Abstract
Curli are functional extracellular amyloid fibers produced by uropathogenic Escherichia coli (UPEC) and other Enterobacteriaceae. Ring-fused 2-pyridones, such as FN075 and BibC6, inhibited curli biogenesis in UPEC and prevented the in vitro polymerization of the major curli subunit protein CsgA. The curlicides FN075 and BibC6 share a common chemical lineage with other ring-fused 2-pyridones termed pilicides. Pilicides inhibit the assembly of type 1 pili, which are required for pathogenesis during urinary tract infection. Notably, the curlicides retained pilicide activities and inhibited both curli-dependent and type 1–dependent biofilms. Furthermore, pretreatment of UPEC with FN075 significantly attenuated virulence in a mouse model of urinary tract infection. Curli and type 1 pili exhibited exclusive and independent roles in promoting UPEC biofilms, and curli provided a fitness advantage in vivo. Thus, the ability of FN075 to block the biogenesis of both curli and type 1 pili endows unique anti-biofilm and anti-virulence activities on these compounds.
Bacterial biofilms are complex microbial communities that exhibit reduced sensitivity to conventional antibiotics, host defenses and external stresses1,2. Multiple determinants contribute to biofilm development and maintenance, and their requirements in biofilm formation may vary depending on environmental conditions. In addition, factors important in biofilm formation are often functionally redundant, making anti-biofilm strategies challenging and complex.
E. coli and other Enterobacteriaceae assemble adhesive amyloid fibers termed curli at the bacterial cell surface that are involved in biofilm formation3,4. Curli mediate cell-cell and cell-surface interactions to promote bacterial adhesion to mammalian and plant cells as well as inert surfaces such as glass, stainless steel and plastic5,6. Curli also serve as an adhesive and structural scaffold to promote biofilm assembly and other community behaviors7–9. Biofilms within the host are implicated in serious and persistent infectious diseases, including cystic fibrosis, chronic otitis media and urinary tract infection (UTI)10,11. In the environment, biofilms can serve as reservoirs for pathogens and can result in surface and water contamination. Amyloid adhesins and amyloid-integrated biofilms, in particular, are prevalent among diverse phyla (for example, Proteobacteria, Bacteroidetes, Chloroflexi, Actinobacteria) that thrive in drinking water reservoirs and other environmental habitats12,13.
Bacterial adhesion and biofilm models emphasize redundant features, and virulent pathogens often harbor multiple adhesive systems that are used in different stages of pathogenesis, supporting the notion that broad-spectrum approaches to prevent biofilm formation may be most attractive6,14. In addition to curli, bacteria can assemble hundreds of extracellular adhesive fibers known as pili, many of which mediate host-cell binding and invasion and biofilm formation, all of which contribute to bacterial pathogenesis in the human host15. Type 1 pili are crucial to UPEC pathogenesis. They contain the FimH adhesin at their tip, which mediates binding to mannosylated receptors present on the luminal surfaces of mammalian bladder epithelial cells—an event that is critical in the pathogenesis of urinary tract infection16–18. Thus, type 1 pili are essential virulence factors representing an excellent target for therapeutic intervention19.
Both curli and type 1 pili have been implicated in mediating biofilm formation by UPEC as well as enterohemorrhagic E. coli7–9,20,21. Recent data suggest that curli and cellulose act synergistically to promote host colonization, biofilm formation and survival in different environments22. Ultimately, strategies that target multiple adhesive structures may be needed to prevent colonization, invasion and biofilm formation in order to have the most potent therapeutic potential23.
Thiazolo ring-fused 2-pyridones are peptidomimetics that can target essential protein-protein interactions in macromolecular assembly24. Previously, we discovered that the dihydro thiazolo ring-fused 2-pyridone provides an excellent scaffold for the rational design and synthesis of pilicides (for example, BibC10 (1), Fig. 1a) that block type 1 pilus biogenesis in E. coli19,24–26. Type 1 pili are assembled by the well-characterized chaperone-usher pathway27,28. Pilicides such as 1 bind to the pilus chaperone and block the targeting of chaperone-subunit complexes to the outer membrane usher, thus preventing the donor strand exchange reaction that is required for assembly of the fiber24. Exchange of the cyclopropyl group of 1 with a CF3-phenyl substituent generated a compound, FN075 (2), that gained anti-amyloid activity and inhibited Alzheimer's-associated β-amyloid (Aβ) polymerization in vitro29. The original cyclopropyl-substituted pyridone, 1, exerted no anti-amyloid activity29.
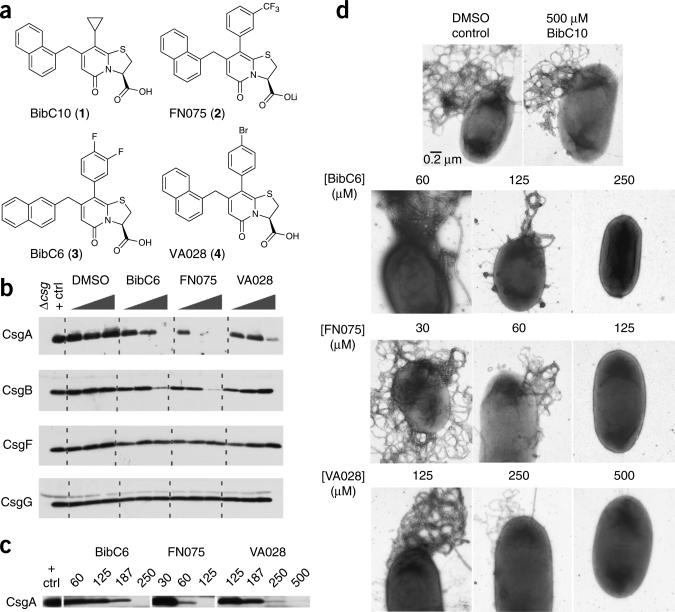
Figure 1.
