Abstract
S-Adenosylhomocysteine hydrolase (AHCY) is the only mammalian enzyme known to catalyze the hydrolysis of S-adenosylhomocysteine (AdoHcy). We have used a genotype-to-phenotype strategy to study this important enzyme by resequencing AHCY in 240 DNA samples from four ethnic groups. Thirty-nine polymorphisms were identified – 28 of which were novel. Functional genomic studies for wild type (WT) AHCY and the three variant allozymes identified showed that two variant allozymes had slight, but significant decreases in enzyme activity, but with no significant differences in levels of immunoreactive protein. Luciferase reporter gene assays for common 5′-flanking region (5′-FR) haplotypes revealed that one haplotype with a frequency of ∼2% in Caucasian-American subjects displayed a decreased ability to drive transcription. The variant nucleotide at 5′-FR SNP (-34) in that haplotype altered the DNA-protein binding pattern during electrophoresis mobility shift assay (EMSA). Finally, an AHCY genotype-phenotype association study for expression in lymphoblastoid cells identified four SNPs that were associated with decreased expression. For the IVS6 (intervening sequence 6, i.e., intron 6) G56>C SNP among those four, EMSA showed that a C>G nucleotide change resulted in an additional shifted band. These results represent a step toward understanding the functional consequences of common genetic variation in AHCY for the regulation of neurotransmitter, drug and macromolecule methylation.
Keywords: S-Adenosylhomocysteine hydrolase, AHCY, functional genomics, gene sequence variation, single nucleotide polymorphisms, SNPs, gene resequencing
Methylation plays a critical role in monoamine neurotransmitter biosynthesis and metabolism (Weinshilboum et al. 1999). Methyltransferase (MT) enzymes involved in these process include catechol O-methyltransferase (COMT (Axelrod & Cohn 1971, Axelrod & Tomchick 1958, Lachman et al. 1996), phenylethanolamine N-methyltransferase (PNMT) (Axelrod 1962a, Axelrod & Cohn 1971, Ji et al. 2005), indolethylamine N-methyltransferase (INMT) (Axelrod 1962b, Thompson et al. 1999), and hydroxyindole O-methyltransferase (HIOMT) (Axelrod et al. 1965, Donohue et al. 1993). All of these MT enzymes utilize S-adenosyl-L-methionine (AdoMet) as a methyl donor (Chiang et al. 1996), and S-adenosylhomocysteine (AdoHcy) is generated during the methyltransferase reaction (Weinshilboum 2006). AdoHcy is a competitive inhibitor of all AdoMet-dependent MT enzymes (Borchardt 1980). The ratio of AdoMet to AdoHcy is sometimes referred to as “methylation potential” (Loehrer et al. 1998). The accumulation of AdoHcy decreases this ratio, resulting in the inhibition of MT enzymes (McKeever et al. 1995a). In the brains of both human and pigs, methylation is significantly reduced with increased AdoHcy concentrations (Goggins et al. 1999, McKeever et al. 1995a, McKeever et al. 1995b). Therefore, it is important that AdoHcy be degraded in vivo to maintain methylation.
S-Adenosylhomocysteine hydrolase (AHCY, EC 3.3.1.1) catalyzes the reversible hydrolysis of AdoHcy to form adenosine (Ado) and homocysteine (Hcy) (Castro et al. 2005, Palmer & Abeles 1976). Although the synthetic direction is favored thermodynamically, the reaction proceeds in the hydrolytic direction in vivo because of the rapid removal of Hcy and Ado (Cantoni 1975, Palmer & Abeles 1976, Castro et al. 2005) (Fig. 1). Reduced AHCY activity can result in elevated AdoHcy levels, with a resultant reduction in the AdoMet to AdoHcy ratio and impaired methylation (Loehrer et al. 1998). As a result, understanding the functional implications of common genetic polymorphism in the gene encoding AHCY could provide insight into individualized variation in methylation.
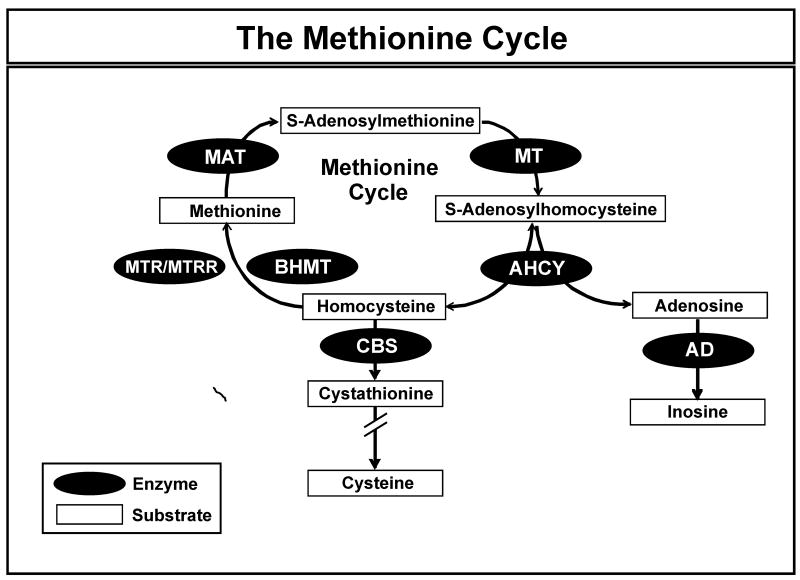
Figure 1.
The Methionine Cycle. AHCY = S-adenosylhomocysteine hydrolase; AD = adenosine deaminase; BHMT = betaine homocysteine methyltransferase; CBS = cystathione β-synthase; MAT = methionine adenosyltransferase; MTR = 5-methyltetrahydrofolate-homocysteine methyltransferase; MTRR = 5-methyltetrahydrofolate homocysteine methyltransferase reductase; MT = methyltransferase.
Human AHCY has 11 exons and maps to the long arm of chromosome 20. Deletion of AHCY is an embryonic lethal in mice (Miller et al. 1994). Mutations in the human AHCY gene that are associated with impaired function result in elevated AdoMet levels and even more dramatically elevated AdoHcy concentrations (Baric et al. 2005, Baric et al. 2004, Buist et al. 2006). All of these patients demonstrated severe myopathy, developmental delay and hypermethionemia. Case-control studies have reported several common AHCY sequence variants (Gellekink et al. 2004), although follow-up functional studies have generally been lacking. In the present study, we set out to resequence the AHCY gene using 240 DNA samples from four different ethnic groups, followed by functional genomic characterization of the polymorphisms observed. Specifically, we resequenced all exons and splice junctions as well as the 5′-flanking region (5′-FR) of the gene, and identified 39 single nucleotide polymorphisms (SNPs) – 28 novel, including 3 nonsynonymous SNPs. The functional implications of the nonsynonymous SNPs were then evaluated as well as the transcriptional activity of common AHCY 5′-FR haplotypes. Finally, microarray analysis was performed with the lymphoblastoid cell lines from which the resequenced DNA had been obtained to investigate the possible relationship between AHCY expression and polymorphisms in the gene. These studies represent a comprehensive attempt to link sequence variation in the human AHCY gene with function as a step toward a more complete understanding of the relationship among common AHCY genetic polymorphisms and variation in methylation.
Materials and Methods
DNA samples
DNA samples from 60 Caucasian-American (CA), 60 African-American (AA), 60 Han-Chinese American (HCA) and 60 Mexican-American (MA) subjects (HD100CAU, HD100AA, HD 100CHI, and HD100MEX) were obtained from the Coriell Cell Repository. These DNA samples had been collected and anonymized by the National Institute of General Medical Sciences, National Institutes of Health. All subjects had provided written informed consent for the use of their DNA for research purpose, and the present studies were reviewed and approved by the Mayo Clinic Institutional Review Board.
