 (−10.7) | |
| Exp.71 | −24.0 | −43.8 (−19.8) | −59.6 (−15.8) | −73.4 (−13.8) | −85.7 (−12.3) | −96.4 (−10.7) | |
|
| |||||||
| K+ | Drude | −17.6 | −33.2 (−15.6) | −47.0 (−13.8) | −58.6 (−11.6) | −67.7 (−9.1) | −76.0 (−8.3) |
| Exp.72 | −18.1 | −34.2 (−16.1) | −47.4 (−13.2) | −59.2 (−11.8) | −69.9 (−10.7) | −79.9 (−10.0) | |
| Exp.71 | −17.9 | −34.0 (−16.1) | −47.2 (−13.2) | −59.0 (−11.8) | −69.7 (−10.7) | −79.7 (−10.0) | |
|
| |||||||
| Rb+ | Drude | −15.3 | −29.0 (−13.7) | −41.2 (−12.2) | −51.9 (−10.7) | −60.3 (−8.4) | −68.5 (−8.2) |
| Exp.72 | −16.0 | −29.6 (−13.6) | −41.8 (−12.2) | −53.0 (−11.2) | −63.5 (−10.5) | ||
| Exp.71 | −15.9 | −29.5 (−13.6) | −41.7 (−12.2) | −52.9 (−11.2) | −63.4 (−10.5) | ||
|
| |||||||
| Cs+ | Drude | −12.4 | −24.3 (−11.9) | −34.7 (−10.3) | −44.0 (−9.3) | −52.0 (−8.0) | −64.0 (−12.0) |
| Exp.71 | −13.7 | −26.2 (−12.5) | −37.4 (−11.2) | −48.0 (−10.6) | |||
|
| |||||||
| F− | Drude | −23.2 | −45.4 (−22.2) | −66.9 (−21.5) | −80.2 (−13.3) | −90.7 (−10.5) | −103.1 (−12.4) |
| Exp.72 | −23.3 | −41.2 (−17.9) | −55.7 (−14.5) | −69.4 (−13.7) | −82.2 (−12.8) | −93.1 (−10.9) | |
|
| |||||||
| Cl− | Drude | −13.7 | −27.6 (−13.9) | −43.9 (−16.3) | −57.4 (−13.5) | −64.4 (−7.0) | −71.7 (−7.3) |
| Exp.72 | −14.4 | −27.2 (−12.8) | −38.9 (−11.7) | −49.8 (−10.9) | −59.3 (−9.5) | −68.1 (−8.8) | |
|
| |||||||
| Br− | Drude | −11.9 | −24.6 (−12.7) | −40.4 (−15.8) | −53.2 (−12.8) | −59.6 (−6.4) | −68.1 (−8.5) |
| Exp.72 | −13.0 | −25.0 (−12.0) | −36.4 (−11.4) | −47.4 (−11.0) | −58.2 (−10.8) | −68.5 (−10.3) | |
|
| |||||||
| I− | Drude | −9.7 | −20.9 (−11.2) | −35.6 (−14.7) | −42.4 (−6.8) | −50.7 (−8.4) | −59.6 (−8.9) |
| Exp.72 | −10.5 | −20.2 (−9.7) | −29.5 (−9.3) | −38.7 (−9.2) | −47.7 (−9.0) | ||
|
| |||||||
| Zn2+ | Drude | −99.4 | −176.9 (−77.5) | −237.2 (−60.3) | −284.4 (−47.2) | −306.2 (−21.8) | −329.0 (−22.8) |
| Theo. or Exp.87 | −103.1 | −191.1 (−88.0) | −246.9 (−55.8) | −289.8 (−42.9) | −314.8 (−25.0) | −339.0 (−24.2) | |
|
| |||||||
| Mg2+ | Drude | −89.0 | −164.6 (−75.6) | −223.3 (−58.7) | −269.8 (−46.5) | −293.0 (−23.2) | −316.5 (−23.5) |
| Theo. or Exp.87 | −81.8 | −153.8 (−72.0) | −210.4 (−56.6) | −254.3 (−43.9) | −282.3 (−28.0) | −306.9 (−24.6) | |
|
| |||||||
| Ca2+ | Drude | −55.1 | −105.2 (−50.1) | −149.5 (−44.2) | −188.1 (−38.6) | −217.8 (−29.7) | −245.3 (−27.5) |
| Theo. or Exp.87 | −56.5 | −105.1 (−48.5) | −148.0 (−42.9) | −183.6 (−35.6) | −211.3 (−27.7) | −236.6 (−25.3) | |
|
| |||||||
| Sr2+ | Drude | −44.7 | −86.2 (−41.5) | −124.0 (−37.8) | −158.4 (−34.4) | −187.0 (−28.6) | −213.6 (−26.6) |
| Exp.86 | − (−22.7) | ||||||
|
| |||||||
| Ba2+ | Drude | −37.3 | −72.0 (−34.7) | −103.7 (−31.7) | −133.3 (−29.6) | −158.7 (−25.4) | −182.6 (−23.9) |
| Exp.86 | − (−23.9) | − (−19.8) | |||||
Numbers in parentheses are for the reaction .
Table 3.
