Abstract
Amyloid formation is characterized by the conversion of soluble proteins into biochemically and structurally distinct fibers. Although amyloid formation is traditionally associated with diseases such as Alzheimer disease, a number of biologically functional amyloids have recently been described. Curli are amyloid fibers produced by Escherichia coli that contribute to biofilm formation and other important physiological processes. We characterized the polymerization properties of the major curli subunit protein CsgA. CsgA polymerizes into an amyloid fiber in a sigmoidal kinetic fashion with a distinct lag, growth, and stationary phase. Adding sonicated preformed CsgA fibers to the polymerization reaction can significantly shorten the duration of the lag phase. We also demonstrate that the conversion of soluble CsgA into an insoluble fiber involves the transient formation of an intermediate similar to that characterized for several disease-associated amyloids. The CsgA core amyloid domain can be divided into five repeating units that share sequence and structural hallmarks. We show that peptides representing three of these repeating units are amyloidogenic in vitro. Although the defining characteristics of CsgA polymerization appear conserved with disease-associated amyloids, these proteins evolved in diverse systems and for different purposes. Therefore, amyloidogenesis appears to be an innate protein folding pathway that can be capitalized on to fulfill normal physiological tasks.
Amyloid formation is the hallmark of medically related disorders such as Alzheimer disease, Huntington disease, Parkinson disease, and the transmissible spongiform encephalopathies (1). The root of these diseases is the uncontrolled conversion of seemingly unrelated soluble proteins into biochemically and structurally related fibers known as amyloids. Despite their diversity in size and amino acid content, all amyloid proteins assemble into 4–12-nm-wide fibers that are β-sheet-rich and exhibit conserved tinctorial properties. Soluble preamyloid species also share common pore-like epitopes, and these globular species may induce cytotoxicity (2–4).
Numerous studies have revealed that amyloidogenic proteins are mostly unstructured or contain mixtures of β-sheets and α-helices in their native state, but when polymerized into fibers, they invariably adopt a characteristic cross β-sheet structure (5–7). This cross β-sheet structure is common to all amyloids and is characterized by β-strands that orient perpendicular to the fiber axis. In vitro, disease-associated amyloids polymerize into fibers with nucleation-dependent kinetics with characteristic lag, growth, and stationary phase. The lag phase is proposed to contain folding intermediates that are key to the toxicity associated with certain amyloidogenic proteins (8, 9). During the lag phase, amyloidogenic proteins adopt a transient folding species that disrupts membrane integrity (3, 8, 10). Loss of membrane integrity is proposed to underlie the cell death and disease associated with many amyloids (3, 10). A conformational-specific antibody has been generated that recognizes a transient intermediate formed during amyloidogenesis of several disease-associated proteins (3).
A new class of amyloids has recently been found that plays important physiological roles for the cell. These so-called “functional” amyloids are found in bacteria (11–13), fungi (14, 15), and mammals (16). The first example of a functional amyloid in bacteria was curli (12). Curli compose part of the complex extracellular matrix that is required for biofilm formation (17–19), host cell adhesion (20), and invasion (21, 22), and they are proposed to be important stimulants of the host inflammatory response (23–25). An intriguing question is whether these functional amyloid proteins polymerize in a manner similar to disease-associated amyloids.
Curli formation is the result of an elegant biosynthetic pathway directed by the Csg proteins in Escherichia coli. The major curli subunit, CsgA, can be secreted to the cell surface as a soluble, unstructured protein (12, 26). CsgA is efficiently nucleated into an insoluble amyloid fiber in the presence of the outer membrane-associated protein, CsgB (27). After nucleation, the fibers are predicted to grow by subsequent CsgA addition to the tip of the amyloid fiber (26).
Both CsgA and CsgB display a remarkable five-fold internal symmetry characterized by conserved polar residues. These five “repeating units” consist of 19–24 amino acids and align along serine, glutamine, and asparagine residues (26, 28). Each repeating unit is predicted to form a strand-loop-strand motif that closely resembles the cross β-spine structure described for many disease-associated amyloids (28 –30).
Here we characterize the folding of purified CsgA and show that its polymerization is similar to that of disease-associated amyloids. CsgA polymerization involves a transient structurally conserved intermediate that implies a common polymerization pathway between functional and disease-associated amyloids. We found that the conserved folding intermediate for CsgA is a monomer or low molecular weight multimer. We demonstrate that at least three of five repeating units of CsgA are amyloidogenic. These results suggest that covalent linkage of multiple amyloidogenic units facilitates efficient fiber formation.
