Abstract
The human immunodeficiency virus type 1 (HIV-1) maturation inhibitor bevirimat disrupts virus replication by inhibiting the cleavage of the capsid-spacer peptide 1 (CA-SP1) Gag processing intermediate to mature CA. The observation that bevirimat delays but does not completely block CA-SP1 processing suggests that the presence of uncleaved CA-SP1 may disrupt the maturation process in trans. In this study, we validate this hypothesis by using a genetic approach to demonstrate that a non-cleavable CA-SP1 mutant exerts a dominant-negative effect on maturation of wild-type HIV-1. In contrast, a mutant in which cleavage can occur internally within SP1 is significantly less potent as a dominant-negative inhibitor. We also show that bevirimat blocks processing at both the major CA-SP1 cleavage site and the internal site. These data underscore the importance of full CA-SP1 processing for HIV-1 maturation and highlight the therapeutic potential of inhibitors that target this Gag cleavage event.
Keywords: HIV-1, Gag, virus maturation, assembly, retrovirus, protease
Introduction
Maturation of human immunodeficiency virus type 1 (HIV-1) virions occurs concomitant with virus budding and is triggered by the proteolytic cleavage of the Gag precursor protein, Pr55Gag, and the Gag-Pol precursor, Pr160Gag-Pol, into the mature Gag proteins and pol-encoded enzymes. Cleavage of Pr55Gag by the viral protease (PR) occurs sequentially to yield matrix (MA; p17), capsid (CA; p24), spacer peptide 1 (SP1), nucleocapsid (NC; p7), spacer peptide 2 (SP2), and p6 (Fig. 1A) (Adamson and Freed, 2007; Adamson, Salzwedel, and Freed, 2009; Freed, 1998; Swanstrom, 1997; Vogt, 1996). In addition to the major Gag cleavage sites mentioned above, a secondary site has also been reported within SP1 (Henderson, 1988; Kräusslich et al., 1995). The ordered nature of Gag cleavage is a consequence of substantial differences in the rates at which individual processing sites are cleaved by PR (Erickson-Viitanen et al., 1989; Kräusslich et al., 1995; Pettit et al., 2002; Pettit et al., 1994; Tritch et al., 1991). Following its liberation from the Gag precursor, the mature CA protein reassembles into a hexameric lattice that forms the mature, conical viral core inside which are localized the viral RNA genome, the mature NC protein, and the viral enzymes reverse transcriptase (RT) and integrase (IN) (Ganser-Pornillos, Yeager, and Sundquist, 2008). Formation of the conical core is essential for virus infectivity. A number of studies have demonstrated that not only the full completion but also the precise ordering of Gag processing is required for proper virion maturation (Kaplan et al., 1993; Pettit et al., 1994).
Fig. 1.
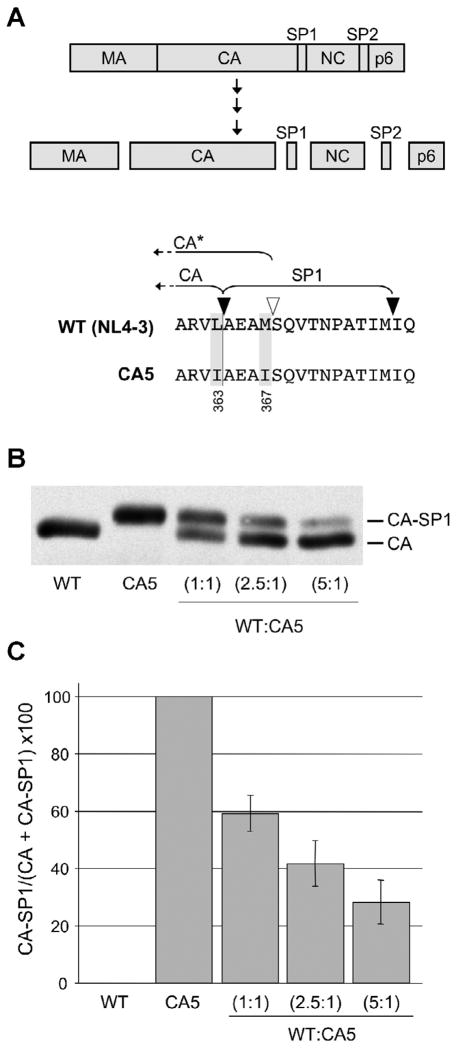
Levels of uncleaved CA and CA-SP1 in WT and WT/CA5 mixed virions. (A, Upper panel). Proteolytic processing of the HIV-1 Gag precursor protein, Pr55Gag. Major Gag domains – matrix (MA), capsid (CA), nucleocapsid (NC), and p6 - and spacer peptides SP1 and SP2 are indicated. Final Gag processing products are shown. (A, Lower panel). WT (NL4-3) and CA5 amino acid sequence spanning the CA-SP1 junction. Mutations in CA5 are highlighted and the Gag amino acid numbers are provided. PR cleavage sites flanking SP1 are shown with black arrowheads. The white arrowhead indicates the secondary, internal cleavage site within SP1. CA* represents CA plus a four-amino-acid extension into SP1, which accumulates in samples containing the L363I mutation. (B) Western blot of CA-SP1 and CA in particles generated by WT, CA5 or a mixture of these clones at the indicated DNA ratios. (C) Quantification of CA and CA-SP1 in virus particles. Percentages of CA-SP1 relative to total CA + CA-SP1 were determined using quantitative Western analysis. Error bars indicate standard deviation (n = 4).