Hydration free energy of neutral salts (in kcal/mol)a.
| Li+ | Na+ | K+ | Rb+ | Cs+ | |
|---|---|---|---|---|---|
| F− | −228.5 (−229.0) | −204.3 (−203.8) | −186.6 (−186.6) | −181.7 (−181.2) | −174.5 (−173.8) |
| ClR− | −198.9 (−199.3) | −174.7 (−174.0) | −157.0 (−156.8) | −152.1 (−151.4) | −144.9 (−144.0) |
| Br− | −192.1 (−192.9) | −167.9 (−167.6) | −150.2 (−150.4) | −145.3 (−145.0) | −138.1 (−137.6) |
| I− | −183.6 (−183.9) | −159.4 (−158.7) | −141.7 (−141.5) | −136.8 (−136.1) | −129.6 (−128.7) |
|
| |||||
| Zn2+ | Mg2+ | Ca2+ | Sr2+ | Ba2+ | |
|
| |||||
| F− | −676.2 (−696.8) | −663.2 | −585.9 (−589.4) | −553.2 | −526.0 (−528.1) |
| Cl− | −617.0 (−637.2) | −604.0 (−607.0) | −526.7 (−529.9) | −494.0 (−499.0) | −466.8 (−468.6) |
| Br− | −603.4 (−624.4) | −590.4 (−594.3) | −513.1 (−517.0) | −480.4 (−486.2) | −453.2 (−455.7) |
| I− | −586.4 (−606.5) | −573.4 (−576.4) | −496.1 (−499.1) | −463.4 (−468.3) | −436.2 (−437.8) |
The experimental solvation free energies reported for alkali halide salts by Tissandier et al.72 and Klots81 (Cs+) are listed in parentheses for comparison. The experimental solvation free energies for salts of divalent cations were derived from the free energies of formation of the salt minus that of the gas-phase ions (in kJ/mol): , with and where M is the molar mass in grams, v* is 1/2 for anions and 1 for cations, S(A*) is the element's absolute entropy in the standard state and S(e−(g)) is the absolute entropy of the gaseous electron (see Ref.72 for details). The experimental thermodynamic data are taken from the Handbook of Chemistry and Physics95 and the 1982 NBS tables.85
3.1 Instabilities and overpolarization
The polarizable Drude models based on the simple scheme presented by Eq. (1) yield numerically stable simulations for all the monovalent cations. As expected, based upon substitution into Eq. (4), no numerical instabilities based on overpolarization were present. In contrast, Eq. (4) can be used to predict overpolarization catastrophes that occur for halide anions (Br− and I−) when ion-proton separations are within physically realistic distances. Such instabilities are not present in our previously published set of alkali-halide ionic models29 because the positive Drude particle and proton do not attract one another. Introducing an anharmonic restoring force to the anionic Drude oscillator removes those instabilities by preventing excessively large excursions of the Drude particle away from the ion. Importantly, as shown in Table 2, Table 3 and Table 4, this construction did not affect our ability to adjust the parameters of the model to accurately fit the target data. In particular, the properties of the small cluster hydrates can be reproduced with the simple anharmonic restoring spring.
A more serious overpolarization problem is encountered with the divalent cations. We first tried the two simple schemes to solve this issue, namely, adding an anharmonic restoring force to the Drude oscillator, or adding a LJ core repulsion between the oxygen-tethered Drude particle and the divalent cations. Experimenting with various parametrizations showed that, while introducing an anharmonic restoring force or a LJ core can prevent the numerical instabilities, it remained difficult to get reasonable monohydrate energies as the large Coulombic repulsion between the bare positive charge carried by the oxygen nucleus and the ion remains unshielded. This Coulombic interaction gives rise to large repulsion energies that must be compensated in order to have reasonable monohydrate energies and geometries with the divalent cations. Ultimately, it was found that introducing a Thole-type screening function49 for the Coulombic electrostatic for the interactions between the divalent ion and the induced dipole component of the water molecules led to the best models. A similar approach was used to construct models of divalent ions consistent with the AMOEBA polarizable force field.30,52 The comparison between the Drude model with Thole-like damping and ab initio calculations are shown in Figure 2 and Figure 3. The Thole screening scheme given by Eq. 7 effectively removes the singularity problems illustrated in Figure 1 and yields divalent monohydrates with reasonable binding energies and total dipole moments.
Figure 3.
Total dipole moments of the divalent monohydrates as a function of the distance between the ion and the oxygen atoms. The ions are located at the origin to compute the dipole moment. See Figure 2 for details.
3.2 Bulk hydration free energies
The total hydration free energies of neutral salts are shown in Table 3, Figure 4 and Figure 5 together with available experimental data. Although the relative hydration free energy difference between different ions in the same series and the total hydration free energy of a neutral salt can generally be measured very accurately, we note that there are noticeable differences among the experimental data from difference sources72,78-83,85 (See Supporting Information Table S1 and S2). Overall, the current Drude models agree very well with the experimental data, with the monovalent neutral salts closer to the experimental estimations by Tissandier et al.,72 Klots,81 and Schmid et al.83 (Figure 4) and the divalent neutral salts closer to the experimental estimations by Schmid et al.83 and Gomer and Tryson80 (Figure 5).
Figure 4.
Hydration free energies of salts with monovalent cations. Tissandier;72 Klots;81 Marcus;82 Noyes;79 Schmid;83 Randles;78 Gomer.80
Figure 5.
Hydration free energies of salts with divalent cations. Schmid:;83 Marcus:;82 Gomer:;80 Noyes:;79 and this work: see the caption of Table 3 for details; The entries for MgF2 and SrF2 in this work were derived based on hydration free energy differences between F− and Cl− and the total hydration free energies of salts MgCl2 and SrCl2.