Experimental Procedures
CsgA Purification
CsgG and CsgA-His were overexpressed in LSR12 (C600::Δcsg) as described previously (12). Following centrifugation for 15 min at 10,000 × g, the supernatant was clarified by filtration through a 0.22-μm polyethersulfone bottle-top filter (Corning, Acton, MA). Filtrates containing CsgA were passed over a HIS-Select™ HF nickel-nitrilotriacetic acid (Sigma) column, washed with 10 volumes of 10 mM potassium phosphate buffer (KPi),2 pH 7.2, and eluted with 10 mM KPi 100 mM imidazole, pH 7.2. CsgA-containing fractions were combined and passed through a 0.02-μm Anotop 10 filter (Whatman, Maidstone, UK). The N-terminal portion of purified CsgA-His was sequenced by mass spectrometry and found to be identical to the predicted CsgA amino acid sequence. A modified protocol using guanidine hydrochloride (GdnHCl) was employed to fully denature CsgA-His. Following the first wash, the column was equilibrated with 5 volumes of 10 mM KPi, 8 m GdnHCl, pH 7.2, and eluted with 50 mM KPi, 8 m GdnHCl, pH 2. Sephadex G25 was used for desalting/buffer exchange. To create CsgA-His seeds, 2-week-old fibers were sonicated using a Fisher Model 100 sonic dismembrator (Fisher) for three 15-s bursts on ice. Where indicated, CsgA samples were filtered through a prewashed Amicon Ultra-4 (Millipore, Bedford, MA) centrifugal filter device. Samples were centrifuged at 4,000 × g for 2 min, and the retentate and filtrate fractions were collected. A plasmid encoding CsgA-His can complement ΔcsgA cells in vivo, and purified CsgA-His polymerizes into an amyloid fiber with similar kinetics as wild-type CsgA (12). Therefore, CsgA-His is referred to as “CsgA” throughout this report.
Thioflavin T (ThT) Assay
Following desalting to remove imidazole or GdnHCl, CsgA was incubated at room temperature. At different time intervals, CsgA samples were mixed with 20 μm ThT, and fluorescence was measured using a Spectramax M2 plate reader (Molecular Devices, Sunnyvale, CA) set to 438 nm excitation and 495 nm emission with a 475 nm cutoff. Alternatively, samples amended with 25 μm ThT were assayed directly in the Spectramax M2 plate reader every 10 min after shaking for 5 s.
CD Spectroscopy
CsgA samples (10 μm CsgA in 50 mm KPi, pH 7.2) were assayed in a Jasco J-810 spectropolarimeter from 190 to 250 nm in a quartz cell with a 1-mm path length at 25 °C.
Blot Assay with A11 and Anti-CsgA
CsgA samples were dripped onto 0.2-μm Transblot nitrocellulose membranes (Bio-Rad) as described (3) and allowed to dry for 5 min. The membrane was blocked in 5% milk in TBS-T (Tris-buffered saline with 0.01% Tween 20) for at least 1 h. The dot blots were washed three times in TBS-T before and after incubating with 1:10,000 dilutions of A11 primary antibody (BioSource, Camarillo, CA) and goat anti-rabbit-horseradish peroxidase (Sigma) in 3% bovine serum albumin/TBS-T. The blots were developed using the SuperSignal® West Dura system (Pierce). Blots were stripped and reprobed with a 1:10,000 dilution of rabbit anti-CsgA antibody (31).
Electron Microscopy
Philips CM12 Scanning Transmission Electron Microscope was used to visualize the fiber aggregates. Samples (10 μl) were placed on Formvar-coated copper grids (Ernest F. Fullam, Inc., Latham, NY) for 2 min, washed with deionized water, and negatively stained with 2% uranyl acetate for 90 s.
Peptide Preparation
Peptides were chemically synthesized by Proteintech Group Inc., Chicago, IL. Purity was greater than 90% by high pressure liquid chromatography, and size was confirmed by mass spectroscopy. To equilibrate the pH of each sample and to remove any potential seed in the peptide preparations, the peptides were denatured using a modified protocol described previously (32). Briefly, peptides were dissolved to 0.5 mg/ml in trifluoroacetic acid/hexafluoroisopropanol (1:1 v/v) and sonicated for 10 min. The suspensions were incubated at room temperature until they visibly cleared. The solvent was then removed by vacuum. Peptides were then dissolved in cold 2 mm HCl and centrifuged at 100,000 × g in a TLA-55 (Beckman Coulter) for 1 h at 4°C. The supernatants were equilibrated to 50 mm KPi, pH 7.2, by 200 mm KPi, pH 7.2, on ice. When the samples were shifted to room temperature, the polymerization was measured by ThT.
Results
CsgA Polymerization Kinetics
To determine the polymerization kinetics of CsgA, an in vitro polymerization assay was developed. The transition of freshly purified, soluble CsgA to amyloid fibers was monitored using ThT, an amyloid-specific dye commonly used to assay amyloid formation (33, 34). The ThT fluorescence of CsgA samples followed a sigmoidal curve with distinguishable lag, growth, and stationary phases (Fig. 1A). The length of the lag phase was concentration-independent when CsgA was incubated at concentrations above 4 μm (Fig. 1A). Concentration-independent lag phases have been reported for other amyloidogenic proteins (35, 36). ThT fluorescence signal did not appreciably change after 8 h, remaining at approximately the same level for over 30 days (data not shown).
