Abstract
Nestled at the tip of a branch of the kinome, protein kinase C (PKC) family members are poised to transduce signals emanating from the cell surface. Cell membranes provide the platform for PKC function, supporting the maturation of PKC through phosphorylation, its allosteric activation by binding specific lipids, and, ultimately, promoting the downregulation of the enzyme. These regulatory mechanisms precisely control the level of signaling-competent PKC in the cell. Disruption of this regulation results in pathophysiological states, most notably cancer, where PKC levels are often grossly altered. This review introduces the PKC family and then focuses on recent advances in understanding the cellular regulation of its diacylglycerol-regulated members.
Keywords: calcium, diacylglycerol, phorbol esters
the family of pkc isozymes transduces the myriad of signals resulting from receptor-mediated hydrolysis of phospholipids, playing critical roles in diverse cellular functions. The discovery in the 1980s that they are the receptors for the potent tumor promoting phorbol esters, coupled with the discovery that they mediate signaling by the lipid second messenger diacylglycerol, secured a center stage position for this family of enzymes in cellular regulation (6, 66). Family members, themselves, are regulated by precise mechanisms that control their structure, function, and subcellular localization. This review discusses our current understanding of the mechanisms controlling protein kinase C signaling.
The PKC Family
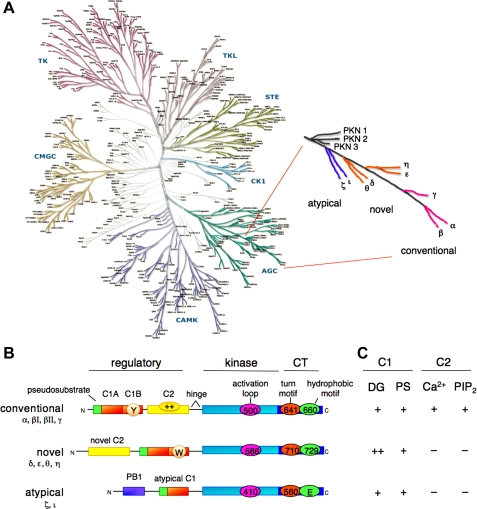
The PKC family occupies the tip of a branch of the AGC kinases from which the related kinases protein kinase N (PKN), Akt/PKB, p70 S6 kinase, and the phosphoinositide-dependent kinase-1 (PDK-1) diverge (Fig. 1A). The 10 members that populate the mammalian family evolved from the single PKC in saccharomyces cerivisiae, PKC1 (57, 98), and are grouped into three classes based on their domain composition (Fig. 1B) (71). This domain composition, in turn, dictates the cofactor regulation of the classes (Fig. 1C). At the very tip of the branch are the four conventional isozymes: PKCα, the first isozyme cloned (76); the alternatively spliced PKCβI and PKCβII (which differ in the last 43 amino acids) and PKCγ; next are the four novel isozymes, PKCδ, -ε, -η, and -θ; closest to the point of divergence are the atypical isozymes PKCζ and PKCι (human; murine isozyme is PKCλ).
Fig. 1.
Protein kinase C (PKC) family members, showing position on branch of AGC kinome, domain composition, and cofactor dependence. A: human kinome (reproduced from www.cellsignal.com/reference/kinase), showing the position of the AGC kinases (bottom right). PKC isozymes (enlarged on right) are poised on a branch that includes Akt, p70 S6 kinase, and PDK-1 (phosphoinositide-dependent kinase-1). The PKN family diverges, then the atypical PKC isozymes, the novel PKC isozymes, and finally, most divergent, the conventional PKC isozymes. B: domain composition of PKC family members, showing pseudosubstrate (green rectangle), C1 domain [orange rectangle; Y/W switch that dictates affinity for diacylglycerol (DG)-containing membranes indicated by circle in C1B domain], C2 domain [yellow rectangle; basic patch that drives binding to PIP2 (phosphatidylinositol-4,5-bisphosphate), indicated by oval with ++], connecting hinge segment, kinase domain (cyan), and carboxyl-terminal tail (CT; dark blue rectangle). Also shown are the 3 priming phosphorylations in the kinase domain and CT, with numbering indicated for PKCβII, PKCε, and PKCζ (note atypical PKC isozymes have Glu at phospho-acceptor position of hydrophobic motif). C: table showing dependence of PKC family members on C1 domain cofactors, DG, and phosphatidylserine (PS) and C2 domain cofactors Ca 2+ and PIP2.
All family members share the same architecture: a carboxyl-terminal kinase domain linked by a flexible hinge segment to an amino-terminal region containing regulatory modules (Fig. 1B) (69, 77). These regulatory modules confer sensitivity to the second messengers diacylglycerol (C1 domain) or Ca2+ (C2 domain), although, importantly, some isozymes have variants of these modules that do not bind ligand (8, 20).
Conventional isozymes contain tandem C1 domains that bind diacylglycerol and a C2 domain that binds anionic lipids in a Ca2+-dependent manner. The small globular C1 domain is also the binding site for the potent tumor-promoting phorbol esters (43, 100), which bind competitively with respect to diacylglycerol (87). The C1 domain stereospecifically binds the anionic phospholipid phosphatidylserine (46, 70). Pioneering work by Blumberg and colleagues, who developed lipophilic analogs of phorbol esters that allowed, for the first time, demonstration of specific and saturable binding of phorbol esters to cells (22), identified the stoichiometry of phorbol ester binding as 1 mol ligand/mol PKC (53); for most isozymes, it is the C1b domain that is the relevant binding module in the context of the full-length protein (49, 80). The C2 domain of conventional PKC isozymes binds anionic phospholipids with modest, but not stereospecific, selectivity for phosphatidylserine (16, 19, 46, 61). More significantly, it has a strong preference for phosphatidylinositol-4,5-bisphosphate (PIP2), and it is this preference for PIP2, which is mediated by a basic patch distal to the Ca2+ binding site (Fig. 1B; oval with ++ indicated in the C2 domain of conventional PKC isozymes), that selectively targets conventional PKC isozymes to the plasma membrane (20, 28, 59).
