Abstract
The loss of HBII-52 and related C/D box small nucleolar RNA (snoRNA) expression units have been implicated as a cause for the Prader–Willi syndrome (PWS). We recently found that the C/D box snoRNA HBII-52 changes the alternative splicing of the serotonin receptor 2C pre-mRNA, which is different from the traditional C/D box snoRNA function in non-mRNA methylation. Using bioinformatic predictions and experimental verification, we identified five pre-mRNAs (DPM2, TAF1, RALGPS1, PBRM1 and CRHR1) containing alternative exons that are regulated by MBII-52, the mouse homolog of HBII-52. Analysis of a single member of the MBII-52 cluster of snoRNAs by RNase protection and northern blot analysis shows that the MBII-52 expressing unit generates shorter RNAs that originate from the full-length MBII-52 snoRNA through additional processing steps. These novel RNAs associate with hnRNPs and not with proteins associated with canonical C/D box snoRNAs. Our data indicate that not a traditional C/D box snoRNA MBII-52, but a processed version lacking the snoRNA stem is the predominant MBII-52 RNA missing in PWS. This processed snoRNA functions in alternative splice-site selection. Its substitution could be a therapeutic principle for PWS.
INTRODUCTION
It has been estimated that 95% of human multi-exon genes undergo alternative splicing (1,2), indicating that this pre-mRNA processing step is central for human gene expression. Unlike promoter activity that is predominantly reflected in the abundance of transcripts, alternative splicing influences the structure of the mRNAs and their encoded proteins. As a result, it influences binding properties, intracellular localization, enzymatic activity, protein stability and post-translational modification of numerous gene products (reviewed in 3).
We recently found that usage of the alternative exon Vb of the serotonin receptor 2C (HTR2C) is regulated by expressing a C/D box snoRNA, HBII-52. SnoRNAs are small nuclear RNAs that can be detected in the nucleolus. They reside in introns from which they are released through nuclease action during the processing of the host pre-mRNA. On the basis of their sequence, snoRNAs can be subdivided into C/D and H/ACA snoRNAs. C/D box snoRNAs have C and D boxes as characteristic sequence elements at the ends of the RNA. The 5′ and 3′ ends of the snoRNA form a short stem that precedes the C and D boxes, which together form a kink-turn (K-turn) structure (4).
A well-understood function attributed to C/D box snoRNAs is the guiding of 2′-O-methylation in ribosomal, transfer and small nuclear RNAs. This activity is achieved through the formation of a specific RNA:RNA duplex between the snoRNA and the target. Most snoRNAs contain two regions to interact with other RNAs, termed the antisense boxes. Each antisense box exhibits sequence complementarity to its target and forms a short, transient double strand with it. On the target RNA, the nucleotide that base pairs with the fifth snoRNA nucleotide upstream of the snoRNA D-box is methylated on the 2′-O-hydroxyl group (reviewed in 5). Several snoRNAs are complementary to pre-rRNA, but the rRNA is not 2′-O-methylated at the predicted positions (6). Recently, numerous C/D box snoRNAs were discovered that show no clear sequence complementarity to other non-mRNAs, suggesting that C/D box snoRNAs might have functions other than 2′-O-methylation (7). One of these ‘orphan’ C/D box snoRNAs is HBII-52 (SNORD 115). It is expressed from the SNURF–SNRPN locus. Loss of expression from this locus is the most likely cause for Prader–Willi syndrome (PWS) (8), which was supported by the recent finding that a microdeletion containing only snoRNAs causes PWS (9). HBII-52 exhibits sequence complementarity to an alternative exon of the human serotonin receptor 2C mRNA and changes alternative splicing of this pre-mRNA (HTR2C) in transfection experiments. This change has also been observed in brain tissue from PWS patients (10) and a mouse model lacking MBII-52 snoRNAs shows differences in pre-mRNA processing of the serotonin receptor (11). Finally, it was reported that an increase of C/D box snoRNA expression from the 15q11–q13 region leads to autistic phenotypes in mice, which further suggests that snoRNAs play an important role in gene regulation (12).
PWS is a congenital disease with an incidence of about 1 in 8000–20 000 live births. PWS is the most common genetic cause of marked obesity in humans. The excess weight makes PWS the most frequent genetic cause for type II diabetes (8). Early PWS is characterized by a failure to thrive, feeding difficulties and hypogonadism. Later, the patients are characterized by short stature and develop mild to moderate mental retardation, behavioral problems and hyperphagia that lead to severe obesity. Children with PWS show low levels of growth hormone, IGF-I and insulin as well as elevated levels of ghrelin (13–15) and often exhibit central adrenal insufficiency (16). Subsequently, growth hormone substitution was approved for the treatment of children with PWS (17).
PWS is caused by the loss of gene expression from a maternally imprinted region on chromosome 15q11–q13 (reviewed in 8). The SNURF–SNRPN locus in the 15q11–q13 region plays a major role in PWS, and its deletion causes PWS-like symptoms in mouse models (18). The SNURF–SNRPN locus spans more than 460 kb and contains at least 148 exons (19). Ten exons in the 5′ part of the gene are transcribed into a bicistronic mRNA that encodes the SNURF (SmN upstream reading frame) and the SmN (small RNP in neurons) protein. The locus harbors a bipartite imprinting center that silences most maternal genes of the PWS critical region. Owing to this imprinting, the SNURF–SNRPN pre-mRNA is expressed only from the paternal allele. The large 3′-UTR region of the SNURF–SNRPN locus harbors clusters of the C/D box snoRNAs HBII-85 and HBII-52 that are present in 24 and 47 copies, respectively. In addition, the region harbors single copies of other C/D box snoRNAs: HBII-13, HBII-436, HBII-437, HBII-438A and HBII-438B. Recent evidence suggests that the HBII-85 and HBII-52 snoRNA clusters are expressed as two transcriptional units (20). The highly conserved snoRNAs are flanked by poorly conserved non-coding exons, suggesting that the functional relevant products of the locus are snoRNAs, not the flanking exons. The expression of these snoRNAs is tissue-specific. HBII-52 could be detected only in brain, whereas other snoRNAs from the SNURF–SNRPN locus are also expressed in non-brain tissues (reviewed in 21).