FIGURE 1. In vitro polymerization of CsgA measured by ThT fluorescence, CD, and TEM.
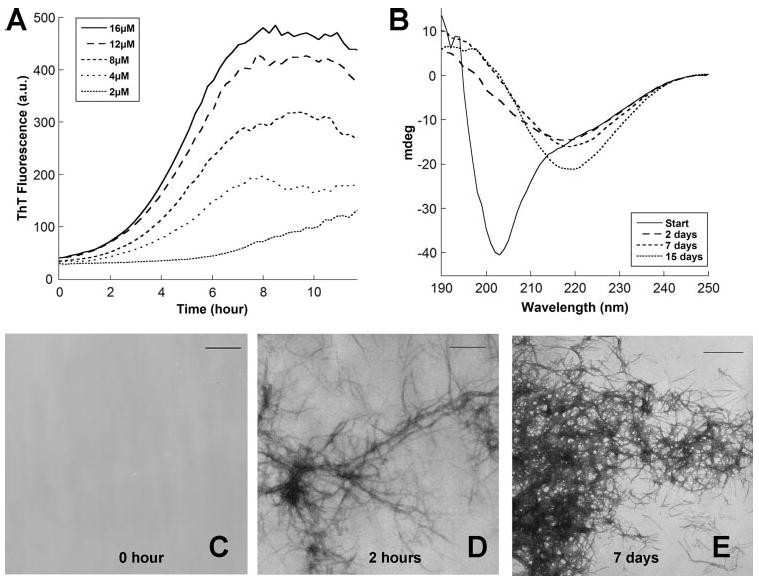
A, the fluorescence of freshly purified CsgA mixed with 25 μm ThT was measured in 10-min intervals at 495 nm after excitation at 438 nm. B, circular dichroism analysis of 10 μm CsgA immediately after purification, 2 days after purification, 7 days after purification, and 15 days after purification. CsgA was incubated at room temperature without shaking after purification. C–E, TEM micrographs of 30 μm CsgA after incubation at room temperature for the indicated times. Scale bar, 500 nm.
Circular dichroism spectroscopy and transmission electron microscopy (TEM) were used to measure the structural changes that occurred during CsgA amyloidogenesis. Circular dichroism spectrum indicated that immediately after purification, CsgA was largely unstructured (Fig. 1B). However, CsgA adopted a β-sheet-rich structure after 2 days of incubation at room temperature (Fig. 1B). Immediately after purification, there was no apparent fiber formation or aggregation by TEM (Fig. 1C). Two hours after purification, regular, unbranched fibers were readily observed (Fig. 1D). Dense fiber aggregates were also observed 7 days after purification (Fig. 1E).
The A11 Antibody Recognizes a Transient CsgA Folding Species
The polymerization of eukaryotic amyloids involves the formation of an intermediate folding species proposed to cause amyloid-associated toxicity to host cells (2, 3). The A11 antibody recognizes an Aβ transient intermediate (3). Remarkably, this antibody also recognizes a transient intermediate formed by islet amyloid polypeptide, polyglutamine, prion peptide 106–126, and Sup35p, among others (3, 37). The A11 antibody recognizes only a transient intermediate species, not soluble monomers or mature amyloid fibers derived from these proteins.
The A11 antibody was used to determine whether CsgA shared a common polymerization intermediate with eukaryotic amyloids. We found that immediately after purification, CsgA was recognized by the A11 antibody (Fig. 2A). As fiber formation proceeded, evidenced by increased ThT fluorescence and the appearance of fiber aggregates by TEM, the A11 antibody lost its affinity for CsgA (Fig. 2A). A polyclonal antibody generated against CsgA recognized purified CsgA independent of its polymerization status (Fig. 2A).
FIGURE 2. Detection of transient conserved intermediate species during CsgA polymerization.
