Abstract
Permanent neonatal diabetes mellitus is a rare form of insulin-requiring diabetes presenting within the first few weeks or months of life. Mutations in the insulin gene are the second most common cause of this form of diabetes. These mutations are located in critical regions of preproinsulin and are likely to prevent normal processing or folding of the preproinsulin/proinsulin molecule. To characterize these mutations, we transiently expressed proinsulin-GFP fusion proteins in MIN6 mouse insulinoma cells. Our study revealed three groups of mutant proteins: 1) mutations that result in retention of proinsulin in the endoplasmic reticulum (ER) and attenuation of secretion of cotransfected wild-type insulin: C43G, F48C, and C96Y; 2) mutations with partial ER retention, partial recruitment to granules, and attenuation of secretion of wild-type insulin: G32R, G32S, G47V, G90C, and Y108C; and 3) similar to (2) but with no significant attenuation of wild-type insulin secretion: A24D and R89C. The mutant insulin proteins do not prevent targeting of wild-type insulin to secretory granules, but most appear to lead to decreased secretion of wild-type insulin. Each of the mutants triggers the expression of the proapoptotic gene Chop, indicating the presence of ER stress.
Keywords: endoplasmic reticulum stress
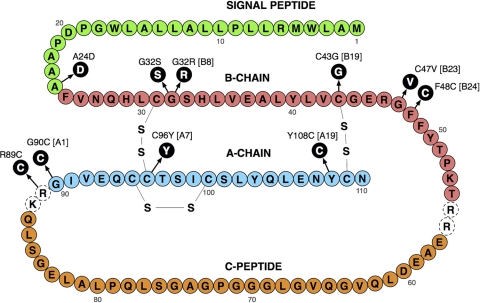
mutations in the heterozygous state in the insulin gene were recently identified as a cause of permanent neonatal diabetes as well as maturity-onset diabetes of the young (MODY) and type 1b diabetes (1, 3, 4, 6, 12, 16). These mutations lead to insulin-requiring diabetes, with onset at various ages, even though all the subjects have a normal insulin gene. The insulin derived from the normal gene is sufficient to maintain normal blood glucose levels for variable periods of time before the effect of the mutant insulin protein leads to cell death. Here, we analyzed the subcellular localization and processing of 10 mutant insulin proteins associated with permanent neonatal diabetes (16). These mutations (Fig. 1) are located in critical regions of preproinsulin and affect signal peptide processing (A24D) or normal folding of the proinsulin molecule (G32S, G32R, C43G, G47V, F48C, R89C, G90C, C96Y, and Y108C).
Fig. 1.
Cartoon of the preproinsulin protein. Residues in black are the mutations studied here that cause permanent neonatal diabetes. Residues in dotted circles represent the C-peptide cleavage sites. Enhanced GFP was introduced between Pro 72 and Gly 73 of the C-peptide (modified from Ref. 16).
To study the subcellular localization, proteolytic processing and glucose-stimulated secretion of these mutant proteins, we used green fluorescent protein (GFP)-proinsulin fusion protein constructs, where GFP was inserted in the C-peptide of proinsulin. We confirmed that in MIN6 mouse insulinoma cells the wild-type insulin-GFP fusion protein is processed, properly targeted to insulin granules, and secreted in response to stimulation with glucose (9, 17). The fusion proteins provided a means to visualize the mutant proteins following transient transfection in live MIN6 cells. Interestingly, whereas some mutants were targeted to secretory granules, others were completely retained in the endoplasmic reticulum (ER) and remained unprocessed. Expression of most of the mutants was found to attenuate secretion of coexpressed wild-type insulin. This may help explain the mechanism by which each mutation impairs β-cell function and suggests that the clinical variability of the onset of diabetes may be mutation specific.
MATERIALS AND METHODS
DNA constructs, mutagenesis, and RT-PCR.
Human preproinsulin was subcloned into pcDNA3.1, and enhanced GFP or mCherry was introduced between proline 72 and glycine 73 of preproinsulin. Mutagenesis was carried out using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). MIN6 mouse insulinoma cells were grown in DMEM supplemented with 15% fetal calf serum and transiently transfected with 4 μg of DNA per 35-mm plate, using Lipofectamine 2000 and following the manufacturer's instructions (Invitrogen, Carlsbad, CA). RNA from the transfected cells was prepared with the RNeasy Mini Kit (Qiagen, Valencia, CA), and cDNA was made by Superscript III reverse transcriptase (Invitrogen).