Curlicide inhibition of in vivo amyloid biogenesis in E. coli. (a) BibC10 (1), FN075 (2), BibC6 (3) and VA028 (4) are ring-fused 2-pyridones that differ in their phenyl ring modifications and, for 3, in the position of naphthyl substitution. (b) CsgA, CsgB, CsgF and CsgG protein profiles obtained by western analysis of MC4100 grown on curlicide-amended agar for 48 h. Each curlicide was tested at 0.1, 1.0 and 2.5 mM, and the positive control sample corresponds to cells grown in DMSO-amended agar. MC4100Δcsg was the negative control. Curli production was abolished at 1.0 mM compound 2, 2.5 mM compound 3. Only a faint CsgA band was observed at 2.5 mM compound 4. (c) Curlicides were effective at the designated micromolar concentrations in blocking curli biogenesis in UTI89 growing in shaking YESCA broth (100 rpm) for 48 h at 28 °C. (d) Representative high-resolution EM images of UTI89 prepared as in c. Titratable reductions in bacterial curliation were observed for cells grown in the presence of curlicides. The scale bar in the first electron micrograph represents 0.2 μm and applies to all images.
Inspired by the anti-amyloid properties of 2 and two additional molecules with a similar substitution pattern as 2 (BibC6 (3) and VA028 (4); Fig. 1 and Supplementary Fig. 1), we found that compounds 2, 3 and 4 interrupted UPEC curli biogenesis and thus prevented curli-dependent biofilm formation. Evaluation of the structure of the binding site identified previously for pilicides24 revealed that compounds with a larger aryl substituent (for example, 2) should still fit well in the chaperone binding site. Thus, we hypothesized that curlicides, such as 2, would retain the pilicide activity harbored by 1. We confirmed this hypothesis and demonstrated the dual ability of 2 to block both curli- and type 1 pili-dependent biofilms in vitro and to block colonization and intracellular bacterial community (IBC) formation in vivo, thus significantly attenuating virulence of UPEC in a mouse model of urinary tract infection. Inspired by this chemical-induced attenuation in virulence by the dual curlicide-pilicide, we examined the potential role of curli in UTI and demonstrated that two different mutants that eliminate curli fiber production, a csgA mutant (lacking the major curli subunit) and a csgB; csgG mutant (lacking the curli fiber nucleator and secretion pore), had reduced fitness in a UTI model. This discovery opens up a new avenue to understand how amyloid production by E. coli influences bladder colonization, and it also reveals a class of compounds that has potential therapeutic value by blocking multiple adhesive organelles important in biofilm formation and colonization.
RESULTS
Curlicides inhibit in vivo amyloid biogenesis
Three compounds, 2, 3 and 4, were tested for their ability to inhibit curli assembly in a whole-cell assay using the laboratory E. coli K-12 strain MC4100 and the UPEC strain UTI89. Curli assembly in E. coli requires the concerted action of several gene products encoded by the divergently transcribed csgB; csgA operon and csgD; csgE; csgF; csgG operon (csg, curli-specific genes)4,30. In vivo polymerization of the major curli subunit CsgA into β-sheet–rich amyloid fibers depends on the nucleating activity of the minor subunit CsgB31. CsgE, CsgF and CsgG are assembly factors required for the stabilization and transport of CsgA and CsgB to the cell surface to mediate fiber formation30,32. Curli protein profiles for MC4100 were obtained from bacteria grown on YESCA agar amended with various concentrations of each compound. Cell-associated CsgA levels in MC4100 were substantially reduced in a dose-dependent manner when bacteria were grown in the presence of 2, 3 and 4 (Fig. 1b). Compound 2 completely inhibited curli biogenesis at 1.0 mM, and compound 3 completely inhibited at 2.5 mM (Fig. 1b). Compound 4 exerted near-complete curli inhibition at 2.5 mM (Fig. 1b). There was less cell-associated CsgB at the highest concentration tested of 2 and 3 compared to cells incubated with no compound (Fig. 1b). CsgF and CsgG levels were approximately the same in cells grown in the presence or absence of compounds, suggesting that the compounds were specifically interfering with CsgA stability or polymerization (Fig. 1b).
Curlicides block UPEC amyloid biogenesis
E. coli curli biogenesis has been characterized most extensively in the avirulent E. coli K12 strain MC4100, for which curli expression is restricted to agar medium and low temperature3,4,33. We sought to (i) develop a broth-based assay for testing curlicides to enable future testing in high-throughput assays and (ii) examine whether curlicides influence curli assembly in pathogenic E. coli, where the regulation and expression of curli and other genes can differ from that in MC4100. We discovered that UTI89, a well-characterized uropathogenic E. coli strain, produced curli in liquid YESCA broth. Thus, we investigated the ability of curlicides to block UTI89 curli formation during growth in YESCA broth. Compounds 2, 3 and 4 blocked UTI89 curli biogenesis, and did so at lower concentrations (60–250 μM) relative to the agar assay (Fig. 1c). The difference may be attributable to uniform and better access of the curlicides to bacteria in broth. The relative potencies of the three curlicides were the same in the liquid and agar assays. Compound 2 was the most effective curli inhibitor in both MC4100 and UTI89, followed by 3 and 4 (Fig. 1b,c). Effects on curli production were also assessed by electron microscopy (Fig. 1d). The effect of each compound was titratable, as intermediate curliation patterns were observed in the electron micrographs at increasing curlicide concentrations (Fig. 1c,d). The original pilicide, 1, had no effect on curli production (Fig. 1d), and none of the compounds affected UTI89 growth (Supplementary Fig. 2).
