Abstract
Ribosomal large subunit protein RPL41 is a basic (positively charged) peptide consisting of only 25 amino acids. An antisense-based functional screening revealed that the down-regulation of RPL41 led to an anchorage-independent growth of NIH3T3 cells in soft agar plates. RPL41 depletion with gene-specific small interfering RNA also resulted in malignant transformation of NIH3T3 cells including increased tumor growth in mice. RPL41 deletion was detected in 59% of tumor cell lines by fluorescence in situ hybridization analyses and RPL41 down-regulation in 75% of primary breast cancers by real-time quantitative reverse transcription-polymerase chain reaction. These studies suggest a tumor suppression role for RPL41. By mass spectrometry, RPL41 was associated with several cytoskeleton components including tubulin β, γ, and myosin IIA, which was confirmed by Western blot analysis on both cellular lysis and individually in vitro-expressed proteins. RPL41 also bound directly to polymerized tubulins. Cells overexpressing a GFP-RPL41 were resistant to nocodazole-induced microtubule depolymerization. A synthetic RPL41 induced cellular α-tubulin acetylation and G2/M cell cycle arrest. These results indicate a stabilizing role of RPL41 on microtubule. Microtubule spindles are essential for chromosome segregation during mitosis. Cells with RPL41 knock-down showed abnormal spindles, frequent failure of cytokinesis, and formation of polynuclear cells. In interphase cells, RPL41-depleted cells had premature splitting of centrosome. Our results provide evidence that RPL41 is a microtubule-associated protein essential for functional spindles and for the integrity of centrosome and that the abnormal mitosis and disrupted centrosome associated with the RPL41 down-regulation may be related to malignant transformation.
Introduction
Ribosomal proteins are a major component of ribosomes where cellular proteins are synthesized. To date, approximately 80 ribosomal proteins have been identified. In addition to their key role in protein synthesis, some ribosomal proteins are involved in extraribosomal functions, such as DNA repair, apoptosis, transcription regulation, and translation regulation [1–6]. For example, the ribosomal protein RPL13a can exit the ribosome on interferon IFN-γ treatment, bind to specific messenger RNA, and translationally silence their expression, suggesting that the ribosome could act as a depot for some releasable regulators [2].
Several ribosomal proteins have been found to be downregulated in tumors, which is inconsistent with increased protein synthesis in tumor cells and suggests that these proteins are more important to cell proliferation and/or transformation than to the translation machinery [7,8]. Inactivation mutation of RPS6 in Drosophila led to overgrowth of the lymph glands, abnormal blood cell differentiation, and melanotic tumor formation [9]. Mutations and deletions of RPL9 and RPL26 are associated with tumor progression in mice [10]. In a zebra fish tumor model, 11 of 12 lines of zebra fish with increased cancer incidence harbored a heterozygous inactivation mutation in different ribosomal protein genes [11]. Several human cancer syndromes are associated with defective ribosomal genes. Dyskeratosis congenita is characterized by premature aging and increased tumors. One of the genes mutated in dyskeratosis congenita is DKC1, which encodes a component of small nucleolar ribonucleoprotein particles and functions in ribosomal RNA processing [12]. Diamond-Blackfan anemia is a congenital disease characterized by defective maturation of erythroid progenitors and an increased risk for several types of tumors. Multiple ribosomal protein genes, including RPS19, RPS24, RPS17, or RPL35A, are inactively mutated [13–17]. Recently, partial loss-of-function of RPS14 was associated with a subgroup of myelodysplastic syndrome with chromosome 5q deletion [18]. These studies all point to the hypothesis that some ribosomal proteins function as tumor suppressors, although the exact mechanisms are unclear [19].
Ribosomal protein RPL41 is a small peptide of 3456 Da consisting of 25 amino acids, 17 of which are basic arginines and lysines. Posttranslational modifications, including N-terminal loss of methionine, acetylation, internal methylation, or hydroxylation, have not been found in mature RPL41 [20]. It is believed that RPL41 is not only the smallest but also the most basic eukaryotic protein [21]. RPL41 is highly conserved in eukaryotes and is present in certain archaea but not in eubacteria [22]. In our attempt to identify cellular transforming genes, we found that RPL41, when expressed in antisense orientation, induced NIH3T3 cell transformation. The RPL41 down-regulation and deletions were frequently detected in human tumors. These studies suggest a tumor suppression role for RPL41. Further studies showed that RPL41 interacted with cytoskeleton components, including tubulin β, γ, and myosin IIA, and a dynamic cellular localization of RPL41 was found in mitotic cells. When RPL41 was downregulated, cells had abnormal mitosis and premature centrosome split-apart. Our studies suggest that RPL41 is yet another ribosomal protein whose deregulation is associated with tumors and that the abnormal mitosis and defective centrosome integrity in cells with RPL41 down-regulation may be related to malignant transformation.
Materials and Methods
Functional Screening for Transforming Genes
NIH3T3 cells and tumor cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA) and cultured according to the ATCC protocol. A functional screening for transforming genes was performed by expressing complementary DNA (cDNA) from a pool of primary tumors including four breast cancers and three prostate cancers in NIH3T3 cells. Transfected cells were cultured in 0.35% Bactoagar in RPMI 1640 with 10% fetal calf serum, and transformed NIH3T3 cells were identified by their capability for anchorage-independent growth [23].