Novel isozymes also contain tandem C1 domains that bind diacylglycerol; these isozymes contain a variant of the C2 domain (novel C2) that lacks key residues that coordinate Ca2+ and, as a result, the novel isozymes are not sensitive to Ca2+. These isoforms are thus regulated only by diacylglycerol. Because these isozymes are regulated by only one membrane-targeting module, their affinity for diacylglycerol is two orders of magnitude higher than that for the conventional PKC isozymes (35). A single residue in the C1b domain tunes the module from a high-affinity diacylglycerol sensor in novel isozymes (which have a Trp at a conserved position on the membrane proximal moiety of the C1b domain) to a low-affinity diacylglycerol sensor for conventional isozymes (which have a Tyr at the equivalent position on their C1b domain) (23). This increased affinity allows novel PKC isozymes to respond to agonist-evoked increases in diacylglycerol. Conventional PKC isozymes depend on membrane recruitment by the Ca2+-regulated C2 domain to sense agonist-evoked diacylglycerol.
Atypical isozymes contain a variant of the C1 domain (atypical C1) with an impaired ligand-binding pocket that binds neither diacylglycerol nor phorbol esters (48, 81). Nor are they regulated by Ca2+; rather, protein-protein interactions provide the major driving force for controlling the function of these isozymes in cells. To this end, they contain a PB1 domain (Fig. 1B) involved in protein interactions (54) as well as a carboxyl-terminal PDZ ligand.
All isozymes have a conserved carboxyl-terminal tail (CT) that serves as a phosphorylation-dependent docking site for key regulatory molecules (Fig. 1B, CT; phosphorylation sites indicated by ovals). In addition, all isozymes have an autoinhibitory pseudosubstrate sequence (Fig. 1B, green rectangle) that maintains PKC in an inactive state by occupying the substrate-binding cavity. Although the structure of full-length PKC has remained refractory to elucidation, structure-function studies reveal that many other contacts maintain the enzyme in a “closed” state in the absence of membrane binding (59, 90).
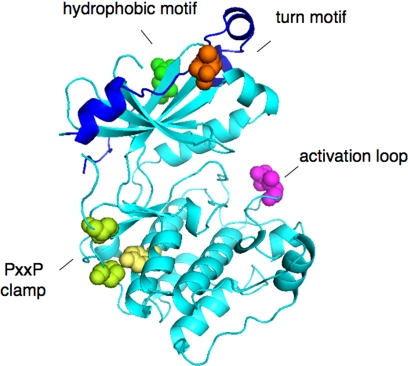
The past few years have seen the determination of the structure of the kinase domain of several PKC isozymes [PKCβII, PKCθ, and PKCι (38, 62, 99)]. Particularly noteworthy in the crystal structure of the PKCβII kinase domain is the clear identification of the three key priming phosphorylations discussed below: the activation loop and the two sites on the CT, which is well ordered in this structure (Fig. 2).
Fig. 2.
Structure of kinase domain of PKCβII showing priming site phosphorylations: activation loop (pink), turn motif (orange), and hydrophobic motif (green) (38). Also shown is the clamp between the PXXP motif (Pro, in green) and the conserved Tyr (yellow) of the αE helix.
Maturation of PKC
PKC isozymes are matured by a series of ordered, tightly coupled, and constitutive phosphorylations that are essential for the stability and catalytic competence of the enzyme (69, 78). Recent studies have identified two new players in this maturation: heat shock protein-90 (HSP90), whose interaction with a specific motif on PKC is essential to allow phosphorylations to occur (36), and the mammalian target of rapamycin (mTOR) complex 2 (mTORC2), a structure comprised of the kinase mTOR, Sin1, Rictor, and mLST8, whose integrity is required to allow the priming phosphorylations (39, 44).
Regulation by HSP90.
The earliest identified step in the maturation of conventional and novel PKC is the binding of the chaperone HSP90 and the co-chaperone Cdc37 to a molecular clamp in the kinase domain formed by a conserved PXXP motif (Fig. 2, Pro in green) in the CT with a conserved Tyr on the αE-helix of the kinase core (Fig. 2, Tyr in yellow) (36). This motif is present throughout the AGC family (47), suggesting that other kinases in this family will be similarly regulated by the binding of HSP90 to the surface aligned by the PXXP clamp. The integrity of this clamp is essential to allow HSP90 binding, an event that is required for the processing of conventional and novel PKC by phosphorylation: either inhibition of HSP90 or mutations of residues that maintain the clamp result in PKC that cannot be phosphorylated and is degraded. Perturbation of this key regulatory step provides one mechanism by which PKC becomes dysregulated in disease: a mutation in PKCα of the first Pro of this motif was recently identified as a cancer-driver mutation in glioblastoma (13a).
Regulation by priming phosphorylations.
Pulse-chase analyses revealing discrete mobility shifts during the “maturation” of PKC provided the first hint that the enzyme is processed by a series of ordered phosphorylation events (10). Mass spectrometric analysis of PKCβII identified three constitutively phosphorylated sites in the kinase domain of PKC that are conserved not only among all the PKC isozymes but among most of the AGC kinases, including PKB (50, 92). These phosphorylations occur on 1) the activation loop site at the entrance to the active site and 2) two carboxyl-terminal sites that were named the turn motif because, by analogy with PKA, the site was predicted to occupy the apex of a turn on the upper lobe of the kinase domain, and the hydrophobic motif because the phosphorylation site is flanked by hydrophobic residues (50, 68). Note that a phospho-mimetic, Glu, occupies the phospho-acceptor position of the hydrophobic motif of the atypical PKC isozymes.
ACTIVATION LOOP.
PDK-1 phosphorylates the activation loop site of all PKC isozymes, an event essential to generate a catalytically competent enzyme (17, 26, 55). Constructs of PKC that have a nonphosphorylatable residue at this site are not processed by phosphorylation and, because phosphorylation stabilizes PKC isozymes, are proteolytically-labile. For this reason, cells that lack PDK-1 have greatly reduced PKC levels (3). Phosphorylation at this site is constitutive for conventional and novel PKC isozymes but displays agonist dependence for atypical PKC isozymes. The activity of PDK-1 toward conventional PKC isozymes has been shown to be independent of phosphoinositides (88). Rather, phosphorylation is controlled by the conformation of PKC: newly synthesized PKC is in an open conformation in which the pseudosubstrate is removed from the substrate-binding cavity, thus unmasking the activation loop site to allow phosphorylation by PDK-1 (Fig. 3) (25). The maturation of PKC requires priming phosphorylation by PDK-1, but once the enzyme is phosphorylated at the two CT sites, phosphorylation at this site, at least for PKCβII, becomes dispensable (25). Indeed, this site is dephosphorylated in a serum-sensitive manner such that only about one-half of the pool of conventional PKC in cells cultured in serum is phosphorylated on the activation loop, yet is quantitatively phosphorylated at the two CT sites (50, 88).