Here, we analyzed the function of the mouse ortholog of HBII-52, MBII-52. We found that it regulates alternative pre-mRNA processing of at least five more genes. The unit expressing MBII-52 expresses smaller RNAs that appear to be nuclease processing products of the full-length MBII-52 snoRNA. We termed these shorter RNAs psnoRNAs for processed snoRNAs. psnoRNAs associate with hnRNPs and not with the known C/D box snoRNA binding proteins. We postulate that psnoRNAs recognize target RNAs by sequence complementarity and influence splice-site selection by interfering with splicing regulatory proteins acting on pre-mRNA.
RESULTS
New targets for MBII-52
The recent finding that HBII-52 regulates alternative splicing of the 5-HT2C receptor (10) raised the question whether there are other targets for this snoRNA. The antisense boxes of the 47 human copies of HBII-52 show up to three sequence variations from their 18 nt consensus sequences (21). We tested HBII-52 variants with one, two, three and five mutations in their antisense box for their ability to change alternative splicing of exon Vb of the serotonin receptor. We found that a snoRNA with three mismatches can still promote exon Vb inclusion (Supplementary Material, Fig. S1). There is no statistically significant change when five mismatches are present in the antisense box. This argues that naturally occurring HBII-52 variants with up to three mismatches between antisense box and target region can influence pre-mRNA processing of the serotonin receptor.
In order to uncover additional targets of HBII-52, we performed a computational screen. Because the mode of interaction between HBII-52 and its targets is not yet known, we based our analysis on the constraints on snoRNA:rRNA interactions leading to ribose methylation in ribosomal targets (22). Concretely, we started by extracting an 18-nt-long antisense element upstream of the D box of MBII-52. We defined as a putative target site of MBII-52 a genomic region that can either form a perfect stem of length at least 10 bp or form a duplex of low free energy (below −15 kcal/mol) with the MBII-52 antisense element, with the duplex satisfying additional constraints. Minimum free energy duplexes were predicted with RNAhybrid (23) allowing G:U wobble in addition to canonical base pairing. The constraints on the duplexes were that (i) loops in the duplex were limited to maximum two nucleotides in either the target sequence or in antisense element and (ii) only up to three unpaired nucleotides in any of the sequences was allowed. Finally, similar to approaches previously employed to predict miRNA targets, we required that the predicted target site be conserved across mammalian species. More specifically, we extracted the regions in the human, rhesus macaque, cow and dog that are orthologous to the predicted HBII-52 target sites in human and we determined whether they would also be predicted as target sites. As the antisense box of HBII-52 is highly conserved in mammals, we compared all orthologous genes to the human antisense box sequence. Our final set of predictions included only putative MBII-52 target sites that were conserved in all of these other species. We obtained 457 such sites, 222 of which are in close proximity (200 nt) or within known exons. The predictions are available at http://www.mirz.unibas.ch/MBII-52/.
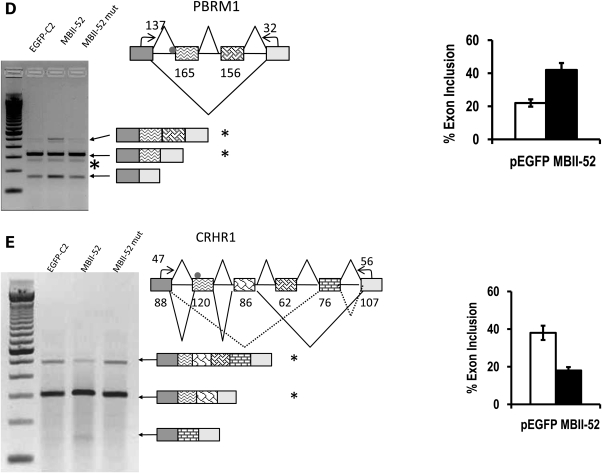
We next tested more than 100 computational predictions experimentally. Neuro2A cells were transfected with either MBII-52 or MBII-52mut, an MBII-52 variant with a scrambled antisense box (10), and the isolated RNA was analyzed by RT–PCR, using primers in the flanking constitutive exons. As shown in Figure 1, we observed a change in alternative splicing patterns in the DPM2, TAF1, RALGPS1, PBRM1 and CRHR1 pre-mRNAs. MBII-52 overexpression promoted either inclusion or skipping of the different exons. Their sequences and the complementarity to MBII-52 are shown in Table 1. These data suggest that MBII-52 expression changes alternative splicing of several endogenous pre-mRNAs.
Figure 1.
MBII-52 changes the alternative splicing pattern of predicted targets. Computationally predicted MBII-52 target genes expressed in Neuro2A cells were analyzed by RT–PCR. Cells were transfected with 1 µg pEGFP-C2, 1 µg of the MBII-52 expressing construct pCMV/MBII-52 (MBII-52) (29) and 1 µg MBII-52 consensus box mutation, MBII-52 cm, (MBII-52 mut) expressing an antisense box mutation of MBII-52 (10). A representative ethidium bromide-stained agarose gel is shown. The adjacent diagram shows the part of the genes that was analyzed. Small arrows indicate the location of the primers used. The MBII-52 complementarity region is indicated by a dot. Numbers in boxes indicate the length of the exons and numbers next to PCR primers indicate the length of the amplified exon fragment. The structure of the PCR products is indicated by similar shading of exons in cDNA and genomic DNA. The statistical analysis of at least four independent experiments is shown on the right. Stars indicate the bands that were used for quantification. The sequences of the regulated exons are shown in Table 1.