Stable Cell Lines with RPL41 Knockdown
Two pairs of mouse RPL41-specific small interfering RNA (siRNA) templates (pair no. 1, 5′-gatccgtggcggaagaagagaatgttcaagagacattctcttcttccgccactta-3′ and 5′-agcttaagtggcggaagaagagaatgtctcttgaacattctcttcttccgccacg-3′; and pair no. 2, 5′-gatccagatgaggcagaggtccaattcaagagattggacctctgcctcatcttta-3′ and 5′-agcttaaagatgaggcagaggtccaatctcttgaattggacctctgcctcatctg-3′) were synthesized (Invitrogen, Carlsbad, CA), annealed, and ligated into pSilencer4.1-CMV neo vector (Ambion, Austin, TX) according to the manufacturer's protocol. Constructs were sequenced to exclude mutations and transfected into NIH3T3 cells using Lipofectamine 2000 (Invitrogen). Cells were selected in G418 (400 µg/ml) for 2 weeks and subjected toWestern blot analysis with antibody specific to RPL41. Both cell lines, which resulted in approximately 50% (RPL41 siRNA no. 1) and 80% (RPL41 siRNA no. 2) decreases in RPL41, were used for the evaluation of transforming capacities by soft agar assays. Cell line RPL41 siRNA no. 2 was used for the studies of abnormal mitosis and premature centrosome split.
Fluorescence In Situ Hybridization
RPL41 BAC clone CTD-2560J16 was purchased from CHORI (Oakland, CA). DNA from BAC clone was isolated, biotin-labeled with a BioprimeDNA Labeling kit (Invitrogen), and purified over a fine Sephadex column. For chromosome preparation, cells were treated with colcemid and hypotonic solution, and fixed in methanol/acetic acid fixative. Chromosome spreads were dropped on glass slides. For hybridization, an RPL41 probe and a chromosome 12 centromere-specific probe (Abbott, Abbott Park, IL) were mixed, added to slides, sealed, and denatured at 80°C for 2 minutes. Hybridization was performed in a humidified oven overnight. After washing, probes were detected with fluorochrome-conjugated antibodies and analyzed under a fluorescence microscope.
Quantitative Real-time Reverse Transcription-Polymerase Chain Reaction for RPL41 Expression
Total RNA was isolated from frozen tumors and matched normal specimens and was reverse-transcribed with iScript reverse transcriptase (Bio-Rad, Hercules, CA). RPL41 transcript was amplified in the presence of SYBR Green on a Bio-Rad iCycler with RPL41-specific primers (F/RPL41: 5′-gccgtagacggaacttcgcc-3′; and R/RPL41: 5′-tctgctcctgtggcctccac-3′). β-Actin ACTB was also amplified as the reference gene (F/ACTB: 5′-ttctacaatgagctgcgtgtg-3′; and R/ACTB: 5′-ggggtgttgaaggtctcaaa-3′). Quantitation of RPL41 expression was performed using the standard curve method. Polymerase chain reactions (PCRs) were performed by initial denaturation at 95°C for 1 minute followed by 35 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds.
Glutathione S-Transferase Pull-down Experiments
Human RPL41 was amplified by reverse transcription (RT)PCR (F/RPL41 cDNA: 5′-cctttctctcggccttagcgcc-3′, and R/RPL41 cDNA: 5′-cttcagctaaaacagcggaagaggtg-3′). First RPL41 PCRproduct was reamplified by a pair of nested primers (F/RPL41/glutathione S-transferase [GST] BamH1: 5′-atccacggatccatgagagagccaagtggaggaag-3′, and R/RPL41/GST EcoRI: 5′-gaattgaattcccagcgtctggcattccatg-3′), digested with BamH1 and EcoRI, and inserted into pGEX-4T-3 (GE Healthcare Life Sciences, Waukesha, WI). Both PCRs were performed by initial denaturation at 95°C for 1 minute followed by 30 cycles of denaturation at 94°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 30 seconds. For the GST/scrambled RPL41 construct that expresses a GST-scrambled RPL41 with all basic amino acids of RPL41 moved to the C-terminal end (NH2-QSMAWLMM-RRRRRRRRRRKKKKKKK-OH), two 86-bp oligos containing a BamH1 site at 5′-end and an EcoRI site at 3′-end (F/ScramL41GST: 5′-gatcccaatctatggcttggctcatgatgcgtcgccgacggagaaggcgccgaagaaggaaaaagaaaaagaaaaagaaatgaccg-3′, and R/ScramL41GST: 5′-aattcggtcatttctttttctttttctttttccttcttcggcgccttctccgtcggcgacgcatcatgagccaagccatagattgg-3′) were annealed and inserted into pGEX-4T-3. For the GST/arginine-rich peptide construct that expresses a GST-arginine-rich peptide with all the nonarginine amino acids in RPL41 replaced by glutamic acid (NH2-EREEEREERERREERERREEREREE-OH), two 86-bp oligos containing a BamH1 site at 5′-end and an EcoRI site at 3′-end (F/ArgrichGST: 5′-gatccgaacgtgaagaagaacgtgaagaacgtgaacgtcgtgaagaacgtgaacgtcgtgaagaacgtgaacgtgaagaatgaccg-3′, and R/ArgrichGST: 5′-aattcggtcattcttcacgttcacgttcttcacgacgttcacgttcttcacgacgttcacgttcttcacgttcttcttcacgttcg-3′) were annealed and inserted into pGEX-4T-3. GST constructs were expressed in Escherichia coli BL21 cells. Recombinant proteins were purified according to a standard protocol. Pull-down assays were performed by preincubating GST proteins with 0.1% bovine serum albumin in NTEN buffer (0.5% NP40, 1 mM EDTA, 20 mM Tris, pH 7.4, and 200 mM NaCl) for 5 minutes at room temperature. To identify the RPL41-interacting proteins by microcapillary liquid chromatography-tandem mass spectrometry (LC/MS/MS), cell lysates from 1 x 107 