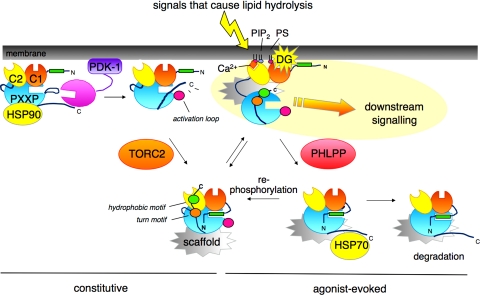
Fig. 3.
Model for the life cycle of PKC, from biosynthesis to degradation. Newly synthesized PKC (leftmost species) associates with a membrane fraction, where it is processed by a series of ordered and tightly coupled phosphorylations. Heat shock protein-90 (HSP90) binds to the PXXP clamp in the kinase domain right before the CT (see text), an event required for priming phosphorylations. Two upstream kinases control priming phosphorylations: PDK-1, bound to the exposed carboxyl terminus of newly synthesized PKC, phosphorylates the activation loop (pink circle); this step appears to be first, and necessary for the processing PKC. The mTORC2 complex promotes the phosphorylation of the turn motif (orange circle), second phosphorylation event, and hydrophobic motif (green circle), the final phosphorylation event. The fully phosphorylated “mature” PKC localizes to the cytosol with the pseudosubstrate (green rectangle) occupying the substrate-binding cavity (bottom species on left). Signals that cause lipid hydrolysis recruit PKC to membranes. For conventional PKC isozymes, binding of Ca2+ to the C2 domain recruits them to the plasma membrane via interaction with PIP2, an event that allows efficient binding of the membrane-embedded ligand DG (top right species). For novel PKC isozymes, the intrinsic affinity of the C1 domain is sufficiently high to allow direct recruitment to membranes by agonist-evoked levels of DG. Membrane-bound PKC adopts an open conformation, in which the pseudosubstrate is released from the kinase domain, allowing downstream signaling (top right species). This open conformation is sensitive to dephosphorylation: the phosphatase PHLPP (PH domain leucine-rich repeat protein phosphatase) dephosphorylates the hydrophobic motif, an event that shunts PKC to the detergent-insoluble fraction where it is further dephosphorylated and degraded (bottom right species). Note: molecular chaperone HSP70 binds the dephosphorylated turn motif (bottom middle species), an event that promotes rephosphorylation of PKC and reentry into the pool of signaling-competent enzyme, thus sustaining the lifetime of the enzyme. Specific protein scaffolds (gray structures shown associated with matured PKC) bind specific isozymes and specific species [e.g., RACKs bind active PKC, but other scaffolds bind inactive PKC] to poise PKC in specific cellular microdomains. The amplitude of the PKC signal and similarly the responsiveness of the cell to low concentrations of agonist are ultimately controlled by how much PKC is in the cell.
Phosphorylation by PDK-1 is likely the first phosphorylation event in the processing of PKC by phosphorylation. First, as noted above, replacement of the Thr at the activation loop of conventional PKC isozymes with neutral, nonphosphorylable residues (Ala or Val) prevents the subsequent phosphorylations on the CT sites, indicating that phosphorylation of the activation loop is necessary to allow CT phosphorylation (15, 74). Second, locking negative charge on either of the COOH-terminal sites by Glu substitution does not significantly alter the rate of processing of PKCβII to the fully phosphorylated species, indicating that phosphorylation of the CT sites is not rate limiting, as would be expected if these were the first sites modified (88). Taken together, the most likely mechanism for the PDK-1 step is that it is first and necessary for the processing of PKC to the fully phosphorylated form. Because perturbation of the PDK-1 step prevents the maturation of PKC by phosphorylation, thus resulting in species of PKC that are highly susceptible to degradation, this step plays a critical role in controlling cellular levels of PKC.
TURN MOTIF.
Structural analysis reveals that phosphorylation of the turn motif stabilizes the structure of mature PKC by anchoring the carboxyl-terminal tail on the upper lobe of the kinase (41). Specifically, the phosphate on the turn motif binds a cluster of basic residues in a pocket above the ATP-binding Gly loop to stablize the active conformation of the kinase. Curiously, mutation of the turn motif residue in PKCβII to Ala results in compensating phosphorylations that accompany the processing of PKC to a functional species; however, mutation of adjacent Thr to Ala prevents these compensating phosphorylations and abolishes the processing of PKCβII (27). These data suggest that the precise presence of phosphate on the actual turn motif Thr is dispensible, supported by the finding that similar mutation in PKCα allows maturation of PKCα, albeit to a more thermally-labile and phosphatase-senstive species (9).
It has recently been shown that phosphorylation of the turn motif depends on the mTORC2 complex (29, 44, 45). Importantly, PKC cannot be processed by phosphorylation in cells lacking this complex, and, because the unphosphorylated species is unstable, it is degraded (39, 44). Whether mTORC2 controls the phosphorylation of this site indirectly, for example by chaperoning or positioning newly synthesized PKC for processing by phosphorylation, or by activating another upstream kinase for this site, or whether it directly phosphorylates PKC is unclear. It is noteworthy that mTORC2 is not able to phosphorylate PKC in vitro at the turn motif (44).
Cells lacking mTORC2 have grossly reduced levels of PKC (39, 44, 84), consistent with the essential role of priming phosphorylations in stabilizing PKC. Thus, steps that interfere with the ability of mTORC2 to control turn motif phosphorylation are another Achilles’ heel in PKC regulation.
HYDROPHOBIC MOTIF.
Kinetic analyses with pure protein have revealed that PKC autophosphorylates by an intramolecular reaction at the hydrophobic motif (7). Whether this is the mechanism of regulation in cells has been difficult to resolve. This site can be phosphorylated in vitro by a number of kinases, including mTORC2 (44). Modulation of this site by the TOR kinase was first reported by Parker et al. (101), who showed that retention of phosphate on the hydrophobic motif of PKCδ was sensitive to the inhibitor rapamycin. More recent studies have established that the hydrophobic motif is not phosphorylated in mTORC2-deficient cells. This may be because TOR directly controls the phosphorylation of this site or because a prerequisite event to allow hydrophobic motif phosphorylation has not taken place (27).
The phosphorylation of the hydrophobic motif is also controlled by the interaction of HSP90 with the PXXP clamp described; inhibitors of HSP90 slow the phosphorylation of this site but not the turn motif site (36). It is noteworthy that PKCζ is not phosphoryated at the hydrophobic motif (constitutive negative charge at this position), nor does its PXXP motif control HSP90 binding. Thus, HSP90 facilitates the phosphorylation of the hydrophobic motif of conventional and novel PKC isozymes by its interaction with the PXXP clamp.