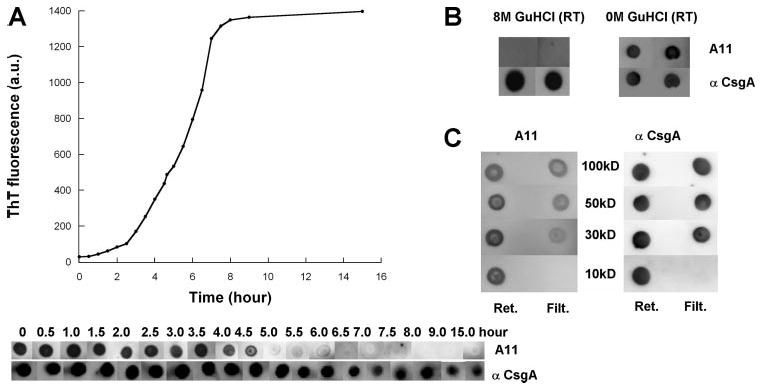
A, ThT fluorescence (top) and immunoblotting (bottom) of 80 μm CsgA incubated for the indicated time after purification. At the indicated times, samples were removed, ThT was added to a final concentration of 20 μm, and fluorescence was measured. Samples were blotted onto a nitrocellulose membrane and probed with the A11 antibody, and after stripping, probed with the anti-CsgA antibody. a.u., arbitrary units. B, CsgA denatured with 8 M GdnHCl was blotted onto nitrocellulose and probed with the A11 and anti-CsgA antibodies (left). GdnHCl (GuHCl) was removed using a Sephadex G25 column (final buffer: 50 mm KPi, pH 7.2) and then immediately blotted onto nitrocellulose and probed with the A11 and anti-CsgA antibodies (right). RT, room temperature; Ret., retentate; Filt., filtrate. C, Amicon ultra filters were used to separate CsgA solutions prior to probing with the A11 and anti-CsgA antibodies. The molecular weight cutoff of the filters is indicated. Retentates and filtrates were immediately blotted onto nitrocellulose and probed with the A11 antibody, and after stripping, probed with the anti-CsgA antibody.
The observation that the A11 antibody recognized CsgA suggested that CsgA polymerization intermediates might be structurally similar to those formed by disease-associated amyloid proteins. It also suggested that immediately after purification, CsgA had already begun its transition to an amyloid fiber. To prevent CsgA from folding during purification, the CsgA-containing fractions were amended with 8 m GdnHCl. Under these strongly denaturing conditions, the A11 antibody did not recognize CsgA; however, denatured CsgA was strongly recognized by the CsgA antibody (Fig. 2B). Immediately after GdnHCl removal with a desalting column, CsgA was recognized by the A11 antibody (Fig. 2B).
To determine the minimum size of the CsgA transient intermediate, freshly purified protein was passed through Amicon Ultra centrifugal membranes with different molecular weight cutoffs. The retentate and filtrate were probed with the A11 antibody (Fig. 2C). The A11 antibody recognized a species in the filtrate of the 30-kDa membrane, suggesting that the smallest reactive species of CsgA is 30 kDa or less. Because CsgA-His has a predicted molecular mass of 13.9 kDa, the species recognized by the A11 antibody is likely either a monomer or a dimer.
CsgA Fibers Can Catalyze Self-polymerization
The approximately sigmoidal ThT fluorescence curve suggests that CsgA polymerizes in a nucleation-dependent mechanism. Therefore, the growing fiber would be expected to direct the polymerization of new CsgA molecules. We tested the hypothesis that preformed CsgA fibers could catalyze CsgA polymerization. The addition of 2.5% (w/w) sonicated CsgA fibers to freshly purified, soluble CsgA resulted in a significant reduction of the lag phase (Fig. 3A). Coincident with the dramatically shorter lag phase in seeded reactions, CsgA was recognized by the A11 antibody for a significantly shorter time when compared with unseeded reactions (Fig. 3B).
FIGURE 3. CsgA fibers can catalyze self-polymerization.
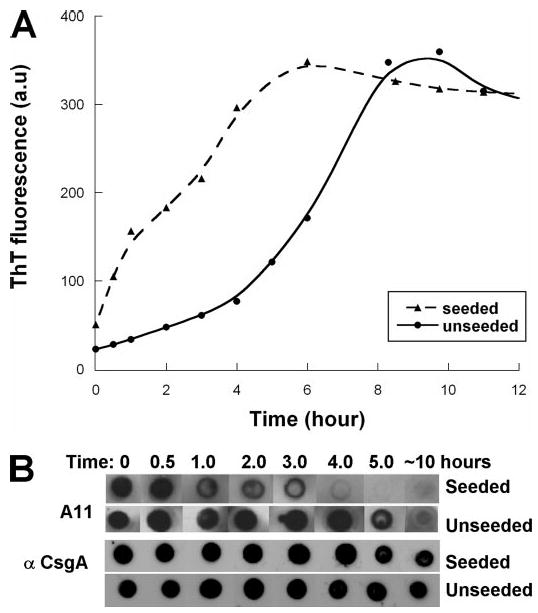
A, CsgA (40 μm) fluorescence in the absence (solid line) or presence of 2.5% by weight of sonicated CsgA fibers (dashed line). Samples were incubated at room temperature, collected at the indicated times, and amended with 20 μm ThT prior to excitation at 438 nm and measurement at 495 nm. a.u., arbitrary units. B, samples were removed at the indicated times and immediately blotted to nitrocellulose. Blots were probed with the A11 antibody, and after stripping, probed with an anti-CsgA antibody.