Table 1.
Sequence properties of MBII-52 targets
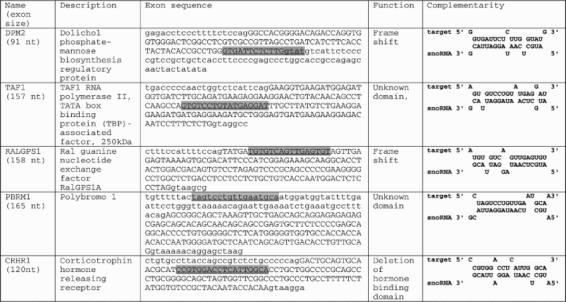 |
Genes that showed a dependency on MBII-52 expression both on endogenous and reporter gene level are listed using their HUGO nomenclature (columns 1 and 2). Numbers in parentheses indicate the exon length. The sequence of the regulated exon and its surrounding sequence is shown in column 3. Introns are in small letters, exons in capital letters. The snoRNA complementary region is highlighted in grey and underlined. Column 5 shows the alignment between the MBII-52 antisense box (snoRNA) and its target region.
MBII-52 changes alternative splicing of targeted pre-mRNAs in reporter gene assays
In the next step of the analysis, we determined whether alternative exons that are influenced by MBII-52 expression show this dependency also in a heterologous system, where they are surrounded by a different RNA context. We cloned the MBII-52 regulated exons into an exon-trap vector, where they were flanked by constitutively spliced insulin exons. All constructs were cloned into pSpliceExpress, a system that we developed previously (24).
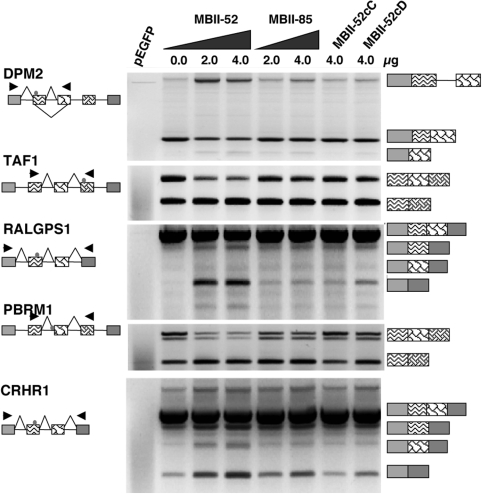
The reporter genes were cotransfected with MBII-52 expression constructs into Neuro2A cells and the splicing patterns were analyzed by RT–PCR. As shown in Figure 2, we observed for the five splicing events identified in endogenous genes a similar dependency on MBII-52 expression. The expression of MBII-85 snoRNA and C and D box mutants of MBII-52 (MBII-52cC,cD) did not show a significant effect on the alternative exons, suggesting that the effect is specific for MBII-52. With the exception of PBRM1, the reporter minigenes followed the splicing pattern of the endogenous genes. In the endogenous PBRM1 gene, MBII-52 promoted both inclusion and skipping of two exons located in a cluster of alternatively spliced cassette exons. In the heterologous system, we observe only the skipping event for PBRM1. This difference is most likely due to the presence of strong insulin exons in pSpliceExpress that interfere with the arrangement of regulatory sequences in this cluster of multiple alternative cassette exons. Finally, we created a series of compensatory mutations in the antisense box of MBII-52 and the snoRNA complementarity regions (snoCR) of its targets. These experiments proved inconclusive, as in most cases mutating the snoCR resulted in strong exon activation that was no longer susceptible to regulation (data not shown).
Figure 2.
Minigene analysis of MBII-52 target genes. The exons harboring the MBII-52 complementary region were subcloned into the exon trap vector pSpliceExpress. The structure of the resulting constructs pSE-RALGPS1, pSE-CRHR1, pSE-DPM2, pSE-PBRM1 and pSE-TAF1 as well as the location of the primers used for RT–PCR analysis is indicated on the left. pEGFP: only an expression construct for GFP is transfected. All other lanes contain 1 µg of pSE-reporter. MBII-52: cotransfection with 2 and 4 µg of MBII-52 expression construct, MBII-85: cotransfection with 2 and 4 µg of an MBII-85 expression construct, MBII52cC: cotransfection with 4 µg of a C-box mutant of MBII-52: MBII52cD: cotransfection with 4 µg of a D-box mutant of MBII-52. The structure of the products is shown schematically on the right, using the same shading scheme as in Figure 1. The usage of alternative exons indicated with a triangle was statistically evaluated. The comparison between MBII-52 and MBII-85 transfected cells showed statistically significant differences, the P-values of the Student's t-test were: DPM2: 0.001, TAF1: 0.023; RALGPS: 0.021; PBRM1: 0.076 and CRHR1: 0.002; (n = 4).
Together, these data suggest that after being transferred into a heterologous gene context at least five alternative exons are influenced by MBII-52 expression.