PR-mediated cleavage at both the N- and C-termini of CA appears to influence CA organization and core condensation. Cleavage at the N-terminus enables the formation of a β-hairpin that is essential for conical core assembly (von Schwedler et al., 1998). The presence of only a few residues of MA at the N-terminus of CA abolishes the assembly of CA cylinders or tubes in vitro (Gross et al., 1998; von Schwedler et al., 1998); CA in these structures is organized in a manner analogous to that observed in mature cores (Ganser et al., 1999; Li et al., 2000). It has been proposed that the residues spanning the CA-SP1 junction form an α-helix that plays an important role in particle formation (Accola, Höglund, and Göttlinger, 1998; Accola, Strack, and Göttlinger, 2000; Kräusslich et al., 1995; Liang et al., 2002; Morellet et al., 2005). In vitro assembly studies demonstrated that the presence of SP1 attached to the C-terminus of CA was important for the formation of Gag spheres that resembled immature virus particles, whereas the removal of SP1 favored the assembly of tubes (Gross et al., 2000).
3-O-(3′,3′-dimethylsuccinyl) betulinic acid (DSB), also known as PA-457 or bevirimat (BVM), is a potent inhibitor of HIV-1 maturation (Li et al., 2003; Zhou et al., 2004). It acts by specifically blocking cleavage of SP1 from the C-terminus of CA, resulting in an accumulation of the Gag processing intermediate, p25 (CA-SP1) (Fig. 1A) (Li et al., 2003; Zhou et al., 2004). Virus particles that assemble in the presence of BVM fail to undergo proper maturation and do not form conical cores. Rather, they exhibit an acentric accumulation of electron density and a crescent-shaped structure adjacent to the viral membrane (Adamson et al., 2006; Li et al., 2003). Previous studies demonstrated that BVM does not completely block CA-SP1 processing but acts as a kinetic inhibitor to delay PR-mediated cleavage at this site (Zhou et al., 2004). These observations suggest that uncleaved CA-SP1 may be capable of acting in trans to block virion maturation.
In this study, we used a genetic approach to investigate the ability of uncleaved CA-SP1 to interfere with HIV-1 maturation and infectivity. We observe that coexpression of WT HIV-1 with a mutant in which the major CA-SP1 cleavage site and the internal SP1 site have been inactivated by mutation markedly interferes with maturation and infectivity. These data highlight the therapeutic potential of inhibitors that, like BVM, block or delay PR-mediated processing at the CA-SP1 cleavage site.
Materials and methods
Cell lines, plasmids, and transfections
Cell lines TZM-bl (obtained from J. Kappes through the NIH AIDS Research and Reference Reagent Program, Bethesda, MD) and HeLa were maintained in Dulbecco’s modified Eagle’s medium (DMEM). Jurkat T cells were maintained in RPMI-1640 medium. All media were supplemented with 10% fetal bovine serum (FBS; HyClone), penicillin, streptomycin, and glutamine (Gibco). Prior to transfection, 6-well cell culture plates (BD Falcon) were seeded with 0.6 × 106 HeLa cells/well. Cells were transfected using Lipofectamine LTX Reagent (Invitrogen) and a total of 3 μg DNA, containing a mixture of an HIV-1 molecular clone (pNL4-3) (Adachi et al., 1986) and the full-length CA5 mutant provirus (kindly provided by Hans-Georg Kräusslich) (Wiegers et al., 1998) or a Gag L363I mutant (kindly provided by S.-K. Lee and R. Swanstrom) (Lee, Harris, and Swanstrom, 2009). The Gag M367I mutant was created by mutagenesis of pNL4-3 DNA with synthetic complementary oligonucleotides (Invitrogen) (5′-CAT AAA GCA AGA GTT TTG GCT GAA GCA ATA AGC CAA GTA ACA AAT-3′) using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene), and verified by DNA sequencing. The change from the WT DNA sequence is underlined.