Gene resequencing
AHCY was resequenced using the 240 DNA samples obtained from the Coriell Cell Repository. Specifically, the PCR was used to amplify all AHCY exons, including exon 1B which is located 8 kb upstream of AHCY exon 1A (see Fig. 2), all intron-exon splice junctions and approximately 1 kb of 5′-FR. The PCR primer sequences used to perform these amplifications are listed in Supplementary Table 1. Amplicons were sequenced on both strands in the Mayo Molecular Biology Core Facility using dye terminator sequencing chemistry. Polymorphisms observed only once, as well any with ambiguous chromatograms, were subjected to a second, independent round of amplification, followed by DNA sequencing. The GeneBank accession number for the AHCY reference sequence used in these studies was NT_028392.5, and the reference sequence for AHCY mRNA was NM_000687.
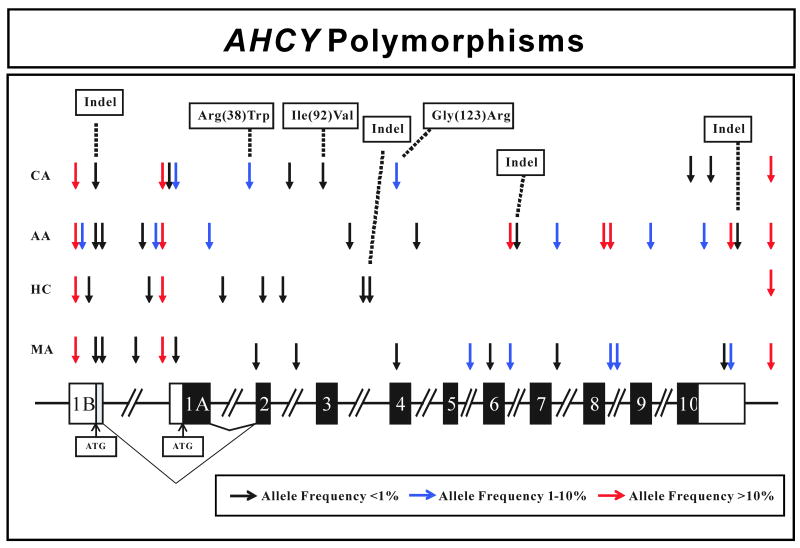
Figure 2.
Human AHCY gene polymorphisms. The figure shows a schematic representation of the human AHCY gene structure, with arrows indicating the locations of polymorphisms. Black rectangles represent exons encoding the open-reading frame (ORF), open rectangles represent portions of exons encoding untranslated region (UTR) sequence, and the grey rectangle represents coding region when exon 1B is transcribed. “Indel” represents an insertion/deletion event. The colors of arrows indicate minor allele frequencies.
Transient expression
An AHCY cDNA clone in pCMV-XL5 was purchased from OriGene (Rockville, MD). This cDNA served as template for site-directed mutagenesis to create expression constructs for each of the AHCY nonsynonymous SNPs. Sequences of all inserts after site-directed mutagenesis were confirmed by sequencing in both directions. These expression constructs were then used to transfect COS-1 cells using Lipofectamine 2000 (Invitrogen, Calsbad, CA). pSV-β-Galactosidase (Promega, Madison WI) was co-transfected to make it possible to correct for possible variation in transfection efficiency (Hall et al. 1983, Rosenthal 1987). Specifically, 10 μg of construct DNA, together with 4 μg of pSV-β-galactosidase DNA, were co-transfected into the COS-1 cells. After incubation at 37°C for 48 hours, the transfected cells were washed with phosphate buffered saline (PBS), resuspended in homogenization buffer and were lysed with a Polytron homogenizer (Brinkmann Instruments, Westbury, NY). The homogenates were centrifuged at 100,000 × g for 1 hour at 4°C, and supernatants were stored at -80°C prior to assay.
Quantitative Western blot analysis
Levels of immunoreactive AHCY protein were determined for each recombinant allozyme by performing quantitative Western blot analysis. A rabbit polyclonal antibody directed against AHCY amino acids 411-432 (Cocalico, Reamstown, PA) was used to perform these study. Specifically, COS-1 cell cytosol was loaded onto 12% SDS mini-gels (Bio-Rad, Hercules, CA) on the basis of levels of β-galactosidase activity to correct for possible variation in transfection efficiency. Electrophoresis was performed for 90 minutes at 120 V, followed by the transfer of proteins to polyvinylidene fluoride membranes (Bio-Rad). After blocking with 5% nonfat milk in Tris buffered saline with Tween 20 (TBST), the membrane was incubated overnight with primary antibody diluted 1:10,000 with 5% milk in TBST. The next morning, goat anti-rabbit horseradish peroxidase antibody (Bio-Rad) was applied at a dilution of 1:5000. The ECL Western Blotting System (Amersham Pharmacia, Piscataway, NJ) was used to detect bound antibody, and results were expressed as a percentage of a WT allozyme control on the same gel.
AHCY enzyme assay
Recombinant allozymes were assayed for AHCY activity in the hydrolase direction using a modification of the adenosine deaminase coupled reaction described by Elrod et al. (Elrod et al. 2002). Briefly, the reaction mixture contained 50 mM potassium phosphate buffer, pH 7.2, 1 mM EDTA, various concentrations of AdoHcy (Sigma-Aldrich Corp., St. Louis, MO), 10 μL of 1 mg/ml adenosine deaminase and recombinant enzyme, in a final volume of 490 μL. The reaction was incubated at 37°C for 10 minutes and was stopped by the addition of 10 μL of 5N HClO4. After centrifugation and filtration, concentrations of inosine (a product of the reaction catalyzed by adenosine deaminase) were determined by HPLC with a C18 reverse-phase column (Shimadzu C18, 3 μm, 150 × 4.5 mm). A binary gradient elution was used, with 25 mM phosphate buffer, pH 3.2, plus 10 mM heptane sulfonic acid as buffer A and with acetonitrile as buffer B. A linear gradient of 2-98% buffer B over 25 min was used to elute the column, with spectrophotometric detection at 260 nm. A standard curve consisting of known quantities of inosine was used to determine inosine concentrations. To determine apparent Km values, 8 concentrations of AdoHcy that varied from 0.78 to 100 μM were assayed. The concentration used to assay “basal” activity for variant allozymes was 50 μM. Cytosol activity for cells tranfected with empty vector controls were used to correct for the presence of endogenous AHCY activity. This activity was never more than 1% of that measured after transfection with WT AHCY.
Reporter gene assays
Luciferase reporter gene constructs were created to study the possible effect on transcription of AHCY 5′-FR polymorphisms. Approximately 500 bp of the AHCY 5′-FR was amplified using as template genomic DNA from subjects with known haplotypes within the area amplified, and the amplicons were ligated into pGL3 basic (Promega). Constructs with variant sequences were created by site-directed mutagenesis using the circular PCR. DNA sequences of the constructs were verified by sequencing in both directions. These constructs were then used to transfect HEK293T and HepG2 cells. Specifically, 2 μg of purified plasmid DNA was co-transfected with 0.2 μg pRL-TK (Promega) DNA. The pRL-TK DNA made it possible to correct for possible variation in transfection efficiency. The Dual-Luciferase Reporter Assay Kit (Promega) was used to assay reporter gene activities, and results were reported as the ratio of firefly to Renilla luciferase light units.
Electrophoresis mobility shift assay (EMSA)
EMSA was performed using the Lightshift Chemiluminescent EMSA Kit (Pierce Biotechnology, Rockford, IL). 5′-Biotin-labeled oligonucleotide probes that included approximately 20 to 22 bp of sequence around the SNP being studied were synthesized for both WT and variant sequences (see Supplementary Table 2). Nuclear extracts were obtained from HEK293T and HepG2 cells during the log growth phase. Probes were annealed to form double stranded DNA, and binding reactions contained 1X binding buffer, 50 ng/μl poly dIdC, 20 fmol Biotin-labeled probe and 9 μg nuclear extract protein. 500-fold excess (10 pmol) unlabeled probe was used in the competition reactions, and reaction mixtures were then run on a 6% non-denaturing polyacrylamide gel, with detection by chemiluminescence (Pierce Biotechnology).