A mouse model of PWS shows changes in the predicted exons
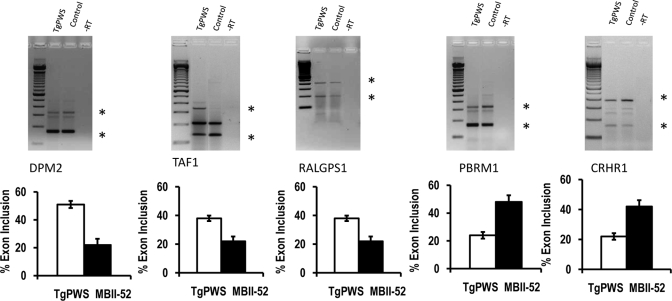
To address the physiological significance of our data, we asked whether MBII-52 influences alternative splicing of the identified target genes in vivo and analyzed RNA samples from the TgPWS mouse model. TgPWS mice have a paternally derived deletion of the PWS critical region that contains the SNURF–SNRPN locus. They show hormonal and metabolic defects resembling those of human newborns with PWS (18). As a larger locus is deleted, in addition to MBII-52, the mice do not express MBII-85 and other snoRNAs from the Prader–Willi critical region.
We compared RNA from newborn TgPWS mice with RNA from littermates expressing the region. As shown in Figure 3, we found that the mouse knockout systems recapitulates a dependency of alternative splicing on the presence of MBII-52. However, the overall splicing patterns of the endogenous genes are different in mouse brain and Neuro2A cells. This most likely reflects the presence of numerous cell types in brain that show different splicing patterns. Despite this limitation, the presence of MBII-52 promotes exon inclusion in the alternative exons with a complementarity to MBII-52 of the DPM2 and PBRM1 pre-mRNAs and promotes skipping of the RALGPS1 and TAF1 exons, similar to the effect seen in Neuro2A cells. The only discrepancy between the MBII-52 effects in brain and Neuro2A cells was an alternative exon of CRHR1 that showed an increase in exon usage in brain tissue, whereas it showed a decrease in response to MBII-52 in Neuro2A cells. The regulated alternative CRHR1 exon is in a cluster of alternative exons and the discrepancy could be due to differences in splicing regulators between brain and Neuro2A cells. Collectively, the data suggest that the loss of MBII-52 expression influences alternative splicing of target genes in a physiological context.
Figure 3.
Comparison of RNA from TgPWS mice lacking MBII-52 expression and control littermates. Total brain samples from TgPWS mice lacking expression of the Prader–Willi critical region were compared with normal littermates expressing all the snoRNAs from the PWS critical region (control). Primers similar to Figure 1 were used. The structure of the gene products is indicated similar to Figures 1 and 2. Stars indicate the bands that were used for comparison. n = 6, other statistical evaluations: P = 0.093, <0.001, 0.0023, 0.001, 0.05 for DPM2, TAF1, RALGPS1, PBRM1 and CRHR1, respectively.
MBII-52 is processed into smaller RNAs
The data indicate that MBII-52 expression influences usage of multiple exons that contain regions with sequence complementarity to the antisense-box of MBII-52. Four recent studies reported that H/ACA snoRNAs give rise to smaller RNAs (25–28). We therefore tested whether the C/D box snoRNA MBII-52 also gives rise to other RNAs by RNase protection analysis.
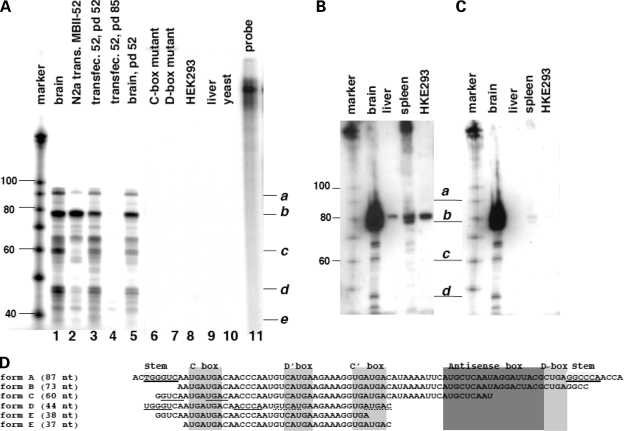
Whereas humans have 47 HBII-52 copies, there are at least 130 copies of MBII-52 snoRNAs in mouse. We used an antisense probe against the MBII-52 copy employed in transfection experiments described above. This isoform is 87 nt in length and its sequence is shown in Figure 4D as form A. In silico analysis shows that this copy shares only an uninterrupted stretch of 20 nt in the antisense box region with other snoRNA isoforms of the MBII-52 cluster. All other regions show single nucleotide differences that prevent longer protected fragments. For the analysis, we used an in vitro transcribed, 32P labeled RNA-antisense probe that detects the 87 nt encompassing the full-length snoRNA. Together with linker and vector sequences, the probe is 175 nt in length. After hybridization, RNase A and T1 digestion, the fragments were separated on 15% acrylamide/TBE/8 m urea gels. As shown in Figure 4A, lane 1, we observed additional fragments when total mouse brain RNA was analyzed with this probe. In agreement with earlier studies, we do not detect expression in liver (29) (Fig. 4A, lane 9). We then asked whether the snoRNA expression construct used in Figure 1 is processed in a similar way. We analyzed total RNA from Neuro2A cells transfected with the pCMV/MBII-52 expression construct (Fig. 4A, lane 2) and found a similar RNA pattern. Importantly, the most abundant RNA species from both the expression construct and brain is shorter than 80 nt (form B, Fig. 4A). SnoRNAs contain C and D boxes that stabilize the snoRNP. Mutation of these RNA elements abolished the effect on splicing (Fig. 2). We therefore tested expression from constructs expressing this mutants and could not detect any RNA expression (Fig. 4A, lanes 6 and 7), suggesting that the smaller RNAs (form B, C, D) derive from a precursor with intact C and D boxes.
Figure 4.