Curlicides prevent CsgA polymerization in vitro
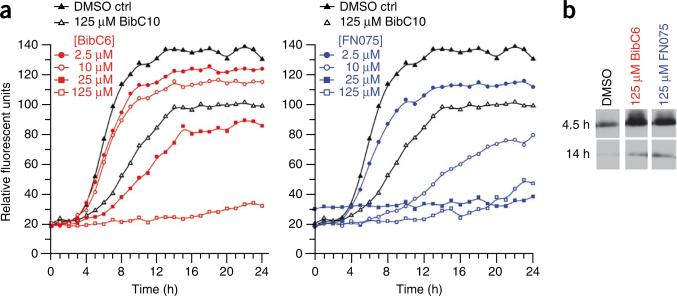
The hypothesis that the curlicides 2 and 3 were directly interfering with CsgA polymerization was investigated using an in vitro thioflavin-T (ThT) amyloid polymerization assay. CsgA was purified in a soluble, unstructured state and allowed to incubate in the presence or absence of curlicides. At a fivefold molar excess (125 μM), 2 and 3 both completely inhibited ThT fluorescence of 25 μM CsgA over time (Fig. 2a). Compound 2 inhibited ThT fluorescence at concentrations as low as 10 μM, representing a 2.5-fold excess of protein to compound (Fig. 2a). Direct comparison of the two compounds at 25 μM indicated partial inhibition of CsgA polymerization by 3 and complete inhibition by 2 at this concentration (Fig. 2a). Thus, 2 was more potent than 3 in this ThT assay and in blocking curli biogenesis in whole cells (Fig. 1). Increased levels of soluble CsgA were observed by immunoblot analysis when the protein was incubated in the presence of 125 μM of either compound (Fig. 2b), demonstrating that the compounds inhibited CsgA polymerization and not ThT binding to the fibers. To determine whether the observed inhibition of amyloid polymerization was specific to the curlicides 2 and 3, we also included the previously described pilicide, 1, which did not inhibit curli assembly. As expected, CsgA was able to polymerize into an insoluble amyloid fiber in the presence of a fivefold molar excess of 1 (Fig. 2a). Compound 4 could not be analyzed in the ThT assay due to high background fluorescence.
Figure 2.
Curlicide inhibition of in vitro CsgA polymerization. (a) The fluorescence of freshly purified CsgA mixed with 25 μM ThT, and respective curlicides, was measured at 495 nm after excitation at 438 nm. Data points corresponding to 60-min intervals are displayed. (b) Reductions in ThT fluorescence corresponded to reduced CsgA polymerization. Soluble CsgA levels were confirmed by SDS-PAGE and western analysis using anti-CsgA antibodies. Polymerized CsgA is SDS insoluble, and migration is impeded during electrophoresis.
Curlicides prevent amyloid-dependent pellicle formation
In order to investigate whether the curlicides inhibited UPEC biofilm formation, we identified a set of curli-dependent biofilm models using the UPEC strain UTI89. Pellicle formation—that is, biofilm formation at the air-liquid interface—requires interbacterial adhesion and the assembly of multicellular architectures that can differ from biofilms formed primarily on a solid surface, such as agar or plastic14. UTI89 pellicle formation during growth in YESCA broth was curli dependent: UTI89ΔcsgA, which is unable to produce curli due to deletion of the major subunit of the fiber, did not form pellicle under these conditions. Pellicle formation was rescued by complementation with plasmid pLR5, encoding csgA (Fig. 3a). Pellicle formation occurred only in YESCA broth and not in LB broth (Fig. 3a).
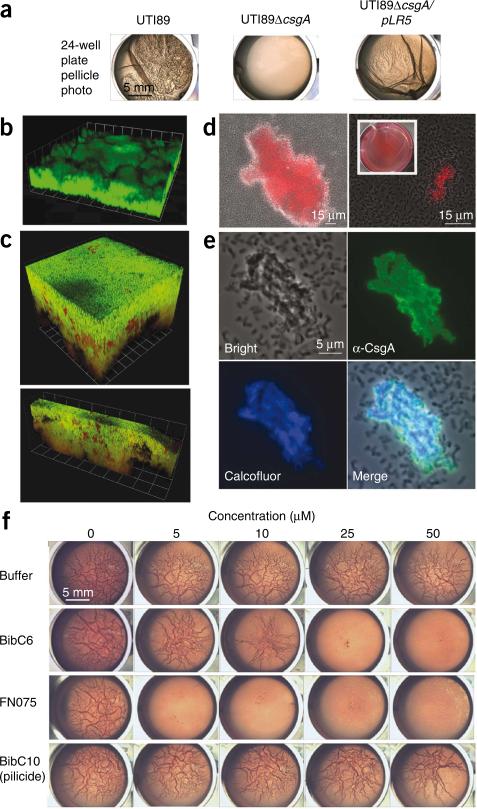
Figure 3.
Curlicide inhibition of curli-dependent UTI89 pellicle formation. (a) Macroscopic observation of curli-dependent pellicles. Pellicle biofilms were visible in wild-type UTI89 culture 48 h post-inoculation in YESCA broth. Deletion of csgA (UTI89ΔcsgA) abolished pellicle formation, and the pellicle was restored by providing csgA in trans (UTI89ΔcsgA/pLR5). (b) Three-dimensional confocal microscopy reconstructions revealed thick pellicles with complex architectures. Pellicles were stained with nucleic acid dye STYO9 (green). Each grid unit is 14.3 μm. (c) Three-dimensional reconstructions of confocal microscopy images showed that GFP reporter gene expression (green) driven by the csgB; csgA promoter is active in UTI89 pellicle. Bacteria were counterstained with nucleic acid dye SYTO83 (red). Grid units were 7.4 μm for the top panel and 14.3 μm for the bottom panel. (d) Curli were detected in the pellicle by Congo red uptake in bright field (DIC) images merged with the red fluorescence channel. Congo red uptake was apparent by macroscopic examination of pellicle (inset). (e) Immunostaining of pellicle segments with anti-CsgA antisera illustrated homogeneous CsgA presence among aggregated cells, whereas planktonic bacteria were unlabeled (top right panel). Calcofluor staining indicated that β-glucans such as cellulose colocalize in the pellicle (bottom panels). (f) Curlicides blocked pellicle formation in a 24-well plate assay in which UTI89 was grown in Congo red–containing YESCA medium at 30 °C (rows 2 and 3). Congo red had no effect on the results but permitted facile display of pellicle roughness. Equivalent carrier volumes of ethanol had no effect on pellicle formation (top row), and pellicle was unaffected by pilicide BibC10, 1 (bottom row).