Confocal microscopy and image analysis.
Transfected cells were imaged on a Leica TCS SP2 laser scanning confocal microscope (Leica Microsystems, Germany) with sequential scanning of GFP and RFP signals to minimize signal bleed-through and cross-talk. A 16-line average was used during image collection to dampen extraneous noise, and photomultiplier tube voltage settings for emission amplification and laser power for excitation were kept constant within and between image acquisition sessions. Before quantification of colocalization, all images were deconvolved with the Huygens Professional software package (Scientific Volume Imaging, Hilversum, The Netherlands) using the classic maximum likelihood estimation method. Deconvolved images were then processed with ImageJ software (National Institutes of Health, Bethesda, MD) to subtract background with the rolling-ball feature (radius 50 pixels) and analyzed for colocalization using the JACoP plug-in (2). We chose a thresholded Mander‘s coefficient as our readout of colocalization, as this is a well-accepted method with low sensitivity to differences in pixel intensity between the two color channels being compared. Thresholds for the two channels were determined empirically to include as much of the remaining pixel intensity in the cell as possible while excluding any remaining non-cell pixels. In an effort to avoid changes in colocalization coefficients based on artifact, threshold levels were kept constant across all images analyzed. Values displayed are means of thresholded Mander’s coefficients for single cells ± SE for n ≥ 7 cells from three independent experiments. Statistical significance was calculated using one-way ANOVA with Dunnett's posttest by Prism software (GraphPad).
Stimulation of insulin secretion.
Transfected MIN6 cells were incubated for 3 h at 37°C in prewarmed Krebs-Ringer HEPES buffer (KRH; in mM: 19 NaCl, 4.7 KCl, 25 HEPES, 1.2 MgSO4, 1.2 KH2PO4, 2.5 CaCl2, pH 7.4) containing 2 mM glucose. The cells were then incubated for 1.5 h in KRH containing 25 mM glucose and 1 mM tetraethylammonium (TEA). The media were collected, and cells were lysed in appropriate lysis buffer for subsequent analysis by Western blot or ELISA.
SDS-PAGE and Western blot analysis.
Cells were lysed in Laemmli sample buffer, sonicated, and heated at 80°C for 10 min. After electrophoresis through a 12% Tris-glycine gel (Invitrogen), proteins were transferred to a nitrocellulose membrane, which was probed with anti-GFP rabbit polyclonal antibody (Invitrogen) and imaged with the ChemiDoc XRS gel documentation system (Bio-Rad, Hercules, CA).
Secreted GFP and C-peptide measurement by ELISA.
MIN6 cells were cotranfected with untagged human preproinsulin and wild-type or mutant insulin-GFP. Seventy-two hours posttransfection, cells were starved, and insulin secretion was stimulated as above. Cells were lysed in lysis buffer (in mM: 20 HEPES, 40 KCl, 100 NaCl, 1 NaEDTA; 10% glycerol, 2% Triton-X-100) with Complete Protease Inhibitor Cocktail tablets (Roche Applied Science, Summerville, NJ). Collected media and cell lysates were analyzed for presence of GFP (GFP ELISA; Cell Biolabs, San Diego, CA) or human C-peptide (ALPCO, Windham, NH). Statistical significance was calculated using one-way ANOVA with Dunnett's posttest by Prism software (GraphPad).
RESULTS
Subcellular localization of mutant insulin-GFP.