Infectivity assays
Virus stocks used for infections were collected by 0.45 μm filtration of supernatant from transfected HeLa cells, then normalized for reverse transcriptase (RT) activity as previously described (Willey et al., 1988). For single-cycle infectivity assays, 5 × 104 TZM-bl cells/well were infected in the presence of 20 μg DEAE-dextran (GE Healthcare) per ml. Infected cells were lysed 48 hr postinfection using Glo Lysis buffer (Promega) and assayed for luciferase activity using Luciferase Assay Kit (Promega) as previously described (Kiernan and Freed, 1998). For analysis of replication kinetics in multiple-round spreading infections, 0.5 × 106 Jurkat cells were infected with 10,000 RT cpm of cell-free virus and passaged at a 1:3 dilution every 2–3 days, at which time culture supernatants were collected to monitor RT activity (Freed and Martin, 1994). For infectivity assays in the presence of BVM, HeLa cells were seeded at 0.25 × 106 cells/well in 12-well cell culture plates (BD Falcon), transfected with 1 μg of DNA (pNL4-3, Gag L363I, or Gag M367I molecular clones) using Lipofectamine LTX, and incubated at 37°C in medium containing 1 μg/ml BVM or an equivalent concentration of DMSO. At ~18 hr posttransfection, cells were washed briefly and incubated at 37°C in medium containing 1 μg/ml BVM or DMSO. After 4 hr incubation, viruses were filtered from the supernatant as described above. TZM-bl cells were then infected with serial dilutions of virus, assayed for luciferase activity as described above, and normalized for virus input by RT activity.
Virus particle purification, Western blotting, and radioimmunoprecipitation analysis
Virus particles were filtered from transfected HeLa cell supernatants at 24 hr posttransfection and pelleted by ultracentrifugation at 100,000 × g for 45 min, then lysed in cell lysis buffer [0.5% Triton X-100, 300 mM NaCl, 50 mM Tris pH 7.5, and Complete protease inhibitor cocktail (Roche)]. Proteins were separated by SDS-PAGE (12% acrylamide) and transferred to nitrocellulose membranes using the iBlot Dry Blotting System (Invitrogen). Gag proteins were detected with HIV-Ig (NIH AIDS Research and Reference Reagent Program) at a 1:50,000 dilution, followed by HRP-conjugated sheep anti-human IgG (GE Healthcare), and visualized by either Western Lightning (Perkin-Elmer) or SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Scientific) and exposed to X-ray film. Levels of p24 (CA) and p25 (CA-SP1) from virus lysates were quantified using a FluorChem SP Imaging System (Alpha Innotech) and Quantity One software (Bio-Rad). The percentage of CA-SP1 was calculated as the amount of CA-SP1 over the sum of CA + CA-SP1. Standard deviations were calculated from at least three independent experiments. For biochemical analysis of CA-SP1 processing in the presence of BVM, radioimmunoprecipitation assays were performed based on methods previously described (Luttge et al., 2008). Briefly, 0.6 × 106 HeLa cells/well in 6-well cell culture plates (BD Falcon) were transfected with 2.5 μg of DNA (pNL4-3, SP1-A1V (Adamson et al., 2006), Gag L363I, or Gag M367I molecular clones) using Lipofectamine LTX reagent and incubated for ~18 hr in media containing 1 μg/ml BVM or an equivalent concentration of DMSO. Cells were washed in labeling medium (RPMI-1640 with 25 mM HEPES Cys−/Met− [Specialty Media]), then metabolically labeled for 2–4 hr at 37°C with 300 μCi [35S] Met/Cys (Express protein labeling mix; Perkin-Elmer) in labeling medium containing 5% FBS and 1 μg/ml BVM or DMSO. Released virions were collected from culture supernatants by 0.45 μm filtration and ultracentrifugation at 125,000 × g for 45 min. Cell and virion samples were lysed in cell lysis buffer [0.5% Triton X-100, 300 mM NaCl, 50 mM Tris (pH 7.5), and Complete protease inhibitor cocktail (Roche)]. Cell lysates were precleared by adsorption with protein A agarose in RIPA buffer [0.1% Triton X-100, 300 mM NaCl, 50 mM Tris (pH 7.5)] and 0.1% bovine serum albumin (BSA). Virion and precleared cell lysates were immunoprecipitated at 4°C with HIV-Ig bound to protein A agarose. Immunoprecipitated cell lysates were washed three times with RIPA buffer and once with SDS-DOC wash buffer [0.1% SDS, 300 mM NaCl, 50 mM Tris (pH 7.5), and 2.5 mM deoxycholic acid]. Immunoprecipitated virus lysates were washed once with RIPA buffer. Immunoprecipitated proteins were eluted in Laemmli sample buffer at 99°C for 5 min and resolved by SDS-PAGE in 15% acrylamide with AcrylAide crosslinker. After fixation and dehydration of the gel, labeled proteins were detected by exposure to storage phosphor screens and quantified using QuantityOne software (BioRad). The percentage of CA-SP1 was calculated as the amount of CA-SP1 over the sum of (CA + CA-SP1) or (CA* + CA-SP1), for the L363I mutant. CA* represents CA plus a four-amino-acid extension into SP1. Standard deviations were calculated from at least three independent experiments.