The macroscopic and microscopic features of the pellicle were analyzed by confocal microscopy. Bacteria assembled a dense three-dimensional multicellular community throughout the pellicle (Fig. 3b). Curli gene expression in the pellicle was monitored by integrating a single-copy chromosomal transcriptional gfp reporter into the λ integration site of strain UTI89 (Fig. 3c). The pellicle was counterstained with the nucleic acid dye SYTO83, which stained the GFP-negative bacteria red. Nearly the entire pellicle was GFP positive, although patches of red-staining bacteria were observed, indicating that curli were expressed throughout most of the pellicle (Fig. 3c). This was characterized further by Congo red staining and immunofluorescence using anti-CsgA antibodies4. Congo red is an amyloid dye that binds to curli and curliated bacteria4. Pellicle bacteria, but not planktonic cells, bound Congo red from the broth medium as visualized both microscopically and macroscopically (Fig. 3d). The immunofluorescence micrographs revealed that CsgA-specific antibodies bound to pellicle-associated bacteria but not to planktonic bacteria (Fig. 3e). Finally, cellulose is known to contribute to multicellular behavior and biofilm formation among the Enterobacteriaceae34. Calcofluor staining and fluorescence confirmed the presence of β-2-glucans such as cellulose in the pellicle (Fig. 3e).
The ability of curlicides to block pellicle formation was tested during growth of UPEC in YESCA broth. UTI89 pellicle formation was examined visually using a 24-well plate assay in the presence and absence of curlicides and the pilicide 1 (Fig. 3f). The curlicides 3 and 2 blocked pellicle formation completely at concentrations of 25 and 5 μM, respectively (Fig. 3f). Compound 4 did not inhibit pellicle formation in this low concentration range and was not tested further in this assay. DMSO carrier and the pilicide 1 had no effect on pellicle formation up to 500 μM (Fig. 3f).
Curlicides prevent biofilm formation on plastic
In another assay, we found that curli were required for biofilm formation on polyvinyl chloride (PVC) when the bacteria were grown in YESCA broth in an assay in which bacteria adhere to and form biofilms in PVC microtiter plates35. In contrast, type 1 pili were required for biofilm formation in this assay when the bacteria were grown in LB in PVC wells24. We confirmed that UTI89ΔfimA, which is unable to produce the major pilus subunit24, and UTI89ΔfimH, which is unable to produce the mannose-binding adhesin36, were both unable to produce biofilms when grown in LB broth in PVC wells (Fig. 4a). Biofilm formation in this LB/PVC condition depended on the expression of type 1 pili but not on curli, since UTI89ΔcsgA formed biofilms comparable to those formed by UTI89 under these conditions (Fig. 4a). In contrast, when grown in YESCA broth in PVC wells, curli, but not type 1 pili, were required for biofilm formation. UTI89ΔcsgA did not form appreciable biofilm, and UTI89ΔfimA and UTI89ΔfimH both formed biofilms comparable to those formed by UTI89 under these YESCA/PVC conditions (Fig. 4a).
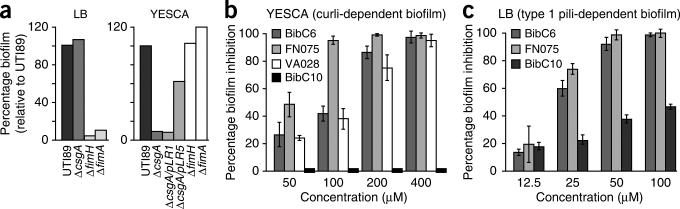
Figure 4.
Curlicide inhibition of UTI89 amyloid-dependent and type 1 pili-dependent biofilm formation on PVC. (a) Curli were dispensable for UTI89 biofilm formation in LB but were required for YESCA-grown biofilms. (b) UTI89 biofilm formation in YESCA in 96-well PVC plates was inhibited in the presence of curlicides, as quantified by crystal violet staining35. (c) FN075 inhibited UTI89 biofilm formation on PVC in LB in a dose-dependent manner. Percent biofilm inhibition represents the decrease in biofilm formation in the presence of curlicide relative to biofilm formation when no compound was present. Bars in b and c represent s.d. of the mean of four experiments, where controls with no compound were included in each experiment on each plate. No effects on bacterial growth rate were observed at these compound concentrations (Supplementary Fig. 2).
Curli-dependent biofilms that formed when UTI89 was grown in YESCA in PVC wells were inhibited from forming by all three curlicides (Fig. 4b). Compound 2 was the most potent inhibitor in this assay and inhibited observable biofilm by 50% at a concentration of 50 μM; over 90% inhibition was achieved at 100 μM (Fig. 4b). Compounds 3 and 4 inhibited YESCA/PVC biofilm formation by nearly 50% at 100 μM and exerted near-complete and complete inhibition at 200 μM and 400 μM, respectively. The pilicide 1 was unable to inhibit the curli-dependent biofilm that formed during growth in YESCA (Fig. 4b). Thus, curli-dependent biofilms (YESCA/PVC) were inhibited from forming by the curlicides, but not by the pilicide 1.