Stabilization of phosphorylation by active site inhibitors.
Constructs of PKC that are catalytically inactive because an essential Lys in the active site that coordinates the α/β phosphates of ATP has been mutated (72) are not processed by phosphorylation (13). It has recently been shown that the binding of active site inhibitors to these constructs stabilizes the retention of phosphate at the priming sites, including the hydrophobic motif (13). This stabilization of phosphate at the hydrophobic motif upon occupancy of the active site also is observed for PKB (73). This suggests that occupancy of the active site by inhibitors stabilizes a conformation that either promotes the phosphorylation of PKC or protects the phosphorylated enzyme from dephosphorylation. It is noteworthy that occupancy of the active site with the pseudo-substrate (i.e., in the inactive conformation of PKC) (24) or with peptide substrates protects pure PKC from dephosphorylation (25). Consistent with active site occupancy “freezing” the CT in a conformation that masks the phosphorylation sites, structural studies with PKA have shown that the CT is highly flexible in the apostructure and highly ordered when active site inhibitors are bound (1, 65). Note the structure of the kinase domain of PKCβII that shows that a highly ordered CT was obtained with bound active-site inhibitor (38). The ability of active site inhibitors to allow accumulation of phosphate on kinase-dead constructs of PKC supports the possibility that other kinases, perhaps TORC2, modify the hydrophobic motif in cells (13); however, mechanistic conclusions await the dissection of phosphorylation versus dephosphorylation steps in promoting phosphate retention.
Regulation by Lipid Second Messengers
The activity of mature conventional and novel PKC isozymes is acutely regulated by binding diacylglycerol, an event that is assisted by Ca2+ for the conventional PKC isozymes (reviewed recently in Refs. 6, 33, 67, 89). For conventional PKC isozymes, the mature enzyme localizes primarily to the cytosol, where it is likely maintained in precise microenvironments by scaffold interactions (86). Agonist-evoked hydrolysis of PIP2 generates two key second messengers for conventional PKC isozymes: Ca2+ and diacylglycerol. Binding of Ca2+ to the C2 domain pretargets PKC to the plasma membrane, where it binds anionic phospholipids, with selectivity for PIP2. Once engaged on the membrane, the C1 domain binds its membrane-embedded ligand diacylglycerol, an interaction that is enhanced by stereospecific binding to phosphatidylserine. The coordinated engagement of both the C1 and C2 domains on the membrane provides the energy to release the autoinhibitory pseudo-substrate sequence from the substrate-binding cavity, allowing substrate phosphorylation. As noted above, novel PKC isozymes are able to respond to agonist-evoked changes in diacylglycerol because their C1b domain has a two-orders-of-magnitude higher affinity for diacylglycerol-containing membranes than the C1b domain of conventional isozymes (35). Note that the higher affinity of novel isozymes for diacylglycerol results in significant basal localization to membranes enriched in diacylglycerol, notably Golgi (14).
The advent of imaging technologies to visualize the generation of second messengers, the subcellular location of PKC, and the activity of PKC at precise cellular locations has allowed much insight into the spatiotemporal dynamics of PKC signaling (12, 32, 33, 96). Fluorescence energy transfer (FRET)-based activity reporters have revealed that Ca2+ oscillations, with or without diacylglycerol oscillations, drive oscillations of the activity of conventional PKC (5, 93, 97). They have also revealed that oscillatory activity does not necessarily require oscillatory membrane translocation: oscillations in Ca2+, under conditions were diacylglycerol levels are sustained and thus retain PKC at the plasma membrane, result in oscillations of activity (97). Live-cell imaging studies have also established that there are different “signatures” of PKC activity at defined microenvironments in cells (32). That is, the level of basal signaling and rate, magnitude, and duration of agonist-evoked signaling vary depending on cellular location and isozyme. Most notably, rapid rises in intracellular Ca2+ drive rapid activation of conventional PKC isozymes at the plasma membrane, where they are likely targeted via unique interactions of their C2 domain with PIP2, a lipid enriched at the plasma membrane (59). Golgi membranes have relatively high levels of diacylglycerol, mediating a “basal” interaction of novel PKC isozymes at the Golgi (14). Agonist-evoked increases in diacylglycerol are highly sustained at the Golgi, resulting in sustained activation of both novel and conventional PKC isozymes at Golgi. In contrast, diacylglycerol at the plasma membrane is transient, resulting in transient activation of conventional PKC isozymes at the plasma membrane. Use of FRET-based activity reporters has recently shown that naturally occurring mutations in PKCγ that are associated with spinocerebellar ataxia type 14 result in reduced signaling output in cells (95).
Regulation by Scaffold Interactions
The importance of protein scaffolds in coordinating PKC activity was first recognized by Mochly-Rosen and colleagues almost 20 years ago (63, 64). Her laboratory went on to identify and characterize a number of structurally unrelated scaffold proteins named receptors for activated C kinase (RACKs) that have in common the ability to bind specific PKC isozymes in a manner that relieves autoinhibition of the isozymes (83). Specifically, her laboratory identified stretches of sequence on scaffold proteins that resemble sequences in specific PKC isozymes and proposed that these sequences participate in intramolecular interactions that clamp PKC in an autoinhibited state. The sequence on the scaffold would compete for these intramolecular interactions, binding PKC and trapping it in an activated conformation. Thus, binding to such scaffolds not only localizes PKC but has the potential to sustain PKC signaling in the absence of second messenger binding. The use of peptides based on these sequences to disrupt the interaction of specific PKC isozymes with their cognate RACKs has been used to modulate the cellular function of PKC isozymes (18). Such peptides have been particularly valuable pharmacological tools to regulate the activity of PKC isozymes in heart disease (75). Note that there are scaffolds for all conformations and species of PKC, from never-phosphorylated, phosphorylated but inactive, phosphorylated and active (e.g., RACKs), to dephosphorylated PKC.
The demonstration by Zuker and colleagues over a decade ago, that disruption of the interaction between the eye-specific PKC in Drosophila melanogaster and the PDZ scaffold InaD impairs light signaling, continues to provide one of the most elegant demonstrations of the role of protein scaffolds in coordinating PKC activity (91). It is now clear that scaffold interactions are an integral part of PKC regulation, poising specific isozymes near key protein and lipid regulators and near substrates.