Transmission electron microscopy (EM)
HeLa cells transfected with HIV-1 molecular clones were fixed 24 hr posttransfection with 2% glutaraldehyde in 0.1 M sodium cacodylate and processed for transmission EM as previously described (Freed et al., 1994). Released virions were scored based on morphology.
Results
The CA-SP1 cleavage mutant CA5 inhibits WT HIV-1 infectivity in trans
To precisely measure the ability of uncleaved CA-SP1 to interfere with HIV-1 infectivity, we used a genetic approach by co-expressing WT Gag and a Gag mutant unable to be cleaved at the CA-SP1 junction. The mutant used in this study, termed CA5 (Wiegers et al., 1998), contains two substitutions in amino acid residues required for CA-SP1 cleavage (L363I and M367I, Fig. 1A, lower panel). CA5 displays defects in Gag processing and virion maturation similar to those induced by BVM treatment (Li et al., 2003; Wiegers et al., 1998). HeLa cells were cotransfected with the wild-type (WT) HIV-1 molecular clone pNL4-3 and the CA5 mutant at varying ratios (1:1, 2.5:1, and 5:1), while the total amount of DNA remained constant. Thus, virus particles formed under these conditions contained a mixture of both p24 (CA) and p25 (CA-SP1) proteins. WT and CA5 virus stocks were generated to serve as controls. The amounts of mature CA and uncleaved CA-SP1 were determined by quantitative Western blotting (Fig. 1B). Transfection of pNL4-3 alone led to the production of virions that contained nearly 100% CA, whereas CA5 virions contained 100% CA-SP1. Cotransfecting WT and CA5 molecular clones at ratios of 1:1, 2.5:1, and 5:1 resulted in the formation of virions that contained ~60%, ~40%, and ~30% CA-SP1, respectively (Fig. 1C).
To determine whether the CA5 mutant imposes a dominant-negative effect on virus infectivity, the TZM-bl indicator cell line (Wei et al., 2002) was infected with the HeLa-derived virus stocks described above. Four viral inputs were used in the TZM-bl infections. Two days postinfection, relative virus infectivity was measured by quantifying the luciferase activity in the infected TZM-bl cultures. A representative graph showing virus infectivity in light units is shown for all virus stocks and controls (Fig. 2A). The percent infectivity was calculated at a viral input of 50,000 RT cpm (Fig. 2B) from several independent experiments. The CA5 mutant is essentially devoid of infectivity (~1% WT levels), whereas the WT/CA5 mixed virions displayed infectivities, relative to those of the WT, of ~16%, ~43%, and ~87% for the 1:1, 2.5:1, and 5:1 mixtures, respectively (n = 4). TZM-bl cells also contain a Tat-inducible β-galactosidase reporter (Wei et al., 2002), allowing infectivity to be quantified either by luciferase activity (as shown in Fig. 2A and B) or by scoring the number of infected cells. Similar defects in infectivity were measured when infectivity was scored by either approach (data not shown). These results demonstrate that uncleaved CA-SP1 disrupts HIV-1 infectivity in a dominant-negative manner.
Fig. 2.
Dominant-negative effect of uncleaved CA-SP1 on HIV-1 infectivity. (A) TZM-bl cells were infected with virus stocks obtained by transfecting HeLa cells with WT (pNL4-3) or CA5 molecular clones, or WT/CA5 mixtures (at indicated DNA ratios). Virus stocks were normalized for RT activity and infection performed with 100,000, 50,000, 25,000, or 12,500 RT cpm. A representative graph indicating light units versus virus input used for each virus stock is shown. (B) Quantitative data showing levels of infectivity with a virus input of 50,000 RT cpm, n = 7. (C) Jurkat cells were infected with virus stocks normalized to 10,000 RT cpm. Supernatants were collected every 2–3 days and monitored for RT activity for a period of 45 days.
To examine the effect of uncleaved CA5 in the context of a multiple-round, spreading viral infection, Jurkat T cells were infected with the HeLa-derived virus stocks described above. Virus replication was monitored by supernatant RT activity for a period of 45 days (Fig. 2C). In the WT-infected cultures, virus replication peaked on day 17. In contrast, replication of the CA5 mutant alone or the virus produced with a 1:1 mix of WT and CA5 was completely blocked. Virus replication in the cultures infected with the 5:1 WT/CA5 mixed virions was similar to that in WT-infected cultures, while the 2.5:1 mixed virus replicated with a delay of 15 days relative to the WT.