As described earlier, evaluation of the structure of the binding site identified previously for pilicides24 revealed that compounds with a larger aryl substituent, such as the best curlicides 2 and 3, should fit well and retain the pilicide activity of the parent compound, 1. Thus, we tested the ability of 2 and 3 to inhibit type 1 pili-dependent biofilms formed by UTI89 when grown in LB broth in PVC wells. The original pilicide 1 was included as a control. Both compounds inhibited the formation of the type 1 pili-dependent LB/PVC biofilm and were more potent than 1. At compound concentrations of 50 μM, 2 and 3 resulted in 99 and 92% reduction of biofilm formation, respectively, whereas biofilm formation in the presence of 50 μM compound 1 was reduced by only 38% (Fig. 4c).
Curli provide fitness advantage early in UTI pathogenesis
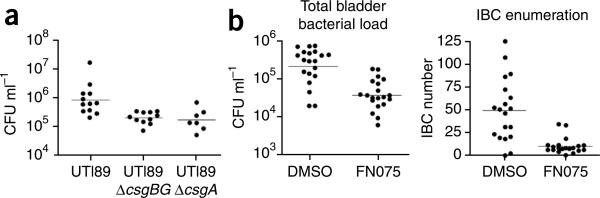
Encouraged by the ability of the best curlicide in this study, 2, to inhibit type 1 pilus-dependent LB/PVC biofilm formation in addition to curli-dependent YESCA/PVC biofilm and pellicle formation, we hypothesized that 2 might have therapeutic efficacy in a mouse model of UTI. The pathogenesis of UTI in the murine model system involves a series of type 1 pili-dependent steps including colonization and invasion of the bladder epithelium and the formation of intracellular bacterial communities. Although type 1 pili are required, some substantial colonization differences have been observed among type 1 pili-expressing UPEC isolates in vivo, suggesting that factors independent of type 1 pili impact adhesion, colonization and virulence37. Many such factors have been identified, including capsule, flagella and siderophores38. We hypothesized that curli, as adhesive amyloid and biofilm-promoting fibers, may aid in bacterial colonization. To test this hypothesis, we investigated the virulence of two mutant strains of UTI89 unable to make curli (UTI89ΔcsgBG and UTI89ΔcsgA) in a murine UTI model. The bladders of C3H/HeN mice were challenged separately with E. coli strains UTI89, UTI89ΔcsgBG and UTI89ΔcsgA, and colony forming units (CFU) from bladder homogenates were determined 6 h post infection (hpi). A significant reduction in CFU was observed for colonization by the curli mutants with respect to wild-type UTI89 (P < 0.007, P < 0.011, respectively), arguing that curli provide a fitness advantage in this model (Fig. 5a).
Figure 5.
FN075 attenuation of virulence in vivo. (a) Curli mutants, UTI89ΔcsgBG and UTI89ΔcsgA, were significantly attenuated in bladder colonization compared to UTI89, as determined by CFU counts (P < 0.007 and P < 0.011, respectively, as determined by a two-tailed Mann Whitney for each mutant–wild-type pair). Geometric means are indicated by solid lines. (b) FN075 (2) reduced virulence of UTI89 in the mouse model of urinary tract infection. Bacteria were grown in the presence of 250 μM FN075 and then introduced into bladders of 7- to 8-week-old female C3H/HeN mice by a single transurethral inoculation of 1–2 × 107 CFU. FN075-treated cells exhibited more than a 10-fold decrease in CFU/bladder at 6 hpi (P < 0.0001, two-tailed Mann-Whitney test) and produced significantly fewer IBCs than nontreated bacteria (P < 0.0001).
Compound 2 attenuates virulence in vivo
We hypothesized that compounds interrupting type 1 pilus assembly and curli biogenesis should attenuate virulence. C3H/HeN mice were challenged with E. coli strain UTI89 grown under type 1 pili-inducing conditions in the absence or presence of 250 μM compound 2. UTI89 that had been pretreated with 2 was significantly (P < 0.0001) attenuated at 6 hpi. The total bacterial load per bladder determined by CFU counts from bladder homogenates was reduced greater than 10-fold at 6 hpi (Fig. 5b). UTI89 grown in the presence of FN075 produced few or no IBCs compared to the wild-type control, as quantified by LacZ staining of whole bladders39 in which IBCs are readily discernable (Fig. 5b).
DISCUSSION
Bacterial pathogens often rely on multiple and redundant virulence determinants23. Biofilm formation is often a key part of the pathogenic cascade, and is implicated in UTI, cystic fibrosis and other chronic infectious diseases10,11,14. Because biofilm formation is multifactorial, considering approaches that simultaneously target multiple factors that contribute to biofilm formation and virulence will facilitate development of anti-virulence compounds40. Curli and type 1 pili are examples of UPEC biofilm determinants that are essential under distinct environmental conditions. Type 1 pili produced by UPEC are essential in UTI pathogenesis, while curli were also found to provide a fitness advantage in vivo. By dissecting the requirements of curli and type 1 pili in UPEC biofilm formation, we have discovered a new class of biofilm inhibitors termed curlicides.