The hydration free energy differences of ions in the same series are well reproduced with some compromise of the monohydrate properties for smaller ions. These present models define an absolute hydration free energy scale in which the real hydration free energy of the proton would be −258.8 kcal/mol. This can be compared with the experimental estimates of −264 kcal/mol by Tissandier et al.,72 −249.5 kcal/mol by Schmid et al.,83 and −253.2 kcal/mol by Gomer and Tryson.80 It is noteworthy that the current value of −258.8 kcal/mol differs by less than 2 percent from the experimental estimates obtained by Tissandier et al.72 This suggests that their analysis, which consisted in monitoring the stepwise changes in ion-cluster free energies, was targeting the absolute real free energy of the proton.72
3.3 Enthalpies of ionic small hydrates
Structural and thermodynamic properties of small clusters offer a rigorous test of the accuracy of the models. As opposed to bulk liquid hydration where induced polarization over different moieties may lead to some compensation of errors, the microscopic interactions are displayed nakedly in small clusters and possible limitations of the models become readily apparent. Furthermore, because the polarizable models are constructed with the intended purpose of responding accurately to different electrostatic environments, the ability to reproduce the properties of small ionic hydrates is very important. Finally, the properties of the ionic monohydrates are used to set the scale for the absolute hydration free energies in bulk solution. The calculated ionic monohydrate energies and geometries are reported in Table 2. The results generally agree well with the target data.73-76 As previously noted,29 the limitations of the models are most apparent for smaller ions. This trend is observed for the divalent cations as well. The less satisfactory performance of the divalent cations is very likely due to the neglect of the important charge transfer effect, which is absent from the current energy function. The enthalpies of hydration for an ion solvated in a small water cluster (1-6) compare favorably with literature values, with the monovalent cations having the best agreement71,72,86,87 (Table 4). In general, the monovalent cations reproduce the experimentally measured properties of ionic hydrate clusters slightly better than do the halide anions. The most likely explanation for this is that the conformational spaces for water molecules in the anion-water clusters are far more complicated than those for the cation-water clusters due to the formation of hydrogen bonding between water molecules coordinating the anion. The hydration enthalpies for the divalent cations have larger deviation from the experimental or ab initio data, consistent with the monohydrate properties.
3.4 Coordination structure in bulk solution
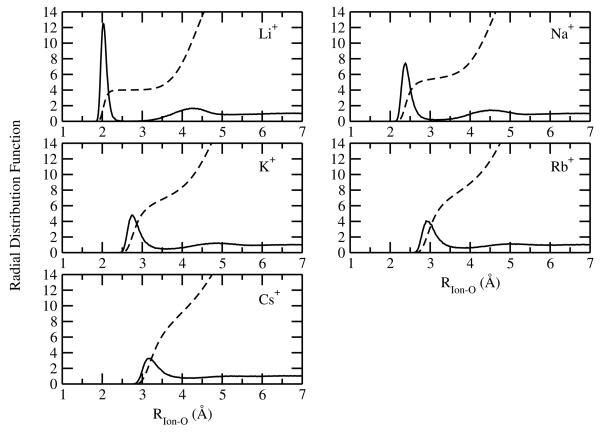
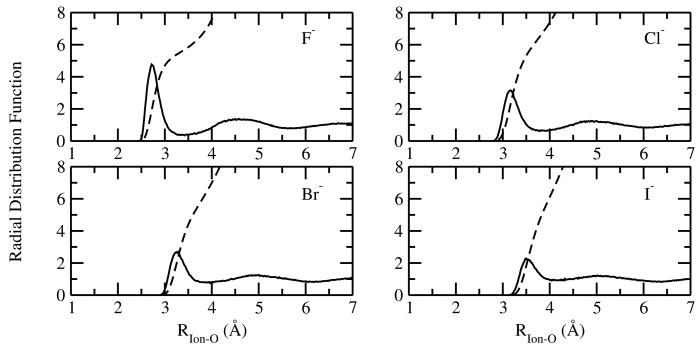
Radial distribution functions (RDF) averaged from ten simulations each of 200 ps for ion-oxygen contacts, g(Rion−O), are presented in Figure 6, Figure 7 and Figure 8 for the alkali cations, halide anions, and divalent ions, respectively. The coordination numbers for each ion, N(Rion−O), defined as the integral of g(Rion−O) from the origin out to the first minimum, rmin, are presented in Table 2. The RDFs and the coordination numbers for the monovalent ions are very similar to those reported from the previous models (Figure 6 and Figure 7) based upon the (positive Drude) SWM4-DP water model.29 Here, we briefly discuss the main findings. In accordance with the most recent computational studies, the current model predicts that the Li+ ion is 4-coordinated within the experimental range obtained from neutron and X-ray diffraction (See Ref.88-90 for a survey). After the first peak, the lithium-oxygen RDF remains very small over a large interval extending from 2.4 to 3.1 Å. This result is consistent with Car-Parrinello molecular dynamics (CPMD) simulation.91 The strong separation between the first and second peak in the RDF implies that the coordination number of Li+ can be defined unambiguously. However, for Na+ and K+, a more populated region is observed between the first and second peaks in in the RDFs. As a result, a broad distribution of coordination numbers within the first solvation shell was observed.22,84 As dissussed in Ref.,29,84,90 both experimental studies and CPMD type simulations have predicted a wide range of coordination numbers for Na+ and K+ and the current models fall well within the reported range. In addition, the coordination number distributions of of Na+ and K+ overlap substantially with one another, indicating that the coordination number alone is not a dominant factor in describing the thermodynamics of ion solvation.22
Figure 6.