cultured cells were precleaned by incubation with 50 µl of glutathione-agarose beads and incubated with 10 µg of GST protein (control) or 10 µg of GST-RPL41 for 2 hours at 4°C with end-over-end mixing. Beads were washed six times with NTEN. Binding proteins were eluted, separated in SDS-PAGE, and analyzed by microcapillary LC/MS/MS. For in vitro expression of tubulin α, β, and γ, and myosin IIA (two overlapping fragments), RT-PCR was performed on human cDNA using gene-specific primers (F/TUBA: 5′-agttctcactgagacctgtcacc-3′, and R/TUBA: 5′-taaccgtgcaaggaaactgtttatttcg-3′; F/TUBB: 5′-ggcgcattccaaccttccagcctgcg-3′, and R/TUBB: 5′-ctactatgtgccaggcactgttctag-3′; F/TUBG: 5′-ctggcgtgcggcgccgttgcgggcg-3′, and R/TUBG: 5′-cagtcaggcagggcttgggccaaccag-3′; F/MYH9: 5′-atccaggttcaggtcctggctataagtcac-3′, and R/MYH9: 5′-gggaggctgtggtgtctgtctgtccatc-3′). PCR products were subjected to reamplification (nested PCR) with gene-specific primers with T7 promoter sequence incorporated (F/TUBA/ATG T7: 5′-ggatcctaatacgactcactataggaacagaccaccatgcgtgaatgcatctcagtcca-3′, and R/TUBA/nest: 5′-aacatagtgaataggctccaggcag-3′; F/TUBB/ATG T7: 5′-ggatcctaatacgactcactataggaacagaccaccatgagggaaatcgtgcacatcca-3′, and R/TUBB/nest: 5′-ggcagttgagtaagacggctaagg-3′; F/TUBG/ATG T7: 5′- ggatcctaatacgactcactataggaacagaccaccatgccgagggaaatcatcaccctac-3′, and R/TUBG for half nested; F/MYH9NT T7: 5′-ggatcctaatacgactcactataggaacagaccaccatggcacagcaagctgccgataagtat-3′, and R/MYH9 NEST: 5′-catctcaggctgcaggagaagag-3′; F/MYH9 CT T7: 5′-ggatcctaatacgactcactataggaacagaccaccatggtcaagccgctgctgcaggtg-3′, and R/MYH9 for half nested). PCR products were visualized in 0.8% agarose gel and used directly for in vitro TNT analysis using the TNT Quick Coupled Transcription/Translation System (Promega, Madison, WI) according to the manufacturer's instructions. TNT products were mixed with 1 µg each of GST or GST-RPL41 beads. The reaction was incubated for 1 hour at 4°C. Beads were washed six times with NTEN. Binding proteins were eluted, separated in 4% to 12% polyacrylamide gel, fixed, visualized by incubating with fluorography Amplify solution (Amersham, Piscataway, NJ), and exposed to X-ray films overnight at room temperature. Note that GST-RPL41 was typically contaminated with heavy DNA/RNA and that GST-RPL41 was pretreated with DNase 4U (Ambion) and RNase 100 µg/ml in 150 µl of reaction buffer at 37°C for 30 minutes.
RPL41 Peptide and Anti-RPL41 Antibody
The RPL41 peptide (NH2-MRAKWRKKRMRRLKRKRRKMRQRSK-OH) was synthesized at Biosyn, Inc. The peptide was HPLC purified to more than 95% and analyzed by mass analysis and analytical HPLC. For generating anti-RPL41 antibody, a full-length RPL41 with the addition of a cysteine and a glycine at N-terminal was used as antigen. The cysteine residue was added for optimal conjugation of the peptide with carrier protein keyhole lympet hemocyanin. The glycine was added to block the amino group on cysteine. Two New Zealand rabbits were immunized with five boosts. Crude serum was collected at 6th, 8th, and 10th weeks and purified over a Melon Gel IgG Purification column (Thermo, Rockford, IL).
RPL41 Immunofluorescence Staining
Human fibroblasts were cultured in chamber slides (Lab-Tek, Scotts Valley, CA), rinsed in PBS, fixed in 4% paraformaldehyde in PBS for 15 minutes, washed twice in PBS, incubated with 1.5% Triton X-100 for 5 minutes, washed twice in PBS, blocked in 10% goat serum in PBS solution for 2 hours, incubated with 5 µl of anti-RPL41 rabbit antibody and 2 µl of anti-B23 mouse antibody (clone FC82291; Sigma, St. Louis,MO) in 500 µl of 2% goat serum for 1 hour or overnight at 4°C, washed two times in PBS, incubated with 1 µl of fluorescein isothiocyanate (FITC)-conjugated goat antirabbit antibody (Santa Cruz Biotech, Santa Cruz, CA) and 1 µl of rhodamine-conjugated goat antimouse antibody (Invitrogen) in 500 µl of 2% goat serumfor 1 hour, washed three times in PBS, and counterstained with 4′,6-diamidino-2-phenylindole. Immunofluorescence staining with α- and γ-tubulin was similar to the procedure described previously except that cells were fixed in -10°C methanol for 5 minutes.
Microtubule Cosedimentation
Purified tubulin (Cytoskeleton, Denver, CO) was dissolved in PEM buffer (100 mM PIPES, pH 6.8, 1 mMEGTA, and 1 mM MgCl2) at a final concentration of 500 ng/ml. Synthetic RPL41 peptide dissolved in distilled water and tubulin solution were centrifuged for 30 minutes at 14,000 rpm in 4°C to remove any aggregates. GTP (1mM) and paclitaxel (20 µM) were added to the tubulin solution, and the solution was incubated at 37°C for 30 minutes for polymerization. RPL41 peptide (10 µg) was mixed with polymerized tubulin (50 µg) and incubated for 10 minutes at 37°C. Samples were layered onto a 10% sucrose cushion containing 20 µM of paclitaxel and centrifuged for 20 minutes at 14,000 rpm in a tabletop centrifuge. Supernatants and sediments were analyzed by SDS-PAGE and Coomassie brilliant blue staining.