Microarray expression analysis
The lymphoblastoid cell lines from which DNA for the gene resequencing studies had been obtained were used to perform expression array analysis. Specifically, total RNA was extracted from these cell lines using the RNeasy Kit (Qiagen, Valencia, CA). Prior to microarray analysis, RNA quality assessment was performed with an Agilent 2100 Bioanalyzer. An RNA Integrity Number (RIN) greater than 9.0 was required for all samples. The RNA was then reverse-transcribed and biotin labeled before hybridization to Affymetrix U133 Plus 2.0 Genechips or to Genechip Human Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA). Microarray images were analyzed in the Mayo Clinic Microarray Core Facility. The array data were normalized with full quantile normalization using XRAY software (Biotique System, Reno, NV). Data from microarray probe set 200903_s_at, corresponding to AHCY, were used to perform the studies described subsequently.
RT-PCR
To verify the expression array results, we performed RT-PCR using total RNA extracted from 12 cell lines with the Qiagen RNeasy kit. These cells were randomly selected from across the range of AHCY expression array values. The primers for RT-PCR were directed to the 3′-UTR and were obtained from Qiagen. The expression array data were then correlated with the RT-PCR results.
Data analysis
DNA sequence data were analyzed using Mutation Surveyor (SoftGenetics, State College, PA). Linkage disequilibrium among AHCY polymorphisms was determined by calculating D′ values for all combinations of polymorphisms (Li et al. 2008). Haplotypes were inferred computationally as described by Schaid et al (Schaid et al. 2002). Apparent Km values were calculated using the Prism program (Graphpad, San Diego, CA). Average levels of AHCY allozyme immunoreactive protein, enzyme activity and reporter gene data were compared by ANOVA using JMP (SAS Institute, Cary, NC). Dunnett's posthoc test was performed to compare group means against one single control group (wild type). The variance measure used consistently in our studies was the standard deviation. The association between SNPs and AHCY mRNA expression array data was determined with PLINK (http://pngu.mgh.harvard.edu/purcell/plink/) (Purcell et al. 2007). Specifically, least square regression was used to determine the association between SNPs and expression. Bonferroni correction was used to correct significance values for associations for multiple testing.
Results
Human AHCY resequencing
The human AHCY gene maps to chromosome 20q13.1, contains 11 exons and is ∼23 kb in length (Fig. 2). A search of GeneBank revealed two different AHCY transcripts encoding proteins that differed at their N-termini. One (mRNA sequence number NM_000687.1) is widely expressed, and is particularly abundant in liver and kidney. The first exon of this transcript will be referred to subsequently as “exon 1A”. The other transcript (mRNA AK097610) had a different initial exon, which we will refer to “exon 1B”. Exon 1B is located 8 kb upstream from exon 1A (Fig. 2). A review of the expressed sequence tag (EST) database showed that exon 1B had only been observed in the testis (DB043400, DB041336 and DB022147).
All AHCY exons, exon-intron splice junctions and approximately 1 kb of 5′-FR sequence upstream of exon 1A were amplified using DNA samples from 60 AA, 60 HCA, 60 CA and 60 MA subjects. A total of 11 PCR amplifications were performed, and both strands were sequenced for all samples. Therefore, a total of ∼2.4 × 106 bp of DNA was sequenced and analyzed. Thirty-nine polymorphisms were observed, including 3 nonsynonymous cSNPs, and four insertion-deletion events (indels) (Table 1 and Fig. 2). SNPs were numbered on the basis of the sequence on the “common” transcript that included exon 1A. Specifically, the ‘A’ in the start codon in exon 1A was assigned the +1 designation. SNPs 5′ to that location were assigned negative and those 3′ were assigned positive numbers. The 3 nonsynonymous SNPs observed resulted in the following changes in encoded amino acid sequence: Arg(38)Trp, Ile(92)Val, and Gly(123)Arg. A 10 bp deletion (TCTCCGCCCC) was observed in exon 1B, located 100 bp upstream of the exon 1B ATG. This indel was observed in one subject each among the AA, CA and MA subjects (Table 1). All polymorphisms observed were in Hardy-Weinberg equilibrium (P>0.05). There were striking ethnic variations in allele frequencies and types, with 20 SNPs in DNA samples from AA, 12 in samples from CA, 10 in DNA from HCA and 18 in samples from MA subjects. Ten polymorphisms were observed only in AA subjects, 4 only in CAs, 7 only in HCA subjects and 4 only in MAs. When compared to the public database dbSNP (www.ncbi.nlm.gov/SNP) (Table 1), twenty-eight of the 39 polymorphisms observed were novel.
Table 1.
Human AHCY polymorphisms. Polymorphism locations, alterations in nucleotide and amino acid sequences, as well as minor allele frequencies (MAFs) for all ethnic groups studied are listed. Numbering of SNPs in exons and the 5′-FR is based on their location in the AHCY cDNA, with the A in start codon (ATG) found in exon 1A designated +1. Polymorphisms 5′ to that nucleotide are assigned negative numbers while those 3′ are assigned positive numbers. Polymorphisms in introns were numbered based on their location relative to the nearest exon-intron splice junction, using negative and positive numbers for splice donor and acceptor sites, respectively. dbSNP designations are listed in the final column if the SNP was already listed in dbSNP. SNPs in exons are boxed.
| AHCY Polymorphisms | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| # | Location | Nucleotide | Nucleotide Sequence Change | Amino Acid Change | MAF | dbSNP designation | |||
| African-American | Caucasian-American | Han Chinese-American | Mexican-American | ||||||
| 1 | 5′-FR | -8598 | C→G | 0.400 | 0.906 | 0.767 | 0.890 | rs 1205349 | |
| 2 | Exon 1B | -8361 | G→C | 0.025 | 0.000 | 0.000 | 0.000 | ||
| 3 | Exon 1B | -8341 | C→T | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 4 | Exon 1B | -8256 to -8247 | deletion of TCTCCGCCCC | 0.008 | 0.008 | 0.000 | 0.008 | ||
| 5 | Exon 1B | -8246 | T→C | 0.008 | 0.000 | 0.000 | 0.008 | ||
| 6 | 5′-FR | -594 | G→A | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 7 | 5′-FR | -528 | T→C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 8 | 5′-FR | -393 | T→G | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 9 | 5′-FR | -225 | C→A | 0.025 | 0.000 | 0.000 | 0.000 | ||
| 10 | 5′-FR | -124 | C→A | 0.392 | 0.892 | 0.792 | 0.892 | rs 819146 | |
| 11 | 5′-FR | -63 | A→G | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 12 | Exon 1A | -34 | C→T | 0.000 | 0.017 | 0.000 | 0.008 | ||
| 13 | Intron 1 | -61 | C→T | 0.033 | 0.000 | 0.000 | 0.000 | ||
| 14 | Intron 1 | -39 | G→T | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 15 | Exon 2 | 112 | C→T | Arg (38) Trp | 0.000 | 0.033 | 0.000 | 0.008 | rs 13043752 |
| 16 | Exon 2 | 171 | C→T | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 17 | Intron 2 | -85 | C→T | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 18 | Intron 2 | -66 | C→T | 0.000 | 0.008 | 0.000 | 0.008 | ||
| 19 | Exon 3 | 274 | A→G | Ile (92) Val | 0.000 | 0.008 | 0.000 | 0.000 | rs 41312290 |
| 20 | Intron 3 | 37 | A→C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 21 | Intron 3 | 57 | C→T | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 22 | Intron 3 | -23 | deletion of C | 0.000 | 0.000 | 0.008 | 0.000 | ||
| 23 | Exon 4 | 367 | G→A | Gly (123) Arg | 0.000 | 0.017 | 0.000 | 0.008 | rs 41301825 |
| 24 | Intron 4 | -21 | G→C | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 25 | Intron 5 | 161 | T→C | 0.000 | 0.000 | 0.000 | 0.042 | ||
| 26 | Exon 6 | 663 | T→C | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 27 | Intron 6 | 56 | C→G | 0.333 | 0.000 | 0.000 | 0.033 | rs 7271501 | |
| 28 | Intron 6 | -7 to -8 | deletion of CT | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 29 | Intron 7 | -29 | A→G | 0.050 | 0.000 | 0.000 | 0.008 | ||
| 30 | Intron 8 | -154 | G→A | 0.333 | 0.000 | 0.000 | 0.033 | rs 34602355 | |
| 31 | Intron 8 | -7 | C→T | 0.333 | 0.000 | 0.000 | 0.033 | rs 17091705 | |
| 32 | Intron 9 | -14 | T→C | 0.025 | 0.000 | 0.000 | 0.000 | ||
| 33 | 3′-UTR | 1341 | C→T | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 34 | 3′-UTR | 1600 | T→G | 0.050 | 0.000 | 0.000 | 0.000 | rs 13245 | |
| 35 | 3′-UTR | 1708 | A→G | 0.000 | 0.008 | 0.000 | 0.000 | ||
| 36 | 3′-UTR | 1902 | A→G | 0.000 | 0.000 | 0.000 | 0.008 | ||
| 37 | 3′-UTR | 1978 | A→T | 0.333 | 0.000 | 0.000 | 0.033 | rs 4239 | |
| 38 | 3′-UTR | 1998 to 1999 | deletion of GA | 0.008 | 0.000 | 0.000 | 0.000 | ||
| 39 | 3′-FR | 2102 | C→T | 0.483 | 0.892 | 0.792 | 0.892 | rs 819169 | |
Two commonly used measures of nucleotide diversity were calculated, π, the average heterozygosity per site, and θ, a population mutation measure that is theoretically equal to the neutral mutation parameter. Tajima's D, a test of the neutral mutation hypothesis, was also calculated. Results for each ethnic group are listed in Table 2. The AA population showed the highest values for both π and θ, suggesting, as anticipated, a greater sequence diversity than that observed in other groups studied.