Uncleaved CA-SP1 disrupts HIV-1 particle maturation in trans
CA5 virions (Wiegers et al., 1998), or WT HIV-1 particles produced in the presence of BVM (Adamson et al., 2006; Li et al., 2003), exhibit a distinct virion morphology characterized by an acentric core accompanied by a crescent-shaped zone of electron density when visualized by transmission electron microscopy (TEM). It was therefore of interest to examine the morphology of virions produced in HeLa cells transfected with the different ratios of WT and CA5 mutant molecular clones described above. To quantify these data, we examined the morphology of >128 virions per sample and classified them into the following categories: conical core, centric core, acentric core, acentric core with crescent, immature, and aberrant (Fig. 3). Mature WT virions have been previously characterized as having primarily a conical or a centric core depending on the angle at which the core is sectioned for EM analysis (Orenstein, 2002). The WT virions mainly contained conical cores (~47%). Centric and acentric cores were also observed (~22% each). In the CA5 mutant, only ~2% and ~8% of all virions contained conical and centric cores, respectively, while ~38% contained acentric cores with a crescent of electron density inside the viral membrane. WT/CA5 mixed virions (1:1, 2.5:1, and 5:1) showed ~13%, ~17%, and 29%, conical cores, respectively, and fewer acentric cores with a crescent, relative to the CA5 mutant. These results demonstrate that increased levels of uncleaved CA-SP1 in virions are accompanied by a reduction in the percentage of virions containing conical cores.
Fig. 3.
Effect of uncleaved CA-SP1 on virion morphology. HeLa cells were transfected with WT (pNL4-3) or CA5 molecular clones, or WT/CA5 mixtures (at indicated DNA ratios). Bar graph shows the quantification of released virions (n = 128–354) based on their morphology and placed into the following categories: conical core, centric core, acentric core, acentric core + crescent, immature, and aberrant. A representative image for each morphology is shown. Percentages for each morphology are shown to the right of each bar.
Cleavage within SP1 mitigates the dominant-negative inhibition imposed by CA-SP1
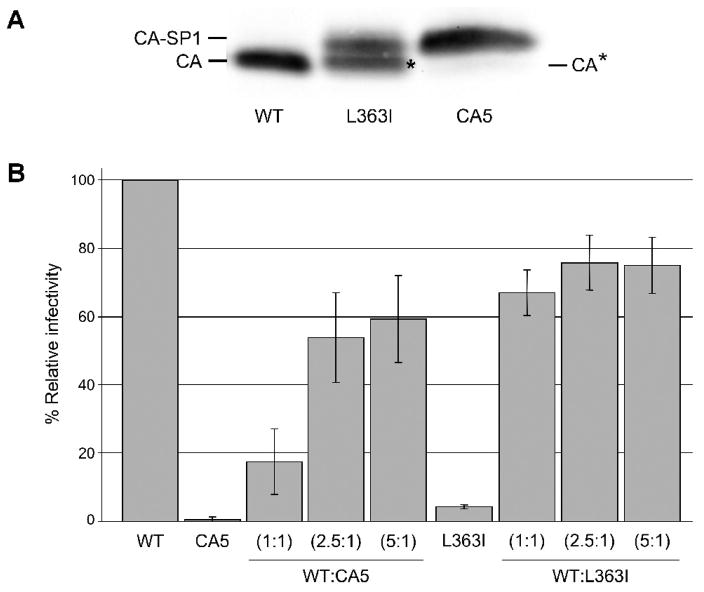
While this manuscript was in preparation, Lee et al. reported that the L363I mutant only weakly inhibits WT HIV-1 infectivity when coexpressed in trans (Lee, Harris, and Swanstrom, 2009). To investigate the difference between CA5 [which bears both L363I and M367I substitutions (Fig. 1)] and the L363I single mutant, we compared the Gag processing pattern of these two mutants. As shown above (Fig. 1B), the CA5 mutant produces exclusively CA-SP1. In contrast, the L363I mutant produces virions containing an approximately 1:1 ratio of CA-SP1 and a smaller species that, as reported earlier (Kräusslich et al., 1995), represents CA plus a four-amino-acid extension into SP1 (Fig. 1A). This species, which we refer to as CA* (Fig. 1 and Fig. 4A) is generated by processing at the internal, cryptic site at residue 367 in SP1 (Kräusslich et al., 1995) (Fig. 1A). This species was also observed in the study by Lee et al. (Lee, Harris, and Swanstrom, 2009). We next compared the ability of CA5 and L363I to inhibit HIV-1 infectivity in trans. As shown above (Fig. 1C), CA5 potently inhibits WT infectivity at a 1:1 input DNA ratio, with a reduction in single-round infectivity in TZM-bl cells of approximately five-fold. In contrast, and in agreement with the data of Lee et al. (Lee, Harris, and Swanstrom, 2009), the L363I has only a weak effect (~30%) in trans despite the fact that this mutation almost completely abolishes infectivity when expressed alone (Fig. 4B). These infectivity data, combined with the analysis of virion-associated CA, CA*, and CA-SP1 present in CA5 and L363I mutant virions, suggest that processing of SP1 at the internal site (residue 367) largely reverses the ability of CA-SP1 cleavage site mutants to dominantly interfere with HIV-1 maturation.
Fig. 4.