CsgA Is Composed of Several Amyloid-forming Units
The observation that CsgA was recognized by the A11 antibody immediately after passing through a 30-kDa cutoff filter (Fig. 2C) was unexpected since the A11 antibody is thought to recognize an oligomeric form of amyloidogenic proteins (3, 9, 37). The number of molecules present in the oligomeric state recognized by A11 varies among amyloidogenic proteins, and Aβ oligomers have been estimated to be larger than tetramers (3, 9). However, CsgA is recognized by A11 as a monomer or at most a dimer as estimated by cutoff filtration. It is possible that a single CsgA molecule includes multiple amyloidogenic domains that collectively contribute to its interaction with the A11 antibody. The primary sequence of CsgA can be divided into three parts: the Sec-dependent signal sequence, the N-terminal 22 amino acids of the mature protein, and a repeat domain that contains five 19–22-amino-acid repeating units (Fig. 4A). The five repeating units form a protease-resistant structure that is proposed to be the amyloid core of CsgA (26, 28). Each repeat has four conserved polar amino acids: serine, glutamine, asparagine, and glutamine (Fig. 4A). The regular arrangement of glutamine and asparagine residues also occurs in CsgA homologs from different Enterobacteriaceae.3
FIGURE 4. Three CsgA intramolecular peptide repeats can assembly into amyloid fibers.
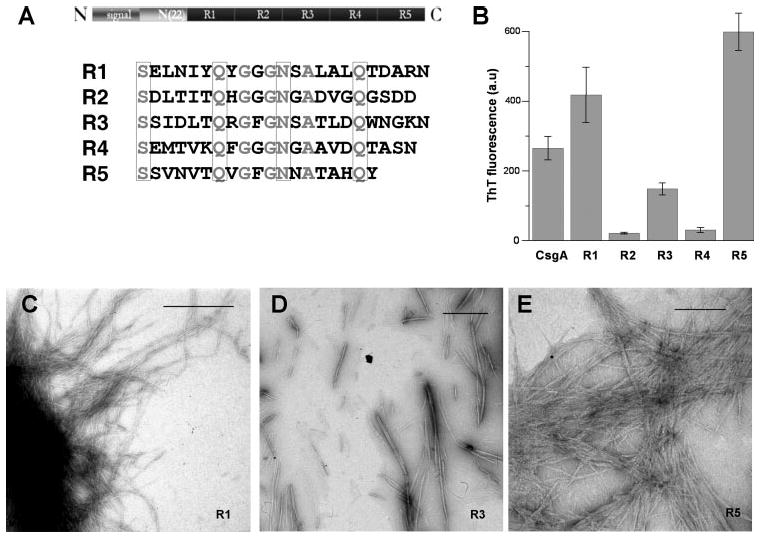
A, alignment of internally conserved residues. The CsgA primary sequence shows the repeated consensus sequences. The identical amino acid residues of five repeats are in gray, and the conserved polar amino acid residues are enclosed in four boxes. N, N terminus; C, C terminus. B, oligopeptides of R1, R2, R3, R4, and R5 at 0.2 mg/ml in KPi were incubated at room temperature for 5 days before ThT fluorescence measurements were taken. The error bar indicates the S.E. for at least three separate experiments. CsgA fibers were diluted to 0.2 mg/ml and assayed for ThT fluorescence. C–E, TEM. 0.5 mg/ml R1, R3, and R5 in pH 7.2 KPi was incubated at room temperature for 5 days. Samples of different peptide solutions were directly applied on Formvar-coated grids and visualized with negative staining electron microscopy. The scale bar is equal to 500 nm.
We hypothesized that these repeating units might represent single amyloid-forming units. Peptides corresponding to each repeating unit were chemically synthesized and tested for their ability to form amyloid fibers. Two independently derived preparations of each peptide were assayed. Peptides corresponding to repeating unit 1, 3 or 5 (R1, R3, or R5) produced a ThT-positive signal and self-assembled into fibers as evidenced by TEM when incubated at 0.2 mg/ml (Fig. 4, B–E). Neither R2 nor R4 showed evidence of amyloidogenesis when resuspended at a concentration of 0.2 mg/ml, although fibers were observed by TEM when R2 or R4 was incubated at 2 mg/ml (Fig. 4B and data not shown). The morphology of R1 fibers was similar to that of those formed by purified CsgA, being generally longer than 1000 nm (compare Fig. 4C with Fig. 1, D and E). R3 fibers were consistently shorter (ranging from 200 to 1000 nm) than those formed by CsgA (compare Fig. 4D with Fig. 1, D and E). R5 fibers appeared more rigid and aggregated than CsgA fibers (Fig. 4E). The morphologies of the fibers did not appreciably change over the course of a 10-day incubation. This analysis suggests that CsgA contains at least three highly amyloidogenic domains, R1, R3, and R5, that likely drive fiber formation in vivo.
Discussion
Amyloid formation is traditionally associated with uncontrolled protein misfolding and aggregation that results in many systemic and neurodegenerative disorders (1, 38). However, there are a growing number of functional amyloids that suggest amyloidogenesis is also a general tenet of normal cellular physiology. In fact, amyloid formation may be a common property of most proteins (39, 40).