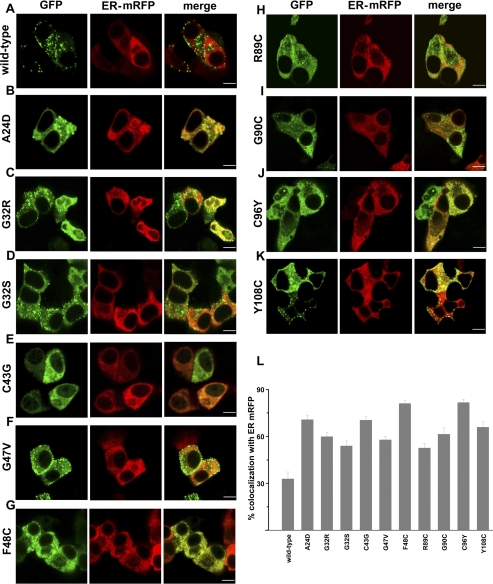
To visualize subcellular localization of insulin proteins, wild-type and mutant insulin-GFP constructs were made by inserting GFP within the C-peptide of proinsulin. These fusion proteins were expressed in MIN6 cells with the ER marker, monomeric red fluorescent protein with a KDEL ER retention sequence (ER-mRFP, gift from Dr. Jennifer Lippincott-Schwartz), by transient transfection, and their subcellular localization was analyzed by confocal microscopy. Degree of colocalization was quantified by calculating the Mander‘s coefficient [(2); Fig. 2L]. Wild-type insulin-GFP was properly targeted to secretory granules and showed 30% colocalization with ER-mRFP (Fig. 2A). Colocalization of each mutant insulin-GFP with ER-mRFP was significantly greater than that of wild-type insulin-GFP. Of the 10 mutants analyzed, three (C43G, F48C, and C96Y) were completely retained in the ER, with 70–85% colocalization with ER-mRFP (Fig. 2, E, G, J, L). Although proinsulin mutants G32R, G32S, G47V, R89C, G90C, and Y108C showed more than 30% colocalization with ER-mRFP, several granules were also visible (Fig. 2, C, D, F, H, I, K). Granules were also present in cells expressing the mutant A24D, despite its high ER retention as calculated by Mander’s coefficient (Fig. 2, B and L). These data demonstrate that degree of retention within the ER and targeting to secretory granules varies between different mutant insulin proteins.
Fig. 2.
Subcellular localization of mutant insulin-GFP. Wild-type or mutant insulin-GFP (green) and mRFP with a KDEL ER retention sequence (ER-mRFP, red) were cotransfected in MIN6 cells and imaged 48 h later by confocal microscopy. Midplane images are shown. A: wild-type insulin-GFP. B–K: mutant insulin-GFP; scale bar, 5 μm. L: %colocalization of insulin-GFP with ER-mRFP, calculated by Mander‘s coefficient. One-way ANOVA with Dunnett’s posttest revealed that %colocalization for each mutant was significantly greater than for wild-type (P < 0.001). Values are means ± SE; for each experiment, n = 7–12 cells. Note that several granules are visible with A24D (B) despite its high colocalization with ER-mRFP.
Proteolytic processing and glucose stimulated secretion of mutant insulin-GFP.
Proteolytic cleavage of proinsulin occurs within maturing secretory granules and yields insulin and free C-peptide, which are subsequently secreted together in equimolar amounts. Having observed varying degrees of mutant proinsulin trafficking to granules, we next determined whether these proteins underwent processing and stimulated secretion.
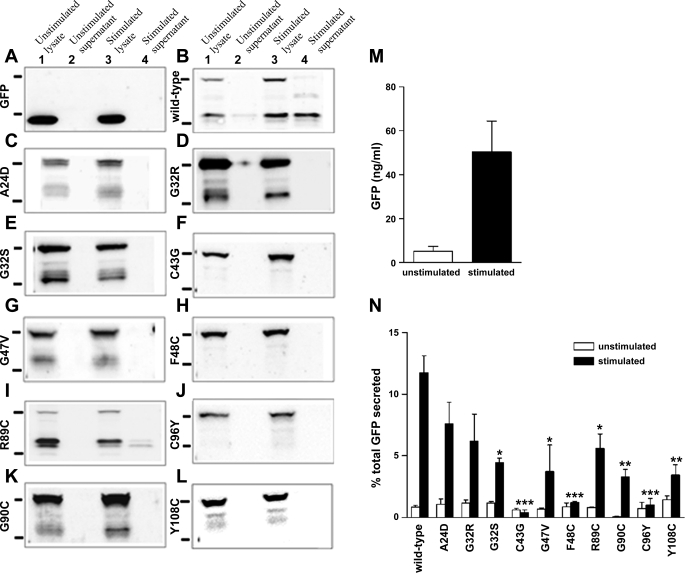
Western blot analysis of the lysate of MIN6 cells expressing wild-type or mutant insulin-GFP with anti-GFP antibody revealed two prominent bands: one at 40 kDa, corresponding to proinsulin-GFP, and one slightly above 30 kDa, corresponding to free GFP-C-peptide (Fig. 3B, lanes 1 and 3). These results indicate that wild-type insulin-GFP was subjected to proper proteolytic processing. Analysis of the mutants C43G, F48C, and C96Y revealed the absence of the GFP-C-peptide band, indicating a complete lack of proinsulin processing (Fig. 3, F, H, J). The signal peptidase cleaves preproinsulin following Ala 24. Under normal circumstances, this process