Radial hydration structure for the alkali cations. Radial distribution functions g(RIon−O) functions are shown in solid line and the coordination numbers N(RIon−O) are shown in dashed line.
Figure 7.
Radial hydration structure for the halide anions. Radial distribution functions g(RIon−O) functions are shown in solid line and the coordination numbers N(RIon−O) are shown in dashed line.
Figure 8.
Radial hydration structure for the divalent cations. Radial distribution functions g(RIon−O) functions are shown in solid line and the coordination numbers N(RIon−O) are shown in dashed line.
For the divalent ions, the first peak in the RDF is generally sharper than those of the monovalent ions, indicating a highly ordered water structure around the ions (Figure 8). In addition, a clear separation exists between the first and the second coordination shells, as indicated by the the small value of the first minimum in the RDFs. The current Drude model for Ca2+ gives a coordination number of 6, smaller than the value of 7.3 based on the AMOEBA force field.30 As a comparison, the coordination number of Ca2+ were estimated to lie between 5.5 and 10 based on various experimental techniques,92 and between 6 and 7 according to the most recent CPMD simulations.92 For Mg2+, the Drude model gives a coordination number of exactly 6, consistent with both the experimental93 and ab initio94 studies. For Zn2+, the coordination number is estimated to be 6, which is within the experimentally estimated range of 5.3 to 6.6.88 Overall, the coordination structure for the ion models is in excellent agreement with experiment, given that no direct adjustments were made to reproduced those features during the parametrization.
3.5 Self-diffusion coefficients
The ability of the polarizable force field to reproduce the dynamic transport properties of ions in aqueous solutions is a critical test of the accuracy of computational models. This ability is best characterized by considering the diffusion of ions in aqueous solutions. The self-diffusion coefficients for the ion models at infinite dilution were computed from the mean-square displacement correlation function. Because there is only a single ion in the atomic system, relatively long simulations are required to obtain converged self diffusion coefficient. To improve convergence, ten simulations of 200 ps each were averaged for each ion. The results are presented in Table 2 and Table 5. Additional simulations indicate that the current estimates from MD are not overly sensitive to finite-sizze effects within PBC.
Table 5.
Effective self-diffusion coefficientsa of the neutral salts in 10−5 cm2/s (the experimentally derived values are in the parentheses95)
| Li+ | Na+ | K+ | Rb+ | Cs+ | |
|---|---|---|---|---|---|
| F− | 1.32 (1.25) | 1.45 (1.40) | 1.77 (1.73) | 1.89 (1.78) | 1.95 (1.77) |
| Cl− | 1.56 (1.53) | 1.70 (1.68) | 2.01 (2.01) | 2.13 (2.05) | 2.19 (2.05) |
| Br− | 1.58 (1.56) | 1.72 (1.71) | 2.02 (2.03) | 2.15 (2.08) | 2.21 (2.07) |
| I− | 1.66 (1.54) | 1.80 (1.69) | 2.11 (2.01) | 2.23 (2.06) | 2.29 (2.06) |
|
| |||||
| Zn2+ | Mg2+ | Ca2+ | Sr2+ | Ba2+ | |
|
| |||||
| F− | 0.85 (0.96) | 0.99 (0.97) | 1.08 (1.02) | 1.08 (1.02) | 1.09 (1.06) |
| Cl− | 1.01 (1.14) | 1.15 (1.15) | 1.25 (1.20) | 1.25 (1.20) | 1.25 (1.25) |
| Br− | 1.02 (1.16) | 1.16 (1.17) | 1.26 (1.22) | 1.26 (1.22) | 1.26 (1.26) |
| I− | 1.08 (1.15) | 1.22 (1.16) | 1.31 (1.21) | 1.31 (1.21) | 1.32 (1.25) |
The effective self-diffusion coefficients for the neutral salts are defined as .
The agreement between the Drude model and available experimental values95 is relatively good. Interestingly, the calculated values appear to be slightly overestimated for the cations while they are underestimated for the anions. This intriguing trend, with the cations diffusing faster and the anions slower than the experimental estimates, has also been noted in a recent study with non-polarizable models17. Limitations in the ions and/or solvent models could perhaps explain these discrepancies. However, it is worth recalling that the polarizable SWM4-NPD water model matches the experimental value of the self-diffusion of water exactly, and the shear viscosity is satisfactorily reproduced as well.35 Alternatively, there is also the possibility that the experimental estimates for the single ions are at fault for the following reason. Experimentally, it is nearly impossible to measure the self-diffusion coefficient of isolated ions in solution. Most of the available values for individual cations and anions have been deduced from conductivity measurements of electrolyte aqueous solutions by invoking additional hypotheses.95 In the limit of very low concentration [c], the conductivity σ of an electrolyte solution varies as . In practice, all the ionic species of a neutral salt contribute to yield the experimentally observed σ, suggesting that a more meaningful comparison is achieved by considering the effective self-diffusion coefficients for neutral salts, (corresponding to the low concentration limit of the whole salt conductivity after removing the dependence on the trivial pre-factor ). In Table 5 the calculated and experimental values of Deff for 40 neutral salts are listed. The agreement between the experimental data and the calculations is striking. With the exclusion of salts involving the smallest anion, F−, the calculated values for the alkali-halides are almost perfect, while the salts with the divalent cations are all less than a few percent off. The remarkable agreement shown in Table 5 strongly argues in favor of the importance of considering the experimental transport properties of neutral salts, rather than reported values for the self-diffusion coefficients of individual ions.