The work presented here, as well as that published previously, demonstrates that both disease-associated and functional amyloids share a common amyloid formation pathway (41). CsgA polymerizes with nucleation-dependent kinetics, and fiber formation is ameliorated by the addition of preformed CsgA fibers. We also found that CsgA polymerization involves the formation of a transient species similar to that produced by other amyloidogenic proteins such as Aβ, synuclein, islet amyloid polypeptide, insulin, lysozyme, and polyglutamine (3).
The transient species that the A11 antibody recognizes during CsgA polymerization is a monomer or low molecular weight multimer (Fig. 2C). It was reported that the A11-recognized species of Aβ and Sup35p were probably large molecular weight oligomers (3, 37). Unlike Aβ and Sup35p (3, 37), CsgA was immediately recognized by the A11 antibody upon removal of strong denaturants such as GdnHCl or after its passage through a 30-kDa Amicon filter. We also found that freshly purified CsgA heated to 95 °C for 5 min was recognized by the A11 (data not shown). At least two hypotheses can be proposed to explain the ability of CsgA to be recognized by A11 immediately after denaturation or passage through a 30-kDa cutoff filter. First, CsgA may adopt an oligomeric conformation so quickly that our ability to measure this transition is lost in the time that it takes to immobilize CsgA on the blotting paper. Another possibility is that the CsgA species recognized by A11 is not an oligomer, but a monomer that contains multiple amyloidogenic units. In support of this hypothesis, we show that CsgA does indeed contain at least three amyloidogenic domains. Nevertheless, these two hypotheses are not mutually exclusive, and there may be other plausible interpretations.
Nonetheless, CsgA contains multiple amyloidogenic domains that may contribute to its ability to efficiently transition from a soluble protein to an amyloid fiber. Many studies have led to the proposal that amyloid fibers themselves are not toxic to cells; instead, toxicity is proposed to be caused by transient folding intermediates (2, 42, 43). Therefore, one mechanism that might be used by functional amyloids to prevent toxicity is to minimize the duration of toxic folding intermediates. This is apparently how Pmel17, an extremely rapidly forming functional amyloid found in mammalian cells, is able to assemble within the cell without eliciting a toxicity cascade (16).
CsgA has a striking primary sequence arrangement (Fig. 4A). The five repeats of CsgA are very similar and share greater than 30% sequence identity. We showed that each repeating unit is potentially a single amyloid domain and that R1, R3, and R5 are the highly amyloidogenic in vitro (Fig. 4, B–E). The covalent linkage of multiple amyloid domains may facilitate amyloid fiber formation by increasing the number of amyloidogenic building blocks. This also results in rapid formation of the intermediate recognized by the A11 antibody. Other amyloidogenic proteins contain repeat sequences that have been postulated to facilitate fiber formation (44, 45). For instance, the N-terminal prion-determining domain of Sup35p has five imperfect oligopeptide repeats, and certain deletions of the repeats are defective in propagation of Sup35p fibrils. Moreover, in vitro, repeat expansion peptides (with two extra repeats) were shown to be more amyloidogenic than wild-type peptides (46). Previous work suggested that the most amyloidogenic domains of CsgA were contained in the hexapeptide GHGGGN and QFGGGN, which are present in R2 and R4, respectively (47). However, our analysis suggests that R1, R3, and R5 contain the more highly amyloidogenic sequences. A thorough mutagenesis study is needed to define the residues that contribute to the highly amyloidogenic nature of CsgA.
The amyloidogenic peptides R1 and R5 contain sequences that contribute significantly to the ability of CsgA to bind human proteins such as fibronectin, plasminogen, tissue plasminogen activator, and β2-microglobulin (48). This correlation suggests that the amyloidogenecity of CsgA may be directly linked to these biological activities. In fact, work by Gebbink et al. (49) suggested that curli contribute to colonization in animal hosts by activating host proteases that are involved in hemostasis.
Curli can also enhance amyloid protein A amyloidosis in mice (50). It is proposed that cross-seeding may play a role in the development of amyloid diseases (50, 51). The in vitro system that we have established here provides an ideal vehicle to test the specificity of curli seeding with other amyloids. Understanding how functional amyloid proteins interact with other host proteins may lead to new ideas about cellular physiology and the processes that promote the toxicity associated with many amyloids.
Most amyloids are known to self-propagate in a process called seeding. In prion diseases such as bovine spongiform encephalopathy, seeding underlies protein infectivity (52). Amyloid self-propagation is also critical to disease development in the non-transmissible amyloid diseases (51, 53). Our demonstration of CsgA seeding suggests that functional amyloids also utilize a controlled self-propagation process to fulfill their biological function. In vivo CsgA polymerization is nucleated by the outer membrane-associated protein CsgB, which shares nearly 49% sequence similarity with CsgA (27). One proposed model of nucleation is that CsgB provides an amyloid-like template that initiates CsgA polymerization (26, 27). The growing fiber tip could then act as a template to direct subsequent CsgA polymerization.