Compounds 2 and 3 block UPEC curli formation in vivo and CsgA polymerization in vitro, and thus are referred to as curlicides. 2 and 3 share a common structural scaffold with previously described pilicides (compound 1) that inhibit UPEC type 1 pili-dependent biofilm formation. Pilicides block pilus biogenesis by preventing chaperone-subunit complexes from interacting with the outer membrane usher24. The crystal structure of a pilicide-chaperone complex, in which the pilicide is a morpholine-substituted analog of 1, indicates that the pilicide binds to a conserved hydrophobic patch on strands F1, C1 and D1 on the back of the N-terminal domain of the pilus chaperone24. This surface of the chaperone is involved in interactions with the outer membrane usher; therefore, pilicides interfere with chaperone-subunit interactions with the usher24. Based on the chaperone-pilicide crystal structure, we rationalized that compounds with larger aryl substituents, such as 2 and 3, might exhibit equal or better structural complementarity in the pilicide binding site. Compounds 2 and 3, which block the formation of curli-dependent biofilms, also retained pilicide activity as measured by their ability to block type 1 pili-dependent biofilms. Therefore, in a multiple-target approach, we identified individual compounds that exhibited a dual mode of action by acting as curlicides and pilicides.
During a UTI, type 1 pili are required for colonization and invasion of the bladder epithelium. Yet bladder infection by UPEC is dependent on the outcome of a series of complex host-pathogen interactions and the molecular cross-talk that occurs as a consequence. Type 1 pili mediate the invasion of UPEC into the bladder epithelium41. This process has been reported to involve lipid raft components such as caveolin-1 (an integral membrane protein found in the inner leaflet of the lipid bilayer), the GTPases Rac1 and dynamin-2 (refs. 42,43), and microtubules44. After invasion, UPEC can be harbored in fusiform vesicles wherein they are expelled from the cell42,45, presumably as part of an innate defense46. When UPEC escape into the cytoplasm of superficial umbrella cells, they are able to rapidly replicate into IBCs16–18,37,46. As the IBC matures, UPEC detach from the biomass and spread to neighboring epithelial cells where further rounds of the IBC perpetuate the massive population expansion of the bacteria as part of a mechanism to subvert host defenses and gain a critical foothold17. Various adhesion systems, toxins, autotransporters, iron acquisition molecules and biofilm-associated factors all contribute to the fitness of UPEC in the urinary tract. The redundancy of these factors provides a competitive advantage to UPEC within the urinary tract. Mutations in any one of these factors result in varying degrees of reduced fitness as measured by luminal colonization, invasion, formation of IBCs and other parameters18,47,48. Fitness advantages conferred in the acute stages of infection are likely to impact the long-term outcome of disease, especially when subjected to severe selective pressure, as is thought to be the case in the course of a natural urinary tract infection. We found that curli are an example of a virulence determinant that provides a fitness advantage in the acute stage of infection, as we showed that curli mutants are attenuated in vivo at 6 hpi.
Pretreatment of UPEC with compound 2, which inhibits both curli and type 1 pili biogenesis, reduced the ability of bacteria to colonize the bladder and prevented the development of IBCs. Thus, the ability of compound 2 to reduce UPEC bladder colonization can be attributed to inhibition of curli and/or type 1 pili. Because biofilm formation is multifactorial and intimately associated with pathogenic cascades of several bacteria, it is attractive to consider the simultaneous targeting of multiple biofilm-promoting factors. Compounds such as 2, with its curlicide-pilicide dual mode of action, accomplish this objective by inhibiting type 1 pili and curli assembly. The dual activity enhances the therapeutic potential of 2, particularly given the crucial role of type 1 pili during urinary tract infection. These results also formulate a new foundation for research that dissects the molecular role of curli during UPEC pathogenesis and, more generally, of bacterial amyloid production in the host.
Since the discovery of curli in 1989 (ref. 3) and the identification of curli as amyloid in 2002 (ref. 4), we now appreciate that functional amyloids are prevalent among microorganisms, and that they are also an integral part of normal mammalian cellular physiology49,50. Assembly of functional amyloids is regulated in order to direct polymerization at the right time and place, and to prevent toxicity. The continued dissection of amyloidogenesis in these systems and our ability to interfere with gene-directed amyloid assembly will enhance our perspective on functional amyloid folding pathways and disease-associated amyloidosis.
METHODS
Synthesis of ring-fused 2-pyridones
Full synthetic schemes and supporting data are described in the Supplementary Methods.
Whole-cell curli biogenesis inhibition assays
BibC10 (1), FN075 (2), BibC6 (3) and VA028 (4) stock solutions were prepared at 70 mM in DMSO. For MC4100 assays (Fig. 1b), appropriate volumes of curlicides were added to 15 ml conical tubes with 6 ml warm YESCA agar to yield desired final concentrations. YESCA agar contains 1 g yeast extract, 10 g casamino acids and 20 g agar per liter. Tubes were inverted three times and poured into 35 × 10 mm petri dishes. After 48 h growth at 26 °C, cells were collected, resuspended in phosphate-buffered saline (PBS) and normalized by cell density (to measured optical density at 600 nm (OD600) = 0.5). 1 ml aliquots of normalized cells were pelleted and resuspended in 100 μl hexafluoroisopropanol (HFIP) to dissociate curli. HFIP was removed by vacuum centrifugation. Pellets were resuspended in 200 μl SDS-PAGE loading buffer and analyzed by immunoblot analysis using curli-specific antibodies. Identical results were obtained for cells subjected to formic acid treatment in place of HFIP. A control sample contained DMSO carrier at the highest equivalent volume tested corresponding to 2.5 mM compound (3.6% DMSO v/v).
In the UTI89 whole-cell curli biogenesis assay, appropriate volumes of compound stock solutions were added to 3 ml YESCA broth in sterile glass test tubes and inoculated with 3 μl of an overnight culture grown in LB. YESCA cultures were incubated overnight, shaking at 100 rpm at 28 °C. Cells were pelleted, resuspended in PBS and processed as above using formic acid treatment for curli dissolution.