3.6 Comparison with other models
Over the years, a number of non-polarizable13-18,96 and polarizable,28-30,84 models of the monovalent ion series have been developed and parametrized to reproduce different sets of target data. Previous non-polarizable models have been parametrized primarily on the basis of the absolute hydration free energies of single ions.13-15,18,96 Recently, Joung and Cheatham pushed the limit of non-polarizable force fields by enforcing a balanced description of crystal and solution properties.16,17 Induced polarization is not a dominant factor for the hydration of a large cation like K+, and satisfactory results can be obtained with nonpolarizable models.84 Nevertheless, adopting nonpolarizable models often leads to some compromises concerning the gas phase properties (e.g. the monohydrate properties15) or the dynamic propeties (e.g. the self-diffusion constant17). Furthermore, the nonpolarizable models are calibrated for bulk solutions and their validity in different environments is uncertain. In contrast, the polarizable force fields improve the transferability by capturing the electronic polarization in different environments.28-30,84 The AMOEBA models, an extension of the induced dipole model, developed by Ponder and co-workers,28,30 accurately reproduce the various gas phase and condensed phase properties of ions, with more complex terms for electrostatics. One important difference of the polarizable models developed here compared to AMOEBA, where multipolar electrostatic terms are required, is that all the interactions are charge-charge Coulombic interactions and the functional form of the potential energy terms can be implemented in standard MD codes without major changes.
Another important difference concerns the parametrization strategy. Here, the parametrization sought to reproduce the well-established experimental data (i.e. the total hydration free energy of neutral salts as well as the relative free energy difference between different ions in the same series). By virtue of this procedure, the monohydrate properties served to lock down the absolute free energy scale. The parameterization of the ion models in AMOEBA followed a somewhat similar route, focusing mainly on the monohydrate properties and the neutral salts.28,30 This is in contrast with other efforts aimed at fitting the actual single-ion hydration free energies presented by a given experimental scale.13-18,96 As noted previously,29 we consider this to be problematic because the absolute hydration free energies measured experimentally depend on an undetermined phase potential, which is reflected in the large spread in the reported values. Coincidentally, the final models presented here happen to be close to the single-ion hydration free energies presented by Noyes for the monovalent cations,79 although this scale was not imposed during parametrization. However, the results from the final models differ markedly from the the single-ion hydration free energies presented by several of the experimental scales, and if those values were imposed as target data, then the unavoidable consequence would be a deterioration of the monohydrate properties. These considerations highlight the pitfalls with using the absolute single-ion hydration free energy reported from experiments as target data for force field parametrization.
4 Conclusion
In the current study, parameters for both cations and anions (Table 1) were developed for the polarizable force field based on classical Drude oscillator in conjunction with the SWM4-NDP water model. The parameters for the ions have been derived based on the gas phase monohydrate properties and the hydration free energies in the bulk phase. The various gas phase and condensed properties (Table 2, Table 3 and Table 4) are in reasonable agreement with the available experimental data and ab initio calculations. The dynamic transport properties of the models are in excellent agreement with experimental estimates based on electrolyte conductivity (Table 5). Overall, the set of models developed here should provide an important tool for accurate studies of a wide range of biological and physical processes involving ions in aqueous solutions.
The models for divalent cations from the present work are somewhat less accurate than those of the monovalent ions. The results from ab initio quantum chemistry calculations and the hydration free energy are not perfectly reproduced. This could be in part due to the neglect of charge transfer effects and to the breakdown of linear response.97 As shown previously, the deviations from linear polarization response become quite severe with a perturbation by a charge of +2.0e.97 In this case, it was necessary to treat the overpolarization that occurs when the ion is in direct contact a water molecule. Analysis of the functional form of the potential function showed that an overpolarization catastrophe is unavoidable when the distance between a polarizable water and the divalent cation is less than 1.97 Å. By contrast, monovalent cations require no special treatment. Several strategies to avoid over-polarization for the divalent ions were considered. The best approach turned out to be the introduction of a Thole-like screening function to modulate the Coulomb electrostatic interactions between the cation and the water molecule. Careful parameterization of this functional form against the results from quantum mechanical calculations led to reasonably accurate models. A similar approach was used to model Mg2+ and Ca2+ in the context of the AMOEBA polarizable force field.30,52
The present effort at parametrizing a complete set of ion models underlies the difficulties that arise when considering single-ion bulk properties deduced from experiments as objective target data. Experimental measurement of ions in bulk liquid phases are performed on globally neutral systems, and deriving single-ion properties requires additional non-thermodynamic hypotheses. The most meaningful comparisons are made with data directly obtained from neutral salts. For example, while the models do not match the single-ion diffusion coefficients deduced from electrolyte conductivity measurements, the effective diffusion coefficients for the neutral salts are in remarkably good agreement with experiments (Table 5). Such issues become particularly accute with respect to single-ion hydration free energies. Experimental estimates often rely on the free energy associated with the solvation of H+ as a reference. However, published experimental scales for the alkali-halides solvation free energy can shift up and down, depending upon the chosen reference. Some of the experimental values may appear to be inconsistent with one another, which increases the difficulties in developing an accurate force field. These problems are further compounded by the fact that absolute free energies of charged species are affected by the air-liquid interfacial potential. While the solvation free energies of neutral salts reported in experimental tables are unambiguous and independent of the interfacial potential, absolute hydration free energies of ions are somewhat ambiguous and cannot be readily utilized as the target data for parametrization. Preserving complete consistency of the solvation free energies of all ions has been a critical aspect of the present effort. To extract a reliable set of target data for the absolute hydration free energy as required to parameterize our models, we previously adopted a strategy based on a “solvation-consistent” scale.29 The consequences of this scale are well illustrated by considering the K+ model. The parameters of K+ have been adjusted to fit the properties of small cluster hydrates in the gas phase (Table 4). Once used in bulk phase simulations, this model yields a (real) hydration free energy of −78.6 kcal/mol (Table 2). Further validating the K+ model was the level of agreement of K+-water oxygen radial distributions for the Drude model with results from both experimental and ab initio Born-Oppenheimer and Car-Parrinello MD simulations.84 Therefore, the absolute free energy scale has been “locked down” by using the K+ model, which then serves as a central reference for other ionic solutes. This suggests that the parametrization of additional charged moieties should be developed to be in accord with the present solvation-consistent scale.