Comparison between CA5 and the L363I mutant in Gag processing patterns and dominant-negative effects on HIV-1 infectivity. (A) Western blot of CA, CA-SP1, and CA*, in WT, L363I, and CA5 particles. CA* represents CA + four amino acids of SP1. (B) TZM-bl cells were infected as described in Fig 2A with virus stocks obtained by transfecting HeLa cells with WT (pNL4-3), CA5, or L363I molecular clones, or WT/CA5 and WT/L363I mixtures at the indicated DNA ratios. Quantitative data showing levels of infectivity with a virus input of 50,000 RT cpm, n=3.
BVM disrupts Gag processing at both the major CA-SP1 cleavage site and the internal cryptic site
To determine whether BVM disrupts CA-SP1 processing in the context of the L363I and M367I mutations, we evaluated the effect of this compound on levels of CA, CA*, and CA-SP1 in virions by quantitative radioimmunoprecipitation analysis. As controls, we included WT NL4-3, which is sensitive to BVM, and the SP1-A1V mutant, which is BVM-resistant (Adamson et al., 2006; Li et al., 2003). As expected, we observed that BVM treatment of virus-producing cells increases the levels of CA-SP1 in WT virions but not in SP1-A1V particles (Fig. 5A and B). The M367I virions showed somewhat more CA-SP1 in virions in the absence of BVM relative to WT or SP1-A1V, but exhibited a clear increase in CA-SP1 levels upon BVM treatment. Interestingly, in L363I virions, a marked increase in CA-SP1 was also observed (Fig. 5A and B), demonstrating that generation of CA* as a result of cleavage at the internal SP1 site is also inhibited by BVM. The effect of BVM on CA-SP1 accumulation was less striking than with the WT, because even in the absence of BVM approximately 50% of the CA-related product was in the form of CA-SP1 (Fig. 5B, right panel). We also evaluated the effect of BVM on virion infectivity. The infectivity of WT and M367I mutants was markedly reduced by BVM treatment (Fig. 5C). The effect of BVM on the infectivity of L363I could not be readily determined, as this mutant is very poorly infectious even in the absence of the compound.
Fig 5.
Effect of BVM on Gag processing and HIV-1 infectivity for the L363I and M367I mutants. (A–B) CA-SP1 processing in the presence or absence of 1 μg/ml BVM. HeLa cells were transfected with WT (pNL4-3), A1V (BVM-resistant mutant), L363I, or M367I molecular clones and incubated in the presence (+) or absence (−) of BVM. One day posttransfection, cells were metabolically radiolabeled with [35S] Met/Cys for 2–4 hr and released virus particles were collected by ultracentrifugation. Gag proteins were detected from virus lysates by immunoprecipitation with HIV-Ig and (A) CA-SP1, CA*, and CA were resolved by SDS-PAGE. CA* is denoted by *. (B) Quantification of CA, CA-SP1, and CA* in virus particles. Percentages of CA-SP1 relative to CA + CA-SP1 [for WT (pNL4-3), A1V, or M367I mutants] or CA* + CA-SP1 (for L363I mutant) were determined by phosphorimager analysis. Error bars indicate standard deviations (n = 3). (C) TZM-bl cells were infected with virus obtained from HeLa cells transfected with WT (pNL4-3), L363I, or M367I molecular clones in the presence (+) or absence (−) of 1 μg/ml BVM. Virus produced one day posttransfection in the course of 4 hr with or without BVM was used for infection at different dilutions ranging from 63,000 to 1,250,000 RT cpm. Quantitative data showing levels of infectivity with a virus input of 250,000 RT cpm, n = 3.
Discussion
The results of this study show that uncleaved CA-SP1 imposes a dominant-negative effect on HIV-1 maturation and infectivity. Also, there is a direct correlation between the levels of mature CA in the mixed virions and the percentage of virus particles containing conical cores. While this manuscript was in preparation, two papers relevant to the current study were published. Müller et al. used low concentrations of PR inhibitors and Gag processing mutants, including CA5, to show that incompletely processed Gag inhibits virus infectivity in trans (Müller et al., 2009). Lee et al. also used a genetic approach to demonstrate dominant-negative effects of HIV-1 Gag processing mutants (Lee, Harris, and Swanstrom, 2009). In contrast to results with CA5, Lee et al. observed that a CA-SP1 cleavage mutant exerted only weak dominant-negative inhibitory activity. In their study, they used the L363I mutant, whereas we and Müller et al. (Müller et al., 2009) used the CA5 mutant (Wiegers et al., 1998), which bears both the L363I mutation at the primary CA-SP1 cleavage site and an Ile substitution at residue M367 at the internal SP1 processing site (Fig. 1). To investigate whether these divergent results were due to the use of two different Gag mutants, we compared the ability of CA5 and the L363I mutant to inhibit WT infectivity. Our results with the L363I mutant were in excellent agreement with those of Lee et al. (Lee, Harris, and Swanstrom, 2009). While the L363I mutant when expressed alone was severely impaired for virus infectivity, it was a very weak inhibitor of the WT when supplied in trans (Fig. 4B) Also consistent with the data of Lee et al., L363I virions contain high levels of a protein close in size to mature CA (Fig. 4A). This species is likely CA with a four-amino-acid C-terminal extension resulting from processing at position M367 (Kräusslich et al., 1995). These results demonstrate that uncleaved CA-SP1 is a strong dominant-negative inhibitor of HIV-1 maturation, whereas CA with the four amino acid SP1 extension is not; i.e., cleavage at the internal SP1 site largely ablates the dominant-negative inhibitory activity of a CA-SP1 cleavage-site mutant.