MBII-52 is processed into smaller RNAs. (A) RNase protection analysis using a probe detecting the MBII-52 copy used in transfection studies in Figures 1 and 2. Five microgram of the following total RNAs was hybridized to an MBII-52 antisense probe: (1): total mouse brain, (2): Neuro2A cells transfected with pCMV/MBII-52. Lanes (3–5) are protections from RNPs captured with oligonucleotides against the antisense box (Fig. 5). (3): Affinity captured RNA from Neuro2A cells expressing MBII-52 using a MBII-52 probe for pull down (pd), (4): affinity captured RNA from Neuro2A expressing MBII-52 using an MBII-85 probe (negative control) and (5): affinity captured RNA from brain using an MBII-52 probe for pull down. Lane 6: RNA from Neuro2A cells transfected with an expression construct containing a C-box mutant, (7): RNA from Neuro2A cells transfected with an expression construct containing a D-box mutant, (8): HEK293 cells non-transfected, (9, 10): RNA from liver and yeast. (11): Undigested probe. The marker is a 100 nt RNA base ladder. (B) Northern blot analysis of MBII-52. Fifteen microgram total RNA from brain, liver, spleen and HEK293 cells was separated on 15% polyacrylamide gels and probed with a 32P labeled probe for MBII-52. After stringent washing, cross-hybridizing bands in liver, spleen and HEK293 cells still remain. Exposure was overnight. (C) The filter from (B) was treated with RNase A/T1 and again washed. The cross-hybridizing bands disappear, but the signals corresponding to smaller bands remain. Exposure was for 3 days. (D) Sequences of the shorter RNAs. The stems and functional boxes are indicated. The clones are ordered according to their length. Form A corresponds to the published snoRNA MBII-52. Underlined nucleotides in forms C and D indicated predicted stems.
It is possible that MBII-52 undergoes nucleotide modifications that would result in mismatching of an RNase protection probe and subsequent generation of smaller fragments. To rule out this possibility, we performed northern blot analysis, using denaturing 15% PAGE gels. Total RNA from brain, liver and spleen was probed with MBII-52 antisense RNA corresponding to the sequence in Figure 4D, form A. Even after stringent washing, we see cross-hybridization of MBII-52 with RNAs from liver, spleen and HEK293 cells (Fig. 4B). This is to be expected, as there are numerous copies of sequence-related snoRNAs expressed in these tissues (29). To detect the specific hybridization between MBII-52 form A and brain RNA, we treated the membrane with RNase A and RNase T1. The RNase treatment reduced the overall signal strength, as we had to use a 3-fold longer exposure time. As shown in Figure 4C, this treatment abolishes the cross-hybridization with non-brain RNAs. However, this treatment does not abolish the signal from brain RNA tissue that corresponds in length to RNA forms B–D. Similar to the RNA protection experiment, the major RNA species is shorter than 80 nt. This indicates that the protection pattern observed in the protection assay is due to shorter RNAs and not the result of nucleotide editing. Unexpectedly, we observe a distinct pattern of shorter RNAs and not a continuous smear of bands. This finding implies that all of the estimated MBII-52 copies are processed in a similar way, giving rise to specific metabolically stable short RNAs.
To determine the identity of the novel short RNAs, we cloned the protected fragments. Total mouse brain RNA was subjected to RNase protection. Subsequently, the RNases were removed by Proteinase K treatment and phenol extraction. The double-stranded RNA was phosphorylated using T4 kinase, and an adenylated-linker was ligated in the absence of ATP (30). After gel purification and isolation, an adapter linker was ligated using T4 DNA ligase. The reaction was subsequently reverse transcribed, amplified and cloned. The positive clones are shown in Figure 4D. All shorter RNAs lack the sequences forming the stem of the snoRNA, but contain the C and C′ box. The stem conveys complementarity between the snoRNA ends and stabilizes the snoRNP. The remaining cloned RNAs are shortened from the 5′ and 3′ ends, indicating that they are generated by 3′→5′ and 5′→3′ exonuclease activity that stops at the C and C′ boxes.
Together, these data suggest that the expression unit consisting of MBII-52 and its flanking intron and exons gives rise to several RNAs. These RNAs include the previously described MBII-52 snoRNA (form A), as well as shorter RNA species. The major RNA species (form B) expressed from the MBII-52 cluster lacks the stem box, but still contain C and D boxes.
MBII-52 derived RNAs do not bind to classical snoRNA-associated proteins
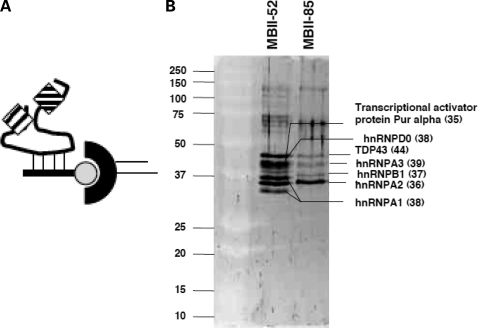
As we found that the MBII-52 locus gives rise to previously not described products, we identified the proteins that associate with these RNAs. We used the affinity between a biotinylated 2′-O-methylated oligonucleotide and the antisense box of MBII-52 to isolate RNAs derived from the MBII-52 locus (Fig. 5A). Using streptavidin beads, we isolated the MBII-52 snoRNA particle (snoRNP) from nuclear extracts generated from cells transfected with the MBII-52 expression construct. Nuclear extract was generated by a scaled-down Dignam procedure (31). After washing with 100 and 200 mm NaCl, the captured material was separated by SDS–PAGE and proteins were identified by mass spectrometry and database matching. An oligonucleotide against the snoCR of MBII-85 was used as the control. As shown in Figure 5B, we found that hnRNPs were associated with the expressed snoRNA. Similar results were seen with samples obtained from mouse brain nuclear extracts (data not shown). Repeated experiments using different washing and isolation methods to find canonical snoRNP proteins, such as fibrillarin or NOP56, in pulled-down material from MBII-52 affinity material failed to identify known snoRNP-associated proteins.