Proteins that are not predicted to form stable globular folds may be prone to aggregation and amyloid formation, and indeed, most functional amyloid proteins have natively disordered segments (29, 54). Consistent with this, some proteins have been shown to form amyloid fibers only after the native, globular fold has been compromised by chemical denaturants or by mutations (39, 40, 55). The circular dichroism studies presented here suggest that CsgA is unstructured after secretion (Fig. 1B). Mature CsgA is also predicted to be natively unfolded (data not shown) by the Uversky and Galzitskaya algorithms (54, 56). The natively unfolded segments of CsgA may facilitate amyloidogenesis indirectly by preventing formation of stable globular structures that would be less likely to aggregate and precipitate into a fiber. Alternatively, the unfolded regions of CsgA may actively direct amyloidogenesis by presenting specific aggregative surfaces to neighboring molecules. In this context, “natively unfolded” would be a transition state during the formation of a stably folded amyloid fiber. Importantly, in the case of functional amyloids, the amyloid fiber would not be the product of protein misfolding but that of protein folding. Certainly, the growing number of functional amyloids suggests that amyloid is an evolutionarily conserved structure. The selective processes that have been employed by functional amyloids to limit cellular toxicity provide a unique context from which to investigate disease-associated amyloidogenesis.
Acknowledgments
We thank members of the Chapman and Hultgren laboratories for helpful discussions. Special thanks to Dr. Glabe and Dr. Kayed for providing the A11 antibody. We thank Dr. Ursula Jakob for technical assistance with the circular dichroism experiments.
Footnotes
This work was supported by National Institutes of Health Grant Award Number K22A1054967 and by the Michigan Alzheimer's Disease Research Center (MADRC) and National Institutes of Health Award P50-A608671 (to M. R. C.).
The abbreviations used are: KPi, potassium phosphate buffer; ThT, thioflavin T; GdnHCl, guanidine hydrochloride; R, repeating unit; TEM, transmission electron microscopy; Aβ, amyloid β.
X. Wang and M. R. Chapman, unpublished observation.
References
- 1.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe DJ. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 3.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 4.Quist A, Doudevski I, Lin H, Azimova R, Ng D, Frangione B, Kagan B, Ghiso J, Lal R. Proc Natl Acad Sci U S A. 2005;102:10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrow CJ, Zagorski MG. Science. 1991;253:179–182. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 6.Barrow CJ, Yasuda A, Kenny PT, Zagorski MG. J Mol Biol. 1992;225:1075–1093. doi: 10.1016/0022-2836(92)90106-t. [DOI] [PubMed] [Google Scholar]
- 7.Hurshman AR, White JT, Powers ET, Kelly JW. Biochemistry. 2004;43:7365–7381. doi: 10.1021/bi049621l. [DOI] [PubMed] [Google Scholar]
- 8.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 9.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 10.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 11.Bieler S, Estrada L, Lagos R, Baeza M, Castilla J, Soto C. J Biol Chem. 2005;280:26880–26885. doi: 10.1074/jbc.M502031200. [DOI] [PubMed] [Google Scholar]
- 12.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–885. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. Genes Dev. 2003;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coustou-Linares V, Maddelein ML, Begueret J, Saupe SJ. Mol Microbiol. 2001;42:1325–1335. doi: 10.1046/j.1365-2958.2001.02707.x. [DOI] [PubMed] [Google Scholar]
- 15.True HL, Lindquist SL. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 16.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. PLoS Biol. 2005;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Austin JW, Sanders G, Kay WW, Collinson SK. FEMS Microbiol Lett. 1998;162:295–301. doi: 10.1111/j.1574-6968.1998.tb13012.x. [DOI] [PubMed] [Google Scholar]
- 18.Zogaj X, Bokranz W, Nimtz M, Romling U. Infect Immun. 2003;71:4151–4158. doi: 10.1128/IAI.71.7.4151-4158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zogaj X, Nimtz M, Rohde M, Bokranz W, Romling U. Mol Microbiol. 2001;39:1452–1463. doi: 10.1046/j.1365-2958.2001.02337.x. [DOI] [PubMed] [Google Scholar]
- 20.Johansson C, Nilsson T, Olsen A, Wick MJ. FEMS Immunol Med Microbiol. 2001;30:21–29. doi: 10.1111/j.1574-695X.2001.tb01545.x. [DOI] [PubMed] [Google Scholar]