Table 2.
Estimates of π, θ and Tajima's D for AHCY in four ethnic groups. Values are parameter estimates of ± SEM. P-values refer to Tajima's D.
| Population | π × 10-4 | θ × 10-4 | Tajima's D-values | P-value |
|---|---|---|---|---|
| African-American | 8.21 ± 4.62 | 6.99 ± 2.32 | 0.49 | 0.64 |
| Caucasian-American | 1.66 ± 1.36 | 4.51 ± 1.69 | -1.61 | 0.11 |
| Han Chinese-American | 2.5 ± 1.81 | 4.08 ± 1.57 | -0.97 | 0.35 |
Linkage disequilibrium and haplotype analysis
It is becoming increasing clear that the determination of haplotype may be more helpful than the assay of single SNPs for use in genetic association studies (Brodde & Leineweber 2005), so haplotype and linkage disequilibrium analyses for AHCY were performed for each ethic group studied. AHCY haplotypes with frequencies ∼1% or greater in at least one ethnic group are listed in Table 3. Twenty-one observed and inferred haplotypes with frequencies of ∼1% were identified in AHCY. Only two of those haplotypes were present in all ethnic groups. Specifically, 12 were present in AA subjects, with eight unique to that population; eight were observed in CA subjects, three of which were unique; four were present in HCA subjects, with one unique to that population; and eight were observed in MA subjects, one of which was unique (Table 3). Haplotype designations were based on the amino acid sequence of the encoded allozyme, with the most common amino acid sequence in AA subjects designated *1. Letter designations were then added in descending order of allele frequency. For example, *2, *3, and *4 represent variant allozymes with Trp38, Val92, and Arg123 variant amino acids.
Table 3.
AHCY haplotypes. AHCY haplotypes with frequencies of ∼1% or greater are listed. Nucleotide positions are numbered as described in Table 1. Variant nucleotides compared to the reference sequence (the most common sequence in the AA population) are highlighted as white type against black. The first column lists observed (o) or inferred (i) haplotypes.
| Allele | Frequency | 5′-FR (-8598) |
5′-FR (-8361) |
5′-FR (-225) |
5′-FR (-124) |
5′-FR (-34) |
Intron 1 (-61) |
Exon 2 (+112) |
Exon 3 (+274) |
Exon 4 (+367) |
Intron 5 (+161) |
Intron 6 (+56) |
Intron 7 (-29) |
Intron 8 (-154) |
Intron 8 (-7) |
Intron 9 (-14) |
3′-UTR (+1600) |
3′-UTR (+1978) |
3′-FR (+2102) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CA | HCA | MA | CEPH | AA | ||||||||||||||||||||
| o | *1 A | 0.814 | 0.708 | 0.800 | 0.804 | 0.344 | G | G | C | A | C | C | C | A | G | T | C | A | G | C | T | T | A | T |
| o | *1 B | 0.025 | 0.274 | C | G | C | C | C | C | C | A | G | T | G | A | A | T | T | T | T | C | |||
| o | *1 C | 0.070 | 0.174 | 0.058 | 0.130 | 0.104 | C | G | C | C | C | C | C | A | G | T | C | A | G | C | T | T | A | C |
| i | *1 D | 0.042 | C | G | C | C | C | C | C | A | G | T | C | A | G | C | T | T | A | T | ||||
| o | *1 E | 0.035 | C | G | C | C | C | C | C | A | G | T | C | G | G | C | T | T | A | C | ||||
| i | *1 F | 0.033 | C | G | C | C | C | T | C | A | G | T | G | A | A | T | T | T | T | C | ||||
| i | *1 G | 0.025 | C | C | C | C | C | C | C | A | G | T | C | A | G | C | C | T | A | T | ||||
| i | *1 H | 0.025 | C | G | A | C | C | C | C | A | G | T | C | A | G | C | T | T | A | T | ||||
| i | *1 I | 0.021 | C | G | C | C | C | C | C | A | G | T | C | A | G | C | T | G | A | C | ||||
| i | *1 J | 0.017 | G | G | C | A | C | C | C | A | G | T | G | A | A | T | T | T | T | C | ||||
| i | *1 K | 0.014 | G | G | C | A | C | C | C | A | G | T | C | A | G | C | T | G | A | T | ||||
| o | *1 L | 0.017 | 0.022 | 0.009 | G | G | C | C | C | C | C | A | G | T | C | A | G | C | T | T | A | C | ||
| i | *1 M | 0.008 | 0.008 | 0.022 | C | G | C | C | T | C | C | A | G | T | C | A | G | C | T | T | A | C | ||
| i | *1 N | 0.008 | 0.022 | G | G | C | C | C | C | C | A | G | T | G | A | A | T | T | T | T | C | |||
| o | *1 O | 0.042 | C | G | C | A | C | C | C | A | G | T | C | A | G | C | T | T | A | T | ||||
| o | *1 P | 0.033 | G | G | C | A | C | C | C | A | G | C | C | A | G | C | T | T | A | T | ||||
| i | *2 A | 0.027 | 0.008 | G | G | C | A | C | C | T | A | G | T | C | A | G | C | T | T | A | T | |||
| i | *2 B | 0.006 | C | G | C | C | C | C | T | A | G | T | C | A | G | C | T | T | A | C | ||||
| o | *3 A | 0.008 | G | G | C | A | C | C | C | G | G | T | C | A | G | C | T | T | A | T | ||||
| o | *4 A | 0.009 | 0.008 | G | G | C | A | C | C | C | A | A | T | C | A | G | C | T | T | A | T | |||
| i | *4 B | 0.008 | C | G | C | C | C | C | C | A | A | T | C | A | G | C | T | T | A | C | ||||
D′ values were also calculated for pairwise combinations of all polymorphisms observed to make it possible to determine linkage disequilibrium. When two polymorphisms are maximally associated, D′ values are equal to 1, and they are zero when the polymorphisms are randomly associated. Ethnic group-specific D′ values for the AHCY gene are depicted graphically in Supplementary Figure 1. No obvious haplotype blocks were observed in the gene in any of the four ethnic groups.