Figure 5.
Analysis of proteins associated with MBII-52. (A) Experimental strategy: a biotinylated oligonucleotide is used to capture the snoRNP complex. The oligonucleotide is shown as a line, the complementarity is dashed, biotin is shown as a circle. The RNP complex (boxes) is isolated by streptavidin (half-circle)-capture from extracts expressing MBII-52 and washed in non-denaturing buffer. The extracts were prepared by transfecting MBII-52 expression constructs and performing Dignam mini-extracts. (B) Silver stain of a gel with affinity purified RNPs. MBII-52: the RNPs were isolated with a capture-oligonucleotide against MBII-52. MBII-85: the RNP was isolated with a capture oligonucleotide against MBII-85. Proteins were determined by mass spectrometry and are indicated. Sizes in kDa are shown in parentheses.
We determined which RNAs are present in the pulled-down material and performed RNase protection. As shown in Figure 4A, lane 4 and 5, we found that the isolates contained the smaller MBII-52-related RNAs, as well as the full-length MBII-52 snoRNA. No MBII-52 RNA was pulled-down with the probe against MBII-85, suggesting the selectivity of the pull-down.
In summary, the findings indicate that the shorter RNAs assemble with hnRNPs, but not with proteins that have previously been described to associate with C/D box snoRNAs. Although the major RNA isoform B contains C and D boxes, structural hallmarks of C/D box snoRNAs, the composition of the RNP formed is different from a C/D box snoRNP.
DISCUSSION
The MBII-52 expression unit generates processed snoRNAs (psnoRNAs)
MBII-52 snoRNAs are expressed from a cluster containing multiple copies of tandemly arranged snoRNA expression units. Each unit contains phylogenetically poorly conserved exons that flank an intron which hosts the snoRNA (19). Humans contain 47 HBII-52 copies and mice at least 130 copies. Using RNase protection assays, we analyzed the mouse MBII-52 copy that is most closely related to the copy 27 of human HBII-52 snoRNA cluster. There is enough sequence heterogeneity between the different MBII-52 snoRNA copies that allows their discrimination in protection assays. Unexpectedly, the RNase protection assay indicates that the snoRNA gives rise to other smaller RNAs and that the full-length C/D box snoRNA is a minor form. The presence of the smaller RNAs could be verified by northern blot analysis, which further rules out that signals corresponding to shorter RNAs are caused by the protection of unrelated RNAs or are caused by RNA editing events that introduce mismatches to the probe. Finally, we tested ectopical expression of MBII-52 in HEK293 cells that do not express this snoRNA. The expression construct gives a similar pattern of shorter RNAs, indicating that they are derived from the transfected single MBII-52 expressing unit. The cloning of the shorter RNAs indicates that the major RNA form expressed from the MBII-52 expression unit is a 73 nt long RNA (form B) that lacks the sequences that form the snoRNA stem. However, this RNA contains other C/D box snoRNA elements, such as the C box, D box and antisense box. This RNA appears to be further shortened by exonuclease trimming, giving rise to smaller RNAs. The shorter RNAs can be detected both by northern blot and RNase protection analyses, indicating that they are metabolically stable. It is possible that these RNAs are protected from further endonuclease action by predicted secondary structures. The RNA form D forms a 5 bp stem on its 5′ and 3′ ends and RNA form C contains a short stem at its 5′ end (Fig. 4D, underlined region). In addition, the formation of protein complexes is likely to stabilize the RNAs.
Ectopic expression of snoRNA mutants suggests that the formation of shorter RNAs depends on intact C and D boxes, which suggests that they derive from a C/D box snoRNA or pre-snoRNA structure. A possible scenario is that an unknown RNase initially removes the stem of the C/D box RNA, which gives rise to the predominant form B. This form is stabilized by the presence of the C and D boxes, most likely by binding to other proteins. Form B is shortened by exonucleases, giving rise to forms C, D and E that are most likely stabilized by another stem-loop structure and/or associated proteins.
To obtain insight into proteins associated with these novel RNAs, we isolated them by affinity purification of RNP complexes, using a probe that is complementary to the antisense box of MBII-52. We used nuclear extract generated by the Dignam procedure as starting material. In this method, most of the nucleolar material is separated in a high-speed centrifugation step. As the MBII-52-derived snoRNAs are present in this fraction, they are most likely present in the nucleoplasm. The major form RNA form B derived from MBII-52 does not contain the characteristic k-turn, which most likely prevents its association to Snu13p/15.5 kDa (4). In agreement with this RNA structure, we could not detect C/D box snoRNA-associated proteins, such as fibrillarin, or NOP56 (5) in the isolated material. In contrast, we identified hnRNPs, including hnRNP A1, A2, TDP-43 and D0 that have been reported to be involved in splice-site selection. Unexpectedly, in the pull-downed material, we could still detect RNA forms C and D. These RNAs lack a complete snoCR that is complementary to the pull-down probe. Relative to the starting brain material, the RNA forms C and D are reduced in the pulled down material (compare Fig. 4A, lanes 1 and 5), but are still detectable. This suggests that the different RNA forms could be present within a complex.
We propose to name these shorter RNAs psnoRNAs for processed small nucleolar RNAs. PsnoRNAs could represent a new class of nuclear small RNAs. The psnoRNAs described here are the first to be derived from C/D box snoRNAs.
