Sialic acids occupy terminal positions on glycan chains of glycoproteins and glycolipids, contributing to a rich diversity of glycan structures that mediate cell surface biology through their recognition as ligands by glycan binding proteins. Pathogens including viruses (influenza virus, polyoma virus), bacteria, and a variety of bacterial toxins (e.g cholera toxin) utilize sialic acid containing glycans as cell surface receptors for attachment to the host cell1. Two families of mammalian glycan binding proteins utilize sialic acid containing glycans as cell surface ligands, including the selectin sub-group of the C-type lectins that mediate leukocyte trafficking2, and the sialic acid binding Ig-like lectins, siglecs, that are differentially expressed on white blood cells of the immune system, and modulate cell signaling receptors involved in innate and adaptive immune responses3.
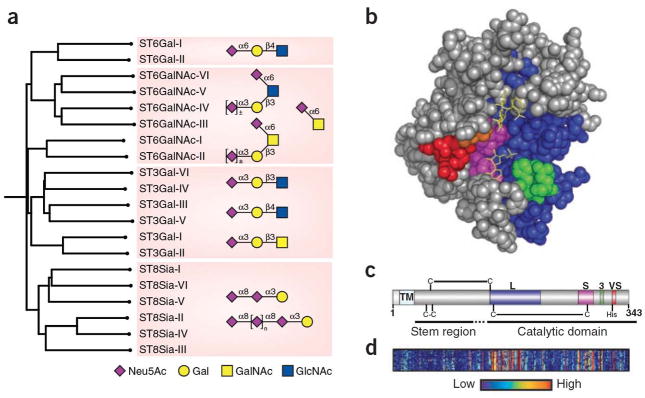
The biosynthesis of sialosides is carried out by a family of twenty sialyltransferases that are highly conserved from mouse to man (Figure 1a)4,5. These enzymes transfer sialic acid from a donor substrate, CMP-sialic acid, to terminal positions of the glycan chains of glycoproteins. In keeping with the role of sialyltransferases as ‘glycan terminators’, they are localized to the trans Golgi network of the secretory pathway, through which glycoproteins and glycolipids pass enroute to the cell surface or secretion from the cell. Each enzyme has a strict specificity for their acceptor glycan, and transfers sialic acid in defined linkage to yield a unique product, completing the non-template mediated synthesis of the glycan chain. They are grouped into four sub-families based on the linkage formed in the product (e.g. Neu5Acα2-6Gal; Figure 1a). Sialyltransferase genes exhibit cell type specific and developmentally regulated expression6 that results in the differential display of the sialic acid containing products of the corresponding enzymes, and contributes to the rich biology mediated by this class of glycan sequences.
Figure 1.
Structural relationships among the human sialyltransferase family. a) Homology dendrogram is shown for twenty members of the human siatlyltransferase family. Sialoside products produced by each of the four major subfamilies are shown in symbol form. b) CPK representation of pST3Gal-I highlighting the four ‘sialyl motifs’: large (L, cyan), small (S, red), 3 (3, green) and very small (VS, yellow). The bound acceptor ligand is highlighted in red. The postulated ‘lid’ domain consisting of a disordered loop located between the motifs 3 and VS is omitted. c) Localization of the sialylmotif regions in the primary sequence of pST3GalI highlighted using the same color-coding for the conserved motifs in panel b. d) Homology heat map highlighting regions of sequence conservation among all 20 members of the sialyltransferase family. relative to hST3GalI. Insertions in other sialyltransferases relative to hST3Gal I are omitted to maintain comparisons relative the sialylmotifs illustrated in panel c.
In this issue Rao et al. (reference) describe the structure of a mammalian sialyltransferase, ST3Gal I (Figure 1b). This enzyme catalyzes the synthesis of the sequence NeuAcα2-3Galβ1-3GalNAc found O-linked to Threonine or Serine on glycoproteins, and as a terminal sequence of the ganglio-series of glycolipids. Although its activity is partially redundant of other members of the ST3Gal subfamily, ablation of St3gal1 in mice causes a profound reduction in CD8+ T cells, revealing a key role of this gene in the development and maintenance of the cytotoxic T cell arm of the immune system7. As the first mammalian sialyltransferase structure to be reported, the structure of ST3Gal I provides an opportunity to examine the relationship of homologous domains in the sialyltransferase family to their function in catalyzing the synthesis of diverse sialoside sequences.
Several structural features of the sialyltransferase family were previously deduced from their primary sequences5. All have N-terminal signal anchor sequences that tether them to the Golgi membrane, and a flexible ‘stem’ region that is not required for catalytic activity (Figure 1c). Within the catalytic domain several homologous regions were identified, termed sialylmotifs, which were postulated to play a role in binding the substrates and catalysis of the transfer of the sialic acid to the acceptor glycan4,5. The reported crystal structure includes the entire catalytic domain of a recombinant ST3Gal I that was produced as an N-terminal truncation missing the signal anchor and stem region. Structures were obtained with bound acceptor and donor substrate analogs that clearly identify the catalytic site (Figure 1c). It is apparent that the conserved sialylmotifs frame the catalytic site, with a stabilizing disulfide bond between two sialylmotifs (L & S) that is conserved across the sialyltransferase family5. A flexible loop missing from the electron density is proposed to form a ‘lid domain’ that participates in binding of the donor substrate.