Variant allozyme activity and protein levels
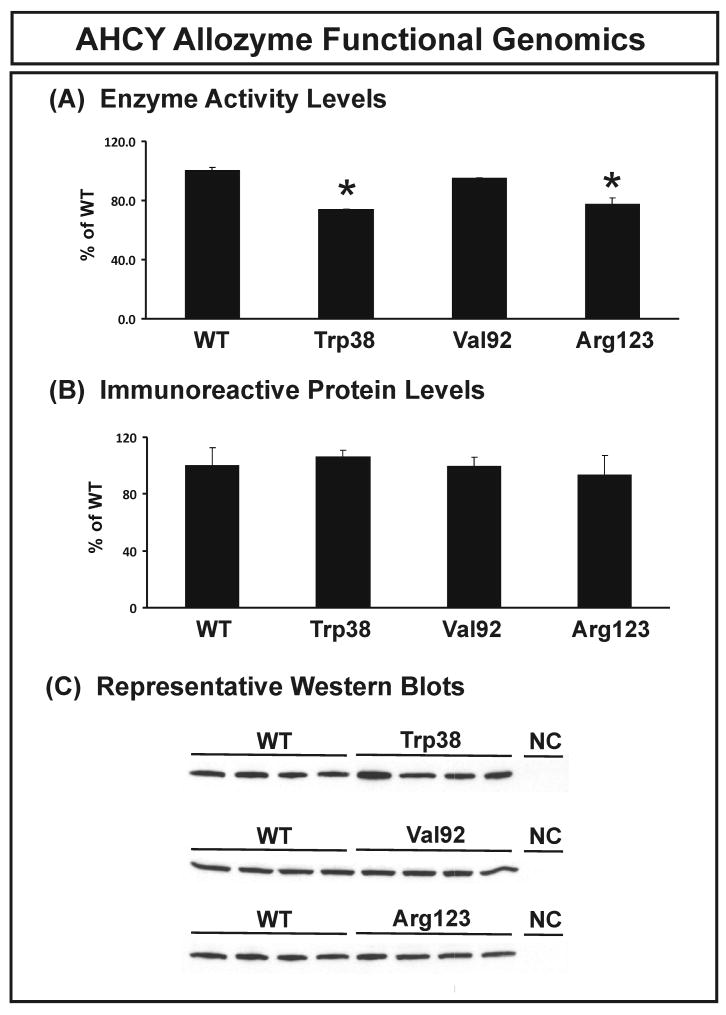
Functional genomic studies were performed to study the possible effects of AHCY nonsynonymous SNPs on function. Expression constructs were created for the wild type (WT) and 3 variants allozymes. These constructs were transiently expressed in a mammalian cell line (COS-1) to ensure that appropriate mammalian post-translational modification and protein degradation systems would be present. pSV-β-galactosidase was co-transfected to make it possible to correct for variation in transfection efficiency. Quantitative Western blot analysis and AHCY enzyme assays were then performed. Levels of immunoreactive protein and AHCY allozyme enzyme activity for all three variant allozymes, expressed as a percentage of WT, are shown in Figure 3. No significant differences from WT were detected for immunoreactive protein (Fig. 3B). Although the results of overexpression experiments must always be interpreted with caution, whenever we have tested the results obtained after COS-1 cell overexpression of variant allozymes with their effects in human tissue biopsy samples, the relative effects have been identical (Honchel et al. 1993, Shield et al. 2004, Szumlanski et al. 1996). The Trp38 and Arg123 variant allozymes showed slight but significant decreases in AHCY activity, to 73.6% and 77.2%, respectively, of that for the WT allozyme measured in the presence of 50 μM AdoHcy (Fig. 3A). Substrate kinetic studies were also performed, and apparent Km values for AdoHcy are listed in Table 4. Trp38 and Arg123 allozymes had significantly lower Km values than did the WT. However, these relatively small differences in levels of enzyme activity or apparent Km values may not be of major physiologic significance. The next set of functional genomic studies focused on possible gene sequence-related variation in transcription.
Figure 3.
AHCY allozyme functional genomics. (A) AHCY allozyme enzyme activity. AHCY activity was assayed in the hydrolase direction with 50 μM AdoHcy as substrate. Activities are expressed as a percentage of WT. Values shown are mean ± SD for 3 determinations, * p<0.001 by ANOVA. (B) AHCY allozyme immunoreactive protein levels. Protein levels are expressed as a percentage of WT. Values shown are mean ± SD for 3 determinations, p=0.11 by ANOVA. (C) A representative Western blot. Cells transfected with empty vector were assayed as a negative control (NC).
Table 4.
Apparent Km values for human AHCY allozymes. Values are mean ± S.D. The P-values listed represent comparison with the WT value using student's t-test. ANOVA was also used to assess differences.
| AdoHcy Km, μM | P-value vs WT | |
|---|---|---|
| Wild Type (WT) | 30.6 ± 5.9 | ------ |
| Trp38 | 13.7 ± 0.2* | 0.01 |
| Val92 | 27.5 ± 5.0 | 0.47 |
| Arg123 | 14.3 ± 0.5* | 0.01 |
P=0.02.
AHCY 5′-FR reporter gene studies
Our gene resequencing studies had identified a total of 7 SNPs located within an area ∼1 kb upstream of the AHCY ATG start codon in exon 1A, i.e., upstream of the most common AHCY transcript that included exon 1A. There were also 5 polymorphism upstream of or within exon 1B, including a 10 bp (TCTCCGCCCC) indel. Therefore, we studied the effect of haplotypes in both areas of the gene on reporter gene activity. Since the AHCY transcript that includes exon 1A has been reported to be highly expressed in liver and kidney, HepG2 and HEK293T cells were chosen for use in reporter gene studies of SNPs located upstream of that exon. However, because the AHCY transcript that includes exon 1B has only been reported in the testis, Hs 181.Tes, a human testicular cell line, was chosen to perform reporter gene studies for polymorphisms upstream of exon 1B.
As a first step, RT-PCR was performed to verify the presence of AHCY transcripts in these three cell lines. One pair of primers was used to amplify the junction of exons 1A and 2, while another was used to detect the junction of exons 1B and 2 (see Supplementary Table 2). Quantitative RT-PCR was performed for all three cell lines, with beta-actin as an internal control. HepG2 and HEK293T cell lines, as expected, showed high expression of AHCY transcripts that included exon 1A (data not shown). However, the predominant AHCY transcript in Hs 181.Tes cells was also amplified using primers for exon 1A, not exon 1B (data not show), so this cell line had no obvious advantage for studies of SNPs located in the region 8 kb upstream of exon 1A and, since the transcript that includes exon 1B has been reported so rarely, we chose to not pursue functional genomic studies of that transcript.
To evaluate the possible influence of exon 1A 5′-FR SNPs on AHCY transcription, haplotype analyses of this region of the gene was performed (Table 5). Luciferase reporter gene constructs were created for haplotypes with frequencies greater than 1% in at least one ethnic group. The SNPs included in these haplotypes were (-225), (-124) and (-34). The CAC haplotype in this region of the gene was designated as “WT” because it was the most common haplotype in all ethnic groups except AAs. In the AA population, the CCC haplotype had a frequency of 58.3%, with a CAC frequency of 38.3%. Each of these common haplotypes was present in all ethnic groups. The ACC haplotype was present only in AA subjects with a frequency of 2.5%, and the CCT haplotype was observed only in CA and MA subjects. Dual luciferase reporter gene assays were performed for these haplotypes and are expressed in Figures 4A and 4B as a percentage of luciferase activity of the pGL3-WT construct. While no significant differences were observed for CCC and ACC when compared to WT, a reduction of approximately 50% was observed for CCT in both HEK293T and HepG2 cell lines. Since a change in nucleotide from C to T at 5′-FR nucleotide (-34) appeared to result in decreased transcription in these two cell lines, we performed EMSA with that polymorphism.