occurs cotranslationally, and the level of preproinsulin in the cell is very low (15). In the case of mutant A24D, we observed minimal processing as well as an additional higher molecular weight band above 40 kDa, probably due to inefficient cleavage of the signal peptide at Asp 24 (Fig. 3C). The mutants G32R, G32S, and G47V showed low but detectable proinsulin processing (Fig. 3, D, E, G). The R89C mutation is predicted to disrupt cleavage by the type II endopeptidase prohormone convertase-2 (5). In addition to the GFP-C-peptide band, we observed a band just above this band, indicating partially incomplete proteolytic processing at this site. Despite the presence of an extra cysteine residue, the G90C and Y108C mutants showed mostly intact proinsulin, with evidence of minimal proinsulin processing (Fig. 3, K and L). Taken together, the data show that mutant insulin proteins that are completely retained in the ER do not undergo proteolytic processing and that all other mutant proteins examined are processed at varying levels.
Fig. 3.
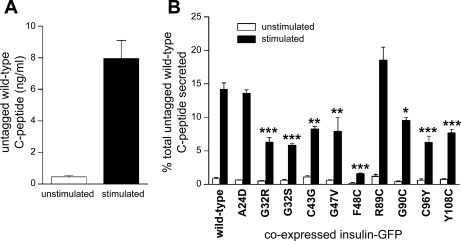
Proteolytic processing and secretion of mutant insulin-GFP. Three days posttransfection with wild-type or mutant insulin-GFP, MIN6 cells were starved for 3 h with 2 mM glucose, and then insulin secretion was stimulated by treatment with 25 mM glucose and 1 mM TEA for 1.5 h. A–L: media and cell lysates were subjected to Western blot analysis with anti-GFP antibody to detect processing of GFP-proinsulin to insulin and GFP-C-peptide. Bands to the left of each blot correspond to protein standards of 40 kDa (top) and 30 kDa (bottom). Lanes 1 and 2, cell lysate and media, respectively, from plates treated with 2 mM glucose; lanes 3 and 4, cell lysate and media, respectively, from plates treated with 25 mM glucose + 1 mM TEA. A and B: GFP without C-peptide runs lower and is distinguishable from GFP-C-peptide. B–L: Western blots showing processing and secretion of wild-type or mutant insulin-GFP, as indicated. While wild-type insulin-GFP underwent proteolytic processing to release GFP-C-peptide, proinsulin mutants C43G, F48C, and C96Y were not processed. However, at least partial processing of the remaining mutants was observed. M: Secretion of GFP-C-peptide under unstimulated or stimulated conditions measured by GFP ELISA. N: secreted GFP was calculated as percentage of total GFP-C-peptide content. Mutants retained in the ER (C43G, F48C, and C96Y) showed minimal secretion, whereas varying quantities of GFP-C-peptide were measured of the remaining mutants, which are partially recruited to the granules. Wild-type, n = 11; each mutant, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we utilized two methods of examining stimulated secretion of each mutant insulin protein. Western blot analysis of the concentrated media collected before and after stimulation with 25 mM glucose and 1 mM TEA showed that, for wild-type insulin-GFP, cleaved GFP-C-peptide is secreted into the media in a glucose dependent manner (Fig. 3B, lanes 2 and 4). For mutant R89C, we also detected secretion of GFP-C-peptide. However, for each remaining mutant insulin protein, the absence of GFP-C-peptide in media collected after stimulation suggests failure to undergo exocytosis in response to glucose.