The structure of ST3Gal-I is also informative with respect to the general fold and catalytic mechanism of the sialyltransferase family. The authors previously reported the structure of a bacterial sialyltransferase8, that shares several features despite only 14% sequence identity. Glycosyltransferases in general fall into two main fold groupings designated GTA and GTB with one and two Rossman-like domains, respectively9. Both ST3Gal I and the bacterial sialyltransferase exhibit variants of the GT-A fold that retains the spatial arrangement of a β-sheet core structure. Sialyltransferases are known to be ‘inverting’ glycosyltransferases since the Neu5Ac in the donor substrate (CMP-β-Neu5Ac) is in the β configuration, while it is in α configuration in the product (Neu5Acα2-3Gal). Based on placement of the donor and acceptor substrates in the catalytic sites of the mammalian and bacterial sialyltransferases, the authors propose that the reaction mechanism of ST3Gal I proceeds through an oxocarbenium-ion transition state, and identify His319 in the conserved VS sialylmotif as the catalytic base9.
While all sialyltransferases use CMP-Neu5Ac as a donor substrate, they vary in their specificity for acceptor substrates. Distinguishing acceptors substrates is the defining property that allows glycosyltransferases to build a glycan of defined sequence in the absence of a template. Indeed, even sialyltransferases of the same ST3Gal sub-family differentiate between disaccharide acceptors with a β1-3 or β1-4 glycosidic linkage and/or disaccharide sequence. The authors credit a single amino acid, namely Tyr269, for the specificity of ST3Gal I for the disaccharide sequence Galβ1-3GalNAc, by making the crucial contacts to the axial C4 positions of both Gal and GalNAc moieties. The authors also note a polar groove adjacent to the acceptor site that can accommodate a peptide or lipid moiety extending from the disaccharide acceptor substrate in the natural ligands of the enzyme. These observations pave the way for a closer look at the basis for substrate specificity of other members of the subfamily as their structures become available.
Genetic ablation of sialyltransferase genes in mice have documented phenotypes that show the importance of the products of these enzymes in modulation of many aspects of immune function and blood coagulation10. Such observations have stimulated interest in these enzymes as targets for development of drugs to treat autoimmune diseases and coagulopathies. The availability of the high resolution ST3Gal I structure reported by Rao et al. is sure to stimulate efforts in the rational design of small molecule inhibitors of this important family of enzymes.
References
- 1.Karlsson KA. Meaning and therapeutic potential of microbial recognition of host glycoconjugates. Mol Microbiol. 1998;29:1–11. doi: 10.1046/j.1365-2958.1998.00854.x. [DOI] [PubMed] [Google Scholar]
- 2.Taylor ME, Drickamer K. Paradigms for glycan-binding receptors in cell adhesion. Curr Opin Cell Biol. 2007;19:572–577. doi: 10.1016/j.ceb.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 3.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 4.Jeanneau C, Chazalet V, Auge C, et al. Structure-function analysis of the human sialyltransferase ST3Gal I: role of n-glycosylation and a novel conserved sialylmotif. J Biol Chem. 2004;279:13461–13468. doi: 10.1074/jbc.M311764200. [DOI] [PubMed] [Google Scholar]
- 5.Datta AK. Comparative sequence analysis in the sialyltransferase protein family: analysis of motifs. Curr Drug Targets. 2009;10:483–498. doi: 10.2174/138945009788488422. [DOI] [PubMed] [Google Scholar]
- 6.Comelli EM, Head SR, Gilmartin T, et al. A focused microarray approach to functional glycomics: transcriptional regulation of the glycome. Glycobiology. 2006;16:117–131. doi: 10.1093/glycob/cwj048. [DOI] [PubMed] [Google Scholar]
- 7.Moody AM, North SJ, Reinhold B, et al. Sialic acid capping of CD8beta core 1-O-glycans controls thymocyte-major histocompatibility complex class I interaction. J Biol Chem. 2003;278:7240–7246. doi: 10.1074/jbc.M210468200. [DOI] [PubMed] [Google Scholar]
- 8.Chiu CP, Watts AG, Lairson LL, et al. Structural analysis of the sialyltransferase CstII from Campylobacter jejuni in complex with a substrate analog. Nat Struct Mol Biol. 2004;11:163–170. doi: 10.1038/nsmb720. [DOI] [PubMed] [Google Scholar]
- 9.Lairson LL, Henrissat B, Davies GJ, Withers SG. Glycosyltransferases: structures, functions, and mechanisms. Annu Rev Biochem. 2008;77:521–555. doi: 10.1146/annurev.biochem.76.061005.092322. [DOI] [PubMed] [Google Scholar]
- 10.Lowe JB, Marth JD. A genetic approach to Mammalian glycan function. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]