Results
Down-regulation of RPL41 Induces Cell Transformation
A functional screening for transforming genes was performed by expressing cDNA from primary tumors in nonneoplastic NIH3T3 cells. Transfected cells were cultured in soft agar plates, and transformed NIH3T3 cells were identified by their capability for anchorage-independent growth. The transforming genes were recovered by cDNA adaptor PCR and sequenced. Genes expressed in sense orientation were considered candidate oncogenes, and those expressed in antisense orientation, which could potentially downregulate the expression of their counterpart in cells, were considered candidate tumor suppressor genes [23]. One of the genes identified was RPL41, which was expressed in antisense orientation. The expression of antisense RPL41 resulted in an approximately 80% decrease of cellular RPL41 in NIH3T3 cells by Western blot analysis with a custom antibody specific to RPL41 (see Online Supplemental Material for specificity evaluation of anti-RPL41 antibody; Figure W1). To further confirm that the down-regulation of RPL41 causes cell transformation, we cloned two mouse RPL41-specific siRNA oligos into pSilencer 4.1-CMV vector and stably expressed them in NIH3T3 cells, which resulted in approximately 50% (RPL41 siRNA no. 1) and 80% (RPL41 siRNA no. 2) decreases in RPL41, respectively (Figure 1A). Multiple foci were formed in cells expressing both RPL41 siRNA but not in control cells (Figure 1B). Soft agar assays showed a significant increase in anchorage-independent colonies in cells expressing both RPL41 siRNA compared with control cells (Figure 1, C and D). Tumorigenesis of NIH3T3 expressing RPL41 siRNA no. 2 was studied in athymic CD-1 nude mice by injecting 2 x 107 cells subcutaneously on both sides of the flanks. After 4 weeks, seven of eight sites injected with RPL41-depleted cells developed palpable tumors, whereas none of the eight sites injected with control cells developed tumors.
Figure 1.
Down-regulation of RPL41 in NIH3T3 cells resulted in malignant transformation. (A) RPL41-specific siRNA effectively knocked down cellular RPL41 expression. NIH3T3 cells were transfected with two RPL41 siRNA expression constructs or a control construct expressing a random hairpin siRNA. Western blot analysis was performed with antibodies specific to RPL41 or β-actin. (B) RPL41-depleted NIH3T3 cells formed foci in liquid culture. No such foci were noted in control cells. (C) RPL41-depleted NIH3T3 cells formed significantly more colonies than did control cells in soft agar plates. (D) Colonies that exceeded 120 µm in diameter in soft agar plates were counted. The number of colonies was the average value based on six wells (35 mm). Both cell lines expressing RPL41 siRNA had significantly more colonies than control cells.
Deletion and Down-regulation of RPL41 in Human Tumors
To evaluate potential RPL41 mutation in human tumors, we performed PCR to amplify a 914-bp genomic region covering the entire RPL41 coding sequence. No mutations or homozygous deletions of RPL41 were identified in 22 tumor cell lines (ATCC; Table W1). Fluorescence in situ hybridization (FISH) analysis with a BAC clone containing RPL41 (CTD-2560J16; CHORI) and a chromosome 12 centromere control probe (Vysis, Inc) showed RPL41 allelic reduction in 13 of 22 tumor cell lines (Figure 2A; Table W1). Real-time quantitative RT-PCR was performed on 12 primary breast cancers and their matched normal tissues in the presence of SYBR Green. Of 12 breast cancers, 9 showed significant RPL41 down-regulation (Figure 2B; Table W2). Interestingly, one case (case no. 9) showed barely detectable RPL41 in both tumor specimen and normal tissue, and a constitutional RPL41 inactivation could not be excluded. Similar RPL41 down-regulation was also revealed by a search of the ONCOMINE GeneChip array database in a large variety of tumors, including breast cancer, lung cancer, head and neck squamous cell carcinoma, malignant glioblastoma, ovarian cancer, glioma, bladder transitional cell carcinoma, melanoma, and adenoid cystic carcinoma of the salivary gland [24].
Figure 2.
Deletion and down-regulation of RPL41 in tumors. (A) RPL41 allelic reduction was detected in human tumors. FISH analysis was performed with an RPL41 BAC clone (red) and a chromosome 12-centromere control probe (green). FISH on normal lymphocytes showed that RPL41 was located at 12q13 as expected (left). Leukemia cells (CCL246) had two copies of centromere and one copy of RPL41 (right). FISH results on 22 tumor cell lines were detailed in Table W1. (B) RPL41 expression was significantly decreased in primary breast cancers. Quantitative real-time RT-PCR was performed with RPL41-specific primers in 12 cases of matched primary breast cancers/normal pairs. Data represent mean ± SD from three reactions. Clinical information of tumors is detailed in Table W2. Constitutional RPL41 inactivation in case 9 could not be excluded.
RPL41 Is a Microtubule-Associated Protein
To identify proteins that interact with RPL41, RPL41 was expressed in E. coli as a fusion to GST, purified by use of glutathione agarose beads, and incubated with human cell lysate. Proteins associated with GST-RPL41 were separated in SDS-PAGE and identified by microcapillary LC/MS/MS. Cytoskeleton components, including tubulin α, β, and γ, and myosin IIA, were found to associate with GST-RPL41. Western blot analysis showed that these cytoskeleton components were all effectively pulled down by GST-RPL41 but not by several controls, including a GST, a GST-scrambled RPL41, or a GST arginine-rich peptide (Figure 3A). The interaction between the endogenous RPL41 and cytoskeleton components was confirmed by a coimmunoprecipitation assay; Cellular RPL41 complex pulled down by the anti-RPL41 antibody contained tubulin α, β, and γ, and myosin IIA (Figure 3B). Each individual protein was then expressed in vitro in a reticulocyte lysate system using gene-specific PCR product containing a T7 promoter and incubated with GST-RPL41. The full-length β- and γ-tubulin as well as N-terminal myosin IIA were readily pulled down by GST-RPL41. However, no interaction between GST-RPL41 and α-tubulin or C-terminal myosin IIA were detected (Figure 3C). The interaction between α-tubulin and GST-RPL41 seen in the pull-down experiment using cellular proteins, therefore, is likely the result of α-tubulin and β-tubulin heterodimerization. We next studied the association of RPL41 with the polymerized tubulin by incubating a synthetic RPL41 peptide with purified tubulin in the presence or absence of Taxol, a known microtubule polymerization inducer. Coprecipitation of RPL41 with polymerized tubulin was detected in the presence of Taxol, consistent with a direct interaction between RPL41 and microtubule (Figure 3D).