The inhibitory activity of CA5 in trans on HIV-1 maturation is evident not only in single-cycle infectivity assays but can also be observed by EM analysis as an effect on virion morphogenesis. In our analysis, WT virions display mature conical cores with a frequency of ~50%. By contrast, the CA5 mutant when expressed alone produces virions that are virtually devoid of conical cores. When CA5 and WT molecular clones are coexpressed, a marked reduction in conical core formation is observed (Fig. 3); for example, at a 1:1 DNA ratio only ~13% of virions contain conical cores. This correlates well with an infectivity defect of ~5-fold under these conditions (Fig. 2). Even at a WT/CA5 ratio of 5:1 some reduction in conical core formation is observed.
We investigated the ability of BVM to disrupt CA-SP1 processing in the context of the L363I and M367I mutations. We found that BVM increases the levels of CA-SP1 in the L363I and M367I mutants (Fig. 5A and B). We also observed that BVM significantly reduces the infectivity of the M367I mutant in single-cycle assays (Fig. 5C). We were not readily able to measure the effect of BVM on the infectivity of the L363I mutant, as this substitution virtually abolishes particle infectivity. The increase in CA-SP1 levels with the L363I mutant indicates that BVM blocks processing not only at the major CA-SP1 cleavage site, but also disrupts cleavage at the internal SP1 site adjacent to residue 367. Related to this observation, we have obtained preliminary mass spectrometry data indicating that BVM treatment does not induce increased processing at the internal site in SP1 (Adamson, Chertova, and Freed, unpublished results).
The data presented here help to explain the potency of BVM in inhibiting HIV-1 maturation. Disruption of CA-SP1 processing not only interferes with virus maturation in cis, but also the uncleaved CA-SP1 fragment exerts a dominant-negative effect on virion maturation. These results imply that drugs targeting the CA-SP1 cleavage site will be most effective if, like the CA5 mutations, they block processing at both the major CA-SP1 site and the internal SP1 site.
Acknowledgments
We thank members of the Freed laboratory for helpful discussion and critical review of the manuscript. We thank H.-G. Kräusslich for providing the CA5 mutant and S.-K. Lee and R. Swanstrom for the L363I mutant. We thank C. Adamson and E. Chertova for sharing unpublished data. The HIV-Ig and TZM-bl cells were obtained from the NIH AIDS Research and Reference Reagent Program. This research was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, NIH, and by the Intramural AIDS Targeted Antiviral Program. This project was funded in part with federal funds from the National Cancer Institute, NIH, under contract N01-CO-12400.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accola MA, Höglund S, Göttlinger HG. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72 (3):2072–8. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accola MA, Strack B, Göttlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74 (12):5395–402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59 (2):284–91. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CS, Ablan SD, Boeras I, Goila-Gaur R, Soheilian F, Nagashima K, Li F, Salzwedel K, Sakalian M, Wild CT, Freed EO. In vitro resistance to the human immunodeficiency virus type 1 maturation inhibitor PA-457 (Bevirimat) J Virol. 2006;80 (22):10957–71. doi: 10.1128/JVI.01369-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson CS, Freed EO. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol. 2007;55:347–87. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- Adamson CS, Salzwedel K, Freed EO. Virus maturation as a new HIV-1 therapeutic target. Expert Opin Ther Targets. 2009;13 (8):895–908. doi: 10.1517/14728220903039714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson-Viitanen S, Manfredi J, Viitanen P, Tribe DE, Tritch R, Hutchison CA, Loeb DD, Swanstrom R. Cleavage of HIV-1 gag polyprotein synthesized in vitro: sequential cleavage by the viral protease. AIDS Res Hum Retroviruses. 1989;5 (6):577–91. doi: 10.1089/aid.1989.5.577. [DOI] [PubMed] [Google Scholar]
- Freed EO. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251 (1):1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- Freed EO, Martin MA. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68 (4):2503–12. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68 (8):5311–20. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Assembly and analysis of conical models for the HIV-1 core. Science. 1999;283 (5398):80–3. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Yeager M, Sundquist WI. The structural biology of HIV assembly. Current Opinion in Structural Biology. 2008;18 (2):203–17. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, Hohenberg H, Huckhagel C, Kräusslich HG. N-Terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72 (6):4798–810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, Hohenberg H, Wilk T, Wiegers K, Grättinger M, Müller B, Fuller S, Kräusslich HG. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000;19 (1):103–13. doi: 10.1093/emboj/19.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson LE, Copeland TD, Sowder RC, Schultz AM, Oroszlan S. Analysis of proteins and peptides from sucrose banded HTLV-III. In: Bolognesi D, editor. Human retroviruses, cancer and AIDS: approaches to prevention and therapy. Alan R. Liss, Inc; New York: 1988. pp. 135–147. [Google Scholar]