To more closely examine and quantify secretion of wild-type and mutant insulin-GFP, we performed secretion assays as above and analyzed secreted GFP, as a marker of insulin secretion, by GFP ELISA. As shown in Fig. 3M, this method reliably measures glucose-stimulated secretion of wild-type GFP-C-peptide. To normalize transfection efficiency, we calculated the percentage of total GFP content that was secreted under unstimulated and stimulated conditions for each mutant [Supplemental Fig. S1 and Fig. 3N (Supplementary materials are found in the online version of this paper)]. Stimulated secretion of mutants C43G, F48C, and C96Y was significantly less than that of wild-type insulin-GFP and not different from basal secretion, confirming the Western blot data. However, by GFP ELISA, which is more sensitive than Western blot, all mutants that appear in granules showed secretion of GFP-C-peptide into the media, and each was decreased relative to wild-type insulin-GFP (Fig. 3N). All together, these results suggest that the mutants that are retained in the ER and do not appear in granules have a more severe effect on the fate of the mutant proinsulin molecule.
Subcellular localization of wild-type insulin-mCherry coexpressed with mutant insulin-GFP.
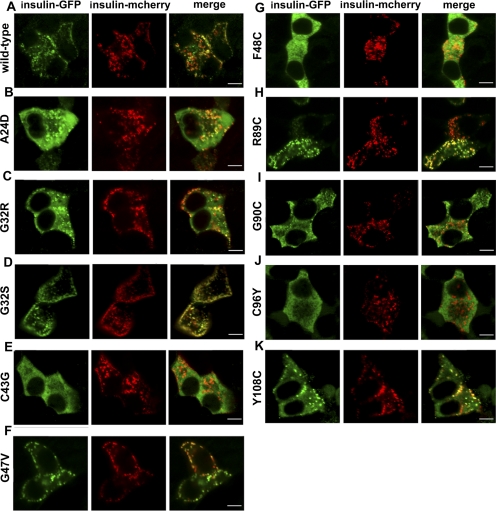
Proinsulin processing and hexamerization occur in secretory granules (5). To monitor trafficking of wild-type proinsulin in the presence of mutant proinsulin, we replaced the GFP in the C-peptide of wild-type insulin-GFP with the fluorescent protein mCherry (14) and cotransfected this construct with wild-type insulin-GFP or mutant insulin-GFP. Similar to wild-type insulin-GFP, wild-type insulin-mCherry was properly targeted to the granules, and it colocalized with the GFP positive granules (Fig. 4A). Interestingly, granules containing insulin-mCherry were visible when this construct was coexpressed with the mutants C43G, F48C, and C96Y, which are completely retained in the ER (Fig. 4, F, G, J). These data suggest that forward trafficking of wild-type insulin-mCherry was not completely inhibited and that the ER retained mutant proteins did not sequester the wild-type insulin-mCherry proteins in the ER. The mutants that are partially retained in the ER (Fig. 4, B, C, D, F, H, I, K) showed both mutant insulin-GFP proteins and wild-type insulin-mCherry proteins in secretory granules. Similar results were observed when wild-type insulin-mCherry was sequentially transfected 24 h after transfection of mutant insulin-GFP (not shown). Unfortunately, differing levels of insulin-GFP retained in the ER across the various mutant insulin constructs made interpretation of thresholded Mander's colocalization coefficients difficult (data not shown). Coexpression of mutant insulin-GFP proteins, as these experiments show, does not alter the subcellular localization of wild-type insulin-mCherry.
Fig. 4.
Coexpression of mutant insulin-GFP with wild-type insulin-mCherry. MIN6 cells were cotransected with wild-type insulin-mCherry (red) and wild-type or mutant insulin-GFP (green) and imaged after 48 h. A: wild-type insulin-GFP colocalizes with wild-type insulin-mCherry. B–K: mutants C43G, F48C, and C96Y, which are retained in the ER, are not colocalized with wild-type insulin-mCherry, suggesting these mutant proteins do not sequester wild-type proinsulin in the ER. Mutants partially targeted to granules (A24D, G32S, G32R, G47V, R89C, G90C, and Y108C) colocalize with wild-type insulin-mCherry in granules. Scale bar, 5 μm.
Secretion of wild-type insulin when coexpressed with mutant insulin-GFP.