Figure 3.
Interaction of RPL41 with cytoskeletal components. (A) Cellular tubulin α, β, and γ and myosin IIA were effectively pulled down by GST-RPL41. Cell lysate was incubated with GST, GST/RPL41, GST/Scrambled RPL41, and GST/Arginine-rich peptide. Western blot analyses of the GST pull-down products were performed with antibodies to tubulin α, β, and γ and myosin IIA. (B) Cell lysate was incubated with an antibody to RPL41 or rabbit preimmunization serum (control), pulled down by incubation with protein A/G agarose, and immunoblotted with antibodies to tubulin α, β, and γ, myosin IIA, and RPL41. (C) Individually expressed tubulin β and γ and N-terminal myosin IIA were effectively pulled down by GST-RPL41. Proteins were expressed in reticulocyte lysate and labeled with 35S methionine. Myosin IIA was expressed in two overlapping fragments (N-myosin and C-myosin) owing to its large size. (D) RPL41 associated with polymerized tubulin. A synthetic RPL41 peptide was incubated with purified tubulin in the presence or absence of Taxol. Reactions were centrifuged, and the supernatant and precipitants were separated in 20% polyacrylamide gel.
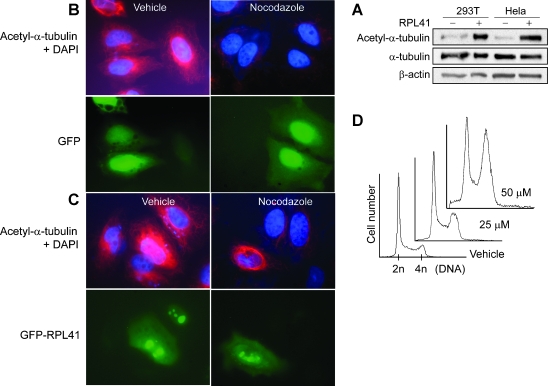
Several microtubule-associated proteins are known to stabilize microtubule by increasing α-tubulin acetylation. To study whether RPL41 affects the acetylation of α-tubulin, cells were treated with a synthetic RPL41, which is self cell-penetrating (unpublished data), and Western blotted with antibodies to α-tubulin and acetyl-α-tubulin. As shown in Figure 4A, RPL41 induced a significant increase in α-tubulin acetylation in both HeLa cells and 293T cells. Immunofluorescence staining on cells overexpressing GFP-RPL41 showed significantly more residual acetylated α-tubulin after nocodazole challenge than those overexpressing a GFP control (Figure 4, B and C). These results are suggesting a stabilizing role of RPL41 on microtubule. Pharmacologic intervention of microtubule dynamics is associated with cell cycle arrest at G2/M. To study whether an exogeneous RPL41 affects cell cycle, 293T cells were cultured with various concentrations of RPL41 for 16 hours and subjected to a flow cytometry assay. A dose-dependent G2/M arrest was detected in cells treated with RPL41 (Figure 4D).
Figure 4.
Acetylation of cellular α-tubulin and G2/M cell cycle arrest induced by RPL41. (A) Significantly increased α-tubulin acetylation was detected in cells treated with a synthetic RPL41. 293T cells and HeLa cells were incubated with 50 µM synthetic RPL41 or vehicle control (saline) for 1 hour and Western blotted with antibodies to α-tubulin, acetyl-α-tubulin, and β-actin. (B and C) Cells overexpressing a GFP-RPL41 showed significantly more residual acetylated α-tubulin after nocodazole challenge than those overexpressing a GFP control. HeLa cells were transfected with GFP (B) or GFP-RPL41 (C) for 24 hours, treated with 10 µM nocodazole or vehicle control (DMSO) for 20 minutes, immunofluorescence-stained with anti-acetyl-α-tubulin antibody (red), and counterstained with DAPI (blue). (D) Cells treated with a synthetic RPL41 accumulated at G2/S phase. 293T cells were incubated with 25 to 50 µM RPL41 or vehicle (saline) for 16 hours and subjected to a flow cytometry assay. Significant increase in G2/M cells was seen in cells treated with RPL41.
Cellular localization of RPL41 was studied by immunofluorescence analysis with the anti-RPL41 antibody. In the interphase, RPL41 is mainly located in nucleoli; diffused signals were also seen in nuclei and cytoplasm (Figure 5A). The nucleolar localization of RPL41 was confirmed by its colocalization with the nucleolar marker B23 (Figure 5A). Colocalization of RPL41 and microtubules was detected in the pseudopodia of cells (Figure 5B). In mitotic cells, RPL41 showed a dynamic cellular localization: RPL41 was evenly distributed in the entire cell in prophase, concentrated on chromosomes in metaphase, enriched on chromosomes and spindle midzone in anaphase, and localized in newly formed nuclei and in midbody in telophase (Figure 5, C and D). Colocalization of RPL41 with microtubules in midbody was confirmed by immunostaining with antibodies to RPL41 and α-tubulin (Figure 5D). The cellular localization of RPL41 was verified by a synthetic FITC-conjugated RPL41 peptide, which showed an identical pattern of localization as described previously (data not shown).
Figure 5.
Cellular localization of RPL41 on human fibroblasts. Immunofluorescence staining of human fibroblasts was performed with antibodies to RPL41, nucleolar marker B23, or α-tubulin, and counterstained with DAPI. (A) In interphase cells, RPL41 is located in nucleoli, nuclei, and cytoplasm. Nucleolar localization of RPL41 was verified by costaining with anti-B23. (B) RPL41 was colocalized with microtubule in the pseudopodia of cells. (C) In mitotic cells, RPL41 was diffused in the entire cell in prophase, concentrated on chromosomes in metaphase, enriched in chromosomes and midzone in anaphase, and localized in newly formed nuclei and midbody in telophase. (D) RPL41 was colocalized with α-tubulin in midbody.