Table 5.
Haplotype analysis for the AHCY exon 1A 5′-FR.
| Construct Names | Haplotype Frequency in Each Ethnic Group | Nucleotide and Location | |||||
|---|---|---|---|---|---|---|---|
| CA | HCA | MA | AA | (-225) | (-124) | (-34) | |
| Wild Type (WT) | 0.883 | 0.783 | 0.883 | 0.383 | C | A | C |
| (-124C) | 0.092 | 0.208 | 0.100 | 0.583 | C | C | C |
| (-225A/-124C) | --- | --- | --- | 0.025 | A | C | C |
| (-124C/-34T) | 0.017 | --- | 0.008 | --- | C | C | T |
Figure 4.
Human AHCY 5′-FR reporter gene and EMS assays. (A) and (B) AHCY 5′-FR reporter gene constructs were used to transfect HEK293T and HepG2 cells. Luciferase activity is expressed as a percentage of the value for WT constructs. Each bar represents the average of 6 independent experiments (mean ± SD), * p<0.001 by ANOVA (C) Human AHCY 5′-FR C(-34)T SNP effect on nuclear protein binding. Nuclear extract from HEK293T and HepG2 cells was incubated with the WT probe containing C(-34), or with the variant probe containing T(-34).
EMSA for SNP C(-34)T
Variation in transcription could result from variation in transcription factor binding (Lamba et al. 2002). Therefore, EMSA was performed with probes that spanned nucleotides (-47) to (-24) of the 5′-FR of the AHCY gene (Supplementary Table 3). For the probe that included (-34)C, the presence of nuclear extract from both HEK293T and HepG2 cell lines resulted in a “shifted” band which could be competitively inhibited by unlabeled probe (Fig. 4C). This same band was not observed with a probe that included (-34)T. This result supported our observations of differences in the ability of 5′-FR reporter gene constructs to drive transcription (Fig. 4A and 4B). We also attempted to determine which transcription factor might bind to this 5′-FR probe. Three proteins, SP1 (Alibaba 2.1, http://darwin.nmsu.edu/∼molb470/fall2003/Projects/solorz/aliBaba_2_1.htm), CREB 1 or CREB 2 (Genomatix, http://www.genomatix.de/), were predicted to possibly bind to DNA sequences spanning SNP C(-34)T. Therefore, we attempted super-shift assays using antibodies against SP1, CREB 1 and CREB 2, but none of those antibodies induced a “supershift” (data not shown).
AHCY expression array analysis
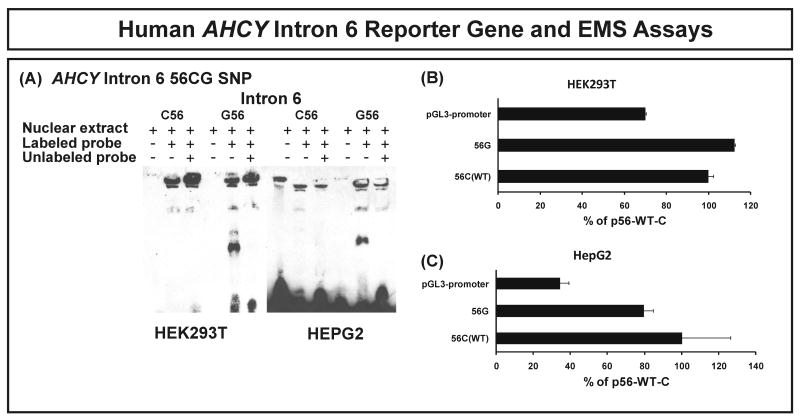
As the next step in our analysis of possible effects of gene sequence variation on expression, we performed microarray studies with Affymetrix U133 2.0 Plus chips for the cell lines from which the DNA used to resequence AHCY had been obtained. We then performed a genotype-phenotype correlation between expression and AHCY SNPs to determine whether there might be a correlation between AHCY SNPs and basal mRNA expression in these lymphoblastoid cells. Since two different AHCY transcripts have been reported, we also obtained exon array data using the same RNA samples that were used to perform the U133 2.0 Plus expression array analysis. Those data showed that in lymphoblastoid cells, as in HEK293T and HepG2 cells, AHCY transcripts that include exon 1A were highly expressed, but not those that include exon 1B (Supplementary Fig. 2). We then performed an association study between AHCY mRNA expression in the lymphoblastoid cell lines using the U133 2.0 Plus data and SNPs. To verify the expression array data, RT-PCR was also performed for 12 samples selected randomly across the distribution of expression array data. Results obtained with the two methods were highly correlated, with a Pearson correlation coefficient of 0.66 (p=0.02). Four SNPs had p-values less than 0.05, C(56)G in intron 6, G(-154)A and C(-7)T in intron 8, and A(1978)T in the 3′-UTR. These four SNPs were tightly linked (D′=1, R2=1). The GATT haplotype for these four SNPs was observed in AA subjects with a frequency of 33.3% and in MA subjects with a frequency of 3.3%. Three of these four SNPs were located in a region of the gene that is highly conserved in many mammalian species. However, these four SNPs were not tightly linked in any species other than humans (data not show). We then performed EMSA using probes for each of the 4 SNPs. No obvious shifted band was observed for intron 8 or 3′-UTR SNPs by using nuclear extract from HEK293T and HepG2 cell line. However, a band shift was observed for the C(56)G SNP for the probe with the G allele after incubation with nuclear extract from both HEK293T and HepG2 cell lines (Fig. 5A). This binding could be inhibited by adding unlabeled “cold” probe. However, the probe for the C(56) allele failed to show the same shift (Fig. 5A). The entire intron 6, with each of the nucleotide substitutions at position 56, was then cloned into the pGL-3-promoter vector. Intron 6 could serve as an enhancer, since luciferase activity increased 1.5-fold in HEK293T cells and 3-fold in HepG2 cell when compared to the empty vector. However, no significant difference was observed between the G56 and C56 alleles (Fig. 5B and 5C).
Figure 5.
Human AHCY intron 6 reporter gene and EMS assays. (A) Effect of human AHCY intron 6 C(56)G SNP on nuclear protein binding. Nuclear extract from HEK293T and HepG2 cells was incubated with a WT C56 intron 6 probe, or with a variant G56 intron 6 probe. (B) and (C) AHCY intron 6 reporter gene studies. Constructs were used to transfect HEK293T and HepG2 cells. Luciferase activity is expressed as a percentage of the value for the WT construct. Each bar represents the average of 3 independent experiments (mean ± SD).
Discussion
Methylation is an important reaction in monoamine neurotransmitter biosynthesis and metabolism. All of the MTs that catalyze these reactions are AdoMet-dependent, and the reaction results in the generation of AdoHcy, a critical compound for the maintenance of appropriate cellular AdoMet/AdoHcy ratios (see Fig. 1) (Finkelstein 2007). AHCY is the only enzyme in mammals known to catalyze the hydrolysis of AdoHcy. Because of its unique role in cellular metabolism and because it is a potential drug target, there have been many studies of the catalytic mechanism of AHCY as well as numerous attempts to develop AHCY inhibitors (Ando et al. 2008, Chiang 1998, Liu et al. 1992). However, little is known with regard to common AHCY DNA sequence variation and there are even fewer reports with regard to the functional consequences of common sequence variation in this gene. In present study, we applied a genotype-to-phenotype strategy to identify common AHCY polymorphisms and to explore their possible effect on function. We began by performing in-depth resequencing of the AHCY gene using 240 DNA samples from four different ethnic groups. In 2004, Gellekink et al. resequenced the coding region of the AHCY gene in 20 cardiovascular patients and reported three polymorphisms, C34T, C112T and C290T (Gellekink et al. 2004). In our studies, we observed the first two of these SNPs, but not the C290T polymorphism. We observed a total of 39 polymorphisms, including 3 nonsynonymous SNPs. Twenty-eight of the polymorphisms that we observed were novel. Linkage disequilibrium and haplotype analysis revealed striking differences in both SNPs and haplotypes among ethnic groups, with the AA population, as anticipated, showing greater DNA sequence diversity than other ethnic groups (Tishkoff & Williams 2002).