Mislocalization likely plays a role in pathophysiological states. Most strikingly, Joubert and coworkers (79, 94) identified a natural mutation in PKCα (D294G in the hinge region) in human pituitary and thyroid tumors, which results in loss of selective targeting to cell-cell contacts. They then went on to show that the sequence around this mutation (GDE in wild-type enzyme) is a targeting sequence that drives PKCα and PKCε to cell-cell contacts (82). This sequence was recently shown to overlap with a 14-3-3 binding region that alternatively targets PKCε from cell-cell contacts (via GDE motif) to cytosolic sequestration (via binding 14-3-3), where a new set of functions is unveiled (21). With respect to the latter, Parker and coworkers reported that PKCε is phosphorylated at three sites in the hinge segment (see Fig. 1B), creating a binding site for 14-3-3 (85). This binding stabilizes an open conformation of PKCε and promotes localized activity at the midbody during cytokinesis. The ability to switch from cytosolic sequestration via 14-3-3 binding to cell-cell contact binding via GDE motif underscores the complex regulation of PKC function by protein interactions.
Downregulation
The life cycle of conventional and novel PKC isozymes is terminated by a process referred to as downregulation (40). In the absence of chronic stimulation, these PKC isozymes have a relatively long half-life (on the order of days for conventional PKC in cells grown in culture). However, sustained activation, as occurs upon treatment of cells with phorbol esters, results in the rapid degradation of PKC. In fact, phorbol ester treatment has been used as a mechanism to deplete cells of the phorbol ester-responsive isozymes, the conventional and novel PKC isozymes (42, 56). Note that atypical isozymes do not bind phorbol esters and are not downregulated in the same manner as their phorbol ester-binding counterparts.
In the inactive conformation, conventional and novel PKC isozymes are relatively resistant to dephosphorylation. However, when they are in the open, membrane-bound conformation, their sensitivity to dephosphorylation increases by two orders of magnitude (24). Thus, prolonged activation of PKC results in dephosphorylation, with the dephosphorylated species being unstable and shunted to degradation (see Fig. 3). The recently discovered PH domain leucine-rich repeat protein phosphatase [PHLPP; (11)] dephosphorylates the hydrophobic motif of conventional and novel PKC isozymes, an event that shunts them to a detergent-insoluble cell fraction, where they are dephosphorylated at the turn motif by an okadaic-sensitive phosphatase (34). The precise mechanisms for degradation remain to be elucidated, and it is likely that different isozymes will be controlled by unique “turn-off” mechanisms.
Summary
The past few years have seen the identification of new key players that regulate the maturation, subcellular location, and downregulation of PKC. HSP90 and mTORC2 are key components of the maturation of PKC, and disruption of either interaction results in the inability of PKC to process into a stable and catalytically competent conformation. Conversely, PHLPP is a key component in the dephosphorylation step preceding downregulation of PKC, and inhibition of this dephosphorylation results in accumulation of PKC. Given that PKC levels are perturbed in so many cancers (2, 31, 37, 52, 60), understanding the mechanisms that control the levels of PKC will have important ramifications for potential therapies. Currently, the use of inhibitors of PKC activity show promise in preventing tumor growth (30, 51), although clinical trials have been somewhat disappointing (58). Because the signaling amplitude of PKC is ultimately dependent on the levels of PKC poised in the cell, targeting mechanisms that control the levels of PKC offer an alternate approach to controlling PKC signaling.
GRANTS
This work was supported by NIH Grant GM-43154.
DISCLOSURES
No conflicts of interest are reported by the author(s).
ACKNOWLEDGMENTS
I thank members of my lab for helpful comments.
REFERENCES
- 1.Akamine P, Madhusudan Wu J, Xuong NH, Ten Eyck LF, Taylor SS. Dynamic features of cAMP-dependent protein kinase revealed by apoenzyme crystal structure. J Mol Biol 327: 159–171, 2003 [DOI] [PubMed] [Google Scholar]
- 2.Aziz MH, Manoharan HT, Church DR, Dreckschmidt NE, Zhong W, Oberley TD, Wilding G, Verma AK. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res 67: 8828–8838, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Balendran A, Hare GR, Kieloch A, Williams MR, Alessi DR. Further evidence that 3-phosphoinositide-dependent protein kinase-1 (PDK1) is required for the stability and phosphorylation of protein kinase C (PKC) isoforms. FEBS Lett 484: 217–223, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Bartlett PJ, Young KW, Nahorski SR, Challiss RA. Single cell analysis and temporal profiling of agonist-mediated inositol 1,4,5-trisphosphate, Ca2+, diacylglycerol, and protein kinase C signaling using fluorescent biosensors. J Biol Chem 280: 21837–21846, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Battaini F, Mochly-Rosen D. Happy birthday protein kinase C: past, present and future of a superfamily. Pharmacol Res 55: 461–466, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behn-Krappa A, Newton AC. The hydrophobic phosphorylation motif of conventional protein kinase C is regulated by autophosphorylation. Curr Biol 9: 728–737, 1999 [DOI] [PubMed] [Google Scholar]
- 8.Blumberg PM, Kedei N, Lewin NE, Yang D, Czifra G, Pu Y, Peach ML, Marquez VE. Wealth of opportunity—the C1 domain as a target for drug development. Curr Drug Targets 9: 641–652, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornancin F, Parker PJ. Phosphorylation of threonine 638 critically controls the dephosphorylation and inactivation of protein kinase C α. Curr Biol 6: 1114–1123, 1996 [DOI] [PubMed] [Google Scholar]
- 10.Borner C, Filipuzzi I, Wartmann M, Eppenberger U, Fabbro D. Biosynthesis and posttranslational modifications of protein kinase C in human breast cancer cells. J Biol Chem 264: 13902–13909, 1989 [PubMed] [Google Scholar]