Finally, it is worth pointing out that at this stage, the models have been optimized to be accurate in the infinite dilution limit. The problem associated with electrolyte solutions at high concentrations, where issues of solubility and ion-ion interactions are important, will be examined in the near future. In particular, the models will be tested by calculating the osmotic pressure of concentrated salt solutions using a new method.98
Supplementary Material
Acknowledgement
T.W.W is supported by a strategic LDRD grant from Argonne National Laboratory. H.Y. is supported by a postdoctoral fellowship from the Joint Theory Institute of the University of Chicago and Argonne Laboratory. This work was in part funded by NIH grants GM072558 and GM051501.
Footnotes
Supporting Information Available
Comparison of the induced dipole moments of Cl− and Br− as a function of external electrostatic field strength and a compilation of the experimentally derived hydration free energy differences between different ions are provided in the supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hille B. Ion channels of excitable membranes. 3rd ed. Sinauer Associates; Sunderland, MA: 2001. [Google Scholar]
- 2.Page MJ, Di Cera E. Physiol. Rev. 2006;86:1049–1092. doi: 10.1152/physrev.00008.2006. [DOI] [PubMed] [Google Scholar]
- 3.Dudev T, Lim C. Chem. Rev. 2003;103:773–787. doi: 10.1021/cr020467n. [DOI] [PubMed] [Google Scholar]
- 4.Torrance JW, MacArthur MW, Thornton JM. Proteins Struct. Funct. Bioinformatics. 2008;71:813–830. doi: 10.1002/prot.21741. [DOI] [PubMed] [Google Scholar]
- 5.Schumacher M, Rivard A, Bachinger H, Adelman J. Nature. 2001;410:1120–1124. doi: 10.1038/35074145. [DOI] [PubMed] [Google Scholar]
- 6.Sakharov D, Lim C. J. Am. Chem. Soc. 2005;127:4921–4929. doi: 10.1021/ja0429115. [DOI] [PubMed] [Google Scholar]
- 7.Bloomfield V. Biopolymers. 1991;31:1471–1481. doi: 10.1002/bip.360311305. [DOI] [PubMed] [Google Scholar]
- 8.Cate J, Gooding A, Podell E, Zhou K, Golden B, Kundrot C, Cech T, Doudna J. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 9.Fang X, Pan T, Sosnick T. Nat. Struct. Biol. 1999;6:1091–1095. doi: 10.1038/70016. [DOI] [PubMed] [Google Scholar]
- 10.Draper D, Grilley D, Soto A. Annu. Rev. Biophys. Biomol. Struct. 2005;34:221–243. doi: 10.1146/annurev.biophys.34.040204.144511. [DOI] [PubMed] [Google Scholar]
- 11.Woodson S. Curr. Opin. Chem. Biol. 2005;9:104–109. doi: 10.1016/j.cbpa.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 12.Qu X, Smith GJ, Lee KT, Sosnick TR, Pan T, Scherer NF. Proc. Natl. Acad. Sci. USA. 2008;105:6602–6607. doi: 10.1073/pnas.0801436105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Åqvist J. J. Phys. Chem. 1990;94:8021–8024. [Google Scholar]
- 14.Beglov D, Roux B. J. Chem. Phys. 1994;100:9050–9063. [Google Scholar]
- 15.Jensen KP, Jorgensen WL. J. Chem. Theory Comput. 2006;2:1499–1509. doi: 10.1021/ct600252r. [DOI] [PubMed] [Google Scholar]
- 16.Joung IS, Cheatham TE. J. Phys. Chem. B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joung IS, Cheatham TE. J. Phys. Chem. B. 2009;113:13279–13290. doi: 10.1021/jp902584c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlsson J, Aqvist J. J. Phys. Chem. B. 2009;113:10255–10260. doi: 10.1021/jp900818z. [DOI] [PubMed] [Google Scholar]
- 19.Roux B. Chem. Phys. Lett. 1993;212:231–240. [Google Scholar]
- 20.Allen TW, Andersen OS, Roux B. Biophys. J. 2006;90:3447–3468. doi: 10.1529/biophysj.105.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bucher D, Guidoni L, Maurer P, Rothlisberger U. J. Chem. Theory Comput. 2009;5:2173–2179. doi: 10.1021/ct9001619. [DOI] [PubMed] [Google Scholar]
- 22.Yu HB, Roux B. Biophys. J. 2009;97:L15–L17. doi: 10.1016/j.bpj.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rick SW, Stuart SJ. Rev. Comp. Chem. 2002:89–146. [Google Scholar]
- 24.Yu HB, van Gunsteren WF. Comput. Phys. Comm. 2005;172:69–85. [Google Scholar]