We also performed functional genomic studies. Expression constructs were created for all of the nonsynonymous cSNPs observed but, after expression in a mammalian cell line, no significant differences in levels of recombinant protein were observed (Fig. 3). However, variant allozymes with Trp38 and Arg123 substitutions displayed significant, but not striking decreases in levels of enzyme activity. The apparent Km value that we observed for the WT allozyme compared favorably with that reported previously (Fumic et al. 2007). We then set out to explore possible relationships between AHCY SNPs and expression. Of two known AHCY transcripts, that which includes exon 1A is expressed in most tissues, including liver and kidney, while the mRNA that includes exon 1B has only been reported in testicular tissue. However, our qRT-PCR analysis failed to demonstrate the expression of the exon 1B transcript in a testicular cell line, and exon array analysis demonstrated that the exon 1A species is the transcript that is predominantly expressed in lymphoblastoid cell lines (Fig. 5), as it is in liver and kidney. We employed luciferase reporter gene constructs to evaluate the ability of different 5′-FR haplotypes for the exon 1A transcript to drive transcription. In both HepG2 and HEK293T cell lines there was a significant decrease of luciferase activity for a reporter gene construct that included both the C(-124) and T(-34) SNPs, but not for a construct with C(-124) – suggesting a potential role for SNP C(-34) in transcription. In subsequent EMSA, a probe spanning this SNP bound to proteins after incubation with nuclear extracts while a change in nucleotide from C to T at that position resulted in the loss of protein binding. A previous epidemiologic study reported that the C(-34)T SNP might be associated with venous thrombosis (Gellekink et al. 2004). We have now demonstrated a functional effect for that SNP.
We also studied the possible influence of AHCY SNPs on expression in the lymphoblastoid cell lines from which the DNA used in our gene resequencing studies had been obtained. An association study performed using all AHCY SNPs and microarray data showed that 4 linked SNPs were associated with lower expression in cells from AA subjects. Those 4 SNPs were tightly linked (D′=1, R2=1), and were present only in AA and MA subjects, with MAFs of 33.3% and 3.3%, respectively. Three of these SNPs were located in a region that shows evolutionary conservation. EMSA was performed with all four of these SNPs and a “shift” was observed for the probe with the intron 6 (G56) SNP, but not the probe with intron 6 (C56). All of AHCY intron 6 was then cloned into the pGL3-promoter vector. Although intron 6 could function as an enhancer when upstream of the luciferase gene, the C(56)G SNP did not alter luciferase activity. In summary, we demonstrated that 4 SNPs near the 3′-end of AHCY are associated with expression in lymphoblastoid cells, and that one of those SNPs, C(56)G, altered the DNA-protein binding pattern (Fig. 5). Since the AHCY nonsynonymous SNPs did not appear to significantly alter function, the major functional effects of AHCY gene sequence variation appeared to result from effects on transcription – specifically involving the exon 1A (-39) and intron 6 (56) SNPs. Whether these DNA sequence variants will also have functional effects in vivo remain to be determined. Therefore, future experiments will be required to extend our in vitro AHCY functional genomic studies to the in vivo situation.
In summary, we have used a genotype-to-phenotype strategy to study AHCY, a gene encoding a critical enzyme in the “methionine cycle” (Fig. 1). We resequenced AHCY in 240 DNA samples from 4 ethnic groups and identified a total of thirty-nine polymorphisms – 28 of which were not present in public databases – as well as twenty-one common haplotypes. Functional characterization was performed for all variant allozymes, but the nonsynonymous SNPs that we observed did not appear to have major functional consequences. However, 5′-FR haplotype-based reporter gene and EMS assays showed that a 5′-FR SNP could significantly alter transcription. Furthermore, a genotype-phenotype correlation analysis showed that four tightly linked SNPs at the 3′-end of the gene were associated with decreased lymphoblastoid cell AHCY expression in AA subjects. One of those SNPs also altered DNA-protein binding patterns during EMSA. These results represent a comprehensive study of common sequence variation in the AHCY gene as well as the functional consequences of that variation, and they will serve as a foundation for future translational and mechanistic studies of this important gene.
Acknowledgments
Supported in part by National Institutes of Health (NIH) grants R01 GM28157, R01 GM35720, R01 CA132780, U01 GM61388 (The Pharmacogenetics Research Network) and a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award. We thank Luanne Wussow for her assistance with the preparation of this manuscript.
Abbreviations used
- AHCY
S-Adenosylhomocysteine hydrolase
- AdoMet
S-adenosylmethionine
- AdoHcy
S-Adenosylhomocysteine
- MT
methyltransferase
- 5′-FR
5′-flanking region
- EMSA
electrophoresis mobility shift assay
- SNP
single nucleotide polymorphism
- WT
wild type
- AA
African-American
- CA
Caucasian-American
- HCA
Han Chinese-American
- MA
Mexican-American
- Ado
adenosine
- Hcy
homocysteine
References
- Ando T, Kojima K, Chahota P, Kozaki A, Milind ND, Kitade Y. Synthesis of 4′-modified noraristeromycins to clarify the effect of the 4′-hydroxyl groups for inhibitory activity against S-adenosyl-L-homocysteine hydrolase. Bioorg Med Chem Lett. 2008;18:2615–2618. doi: 10.1016/j.bmcl.2008.03.029. [DOI] [PubMed] [Google Scholar]
- Axelrod J. The enzymatic N-methylation of serotonin and other amines. J Pharmacol Exp Ther. 1962a;138:28–33. [PubMed] [Google Scholar]
- Axelrod J. Purification and properties of phenylethanolamine-N-methyl transferase. J Biol Chem. 1962b;237:1657–1660. [PubMed] [Google Scholar]
- Axelrod J, Cohn CK. Methyltransferase enzymes in red blood cells. J Pharmacol Exp Ther. 1971;176:650–654. [PubMed] [Google Scholar]
- Axelrod J, Tomchick R. Enzymatic O-methylation of epinephrine and other catechols. J Biol Chem. 1958;233:702–705. [PubMed] [Google Scholar]
- Axelrod J, Wurtman RJ, Snyder SH. Control of Hydroxyindole O-Methyltransferase Activity in the Rat Pineal Gland by Environmental Lighting. J Biol Chem. 1965;240:949–954. [PubMed] [Google Scholar]
- Baric I, Cuk M, Fumic K, et al. S-Adenosylhomocysteine hydrolase deficiency: a second patient, the younger brother of the index patient, and outcomes during therapy. J Inherit Metab Dis. 2005;28:885–902. doi: 10.1007/s10545-005-0192-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baric I, Fumic K, Glenn B, et al. S-adenosylhomocysteine hydrolase deficiency in a human: a genetic disorder of methionine metabolism. Proc Natl Acad Sci U S A. 2004;101:4234–4239. doi: 10.1073/pnas.0400658101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchardt RT. S-Adenosyl-L-methionine-dependent macromolecule methyltransferases: potential targets for the design of chemotherapeutic agents. J Med Chem. 1980;23:347–357. doi: 10.1021/jm00178a001. [DOI] [PubMed] [Google Scholar]