RPL41 Down-regulation Leads to Abnormal Mitosis
Although no significant morphologic difference was seen between NIH3T3 cells expressing an RPL41 siRNA and control cells, RPL41-depleted cells showed sparse cellular microtubules and lack of typical aster structure in interphase cells (Figure 6A). In mitotic cells, abnormal mitotic spindles, including sparse spindles, lack of centrosome foci, and decreased midzone spindles, were found in RPL41-depleted NIH3T3 cells (Figure 6B). Immunostaining with anti-γ-tubulin antibody on mitotic cells showed an apparent normal pattern of signals in RPL41-depleted NIH3T3 cells. However, by measuring the distance between the spindle poles at the end of anaphase, significantly shortened mitotic spindles were found in RPL41-depleted cells, with an average of 9.1 ± 1.89 µm in RPL41-depleted cells compared with 13.6 ± 1.5 µm in control cells (P = 7.4e - 05; n = 20). Similarly, the two groups of newly segregated chromosomes in telophase stayed noticeably closer in RPL41-depleted NIH3T3 cells. RPL41-depleted cells also showed a shorter and twisted midbody, with some containing lagging DNA material (Figure 6C), probably due to the lack of enough chromosome separation before cytokinesis. We counted 100 telophase cells with RPL41 depletion; 16 of them had visible DNA material (DAPI-positive) in midbody; only 2 of 100 control cells had similar DNA material in midbody. By time-lapse image analysis under contrast microscopy, RPL41-depleted NIH3T3 cells showed large membrane bulges at the beginning of cytokinesis; no such large bulges were seen in control NIH3T3 cells. Cleavage furrow did occur, although it often regressed, which led to failed cytokinesis (Video W1 and Video W2). A significant increase in polynuclear cells, including some giant cells with up to eight nuclei per cell, was found in RPL41-depleted cells (Figure 6D). Of 500 cells counted under contrast microscope, 115 (23%) were polynuclear cells in RPL41-depleted cells, whereas only 10 (2%) of 500 were polyploidy cells in control cells.
Figure 6.
Abnormal microtubules in NIH3T3 cells with RPL41 down-regulation. (A) RPL41-depleted interphase cells showed sparse cellular microtubules and lack of typical aster structure. NIH3T3 stable cell lines expressing an RPL41-specific siRNA (RPL41 KD) or a control random siRNA were immunostained with an antibody to α-tubulin and counterstained with DAPI. (B) RPL41-depleted cells showed abnormal mitotic spindles. Mitotic cells of control and RPL41 KD cells were immunostained with an antibody to α-tubulin and counterstained with DAPI. Control cells had spindles focused on centrosomes, seen as prominent bright foci (arrowheads). These foci were undetectable in RPL41-depleted cells. RPL41-depleted cells also showed lack of midzone spindles in anaphase. (C) Lagged DNA materials were commonly seen in the midbodies in RPL41-depleted cells. (D) Increased polynuclear cells were seen in RPL41-depleted NIH3T3 cells. Approximately 80% decrease in cellular RPL41 in cells expressing a RPL41-specific siRNA was verified by Western blot analysis.
RPL41 Down-regulation Leads to Centrosome Splitting
Immunofluorescence staining with anti-γ-tubulin antibody on interphase cells showed a different pattern of signals between RPL41-depleted NIH3T3 cells and control NIH3T3 cells; whereas most control cells had one signal, most RPL41-depleted cells had two signals (Figure 7A). Some of the lone signal in control cells showed a doublet, presumably representing the two centrioles, at higher magnification. No such doublet was seen in either of the two signals in RPL41-depleted cells. This result suggests a possible premature centrosome split-apart of the two centrioles in RPL41-depleted cells, although an arrest of cell cycle at G2, when centrosome normally separates, could not be excluded. A flow cytometry analysis detected no such G2 arrest; in fact, a slightly but consistently decreased G2 and S cells and slightly increased G1 cells was seen in RPL41-depleted cells (Table W3). Immunofluorescence staining with antibodies to γ-tubulin and centrosome protein 170 kDa (Cep170) was then performed. Cep170 is present in the mother centriole, but not in the daughter centriole in G1 cells, and in both matured centrosomes at late G2 [25]. RPL41-depleted cells showed colocalization of γ-tubulin and Cep170 in one centrosome and only of γ-tubulin in the other centrosome (Figure 7B). Considering that most RPL41-depleted cells are in G1, the observed pattern of γ-tubulin and Cep170 signals in RPL41-depleted NIH3T3 cells is consistent with a premature centrosome splitting. To study the nucleation function of the split centrosomes, a microtubules regrowth assay was performed. Cells were incubated on ice to depolymerize microtubules, immersed immediately in prewarmed complete culture medium (37°C) for repolymerization, fixed in methanol, and immunofluorescence-stained with antibodies to α- and γ-tubulin. Whereas control cells showed a nice aster microtubule structure, no such microtubule regrowth was found in cells with RPL41 depletion (Figure 7C).
Figure 7.
Centrosome abnormality in NIH3T3 cells with RPL41 down-regulation. (A) Majority of RPL41-depleted cells had premature centrosome splitting. Interphase cells of NIH3T3 cells expressing an RPL41-specific siRNA (RPL41 KD) or a control random siRNA were immunostained with an antibody to γ-tubulin and counterstained with DAPI. Control cells had one signal with some consisting of a doublet (insert). Most RPL41-depleted cells had two signals (arrowhead) without doublet (insert). (B) Only one of the split centrioles was-positive to Cep170 in RPL41-depleted cells. Control and RPL41 KD cells were immunostained with antibodies to γ-tubulin and Cep170 (red) and counterstained with DAPI. (C) Defective microtubule regrowth in cells with RPL41 down-regulation. RPL41 KD cells and control cells were subjected to a microtubule regrowth assay followed by immunostaining with antibodies to α-tubulin and γ-tubulin. Typical microtubule asters were seen in control cells but not in RPL41-depleted cells with split centrosomes.