- 11.Brognard J, Newton CA. PHLPPing the switch on Akt and protein kinase C signaling. Trends Endocrinol Metab in press, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brumbaugh J, Schleifenbaum A, Gasch A, Sattler M, Schultz C. A dual parameter FRET probe for measuring PKC and PKA activity in living cells. J Am Chem Soc 128: 24–25, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Cameron AJ, Escribano C, Saurin AT, Kostelecky B, Parker PJ. PKC maturation is promoted by nucleotide pocket occupation independently of intrinsic kinase activity. Nat Struct Mol Biol 16: 624–630, 2009 [DOI] [PubMed] [Google Scholar]
- 13a.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrasco S, Merida I. Diacylglycerol-dependent binding recruits PKCtheta and RasGRP1 C1 domains to specific subcellular localizations in living T lymphocytes. Mol Biol Cell 15: 2932–2942, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cazaubon S, Bornancin F, Parker PJ. Threonine-497 is a critical site for permissive activation of protein kinase C alpha. Biochem J 301: 443–448, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cho W, Stahelin RV. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta 1761: 838–849, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Chou MM, Hou W, Johnson J, Graham LK, Lee MH, Chen CS, Newton AC, Schaffhausen BS, Toker A. Regulation of protein kinase C zeta by PI 3-kinase and PDK-1. Curr Biol 8: 1069–1077, 1998 [DOI] [PubMed] [Google Scholar]
- 18.Churchill EN, Qvit N, Mochly-Rosen D. Rationally designed peptide regulators of protein kinase C. Trends Endocrinol Metab 20: 25–33, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conesa-Zamora P, Lopez-Andreo MJ, Gomez-Fernandez JC, Corbalan-Garcia S. Identification of the Phosphatidylserine Binding Site in the C2 Domain that Is Important for PKCalpha Activation and in Vivo Cell Localization. Biochemistry 40: 13898–13905, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Corbalan-Garcia S, Guerrero-Valero M, Marin-Vicente C, Gomez-Fernandez JC. The C2 domains of classical/conventional PKCs are specific PtdIns(4,5)P(2)-sensing domains. Biochem Soc Trans 35: 1046–1048, 2007 [DOI] [PubMed] [Google Scholar]
- 21.Diouf B, Collazos A, Labesse G, Macari F, Choquet A, Clair P, Gauthier-Rouviere C, Guerineau NC, Jay P, Hollande F, Joubert D. A 20-amino acid module of protein kinase C(epsilon) involved in translocation and selective targeting at cell-cell contacts. J Biol Chem 284: 18808–18815, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Driedger PE, Blumberg PM. Specific binding of phorbol ester tumor promoters. Proc Natl Acad Sci USA 77: 567–571, 1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dries DR, Gallegos LL, Newton AC. A single residue in the C1 domain sensitizes novel protein kinase C isoforms to cellular diacylglycerol production. J Biol Chem 282: 826–830, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Dutil EM, Keranen LM, DePaoli-Roach AA, Newton AC. In vivo regulation of protein kinase C by trans-phosphorylation followed by autophosphorylation. J Biol Chem 269: 29359–29362, 1994 [PubMed] [Google Scholar]
- 25.Dutil EM, Newton AC. Dual role of pseudosubstrate in the coordinated regulation of protein kinase C by phosphorylation and diacylglycerol. J Biol Chem 275: 10697–10701, 2000 [DOI] [PubMed] [Google Scholar]
- 26.Dutil EM, Toker A, Newton AC. Regulation of conventional protein kinase C isozymes by phosphoinositide-dependent kinase 1 (PDK-1). Curr Biol 8: 1366–1375, 1998 [DOI] [PubMed] [Google Scholar]
- 27.Edwards AS, Faux MC, Scott JD, Newton AC. Carboxyl-terminal phosphorylation regulates the function and subcellular localization of protein kinase C betaII. J Biol Chem 274: 6461–6468, 1999 [DOI] [PubMed] [Google Scholar]
- 28.Evans JH, Murray D, Leslie CC, Falke JJ. Specific translocation of protein kinase Calpha to the plasma membrane requires both Ca2+ and PIP2 recognition by its C2 domain. Mol Biol Cell 17: 56–66, 2006. 16236797 [Google Scholar]
- 29.Facchinetti V, Ouyang W, Wei H, Soto N, Lazorchak A, Gould C, Lowry C, Newton AC, Mao Y, Miao RQ, Sessa WC, Qin J, Zhang P, Su B, Jacinto E. The mammalian target of rapamycin complex 2 controls folding and stability of Akt and protein kinase C. EMBO J 27: 1932–1943, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fields AP, Calcagno SR, Krishna M, Rak S, Leitges M, Murray NR. Protein kinase Cbeta is an effective target for chemoprevention of colon cancer. Cancer Res 69: 1643–1650, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fields AP, Frederick LA, Regala RP. Targeting the oncogenic protein kinase Ciota signalling pathway for the treatment of cancer. Biochem Soc Trans 35: 996–1000, 2007 [DOI] [PubMed] [Google Scholar]
- 32.Gallegos LL, Kunkel MT, Newton AC. Targeting protein kinase C activity reporter to discrete intracellular regions reveals spatiotemporal differences in agonist-dependent signaling. J Biol Chem 281: 30947–30956, 2006 [DOI] [PubMed] [Google Scholar]
- 33.Gallegos LL, Newton AC. Spatiotemporal dynamics of lipid signaling: protein kinase C as a paradigm. IUBMB Life 60: 782–789, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao T, Brognard J, Newton AC. The phosphatase PHLPP controls the cellular levels of protein kinase C. J Biol Chem 283: 6300–6311, 2008 [DOI] [PubMed] [Google Scholar]
- 35.Giorgione JR, Lin JH, McCammon JA, Newton AC. Increased membrane affinity of the C1 domain of protein kinase Cdelta compensates for the lack of involvement of its C2 domain in membrane recruitment. J Biol Chem 281: 1660–1669, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gould CM, Kannan N, Taylor SS, Newton AC. The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem 284: 4921–4935, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griner EM, Kazanietz MG. Protein kinase C and other diacylglycerol effectors in cancer. Nat Rev Cancer 7: 281–294, 2007 [DOI] [PubMed] [Google Scholar]
- 38.Grodsky N, Li Y, Bouzida D, Love R, Jensen J, Nodes B, Nonomiya J, Grant S. Structure of the catalytic domain of human protein kinase C beta II complexed with a bisindolylmaleimide inhibitor. Biochemistry 45: 13970–13981, 2006 [DOI] [PubMed] [Google Scholar]