- Brodde OE, Leineweber K. Beta2-adrenoceptor gene polymorphisms. Pharmacogenet Genomics. 2005;15:267–275. doi: 10.1097/01213011-200505000-00001. [DOI] [PubMed] [Google Scholar]
- Buist NR, Glenn B, Vugrek O, et al. S-adenosylhomocysteine hydrolase deficiency in a 26-year-old man. J Inherit Metab Dis. 2006;29:538–545. doi: 10.1007/s10545-006-0240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantoni GL. Biological methylation: selected aspects. Annu Rev Biochem. 1975;44:435–451. doi: 10.1146/annurev.bi.44.070175.002251. [DOI] [PubMed] [Google Scholar]
- Castro R, Rivera I, Martins C, et al. Intracellular S-adenosylhomocysteine increased levels are associated with DNA hypomethylation in HUVEC. J Mol Med. 2005;83:831–836. doi: 10.1007/s00109-005-0679-8. [DOI] [PubMed] [Google Scholar]
- Chiang PK. Biological effects of inhibitors of S-adenosylhomocysteine hydrolase. Pharmacol Ther. 1998;77:115–134. doi: 10.1016/s0163-7258(97)00089-2. [DOI] [PubMed] [Google Scholar]
- Chiang PK, Gordon RK, Tal J, Zeng GC, Doctor BP, Pardhasaradhi K, McCann PP. S-Adenosylmethionine and methylation. Faseb J. 1996;10:471–480. [PubMed] [Google Scholar]
- Donohue SJ, Roseboom PH, Illnerova H, Weller JL, Klein DC. Human hydroxyindole-O-methyltransferase: presence of LINE-1 fragment in a cDNA clone and pineal mRNA. DNA Cell Biol. 1993;12:715–727. doi: 10.1089/dna.1993.12.715. [DOI] [PubMed] [Google Scholar]
- Elrod P, Zhang J, Yang X, Yin D, Hu Y, Borchardt RT, Schowen RL. Contributions of active site residues to the partial and overall catalytic activities of human S-adenosylhomocysteine hydrolase. Biochemistry. 2002;41:8134–8142. doi: 10.1021/bi025771p. [DOI] [PubMed] [Google Scholar]
- Finkelstein JD. Metabolic regulatory properties of S-adenosylmethionine and S-adenosylhomocysteine. Clin Chem Lab Med. 2007;45:1694–1699. doi: 10.1515/CCLM.2007.341. [DOI] [PubMed] [Google Scholar]
- Fumic K, Beluzic R, Cuk M, Pavkov T, Kloor D, Baric I, Mijic I, Vugrek O. Functional analysis of human S-adenosylhomocysteine hydrolase isoforms SAHH-2 and SAHH-3. Eur J Hum Genet. 2007;15:347–351. doi: 10.1038/sj.ejhg.5201757. [DOI] [PubMed] [Google Scholar]
- Gellekink H, den Heijer M, Kluijtmans LA, Blom HJ. Effect of genetic variation in the human S-adenosylhomocysteine hydrolase gene on total homocysteine concentrations and risk of recurrent venous thrombosis. Eur J Hum Genet. 2004;12:942–948. doi: 10.1038/sj.ejhg.5201237. [DOI] [PubMed] [Google Scholar]
- Goggins M, Scott JM, Weir DG. Methylation of cortical brain proteins from patients with HIV infection. Acta Neurol Scand. 1999;100:326–331. doi: 10.1111/j.1600-0404.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- Hall CV, Jacob PE, Ringold GM, Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- Honchel R, Aksoy I, Szumlanski C, Wood TC, Otterness DM, Wieben ED, Weinshilboum RM. Human thiopurine methyltransferase: molecular cloning and expression of T84 colon carcinoma cell cDNA. Mol Pharmacol. 1993;43:878–887. [PubMed] [Google Scholar]
- Ji Y, Salavaggione OE, Wang L, Adjei AA, Eckloff B, Wieben ED, Weinshilboum RM. Human phenylethanolamine N-methyltransferase pharmacogenomics: gene re-sequencing and functional genomics. J Neurochem. 2005;95:1766–1776. doi: 10.1111/j.1471-4159.2005.03453.x. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Papolos DF, Saito T, Yu YM, Szumlanski CL, Weinshilboum RM. Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders. Pharmacogenetics. 1996;6:243–250. doi: 10.1097/00008571-199606000-00007. [DOI] [PubMed] [Google Scholar]
- Lamba JK, Lin YS, G SE, Thummel KE. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–1294. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Li F, Feng Q, Lee C, et al. Human betaine-homocysteine methyltransferase (BHMT) and BHMT2: common gene sequence variation and functional characterization. Mol Genet Metab. 2008;94:326–335. doi: 10.1016/j.ymgme.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wolfe MS, Borchardt RT. Rational approaches to the design of antiviral agents based on S-adenosyl-L-homocysteine hydrolase as a molecular target. Antiviral Res. 1992;19:247–265. doi: 10.1016/0166-3542(92)90083-h. [DOI] [PubMed] [Google Scholar]
- Loehrer FM, Angst CP, Brunner FP, Haefeli WE, Fowler B. Evidence for disturbed S-adenosylmethionine: S-adenosylhomocysteine ratio in patients with end-stage renal failure: a cause for disturbed methylation reactions? Nephrol Dial Transplant. 1998;13:656–661. doi: 10.1093/ndt/13.3.656. [DOI] [PubMed] [Google Scholar]
- McKeever M, Molloy A, Weir DG, Young PB, Kennedy DG, Kennedy S, Scott JM. An abnormal methylation ratio induces hypomethylation in vitro in the brain of pig and man, but not in rat. Clin Sci (Lond) 1995a;88:73–79. doi: 10.1042/cs0880073. [DOI] [PubMed] [Google Scholar]
- McKeever M, Molloy A, Young P, Kennedy S, Kennedy DG, Scott JM, Weir DG. Demonstration of hypomethylation of proteins in the brain of pigs (but not in rats) associated with chronic vitamin B12 inactivation. Clin Sci (Lond) 1995b;88:471–477. doi: 10.1042/cs0880471. [DOI] [PubMed] [Google Scholar]
- Miller MW, Duhl DM, Winkes BM, Arredondo-Vega F, Saxon PJ, Wolff GL, Epstein CJ, Hershfield MS, Barsh GS. The mouse lethal nonagouti (a(x)) mutation deletes the S-adenosylhomocysteine hydrolase (Ahcy) gene. Embo J. 1994;13:1806–1816. doi: 10.1002/j.1460-2075.1994.tb06449.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JL, Abeles RH. Mechanism for enzymatic thioether formation. Mechanism of action of S-adenosylhomocysteinase. J Biol Chem. 1976;251:5817–5819. [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal N. Identification of regulatory elements of cloned genes with functional assays. Methods Enzymol. 1987;152:704–720. doi: 10.1016/0076-6879(87)52075-4. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shield AJ, Thomae BA, Eckloff BW, Wieben ED, Weinshilboum RM. Human catechol O-methyltransferase genetic variation: gene resequencing and functional characterization of variant allozymes. Mol Psychiatr. 2004;9:151–160. doi: 10.1038/sj.mp.4001386. [DOI] [PubMed] [Google Scholar]
- Szumlanski C, Otterness D, Her C, et al. Thiopurine methyltransferase pharmacogenetics: human gene cloning and characterization of a common polymorphism. DNA Cell Biol. 1996;15:17–30. doi: 10.1089/dna.1996.15.17. [DOI] [PubMed] [Google Scholar]
- Thompson MA, Moon E, Kim UJ, Xu J, Siciliano MJ, Weinshilboum RM. Human indolethylamine N-methyltransferase: cDNA cloning and expression, gene cloning, and chromosomal localization. Genomics. 1999;61:285–297. doi: 10.1006/geno.1999.5960. [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM. Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet. 2002;3:611–621. doi: 10.1038/nrg865. [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM. Pharmacogenomics: catechol O-methyltransferase to thiopurine S-methyltransferase. Cell Mol Neurobiol. 2006;26:539–561. doi: 10.1007/s10571-006-9095-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Szumlanski CL. Methylation pharmacogenetics: catechol O-methyltransferase, thiopurine methyltransferase, and histamine N-methyltransferase. Annu Rev Pharmacol Toxicol. 1999;39:19–52. doi: 10.1146/annurev.pharmtox.39.1.19. [DOI] [PubMed] [Google Scholar]