Discussion
Several studies have linked the deregulated ribosome proteins with tumors, although their oncogenic mechanisms remain to be determined. The involvement of multiple ribosomal proteins in tumors indicates that they may use a common oncogenic pathway. Deregulation of translation is an obvious possibility. However, it is puzzling that some ribosomal proteins involved in tumors are downregulated, whereas the cellular translation is presumably increased in tumors. Several studies provided evidence that many ribosomal proteins have extraribosomal functions unrelated to cellular translation. Our studies showed that RPL41 is a microtubule-associated protein, and cells with defective RPL41 had abnormal mitosis with frequent cytokinesis failure and increased polynuclear cells. Abnormal mitosis could lead to genome instability and facilitate malignant transformation [26]. Polynuclear cells are also more sensitive to carcinogens [27]. It is likely that the abnormal mitosis and polynuclear cells in RPL41 defective cells contribute to malignant transformation. An interesting question is whether other ribosomal protein defects use a similar hypothetical oncogenic pathway. Microtubule-associated proteins are typically basic proteins that interact with the acid region of tubulin [28]; most ribosomal proteins are basic proteins [29] and therefore potential microtubule-binding proteins. A proteomic study on isolated midbodies, a microtubulebased structure important for cytokinesis, showed that 13% of 577 proteins are ribosomal proteins [30]. The possibility that other ribosomal proteins may play roles in mitosis, therefore, is worth further study. Interestingly, RPL41 was not one of the midbody-associated proteins in the proteomic study of midbody, although our results clearly showed a midbody localization. RPL41 is unique in that 17 of 25 amino acids are arginines and lysines, which are cleaved by trypsin digestion before peptide mass fingerprinting. Small fragments of RPL41 are likely beyond the resolution of mass spectrum.
In addition to failed cytokinesis, we frequently observed lagged chromatids inside cleavage furrow in RPL41-depleted cells, which are probably caused by the shortened mitotic spindles and the lack of enough chromosome separation before cytokinesis. A potential detrimental effect of this abnormality is that the force of contractile ring could fracture these lagged DNA. A loss of DNA fragments encoding tumor suppressors will contribute to malignant transformation. For example, chromosome 1, the longest chromosome of the cell that may be more likely to be “stuck” in midbody, is suspected to harbor multiple tumor suppressors [31].
Our results showed that cells with RPL41 down-regulation had premature centrosome splitting. Centrosome is an organelle that serves as the main microtubule-organizing center. Centrosome abnormalities have been detected in early tumors [32] and in human cells infected with tumor-causing virus [33]. Centrosome at G1 is consisted of two centrioles held together by a dynamic linker. A procentriole is formed adjacent to each of the two parental centrioles during S and G2, and the two matured centrosomes are separated just before the onset of mitosis to form the poles of the bipolar spindle apparatus. Centrosome cohesion is regulated by a balance of centrosome-associated kinase and phosphatase activity; both overexpression of centrosome-associated kinases or inhibition of phosphatases induced centrosome splitting [34]. It is proposed that the phosphorylation status of the docking site for the centriole linker regulates the centrosome cohesion [35]. Microtubule-destabilizing agents are known to induce centrosome splitting likely through an imbalance of centrosome-associated kinases and phosphatases caused by the defective microtubule flux. The mechanism of centrosome splitting in RPL41-depleted cells is unclear. RPL41 is a microtubule-associated protein that stabilizes the microtubule. It is possible that the lack of RPL41 in cells renders microtubule unstable and therefore premature centrosome splitting.
Online Supplemental Material
Evaluation of Specificity of RPL41 Antibody
The specificity of the antibody to RPL41 was evaluated in the following studies: (1) Immunofluorescence staining of human fibroblasts with anti-RPL41 antibody showed a predominant nuclei and nucleoli localization with weak cytoplasmic staining, consistent with the results obtained from a penetrating FITC-conjugated synthetic RPL41 and the expression of a GFP/RPL41 construct (Figure 5, A and B; manuscript submitted); (2) Western blot analysis of human cell lysis showed a single band that comigrated with the synthetic RPL41 (Figure W1); RPL41 needed to be transferred onto membranes in a reversed electronic field, that is, from anode to cathode, consistent with the extremely basic nature of RPL41; and (3) Western blot analysis of NIH3T3 cells expressing an RPL41 siRNA oligo showed that the RPL41 was specifically knocked down (Figure 1A).
Acknowledgment
The authors thank Stefan and Anette Duensing (University of Pittsburgh Cancer Institute) for critical reading of the manuscript.