Soluble CsgA purification
C-terminal His6-tagged CsgA4 was purified using a modified denaturing protocol. NEB 3016 slyD cells harboring a pET11d vector containing a sec signal sequence minus CsgA were grown to OD600 0.9, induced with 0.5 mM IPTG, and incubated for 1 h at 37 °C. Cells were collected by centrifugation and stored at −80 °C. Cells were lysed by addition of an 8 M guanidine hydrochloride, 50 mM potassium phosphate solution. 50 ml of lysis solution was used per pellet generated from a 500 ml culture. The lysate was incubated, with stirring, at 4 °C for 48 h. The insoluble portion of the lysate was removed by centrifuging at 10,000g at 4 °C for 15 min, and the resulting supernatant was incubated with NiNTA resin (Sigma) for 1 h at room temperature (18–22 °C) with rocking. CsgA was purified using nickel affinity beads in a Kontes column. The beads were washed with 50 mM potassium phosphate. CsgA was eluted from the column using a 0.5 M imidazole 50 mM potassium phosphate solution. Fractions were collected and analyzed for the presence of protein using the BCA assay (Thermo) and SDS-PAGE.
Biofilm assay
Bacteria were grown in YESCA or LB broth in wells of PVC microtiter plates in the presence of individual curlicides. After 48 h of growth, wells were rinsed and stained with crystal violet, and biofilms were quantified as described35.
Bacterial strains
UTI89ΔcsgA, UTI89ΔcsgA/pLR1, UTI89ΔcsgA/pLR5 and the curli promoter–gfp transcriptional fusion construction are described in the Supplementary Methods.
Mouse infection and bladder tissue bacterial titer determinations
All animal studies using mice were approved by the Animal Studies Committee of Washington University (Animal Protocol Number 20070029). Bacteria were grown under type 1 pili-inducing conditions, in the presence or absence of 250 μM FN075 (2). The bacteria were harvested and resuspended to an OD600 of 0.5 in PBS. 50 μl of this suspension, resulting in a 1–2 × 107 inoculum, was instilled transurethrally into the mouse bladder. After 6 h, mice were killed, and bladders were aseptically removed and homogenized in 1 ml PBS, serially diluted and plated onto LB agar plates. CFU was enumerated after 16 h of growth at 37 °C.
Enumeration of bladder IBCs
Bacteria were grown under type 1 pili-inducing conditions, in the presence or absence of 250 μM FN075. The bacteria were harvested and resuspended to an OD600 of 0.5 in PBS. 50 μl of this suspension, resulting in a 1–2 × 107 inoculum, was instilled transurethrally into the bladder. After 6 h, mice were killed, and bladders were aseptically removed, bisected, splayed on silicone plates and fixed in 2% paraformaldehyde (v/v). IBCs were quantified by LacZ staining of whole bladders. IBCs are readily discernable as punctate violet spots39.
Supplementary Material
ACKNOWLEDGMENTS
We gratefully acknowledge the expertise of W. Beatty (Imaging Facility, Washington University School of Medicine) and of A. Olofsson (Umeå Centre for Molecular Pathogenesis, Umeå University). This study was supported in part by the Swedish Natural Science Research Council and the Knut and Alice Wallenberg Foundation. The authors acknowledge funding from the US National Institutes of Health to S.J.H. (AI02549, AI048689, AI049950 and P50 DK64540), M.R.C. (AI073847), P.C.S. (K12HD00850 and K08DK074443) and L.C. (T32A107172). L.C. holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Footnotes
Note: Supplementary information and chemical compound information is available on the Nature Chemical Biology website.
Published online at http://www.nature.com/naturechemicalbiology/.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Ryu JH, Beuchat LR. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and Curli production on its resistance to chlorine. Appl. Environ. Microbiol. 2005;71:247–254. doi: 10.1128/AEM.71.1.247-254.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsen A, Jonsson A, Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 4.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cegelski L, Smith CL, Hulgren SJ. Microbial adhesion. In: Schaechter M, editor. Encyclopedia of Microbiology. Academic Press; New York: 2009. pp. 1–10. [Google Scholar]
- 7.Uhlich GA, Cooke PH, Solomon EB. Analyses of the red-dry-rough phenotype of an Escherichia coli O157:H7 strain and its role in biofilm formation and resistance to antibacterial agents. Appl. Environ. Microbiol. 2006;72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 9.Zogaj X, Bokranz W, Nimtz M, Romling U. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsek MR, Singh PK. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 11.Donlan RM, Costerton JW. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larsen P, et al. Amyloid adhesins are abundant in natural biofilms. Environ. Microbiol. 2007;9:3077–3090. doi: 10.1111/j.1462-2920.2007.01418.x. [DOI] [PubMed] [Google Scholar]
- 13.Larsen P, Nielsen JL, Otzen D, Nielsen PH. Amyloid-like adhesins produced by floc-forming and filamentous bacteria in activated sludge. Appl. Environ. Microbiol. 2008;74:1517–1526. doi: 10.1128/AEM.02274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Branda SS, Vik S, Friedman L, Kolter R. Biofilms: the matrix revisited. Trends Microbiol. 2005;13:20–26. doi: 10.1016/j.tim.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 15.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 16.Anderson GG, et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science. 2003;301:105–107. doi: 10.1126/science.1084550. [DOI] [PubMed] [Google Scholar]
- 17.Justice SS, et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. USA. 2004;101:1333–1338. doi: 10.1073/pnas.0308125100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright KJ, Seed PC, Hultgren SJ. Development of intracellular bacterial communities of uropathogenic Escherichia coli depends on type 1 pili. Cell. Microbiol. 2007;9:2230–2241. doi: 10.1111/j.1462-5822.2007.00952.x. [DOI] [PubMed] [Google Scholar]