MBII-52-derived psnoRNAs regulate splicing of several pre-mRNAs
We previously found that the expression of the snoRNA HBII-52 promotes inclusion of exon Vb of the serotonin receptor 5-HT2C. To investigate whether this represents a special, unique case or is part of a new regulatory mechanism, we developed a computational screen that predicted more than 400 putative snoRNA targets. We tested some of these predicted targets by RT–PCR in transfection assays and further concentrated on five splicing events that showed consistent dependency on MBII-52 expression. In contrast to the 5-HT2C receptor pre-mRNA, the pre-mRNAs harboring the MBII-52-dependent exons are expressed in Neuro2A and HEK293 cells, which allows us to determine the influence of MBII-52 expression on the endogenous genes. Also in contrast to the neuron-specific 5-HT2C system where a splice site had to be optimized to detect the dependency on MBII-52 (10), the new alternative exons showed the dependency on MBII-52 expression when analyzed in their endogenous gene context.
The alternative exons were next tested in a heterologous exon trap system and showed the dependency of MBII-52 when flanked by insulin exons that are controlled by a CMV promoter. These experiments suggest that MBII-52 RNAs act on defined parts of the pre-mRNA, in a mechanism that is independent of the promoter usage and genomic context. Together, these data strongly suggest that MBII-52 expression influences alternative pre-mRNA splicing events.
Expression of MBII-52 causes a small, but statistically significant changes in multiple targets. This modest influence on numerous targets has been observed for other splicing factors, such as SMN (32) and NOVA (33). Detailed work in the NOVA system (33) suggested that a splicing factor can control biological processes by coordinating numerous small changes and it is possible that MBII-52 fulfills a similar function. An alignment of the antisense box of MBII-52 and its experimentally confirmed targets is shown in Supplementary Data 2. The complementarity between the MBII-52 antisense box and its targets can be interrupted in multiple positions. With the exception of the serotonin receptor 5HT2C, there are always three mismatches in the alignment of the snoCR and the MBII-52 antisense box. It is interesting that the serotonin pre-mRNA can be edited at three positions within the snoCR. Taking these editing events into account, the data suggest that preferably 15 of the 18 nucleotides of the antisense box show complementarity towards its target. It is noteworthy that we initially concentrated on targets with only one or two mismatches, but did not find a dependency of these exons on MBII-52 expression. The data indicate that MBII-52-derived RNAs need a defined degree of sequence complementarity towards their targets. This scenario is reminiscent of the action of U1 snRNP on the 5′ splice site, where natural occurring exons rarely show 100% complementarity towards the U1 snRNA, but usually have several mismatches, which cluster in certain position of the 5′ splice site (34).
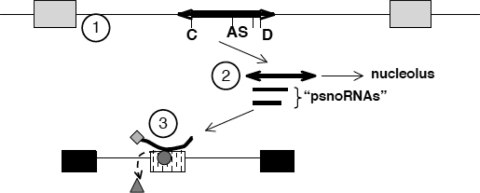
The existence of psnoRNAs could explain the influence of MBII-52 expression on splice-site selection in a model illustrated in Figure 6. We postulate that the MBII-52 expressing unit consisting of two non-coding exons flanking an intron that hosts an snoRNA gives rise to several RNAs. The major form derived from the expression unit is form B that lacks the snoRNA stem-structure and is associated with hnRNPs, but not C/D box snoRNA-typical proteins. Form B contains the antisense box that targets it to other RNAs, including pre-mRNAs identified in this study. Form B RNA can influence splice-site selection by competing with existing splicing regulatory factors on the pre-mRNA or by bringing the associated hnRNPs to the targets, similar to a bifunctional oligonucletide. The longest RNA (form A) has all the hallmarks of a traditional C/D box snoRNA, but is only a minor fraction of the RNA expressed. It is likely that this RNA is transported into the nucleolus, where it can be detected by in situ hybridization (35). It is not clear what function this RNA has in the nucleolus, but it could represent a storage form for the formation of the active RNA form B that is released from the nucleolus according to the physiological needs.
Figure 6.
Model for MBII-52 action on RNA processing. (1) The PWS critical region contains snoRNAs (thick line) located in introns between non-coding exons (grey boxes). The snoRNA is characterized by a C box (C), D box (D) and an antisense box (AS), as well as stem-forming sequences (arrows). (2) This unit generates several RNAs, including the full-length snoRNA that shows its highest concentration in the nucleolus and Cajal bodies (35) as well as several shorter psnoRNAs (for processed snoRNAs). PsnoRNAs are present in the nucleoplasm where they associate with hnRNPs. (3) PsnoRNAs can change splice-site selection, most likely by binding to complementary sequences. We postulate that they either remove regulatory proteins from their targets (triangle) or bring in associated proteins to the exon recognition complex (diamond associated with the small RNA).
Relevance for PWS
The loss of C/D box snoRNA expression has been postulated as the underlying mechanism for the development of PWS (36). This hypothesis was recently supported by a patient with a 174 584 bp large microdeletion that encompassed only snoRNAs and their flanking hosting introns and exons. The deletion led to a Prader–Willi phenotype (9). To date, the only published RNAs expressed from the 174 584 bp region are snoRNAs and fragments of their surrounding non-coding exons.