To investigate the effect of mutant insulin protein expression on secretion of wild-type insulin, we cotransfected MIN6 cells with mutant insulin-GFP together with untagged wild-type proinsulin. Secretion of untagged wild-type C-peptide was measured by human C-peptide-specific ELISA, which did not recognize endogenous mouse C-peptide or GFP-tagged C-peptide (Supplemental Fig. S2). When untagged wild-type proinsulin was coexpressed with wild-type insulin-GFP in MIN6 cells, untagged wild-type C-peptide was secreted in a glucose-dependent manner (Fig. 5A). To normalize for transfection efficiency, we calculated the percentage of total C-peptide content that was secreted in response to glucose stimulation for each mutant (Supplemental Fig. S3 and Fig. 5B). Stimulated secretion of wild-type C-peptide was significantly decreased with coexpression of each of the mutants C43G, F48C, C96Y, G32S, G32R, G47V, G90C, and Y108C (Fig. 5B). In these cases, secretion of wild-type C-peptide was inhibited by ∼30–90% compared with stimulated secretion of wild-type C-peptide coexpressed with wild-type insulin-GFP. Expression of mutant A24D, which disrupts the signal peptide cleavage site, and R89C, which disrupts one of the C-peptide cleavage sites, did not result in attenuation of secretion of the wild-type insulin. These results show that expression of mutant insulin proteins, with the exception of A24D and R89C, attenuates the secretion of wild-type insulin, as previously shown for the C96Y (Akita) mutation (9).
Fig. 5.
Expression of proinsulin mutant proteins attenuates secretion of wild-type insulin. MIN6 cells were transfected with untagged wild-type proinsulin and wild-type or mutant insulin-GFP. Three days posttransfection, cells were incubated in 2 mM glucose for 3 h, and then insulin secretion was stimulated by incubation with 25 mM glucose and 1 mM TEA for 1.5 h. Supernatant and cell lysate were analyzed for untagged wild-type C-peptide by ELISA. A: MIN6 cells expressing untagged wild-type proinsulin exhibit secretion of human C-peptide upon glucose stimulation. B: secreted wild-type C-peptide was calculated as percentage of total C-peptide content. Note that with expression of each mutant insulin-GFP (not including A24D or R89C), secretion of untagged wild-type human C-peptide was significantly decreased. Wild-type, n = 12; each mutant, n = 3. *P < 0.05, **P < 0.01, ***P < 0.001.
Expression of the proapoptotic marker Chop in cells expressing mutant insulin-GFP.
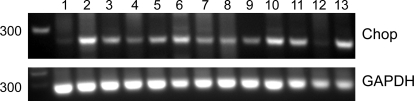
To test whether expression of mutant proinsulin proteins results in ER stress, we estimated the degree of Chop (C/EBP-homologous protein) expression by RT-PCR (Fig. 6). Low basal expression of Chop was observed in cells transfected with wild-type insulin-GFP and in untransfected cells (Fig. 6, lanes 1 and 12). In contrast, cells transfected with mutant proinsulin constructs or treated with thapsigargin showed increased expression of Chop (Fig. 6, lanes 2–11 and 13), suggesting that their expression results in ER stress.
Fig. 6.
Expression of proapoptotic marker Chop (C/EBP-homologous protein). MIN6 cells were transfected with wild-type or mutant insulin-GFP. RNA and cDNA were prepared 72 h posttransfection, and expression of Chop mRNA was analyzed by RT-PCR. A higher level of Chop expression was observed with expression of each of the mutants vs. that seen with wild-type or in untransfected cells. Lanes: 1) wild-type insulin-GFP, 2) A24D, 3) G32R, 4) G32S, 5) C43G, 6) G47V, 7) F48C, 8) R89C, 9) G90C, 10) C96Y, 11) Y108C, 12) untransfected, and 13) treated with 500 nM thapsigargin for 6 h.
DISCUSSION
Here, we have analyzed subcellular localization, processing, and secretion of 10 insulin mutant proteins that cause permanent neonatal diabetes (1, 3, 4, 6, 12, 16). On the basis of our results, we have classified the mutations into three groups. Group A are those that are completely retained in the ER: C43G, F48C, and C96Y; group B includes mutations with partial ER retention, partial recruitment to granules, and those that attenuate secretion of cotransfected wild-type insulin: G32R, G32S, G47V, G90C, and Y108C; and group C, which has the features of B but with no significant attenuation of wild-type insulin secretion, includes mutants A24D and R89C.