- 25.Warshel A, Kato M, Pisliakov AV. J. Chem. Theory Comput. 2007;3:2034–2045. doi: 10.1021/ct700127w. [DOI] [PubMed] [Google Scholar]
- 26.Lopes PEM, Roux B, MacKerell AD. Theo. Chem. Acc. 2009;124:11–28. doi: 10.1007/s00214-009-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dang LX, Rice JE, Caldwell J, Kollman PA. J. Am. Chem. Soc. 1991;113:2481–2486. [Google Scholar]
- 28.Grossfield A, Ren P, Ponder JW. J. Am. Chem. Soc. 2003;125:15671–15682. doi: 10.1021/ja037005r. [DOI] [PubMed] [Google Scholar]
- 29.Lamoureux G, Roux B. J. Phys. Chem. B. 2006;110:3308–3322. doi: 10.1021/jp056043p. [DOI] [PubMed] [Google Scholar]
- 30.Jiao D, King C, Grossfield A, Darden T, Ren P. J. Phys. Chem. B. 2006;110:18553–18559. doi: 10.1021/jp062230r. [DOI] [PubMed] [Google Scholar]
- 31.Drude P. Lehrbuch der Optik. S. Hirzel; Leipzig: 1900. [Google Scholar]
- 32.Sangster MJL, Dixon M. Adv. Phys. 1976;25:247–342. [Google Scholar]
- 33.Lamoureux G, Roux B. J. Chem. Phys. 2003;119:3025–3039. [Google Scholar]
- 34.Lamoureux G, MacKerell A, Roux B. J. Chem. Phys. 2003;119:5185–5197. [Google Scholar]
- 35.Lamoureux G, Harder E, Vorobyov IV, Roux B, MacKerell AD., Jr. Chem. Phys. Lett. 2006;418:245–249. [Google Scholar]
- 36.Dick BG, Jr., Overhauser AW. Phys. Rev. 1958;112:90. [Google Scholar]
- 37.Cochran W. Crit. Rev. Solid. State. Mater. Sci. 1971;2:1–44. [Google Scholar]
- 38.van Maaren PJ, van der Spoel D. J. Phys. Chem. B. 2001;105:2618–2626. [Google Scholar]
- 39.Kunz AP, van Gunsteren WF. J Phys Chem A. 2009;113:11570–11579. doi: 10.1021/jp903164s. [DOI] [PubMed] [Google Scholar]
- 40.Vorobyov I, Anisimov VM, Greene S, Venable RM, Moser A, Pastor RW, MacKerell AD. J. Chem. Theo. Comput. 2007;3:1120–1133. doi: 10.1021/ct600350s. [DOI] [PubMed] [Google Scholar]
- 41.Lopes PEM, Lamoureux G, Roux B, MacKerell AD. J. Phys. Chem. B. 2007;111:2873–2885. doi: 10.1021/jp0663614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anisimov VM, Vorobyov IV, Roux B, MacKerell AD. J. Chem. Theo. Comput. 2007;3:1927–1946. doi: 10.1021/ct700100a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harder E, Anisimov VM, Whitfield TW, MacKerell AD, Jr., Roux B. J. Phys. Chem. B. 2008;112:3509–3521. doi: 10.1021/jp709729d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopes PEM, Lamoureux G, Mackerell AD. J. Comput. Chem. 2009;30:1821–1838. doi: 10.1002/jcc.21183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harder E, MacKerell AD, Roux B. J. Am. Chem. Soc. 2009;131:2760–2761. doi: 10.1021/ja806825g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University Press; Oxford: 1987. [Google Scholar]
- 47.Tuckerman M, Berne BJ, Martyna GJ. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 48.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 49.Thole BT. Chem. Phys. 1981;59:341–350. [Google Scholar]
- 50.van der Hoef MA, Madden PA. Mol. Phys. 1998;94:417–433. [Google Scholar]
- 51.Whitfield TW, Martyna GJ. Chem. Phys. Lett. 2006;424:409–413. [Google Scholar]
- 52.Piquemal JP, Perera L, Cisneros GA, Ren P, Pedersen LG, Darden TA. J Chem Phys. 2006;125:054511. doi: 10.1063/1.2234774. [DOI] [PubMed] [Google Scholar]
- 53.Deng Y, Roux B. J. Phys. Chem. B. 2004;108:16567–16576. [Google Scholar]
- 54.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. J. Chem. Phys. 1995;103:8577–8593. [Google Scholar]
- 55.Nosé S. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 56.Hoover W. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 57.Martyna G, Tobias D, Klein M. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 58.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 59.Martyna GJ, Tuckerman ME, Tobias DJ, Klein ML. Mol. Phys. 1996;87:1117–1157. [Google Scholar]
- 60.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J. Comput. Chem. 1992;13:1011–1021. [Google Scholar]
- 61.Asthagiri D, Pratt LR, Ashbaugh HS. J. Chem. Phys. 2003;119:2702–2708. [Google Scholar]