- 39.Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 11: 859–871, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Hansra G, Garcia-Paramio P, Prevostel C, Whelan RD, Bornancin F, Parker PJ. Multisite dephosphorylation and desensitization of conventional protein kinase C isotypes. Biochem J 342: 337–344, 1999 [PMC free article] [PubMed] [Google Scholar]
- 41.Hauge C, Antal TL, Hirschberg D, Doehn U, Thorup K, Idrissova L, Hansen K, Jensen ON, Jorgensen TJ, Biondi RM, Frodin M. Mechanism for activation of the growth factor-activated AGC kinases by turn motif phosphorylation. EMBO J 26: 2251–2261, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang FL, Yoshida Y, Cunha-Melo JR, Beaven MA, Huang KP. Differential down-regulation of protein kinase C isozymes. J Biol Chem 264: 4238–4243, 1989 [PubMed] [Google Scholar]
- 43.Hurley JH. Membrane binding domains. Biochim Biophys Acta 1761: 805–811, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. EMBO J 27: 1919–1931, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jacinto E, Lorberg A. TOR regulation of AGC kinases in yeast and mammals. Biochem J 410: 19–37, 2008 [DOI] [PubMed] [Google Scholar]
- 46.Johnson JE, Giorgione J, Newton AC. The C1 and C2 domains of protein kinase C are independent membrane targeting modules, with specificity for phosphatidylserine conferred by the C1 domain. Biochemistry 39: 11360–11369, 2000 [DOI] [PubMed] [Google Scholar]
- 47.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci USA 104: 1272–1277, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kazanietz MG, Bustelo XR, Barbacid M, Kolch W, Mischak H, Wong G, Pettit GR, Bruns JD, Blumberg PM. Zinc finger domains and phorbol ester pharmacophore: analysis of binding to mutated form of protein kinase C ζ and the vav and c-raf proto-oncogene products. J Biol Chem 269: 11590–11594, 1994 [PubMed] [Google Scholar]
- 49.Kazanietz MG, Wang S, Milne GWA, Lewin NE, Liu HL, Blumberg PM. Residues in the second cysteine-rich region of protein kinase c δ relevant to phorbol ester binding as revealed by site-directed mutagenesis. J Biol Chem 270: 21852–21859, 1995 [DOI] [PubMed] [Google Scholar]
- 50.Keranen LM, Dutil EM, Newton AC. Protein kinase C is regulated in vivo by three functionally distinct phosphorylations. Curr Biol 5: 1394–1403, 1995 [DOI] [PubMed] [Google Scholar]
- 51.Kim J, Choi YL, Vallentin A, Hunrichs BS, Hellerstein MK, Peehl DM, Mochly-Rosen D. Centrosomal PKCbetaII and pericentrin are critical for human prostate cancer growth and angiogenesis. Cancer Res 68: 6831–6839, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Koivunen J, Aaltonen V, Peltonen J. Protein kinase C (PKC) family in cancer progression. Cancer Lett 235: 1–10, 2006 [DOI] [PubMed] [Google Scholar]
- 53.König B, DiNitto PA, Blumberg PM. Stoichiometric binding of diacylglycerol to the phorbol ester receptor. J Cell Biochem 29: 37–44, 1985 [DOI] [PubMed] [Google Scholar]
- 54.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem 278: 34568–34581, 2003 [DOI] [PubMed] [Google Scholar]
- 55.Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science 281: 2042–2045, 1998 [DOI] [PubMed] [Google Scholar]
- 56.Leontieva OV, Black JD. Identification of two distinct pathways of protein kinase Calpha down-regulation in intestinal epithelial cells. J Biol Chem 279: 5788–5801, 2004 [DOI] [PubMed] [Google Scholar]
- 57.Levin DE, Fields FO, Kunisawa R, Bishop JM, Thorner J. A candidate protein kinase C gene, pkc1, is required for the S. cerevisiae cell cycle. Cell 62: 213–224, 1990 [DOI] [PubMed] [Google Scholar]
- 58.Mackay HJ, Twelves CJ. Targeting the protein kinase C family: are we there yet? Nat Rev Cancer 7: 554–562, 2007 [DOI] [PubMed] [Google Scholar]
- 59.Marin-Vicente C, Nicolas FE, Gomez-Fernandez JC, Corbalan-Garcia S. The PtdIns(4,5)P2 ligand itself influences the localization of PKCalpha in the plasma membrane of intact living cells. J Mol Biol 377: 1038–1052, 2008 [DOI] [PubMed] [Google Scholar]
- 60.Martiny-Baron G, Fabbro D. Classical PKC isoforms in cancer. Pharmacol Res 55: 477–486, 2007 [DOI] [PubMed] [Google Scholar]
- 61.Medkova M, Cho W. Interplay of C1 and C2 domains of protein kinase C-alpha in its membrane binding and activation. J Biol Chem 274: 19852–19861, 1999 [DOI] [PubMed] [Google Scholar]
- 62.Messerschmidt A, Macieira S, Velarde M, Badeker M, Benda C, Jestel A, Brandstetter H, Neuefeind T, Blaesse M. Crystal structure of the catalytic domain of human atypical protein kinase C-iota reveals interaction mode of phosphorylation site in turn motif. J Mol Biol 352: 918–931, 2005 [DOI] [PubMed] [Google Scholar]
- 63.Mochly-Rosen D, Gordon AS. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J 12: 35–42, 1998 [PubMed] [Google Scholar]
- 64.Mochly-Rosen D, Khaner H, Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc Natl Acad Sci USA 88: 3997–4000, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Narayana N, Diller TC, Koide K, Bunnage ME, Nicolaou KC, Brunton LL, Xuong NH, Ten Eyck LF, Taylor SS. Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase. Biochemistry 38: 2367–2376, 1999 [DOI] [PubMed] [Google Scholar]
- 66.Newton AC. Diacylglycerol's affair with protein kinase C turns 25. Trends Pharmacol Sci 25: 175–177, 2004 [DOI] [PubMed] [Google Scholar]
- 67.Newton AC. Lipid activation of protein kinases. J Lipid Res 50Suppl: S266–S271, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Newton AC. Protein kinase C: structural and spatial regulation by phosphorylation, cofactors, and macromolecular interactions. Chem Rev 101: 2353–2364, 2001 [DOI] [PubMed] [Google Scholar]
- 69.Newton AC. Regulation of the ABC kinases by phosphorylation: protein kinase C as a paradigm. Biochem J 370: 361–371, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Newton AC, Johnson JE. Protein kinase C: a paradigm for regulation of protein function by two membrane-targeting modules. Biochim Biophys Acta 1376: 155–172, 1998 [DOI] [PubMed] [Google Scholar]
- 71.Nishizuka Y. Protein kinase C and lipid signaling for sustained cellular responses. FASEB J 9: 484–496, 1995 [PubMed] [Google Scholar]