- 19.Aberg V, Almqvist F. Pilicides-small molecules targeting bacterial virulence. Org. Biomol. Chem. 2007;5:1827–1834. doi: 10.1039/b702397a. [DOI] [PubMed] [Google Scholar]
- 20.Reisner A, Haagensen JA, Schembri MA, Zechner EL, Molin S. Development and maturation of Escherichia coli K-12 biofilms. Mol. Microbiol. 2003;48:933–946. doi: 10.1046/j.1365-2958.2003.03490.x. [DOI] [PubMed] [Google Scholar]
- 21.Pratt LA, Kolter R. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type 1 pili. Mol. Microbiol. 1998;30:285–293. doi: 10.1046/j.1365-2958.1998.01061.x. [DOI] [PubMed] [Google Scholar]
- 22.Saldana Z, et al. Synergistic role of curli and cellulose in cell adherence and biofilm formation of attaching and effacing Escherichia coli and identification of Fis as a negative regulator of curli. Environ. Microbiol. 2009;11:992–1006. doi: 10.1111/j.1462-2920.2008.01824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. The biology and future prospects of antivirulence therapies. Nat. Rev. Microbiol. 2008;6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pinkner JS, et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl. Acad. Sci. USA. 2006;103:17897–17902. doi: 10.1073/pnas.0606795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emtenas H, Alderin L, Almqvist F. An enantioselective ketene-imine cycloaddition method for synthesis of substituted ring-fused 2-pyridinones. J. Org. Chem. 2001;66:6756–6761. doi: 10.1021/jo015794u. [DOI] [PubMed] [Google Scholar]
- 26.Emtenas H, Taflin C, Almqvist F. Efficient microwave assisted synthesis of optically active bicyclic 2-pyridinones via delta2-thiazolines. Mol. Divers. 2003;7:165–169. doi: 10.1023/b:modi.0000006800.46154.99. [DOI] [PubMed] [Google Scholar]
- 27.Jones CH, et al. FimC is a periplasmic PapD-like chaperone that directs assembly of type 1 pili in bacteria. Proc. Natl. Acad. Sci. USA. 1993;90:8397–8401. doi: 10.1073/pnas.90.18.8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung DL, Hultgren SJ. Pilus biogenesis via the chaperone/usher pathway: an integration of structure and function. J. Struct. Biol. 1998;124:201–220. doi: 10.1006/jsbi.1998.4049. [DOI] [PubMed] [Google Scholar]
- 29.Aberg V, et al. Microwave-assisted decarboxylation of bicyclic 2-pyridone scaffolds and identification of Abeta-peptide aggregation inhibitors. Org. Biomol. Chem. 2005;3:2817–2823. doi: 10.1039/b503294f. [DOI] [PubMed] [Google Scholar]
- 30.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol. Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc. Natl. Acad. Sci. USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc. Natl. Acad. Sci. USA. 2009;106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen A, Arnqvist A, Hammar M, Normark S. Environmental regulation of curli production in Escherichia coli. Infect. Agents Dis. 1993;2:272–274. [PubMed] [Google Scholar]
- 34.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 36.Rosen DA, et al. Molecular variations in Klebsiella pneumoniae and Escherichia coli FimH affect function and pathogenesis in the urinary tract. Infect. Immun. 2008;76:3346–3356. doi: 10.1128/IAI.00340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garofalo CK, et al. Escherichia coli from urine of female patients with urinary tract infections is competent for intracellular bacterial community formation. Infect. Immun. 2007;75:52–60. doi: 10.1128/IAI.01123-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beloin C, Roux A, Ghigo JM. Escherichia coli biofilms. Curr. Top. Microbiol. Immunol. 2008;322:249–289. doi: 10.1007/978-3-540-75418-3_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Justice SS, Lauer SR, Hultgren SJ, Hunstad DA. Maturation of intracellular Escherichia coli communities requires SurA. Infect. Immun. 2006;74:4793–4800. doi: 10.1128/IAI.00355-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keith CT, Borisy AA, Stockwell BR. Multicomponent therapeutics for networked systems. Nat. Rev. Drug Discov. 2005;4:71–78. doi: 10.1038/nrd1609. [DOI] [PubMed] [Google Scholar]
- 41.Martinez JJ, Mulvey MA, Schilling JD, Pinkner JS, Hultgren SJ. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 2000;19:2803–2812. doi: 10.1093/emboj/19.12.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan MJ, Li G, Shin JS, Carson JL, Abraham SN. Bacterial penetration of bladder epithelium through lipid rafts. J. Biol. Chem. 2004;279:18944–18951. doi: 10.1074/jbc.M400769200. [DOI] [PubMed] [Google Scholar]
- 43.Terada N, et al. Involvement of dynamin-2 in formation of discoid vesicles in urinary bladder umbrella cells. Cell Tissue Res. 2009;337:91–102. doi: 10.1007/s00441-009-0804-z. [DOI] [PubMed] [Google Scholar]
- 44.Dhakal BK, Mulvey MA. Uropathogenic Escherichia coli invades host cells via an HDAC6-modulated microtubule-dependent pathway. J. Biol. Chem. 2009;284:446–454. doi: 10.1074/jbc.M805010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bishop BL, et al. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nat. Med. 2007;13:625–630. doi: 10.1038/nm1572. [DOI] [PubMed] [Google Scholar]
- 46.Mulvey MA, et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science. 1998;282:1494–1497. doi: 10.1126/science.282.5393.1494. [DOI] [PubMed] [Google Scholar]
- 47.Lloyd AL, Henderson TA, Vigil PD, Mobley HL. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 2009;191:3469–3481. doi: 10.1128/JB.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright KJ, Seed PC, Hultgren SJ. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect. Immun. 2005;73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hammer ND, Wang X, McGuffie BA, Chapman MR. Amyloids: friend or foe? J. Alzheimers Dis. 2008;13:407–419. doi: 10.3233/jad-2008-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Badtke MP, Hammer ND, Chapman MR. Functional amyloids signal their arrival. Sci. Signal. 2009;2:pe43. doi: 10.1126/scisignal.280pe43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.