Two mutations, C43G and C96Y, disrupt interchain disulfide bonds that are required for the ordered secondary structure of proinsulin (7). These two mutants failed to exit from the ER and did not undergo proteolytic processing. The mutation F48C, which causes ablation of a structural motif (10) formed by the F49 and Y108 residues, was also retained in the ER and not processed to release C-peptide. The Y108C mutation resulted in increased ER retention and minimal processing, even though partial granule localization and a low level of exocytosis were observed.
Another mutation that replaces a Gly with a Cys results in an unpaired Cys residue, G90C, which was partially localized in the granules and showed low levels of processing and exocytosis. Three mutations that displace highly conserved Gly residues (G32S, G32R, and G47V) did not completely impair trafficking to granules or proteolytic cleavage of proinsulin. These misfolded proteins are unlikely to form hexamers. Their appearance in granules is consistent with previous studies showing that hexamerization is not required for proinsulin targeting to secretory granules (13). Variable degrees of exocytosis were observed with mutants that are not retained in the ER.
The group C mutations (A24D and R89C) resulted in aberrant processing of proinsulin to insulin. The A24D mutation at the signal peptide cleavage site showed partial inhibition of preproinsulin to proinsulin processing, ER accumulation, and partial granule targeting. The R89C mutation replaces an Arg with a Cys at the A chain-C-peptide cleavage site (8, 15). Despite their aberrant processing, we observed trafficking to secretory granules and secretion of these mutant insulin proteins.
Surprisingly, expression of mutant insulin-GFP proteins retained in the ER did not completely prevent trafficking of wild-type insulin-mCherry in cotransfection experiments. Wild-type proinsulin was reported to coimmunoprecipitate with C96Y (Akita) mutant insulin (9), suggesting that some degree of heteromerization of the wild-type and the mutant occurs in the ER; however, this was not detectable in our optical assay. Moreover, expression of mutant insulin proteins attenuated secretion of coexpressed, untagged wild-type C-peptide, probably as a result of decreased translation of wild-type insulin in response to ER stress. This has also been suggested by Colombo et al. (4), when mutant insulin proteins were expressed in HEK cells. Interestingly, we found no significant inhibition of wild-type insulin secretion with expression of the two mutants A24D and R89C. Unlike other mutations that reside in the A and B chains of the insulin protein, these mutations reside at the enzymatic cleavage sites of the preproinsulin. With these mutations, it remains possible that induction of translational inhibition in response to ER stress requires a longer time course that may vary with each specific mutation. These mutations may have a greater effect on β-cell dysfunction in a stable expression system, in which experiments are performed on a longer time course.
β-Cells in Akita mice (Ins2C96Y) express high levels of ER chaperones and Chop (11). We found elevated Chop expression in insulinoma cells transfected with each of the 10 mutants, also indicating ER stress. Although all of the mutations studied here resulted in elevated Chop expression, several mutants showed some degree of granule localization and exocytosis.
These studies provide a foundation for further studies focused on modulating the ER stress response to maintain ER homeostasis rather than lead to apoptosis. A better understanding the response of the β-cell to ER stress could lead to new therapies for the treatment of diabetes due to the expression of a mutant insulin protein.
GRANTS
This work was partially supported by the National Institute of Diabetes and Digestive and Kidney Diseases (DK-075706 to S. Rajan, DK-48494 to L. H. Philipson, DK-064973 to V. E. Prince), the University of Chicago Diabetes Research and Training Center (P60-DK-20595), and the Juvenile Diabetes Research Foundation International (5-2007-970 to V. E. Prince).
DISCLOSURES
No conflicts of interest are reported by the author(s).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Donald F. Steiner, James P. Lopez, and Adam Shaw for helpful discussions. We thank Dr. Peter Drain for the C-peptide-GFP construct and Dr. Jennifer Lippincott-Schwartz for the ER-mRFP construct.