Footnotes
References
- 1.Zimmermann RA. The double life of ribosomal proteins. Cell. 2003;115:130–132. doi: 10.1016/s0092-8674(03)00804-3. [DOI] [PubMed] [Google Scholar]
- 2.Mazumder B, Sampath P, Seshadri V, Maitra RK, DiCorleto PE, Fox PL. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell. 2003;115:187–198. doi: 10.1016/s0092-8674(03)00773-6. [DOI] [PubMed] [Google Scholar]
- 3.Chavez-Rios R, Arias-Romero LE, Almaraz-Barrera MJ, Hernandez-Rivas R, Guillen N, Vargas M. L10 ribosomal protein from Entamoeba histolytica share structural and functional homologies with QM/Jif-1: proteins with extraribosomal functions. Mol Biochem Parasitol. 2003;127:151–160. doi: 10.1016/s0166-6851(02)00332-8. [DOI] [PubMed] [Google Scholar]
- 4.Neumann F, Krawinkel U. Constitutive expression of human ribosomal protein L7 arrests the cell cycle in G1 and induces apoptosis in Jurkat T-lymphoma cells. Exp Cell Res. 1997;230:252–261. doi: 10.1006/excr.1996.3417. [DOI] [PubMed] [Google Scholar]
- 5.Khanna N, Reddy VG, Tuteja N, Singh N. Differential gene expression in apoptosis: identification of ribosomal protein S29 as an apoptotic inducer. Biochem Biophys Res Commun. 2000;277:476–486. doi: 10.1006/bbrc.2000.3688. [DOI] [PubMed] [Google Scholar]
- 6.Naora H, Takai I, Adachi M, Naora H. Altered cellular responses by varying expression of a ribosomal protein gene: sequential coordination of enhancement and suppression of ribosomal protein S3a gene expression induces apoptosis. J Cell Biol. 1998;141:741–753. doi: 10.1083/jcb.141.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasai H, Nadano D, Hidaka E, Higuchi K, Kawakubo M, Sato TA, Nakayama J. Differential expression of ribosomal proteins in human normal and neoplastic colorectum. J Histochem Cytochem. 2003;51:567–574. doi: 10.1177/002215540305100502. [DOI] [PubMed] [Google Scholar]
- 8.Choi P, Chen C. Genetic expression profiles and biologic pathway alterations in head and neck squamous cell carcinoma. Cancer. 2005;104:1113–1128. doi: 10.1002/cncr.21293. [DOI] [PubMed] [Google Scholar]
- 9.Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc Natl Acad Sci USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck-Engeser GB, Monach PA, Mumberg D, Yang F, Wanderling S, Schreiber K, Espinosa R, 3rd, Le Beau MM, Meredith SC, Schreiber H. Point mutation in essential genes with loss or mutation of the second allele: relevance to the retention of tumor-specific antigens. J Exp Med. 2001;194:285–300. doi: 10.1084/jem.194.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, Lees JA, Hopkins N. Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol. 2004;2:E139–??. doi: 10.1371/journal.pbio.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet. 1998;19:32–38. doi: 10.1038/ng0598-32. [DOI] [PubMed] [Google Scholar]
- 13.Dianzani I, Garelli E, Ramenghi U. Diamond-Blackfan anaemia: an overview. Paediatr Drugs. 2000;2:345–355. doi: 10.2165/00128072-200002050-00002. [DOI] [PubMed] [Google Scholar]
- 14.Draptchinskaia N, Gustavsson P, Andersson B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J, Matsson H, et al. The gene encoding ribosomal protein S19 is mutated in Diamond-Blackfan anaemia. Nat Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 15.Gazda HT, Grabowska A, Merida-Long LB, Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball SE, Orfali KA, et al. Ribosomal protein S24 gene is mutated in Diamond-Blackfan anemia. Am J Hum Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farrar JE, Nater M, Caywood E, McDevitt MA, Kowalski J, Takemoto CM, Talbot CC, Jr, Meltzer P, Esposito D, Beggs AH, et al. Abnormalities of the large ribosomal subunit protein, Rpl35A, in Diamond-Blackfan anemia. Blood. 2008;112:1582–1592. doi: 10.1182/blood-2008-02-140012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cmejla R, Cmejlova J, Handrkova H, Petrak J, Pospisilova D. Ribosomal protein S17 gene (RPS17) is mutated in Diamond-Blackfan anemia. Hum Mutat. 2007;28:1178–1182. doi: 10.1002/humu.20608. [DOI] [PubMed] [Google Scholar]
- 18.Ebert BL, Pretz J, Bosco J, Chang CY, Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al. Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer. 2003;3:179–192. doi: 10.1038/nrc1015. [DOI] [PubMed] [Google Scholar]
- 20.Odintsova TI, Müller EC, Ivanov AV, Egorov TA, Bienert R, Vladimirov SN, Kostka S, Otto A, Wittmann-Liebold B, Karpova GG. Characterization and analysis of posttranslational modifications of the human large cytoplasmic ribosomal subunit proteins by mass spectrometry and Edman sequencing. J Protein Chem. 2003;22:249–258. doi: 10.1023/a:1025068419698. [DOI] [PubMed] [Google Scholar]
- 21.Yu X, Warner JR. Expression of a micro-protein. J Biol Chem. 2001;276:33821–33825. doi: 10.1074/jbc.M103772200. [DOI] [PubMed] [Google Scholar]
- 22.Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, FitzGerald LM, Clayton RA, Gocayne JD, et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 23.Li Y, Huang J, Zhao YL, He J, Wang W, Davies KE, Nosé V, Xiao S. UTRN on chromosome 6q24 is mutated in multiple tumors. Oncogene. 2007;26:6220–6228. doi: 10.1038/sj.onc.1210432. [DOI] [PubMed] [Google Scholar]
- 24.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duensing A, Chin A, Wang L, Kuan SF, Duensing S. Analysis of centrosome overduplication in correlation to cell division errors in high-risk human papillomavirus (HPV)-associated anal neoplasms. Virology. 2008;372:157–164. doi: 10.1016/j.virol.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Duesberg P, Rasnick D, Li R, Winters L, Rausch C, Hehlmann R. How aneuploidy may cause cancer and genetic instability. Anticancer Res. 1999;19:4887–4906. [PubMed] [Google Scholar]
- 27.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- 28.Littauer UZ, Giveon D, Thierauf M, Ginzburg I, Ponstingl H. Common and distinct tubulin binding sites for microtubule-associated proteins. Proc Natl Acad Sci USA. 1986;83:7162–7166. doi: 10.1073/pnas.83.19.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Otaka E, Kobata K. Yeast ribosomal proteins: I. Characterization of cytoplasmic ribosomal proteins by two-dimensional gel electrophoresis. Mol Gen Genet. 1978;162:259–268. doi: 10.1007/BF00268851. [DOI] [PubMed] [Google Scholar]
- 30.Skop AR, Liu H, Yates J, III, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mertens F, Johansson B, Hoglund M, Mitelman F. Chromosomal imbalance maps of malignant solid tumors: a cytogenetic survey of 3185 neoplasms. Cancer Res. 1997;57:2765–2780. [PubMed] [Google Scholar]
- 32.Pihan GA, allace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- 33.Duensing S, Duensing A, Flores ER, Do A, Lambert PF, Munger K. Centrosome abnormalities and genomic instability by episomal expression of human papillomavirus type 16 in raft cultures of human keratinocytes. J Virol. 2001;75:7712–7716. doi: 10.1128/JVI.75.16.7712-7716.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114:3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 35. Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.