- Kaplan AH, Zack JA, Knigge M, Paul DA, Kempf DJ, Norbeck DW, Swanstrom R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J Virol. 1993;67 (7):4050–5. doi: 10.1128/jvi.67.7.4050-4055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiernan RE, Freed EO. Cleavage of the murine leukemia virus transmembrane env protein by human immunodeficiency virus type 1 protease: transdominant inhibition by matrix mutations. J Virol. 1998;72 (12):9621–7. doi: 10.1128/jvi.72.12.9621-9627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kräusslich HG, Fäcke M, Heuser AM, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69 (6):3407–19. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Harris J, Swanstrom R. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J Virol. 2009;83 (17):8536–43. doi: 10.1128/JVI.00317-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Goila-Gaur R, Salzwedel K, Kilgore NR, Reddick M, Matallana C, Castillo A, Zoumplis D, Martin DE, Orenstein JM, Allaway GP, Freed EO, Wild CT. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc Natl Acad Sci USA. 2003;100(23):13555–60. doi: 10.1073/pnas.2234683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Hill CP, Sundquist WI, Finch JT. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature. 2000;407 (6802):409–13. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]
- Liang C, Hu J, Russell RS, Roldan A, Kleiman L, Wainberg MA. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 gag. J Virol. 2002;76 (22):11729–37. doi: 10.1128/JVI.76.22.11729-11737.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge BG, Shehu-Xhilaga M, Demirov DG, Adamson CS, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Molecular characterization of feline immunodeficiency virus budding. J Virol. 2008;82 (5):2106–19. doi: 10.1128/JVI.02337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morellet N, Druillennec S, Lenoir C, Bouaziz S, Roques BP. Helical structure determined by NMR of the HIV-1 (345–392)Gag sequence, surrounding p2: implications for particle assembly and RNA packaging. Protein Sci. 2005;14 (2):375–86. doi: 10.1110/ps.041087605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B, Anders M, Akiyama H, Welsch S, Glass B, Nikovics K, Clavel F, Tervo H, Keppler O, Kräusslich H. Human immunodeficiency virus (HIV-1) Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J Biol Chem. 2009 doi: 10.1074/jbc.M109.027144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orenstein JM. Ultrastructure of HIV/AIDS. Ultrastruct Pathol. 2002;26 (4):245–50. doi: 10.1080/01913120290104502. [DOI] [PubMed] [Google Scholar]
- Pettit SC, Henderson GJ, Schiffer CA, Swanstrom R. Replacement of the P1 amino acid of human immunodeficiency virus type 1 Gag processing sites can inhibit or enhance the rate of cleavage by the viral protease. J Virol. 2002;76 (20):10226–33. doi: 10.1128/JVI.76.20.10226-10233.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit SC, Moody MD, Wehbie RS, Kaplan AH, Nantermet PV, Klein CA, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68 (12):8017–27. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanstrom R, Wills JW. Synthesis, assembly, and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. pp. 263–334. [PubMed] [Google Scholar]
- Tritch RJ, Cheng YE, Yin FH, Erickson-Viitanen S. Mutagenesis of protease cleavage sites in the human immunodeficiency virus type 1 gag polyprotein. J Virol. 1991;65 (2):922–30. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt VM. Proteolytic processing and particle maturation. Curr Top Microbiol Immunol. 1996;214:95–131. doi: 10.1007/978-3-642-80145-7_4. [DOI] [PubMed] [Google Scholar]
- von Schwedler UK, Stemmler TL, Klishko VY, Li S, Albertine KH, Davis DR, Sundquist WI. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17 (6):1555–68. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46 (6):1896–905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegers K, Rutter G, Kottler H, Tessmer U, Hohenberg H, Kräusslich HG. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J Virol. 1998;72 (4):2846–54. doi: 10.1128/jvi.72.4.2846-2854.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey RL, Smith DH, Lasky LA, Theodore TS, Earl PL, Moss B, Capon DJ, Martin MA. In vitro mutagenesis identifies a region within the envelope gene of the human immunodeficiency virus that is critical for infectivity. J Virol. 1988;62 (1):139–47. doi: 10.1128/jvi.62.1.139-147.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Yuan X, Dismuke D, Forshey BM, Lundquist C, Lee KH, Aiken C, Chen CH. Small-molecule inhibition of human immunodeficiency virus type 1 replication by specific targeting of the final step of virion maturation. J Virol. 2004;78 (2):922–9. doi: 10.1128/JVI.78.2.922-929.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]