- 21.Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. Infect Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gophna U, Oelschlaeger TA, Hacker J, Ron EZ. FEMS Microbiol Lett. 2002;212:55–58. doi: 10.1111/j.1574-6968.2002.tb11244.x. [DOI] [PubMed] [Google Scholar]
- 23.Bian Z, Brauner A, Li Y, Normark S. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 24.Bian Z, Yan ZQ, Hansson GK, Thoren P, Normark S. J Infect Dis. 2001;183:612–619. doi: 10.1086/318528. [DOI] [PubMed] [Google Scholar]
- 25.Tukel C, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akcelik M, Adams LG, Baumler AJ. Mol Microbiol. 2005;58:289–304. doi: 10.1111/j.1365-2958.2005.04825.x. [DOI] [PubMed] [Google Scholar]
- 26.Barnhart MM, Chapman MR. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammar M, Bian Z, Normark S. Proc Natl Acad Sci U S A. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collinson SK, Parker JM, Hodges RS, Kay WW. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 29.Nelson R, Eisenberg D. Curr Opin Struct Biol. 2006;16:260–265. doi: 10.1016/j.sbi.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Nelson R, Sawaya MR, Balbirnie M, Madsen AO, Riekel C, Grothe R, Eisenberg D. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barnhart MM, Lynem J, Chapman MR. J Bacteriol. 2006;188:5212–5219. doi: 10.1128/JB.00234-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen S, Wetzel R. Protein Sci. 2001;10:887–891. doi: 10.1110/ps.42301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.LeVine H., III Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeVine H., III Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 35.Lomakin A, Teplow DB, Kirschner DA, Benedek GB. Proc Natl Acad Sci U S A. 1997;94:7942–7947. doi: 10.1073/pnas.94.15.7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rhoades E, Gafni A. Biophys J. 2003;84:3480–3487. doi: 10.1016/S0006-3495(03)70068-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shorter J, Lindquist S. Science. 2004;304:1793–1797. doi: 10.1126/science.1098007. [DOI] [PubMed] [Google Scholar]
- 38.Shorter J, Lindquist S. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- 39.Fandrich M, Fletcher MA, Dobson CM. Nature. 2001;410:165–166. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 40.Guijarro JI, Sunde M, Jones JA, Campbell ID, Dobson CM. Proc Natl Acad Sci U S A. 1998;95:4224–4228. doi: 10.1073/pnas.95.8.4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Glover JR, Kowal AS, Schirmer EC, Patino MM, Liu JJ, Lindquist S. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 42.Demuro A, Mina E, Kayed R, Milton SC, Parker I, Glabe CG. J Biol Chem. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 43.Glabe CG, Kayed R. Neurology. 2006;66:S74–8. doi: 10.1212/01.wnl.0000192103.24796.42. [DOI] [PubMed] [Google Scholar]
- 44.Ross ED, Minton A, Wickner RB. Nat Cell Biol. 2005;7:1039–1044. doi: 10.1038/ncb1105-1039. [DOI] [PubMed] [Google Scholar]
- 45.Wright CF, Teichmann SA, Clarke J, Dobson CM. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 46.Liu JJ, Lindquist S. Nature. 1999;400:573–576. doi: 10.1038/23048. [DOI] [PubMed] [Google Scholar]
- 47.Cherny I, Rockah L, Levy-Nissenbaum O, Gophna U, Ron EZ, Gazit E. J Mol Biol. 2005;352:245–252. doi: 10.1016/j.jmb.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 48.Olsen A, Herwald H, Wikstrom M, Persson K, Mattsson E, Bjorck L. J Biol Chem. 2002;277:34568–34572. doi: 10.1074/jbc.M206353200. [DOI] [PubMed] [Google Scholar]
- 49.Gebbink MF, Claessen D, Bouma B, Dijkhuizen L, Wosten HA. Nat Rev Microbiol. 2005;3:333–341. doi: 10.1038/nrmicro1127. [DOI] [PubMed] [Google Scholar]
- 50.Lundmark K, Westermark GT, Olsen A, Westermark P. Proc Natl Acad Sci U S A. 2005;102:6098–6102. doi: 10.1073/pnas.0501814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kisilevsky R. J Struct Biol. 2000;130:99–108. doi: 10.1006/jsbi.2000.4222. [DOI] [PubMed] [Google Scholar]
- 52.Prusiner SB. Brain Pathol. 1998;8:499–513. doi: 10.1111/j.1750-3639.1998.tb00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lansbury PT., Jr Proc Natl Acad Sci U S A. 1999;96:3342–3344. doi: 10.1073/pnas.96.7.3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uversky VN, Gillespie JR, Fink AL. Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 55.Booth DR, Sunde M, Bellotti V, Robinson CV, Hutchinson WL, Fraser PE, Hawkins PN, Dobson CM, Radford SE, Blake CC, Pepys MB. Nature. 1997;385:787–793. doi: 10.1038/385787a0. [DOI] [PubMed] [Google Scholar]
- 56.Galzitskaya OV, Garbuzynskiy SO. Proteins. 2006;63:144–154. doi: 10.1002/prot.20851. [DOI] [PubMed] [Google Scholar]