- 72.Ohno S, Konno Y, Akita Y, Yano A, Suzuki K. A point mutation at the putative ATP-binding site of protein kinase C alpha abolishes the kinase activity and renders it down-regulation-insensitive. A molecular link between autophosphorylation and down-regulation. J Biol Chem 265: 6296–6300, 1990 [PubMed] [Google Scholar]
- 73.Okuzumi T, Fiedler D, Zhang C, Gray DC, Aizenstein B, Hoffman R, Shokat KM. Inhibitor hijacking of Akt activation. Nat Chem Biol 5: 484–493, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orr JW, Newton AC. Requirement for negative charge on “activation loop” of protein kinase C. J Biol Chem 269: 27715–27718, 1994 [PubMed] [Google Scholar]
- 75.Palaniyandi SS, Sun L, Ferreira JC, Mochly-Rosen D. Protein kinase C in heart failure: a therapeutic target? Cardiovasc Res 82: 229–239, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Parker PJ, Coussens L, Totty N, Rhee L, Young S, Chen E, Stabel S, Waterfield MD, Ullrich A. The complete primary structure of protein kinase C—the major phorbol ester receptor. Science 233: 853–859, 1986 [DOI] [PubMed] [Google Scholar]
- 77.Parker PJ, Murray-Rust J. PKC at a glance. J Cell Sci 117: 131–132, 2004 [DOI] [PubMed] [Google Scholar]
- 78.Parker PJ, Parkinson SJ. AGC protein kinase phosphorylation and protein kinase C. Biochem Soc Trans 29: 860–863, 2001 [DOI] [PubMed] [Google Scholar]
- 79.Prevostel C, Alvaro V, de Boisvilliers F, Martin A, Jaffiol C, Joubert D. The natural protein kinase C alpha mutant is present in human thyroid neoplasms. Oncogene 11: 669–674, 1995 [PubMed] [Google Scholar]
- 80.Pu Y, Garfield SH, Kedei N, Blumberg PM. Characterization of the differential roles of the twin C1a and C1b domains of protein kinase C-delta. J Biol Chem 284: 1302–1312, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pu Y, Peach ML, Garfield SH, Wincovitch S, Marquez VE, Blumberg PM. Effects on ligand interaction and membrane translocation of the positively charged arginine residues situated along the C1 domain binding cleft in the atypical protein kinase C isoforms. J Biol Chem 281: 33773–33788, 2006 [DOI] [PubMed] [Google Scholar]
- 82.Quittau-Prevostel C, Delaunay N, Collazos A, Vallentin A, Joubert D. Targeting of PKCalpha and epsilon in the pituitary: a highly regulated mechanism involving a GD(E)E motif of the V3 region. J Cell Sci 117: 63–72, 2004 [DOI] [PubMed] [Google Scholar]
- 83.Ron D, Mochly-Rosen D. An autoregulatory region in protein kinase C: the pseudoanchoring site. Proc Natl Acad Sci USA 92: 492–496, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol 14: 1296–1302, 2004 [DOI] [PubMed] [Google Scholar]
- 85.Saurin AT, Brownlow N, Parker PJ. Protein kinase C epsilon in cell division: control of abscission. Cell Cycle 8: 549–555, 2009 [DOI] [PubMed] [Google Scholar]
- 86.Schechtman D, Mochly-Rosen D. Adaptor proteins in protein kinase C-mediated signal transduction. Oncogene 20: 6339–6347, 2001 [DOI] [PubMed] [Google Scholar]
- 87.Sharkey NA, Leach KL, Blumberg PM. Competitive inhibition by diacylglycerol of specific phorbol ester binding. Proc Natl Acad Sci USA 81: 607–610, 1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sonnenburg ED, Gao T, Newton AC. The phosphoinositide dependent kinase, PDK-1, phosphorylates conventional protein kinase C isozymes by a mechanism that is independent of phosphoinositide-3-kinase. J Biol Chem 28: 45289–45297, 2001 [DOI] [PubMed] [Google Scholar]
- 89.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev 88: 1341–1378, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stensman H, Larsson C. Identification of acidic amino acid residues in the protein kinase C alpha V5 domain that contribute to its insensitivity to diacylglycerol. J Biol Chem 282: 28627–28638, 2007 [DOI] [PubMed] [Google Scholar]
- 91.Tsunoda S, Sierralta J, Sun Y, Bodner R, Suzuki E, Becker A, Socolich M, Zuker CS. A multivalent PDZ-domain protein assembles signalling complexes in a G-protein-coupled cascade. Nature 388: 243–249, 1997 [DOI] [PubMed] [Google Scholar]
- 92.Tsutakawa SE, Medzihradszky KF, Flint AJ, Burlingame AL, Koshland DE., Jr Determination of in vivo phosphorylation sites in protein kinase C. J Biol Chem 270: 26807–26812, 1995 [DOI] [PubMed] [Google Scholar]
- 93.Uchino M, Sakai N, Kashiwagi K, Shirai Y, Shinohara Y, Hirose K, Iino M, Yamamura T, Saito N. Isoform-specific phosphorylation of metabotropic glutamate receptor 5 by protein kinase C (PKC) blocks Ca2+ oscillation and oscillatory translocation of Ca2+-dependent PKC. J Biol Chem 279: 2254–2261, 2004 [DOI] [PubMed] [Google Scholar]
- 94.Vallentin A, Lo TC, Joubert D. A single point mutation in the V3 region affects protein kinase Calpha targeting and accumulation at cell-cell contacts. Mol Cell Biol 21: 3351–3363, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Verbeek DS, Goedhart J, Bruinsma L, Sinke RJ, Reits EA. PKC gamma mutations in spinocerebellar ataxia type 14 affect C1 domain accessibility and kinase activity leading to aberrant MAPK signaling. J Cell Sci 121: 2339–2349, 2008 [DOI] [PubMed] [Google Scholar]
- 96.Violin JD, Newton AC. Pathway illuminated: visualizing protein kinase C signaling. IUBMB Life 55: 653–660, 2003 [DOI] [PubMed] [Google Scholar]
- 97.Violin JD, Zhang J, Tsien RY, Newton AC. A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol 161: 899–909, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Watanabe M, Chen CY, Levin DE. Saccharomyces cerivisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J Biol Chem 269: 16829–16836, 1994 [PubMed] [Google Scholar]
- 99.Xu ZB, Chaudhary D, Olland S, Wolfrom S, Czerwinski R, Malakian K, Lin L, Stahl ML, Joseph-McCarthy D, Benander C, Fitz L, Greco R, Somers WS, Mosyak L. Catalytic domain crystal structure of protein kinase C-theta (PKCtheta). J Biol Chem 279: 50401–50409, 2004 [DOI] [PubMed] [Google Scholar]
- 100.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell 81: 917–924, 1995 [DOI] [PubMed] [Google Scholar]
- 101.Ziegler WH, Parekh DB, Le Good JA, Whelan RD, Kelly JJ, Frech M, Hemmings BA, Parker PJ. Rapamycin-sensitive phosphorylation of PKC on a carboxy-terminal site by an atypical PKC complex. Curr Biol 9: 522–529, 1999 [DOI] [PubMed] [Google Scholar]