REFERENCES
- 1.Ahamed A, Unnikrishnan AG, Pendsey SS, Nampoothiri S, Bhavani N, Praveen VP, Kumar H, Jayakumar RV, Nair V, Ellard S, Edghill EL. Permanent neonatal diabetes mellitus due to a C96Y heterozygous mutation in the insulin gene. A case report. JOP 9: 715–718, 2008 [PubMed] [Google Scholar]
- 2.Bolte S, Cordelieres FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224: 213–232, 2006 [DOI] [PubMed] [Google Scholar]
- 3.Bonfanti R, Colombo C, Nocerino V, Massa O, Lampasona V, Iafusco D, Viscardi M, Chiumello G, Meschi F, Barbetti F. Insulin gene mutations as cause of diabetes in children negative for five type 1 diabetes autoantibodies. Diabetes Care 32: 123–125, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colombo C, Porzio O, Liu M, Massa O, Vasta M, Salardi S, Beccaria L, Monciotti C, Toni S, Pedersen O, Hansen T, Federici L, Pesavento R, Cadario F, Federici G, Ghirri P, Arvan P, Iafusco D, Barbetti F. Seven mutations in the human insulin gene linked to permanent neonatal/infancy-onset diabetes mellitus. J Clin Invest 118: 2148–2156, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodson G, Steiner D. The role of assembly in insulin's biosynthesis. Curr Opin Struct Biol 8: 189–194, 1998 [DOI] [PubMed] [Google Scholar]
- 6.Edghill EL, Flanagan SE, Patch AM, Boustred C, Parrish A, Shields B, Shepherd MH, Hussain K, Kapoor RR, Malecki M, MacDonald MJ, Stoy J, Steiner DF, Philipson LH, Bell GI, Hattersley AT, Ellard S. Insulin mutation screening in 1,044 patients with diabetes: mutations in the INS gene are a common cause of neonatal diabetes but a rare cause of diabetes diagnosed in childhood or adulthood. Diabetes 57: 1034–1042, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hua QX, Mayer JP, Jia W, Zhang J, Weiss MA. The folding nucleus of the insulin superfamily: a flexible peptide model foreshadows the native state. J Biol Chem 281: 28131–28142, 2006 [DOI] [PubMed] [Google Scholar]
- 8.Lipkind G, Steiner DF. Predicted structural alterations in proinsulin during its interactions with prohormone convertases. Biochemistry 38: 890–896, 1999 [DOI] [PubMed] [Google Scholar]
- 9.Liu M, Hodish I, Rhodes CJ, Arvan P. Proinsulin maturation, misfolding, proteotoxicity. Proc Natl Acad Sci USA 104: 15841–15846, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray-Rust J, McLeod AN, Blundell TL, Wood SP. Structure and evolution of insulins: implications for receptor binding. Bioessays 14: 325–331, 1992 [DOI] [PubMed] [Google Scholar]
- 11.Oyadomari S, Koizumi A, Takeda K, Gotoh T, Akira S, Araki E, Mori M. Targeted disruption of the Chop gene delays endoplasmic reticulum stress-mediated diabetes. J Clin Invest 109: 525–532, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polak M, Dechaume A, Cave H, Nimri R, Crosnier H, Sulmont V, de Kerdanet M, Scharfmann R, Lebenthal Y, Froguel P, Vaxillaire M. Heterozygous missense mutations in the insulin gene are linked to permanent diabetes appearing in the neonatal period or in early infancy: a report from the French ND (Neonatal Diabetes) Study Group. Diabetes 57: 1115–1119, 2008 [DOI] [PubMed] [Google Scholar]
- 13.Quinn D, Orci L, Ravazzola M, Moore HP. Intracellular transport and sorting of mutant human proinsulins that fail to form hexamers. J Cell Biol 113: 987–996, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shu X, Shaner NC, Yarbrough CA, Tsien RY, Remington SJ. Novel chromophores and buried charges control color in mFruits. Biochemistry 45: 9639–9647, 2006 [DOI] [PubMed] [Google Scholar]
- 15.Steiner DF. The proinsulin C-peptide—a multirole model. Exp Diabesity Res 5: 7–14, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stoy J, Edghill EL, Flanagan SE, Ye H, Paz VP, Pluzhnikov A, Below JE, Hayes MG, Cox NJ, Lipkind GM, Lipton RB, Greeley SA, Patch AM, Ellard S, Steiner DF, Hattersley AT, Philipson LH, Bell GI. Insulin gene mutations as a cause of permanent neonatal diabetes. Proc Natl Acad Sci USA 104: 15040–15044, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watkins S, Geng X, Li L, Papworth G, Robbins PD, Drain P. Imaging secretory vesicles by fluorescent protein insertion in propeptide rather than mature secreted peptide. Traffic 3: 461–471, 2002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.