- 62.Harder E, Roux B. J Chem Phys. 2008;129:234706. doi: 10.1063/1.3027513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kastenholz M, Hunenberger P. J. Chem. Phys. 2006;124:124106. doi: 10.1063/1.2172593. [DOI] [PubMed] [Google Scholar]
- 64.Kastenholz MA, Huenenberger PH. J. Chem. Phys. 2006;124:224501. doi: 10.1063/1.2201698. [DOI] [PubMed] [Google Scholar]
- 65.Mahan G. Phys. Rev. A. 1980;22:1780–1785. [Google Scholar]
- 66.Hattig C, Hess BA. J. Chem. Phys. 1998;108:3863–3870. [Google Scholar]
- 67.Coker H. J. Phys. Chem. 1976;80:2078–2084. [Google Scholar]
- 68.Coker H. J. Phys. Chem. 1976;80:2084–2091. [Google Scholar]
- 69.Pyper NC, Pike CG, Edwards PP. Mol. Phys. 1992;76:353–372. [Google Scholar]
- 70.Jemmer P, Fowler PW, Wilson M, Madden PA. J. Phys. Chem. A. 1998;102:8377–8385. [Google Scholar]
- 71.Džidić I, Kebarle P. J. Phys. Chem. 1970;74:1466–1474. [Google Scholar]
- 72.Tissandier MD, Cowen KA, Feng WY, Gundlach E, Cohen MH, Earhart AD, Coe JV, Tuttle TR., Jr. J. Phys. Chem. A. 1998;102:7787–7794. [Google Scholar]
- 73.Feller D, Glendening ED, Kendall RA, Peterson KA. J. Chem. Phys. 1994;100:4981–4997. [Google Scholar]
- 74.Glendening ED, Feller D. J. Phys. Chem. 1995;99:3060–3067. [Google Scholar]
- 75.Borodin O, Bell RL, Li Y, Bedrov D, Smith GD. Chem. Phys. Lett. 2001;336:292–302. [Google Scholar]
- 76.Kim J, Lee HM, Suh SB, Majumdar D, Kim KS. J. Chem. Phys. 2000;113:5259. [Google Scholar]
- 77.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery J, Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision C.02. Gaussian, Inc.; Wallingford, CT: 2004. [Google Scholar]
- 78.Randles J. Trans. Faraday Soc. 1956;52:1573–1581. [Google Scholar]
- 79.Noyes RM. J. Am. Chem. Soc. 1962;84:513–522. [Google Scholar]
- 80.Gomer R, Tryson G. J. Chem. Phys. 1977;66:4413–4424. [Google Scholar]
- 81.Klots CE. J. Phys. Chem. 1981;85:3585–3588. [Google Scholar]
- 82.Marcus Y. J. Chem. Soc.-Faraday Trans. 1991;87:2995–2999. [Google Scholar]
- 83.Schmid R, Miah AM, Sapunov VN. Phys. Chem. Chem. Phys. 2000;2:97–102. [Google Scholar]
- 84.Whitfield TW, Varma S, Harder E, Lamoureux G, Rempe SB, Roux B. J. Chem. Theory Comput. 2008;3:2068. doi: 10.1021/ct700172b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wagman DD, Evans WH, Parker VB, Schumm RH, Halow I, Bailey SM, Churney KL, Nuttall RL. J. Phys. Chem. Ref. Data. 1982;11(Supplement No 2) [Google Scholar]
- 86.Peschke M, Blades AT, Kebarle P. J. Phys. Chem. A. 1998;102:9978–9985. [Google Scholar]
- 87.Peschke M, Blades AT, Kebarle P. J. Am. Chem. Soc. 2000;122:10440–10449. [Google Scholar]
- 88.Ohtaki H, Radnai T. Chem. Rev. 1993;93:1157–1204. [Google Scholar]
- 89.Spangberg D, Rey R, Hynes JT, Hermansson K. J. Phys. Chem. B. 2003;107:4470–4477. [Google Scholar]
- 90.Varma S, Rempe SB. Biophys. Chem. 2006;124:192–199. doi: 10.1016/j.bpc.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 91.Lyubartsev AP, Laasonen K, Laaksonen A. J. Chem. Phys. 2001;114:3120–3126. [Google Scholar]
- 92.Todorova T, Hunenberger PH, Hutter J. J. Chem. Theory Comput. 2008;4:779–789. doi: 10.1021/ct700302m. [DOI] [PubMed] [Google Scholar]
- 93.Caminiti R, Licheri G, Piccaluga G, Pinna G. Chem. Phys. Lett. 1977;47:275–278. [Google Scholar]
- 94.Martinez JM, Pappalardo RR, Marcos ES. J. Am. Chem. Soc. 1999;121:3175–3184. [Google Scholar]
- 95.Lide DR, editor. CRC Handbook of Chemistry and Physics. 87th ed. Taylor and Francis; Boca Raton, FL: 2007. [Google Scholar]
- 96.Roux B. Biophys. J. 1996;71:3177–3185. doi: 10.1016/S0006-3495(96)79511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Harder E, Anisimov VM, Vorobyov IV, Lopes PEM, Noskov SY, MacKerell AD, Roux B. J. Chem. Theory Comput. 2006;2:1587–1597. doi: 10.1021/ct600180x. [DOI] [PubMed] [Google Scholar]
- 98.Luo Y, Roux B. J. Phys. Chem. Lett. 2010;1:183–189. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.