The idea that the loss of snoRNA expression is central to PWS is further supported by genetic evidence that ruled out proteins encoded by MKRN3, MAGEL2 and NDN genes expressed in the Prader–Willi critical region (37). The 174 584 bp microdeletion removes the snoRNAs HBII-438A, -85 and 23 of the 47 HBII-52 copies from the paternally expressed allele. The only snoRNA that was totally removed by the microdeletion was MBII-85, which let to the suggestion that MBII-85 loss is the major reason for PWS. However, there is evidence that HBII-85 and HBII-52 are expressed by two transcriptional units (20). As the 174 584 bp micordeletion contains the 5′ end of the HBII-52 cluster, it could harbor transcriptional elements necessary for proper HBII-52 expression. Furthermore, in the majority of cases, the complete SNURF–SNRPN locus is deleted (8). The contribution of HBII-85 and HBII-52 loss to PWS is therefore not clear.
Our findings indicate that the SNURF–SNRPN locus not only gives rise to typical C/D box snoRNAs, but generates shorter psnoRNAs. The northern blot analysis indicates that all of the at least 130 MBII-52 copies are processed in a similar manner. The major RNA form from the MBII-52 cluster is not the canonical C/D box snoRNA, but a shorter RNA form, most likely similar to psnoRNA form B. psnoRNAs are associated with hnRNPs and could have multiple functions by targeting these proteins to other RNAs. It is not clear whether several psnoRNAs lacking the antisense box use other RNA parts for targeting or have non-related functions.
The loss of the regulatory psnoRNAs could be a significant contribution to the etiology of PWS and substitution of the short psnoRNAs could be a therapeutic principle for the disease.
MATERIALS AND METHODS
Transfection experiments were performed using Ca-phosphate method as described (38).
The construction of reporter minigenes was done by using recombination between pSpliceExpress, an exon-trap vector and BAC-derived PCR fragments that encompassed the alternative exons, as previously described (24).
Pull-down experiments were performed using Dignam-derived miniextracts (31).
RNase protection analysis was performed using the Ambion RNase protection kit using uniformly 32P-labeled probes.
Cloning of the psnoRNAs was performed as follows: 100 µg of total brain RNA, isolated by the Trizol method was protected using 3 × 106 cpm of an MBII-52 antisense probe. Hybridization was overnight. Single-stranded RNA was digested with RNase A/T1 (Ambion, dilution 1:100) for 1 h at 37°C and RNases were subsequently removed by 100 µg/ml Proteinase K treatment for 1 h. Following phenol extraction and precipitation, RNAs were separated on a 15% acrylamide, 8 m urea gel, exposed overnight and the appropriate bands were excised, crushed, eluted overnight in 3 m ammonium acetate/1% SDS and recovered by precipitation. The first RNA linker was 5′rAppCTGTAGGCACCATCAAT/3ddC. It was ligated for 2 h in a 20 µl reaction in 50 mm HEPES pH 8.3, 10 mm MgCl2, 3.3 mm DTT, 10 µg/ml BSA, 8.3 v/v glycerol and 20 U RNA ligase. The reaction was again separated by a 15% acrylamide, 8 m urea gel, bands excised, crushed and eluted overnight. The second RNA linker was 5′-AmMC6/GCTCCAGAATTCGGACCCGArGrUrGrCrCrUrArCrArG. It was ligated at 18°C in 1× ligase buffer using T4 DNA ligase overnight. The reaction was reverse transcribed, PCR amplified and subcloned. Positive clones were isolated using colony hybridization and sequenced.
Primers for PCR detection were: CRHR1: MmNEWCRHR1F CCAGGATCAGCAGTGTGAGA; MmNEWCRHR1R AGTGGCCCAGGTAGTTGATG; TAF1: TAF1NewF TCTGCGATGAAAAACTCAAAGA; TAF1NewR TCCACATCAGAGTCACTTCCA; DPM2: F CAGACCAAGCAGTAGGATTT; R ACAAACAGGAGCAGCAGGAG; RALGPS1: F AGTCCCCAGACACAGGAAGA; R TCTCAGAGGCCCCTCCAT; PB1: F TGGCTACATTTTGTTCAGCAG; R ATGGGGGCTACTCCTTGATT.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the NIH (GM083187 to S.S.), the DFG (Sta399/6-3), the Foundation for Prader–Willi Research, the Prader–Willi Research Foundation, EURASNET (S.S., M.Z.), and in part from a CHP Scientific Program Innovation Award (R.D.N.). The mass spectrometric analysis was performed at the University of Kentucky, Center for Structural Biology Protein Core Facility. This core facility is supported in part by funds from NIH National Center for Research Resources (NCRR) grant P20 RR020171.
Supplementary Material
ACKNOWLEDGEMENTS
Conflict of Interest statement. None declared.
REFERENCES
- 1.Pan Q., Shai O., Lee L.J., Frey B.J., Blencowe B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 2.Wang E.T., Sandberg R., Luo S., Khrebtukova I., Zhang L., Mayr C., Kingsmore S.F., Schroth G.P., Burge C.B. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stamm S., Ben-Ari S., Rafalska I., Tang Y., Zhang Z., Toiber D., Thanaraj T.A., Soreq H. Function of alternative splicing. Gene. 2005;344C:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 4.Watkins N.J., Segault V., Charpentier B., Nottrott S., Fabrizio P., Bachi A., Wilm M., Rosbash M., Branlant C., Luhrmann R. A common core RNP structure shared between the small nucleoar box C/D RNPs and the spliceosomal U4 snRNP. Cell. 2000;103:457–466. doi: 10.1016/s0092-8674(00)00137-9. [DOI] [PubMed] [Google Scholar]
- 5.Matera A.G., Terns R.M., Terns M.P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 6.Steitz J.A., Tycowski K.T. Small RNA chaperones for ribosome biogenesis. Science. 1995;270:1626–1627. doi: 10.1126/science.270.5242.1626. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz W., Pogacic V. Biogenesis of small nucleolar ribonucleoproteins. Curr. Opin. Cell Biol. 2002;14:319–327. doi: 10.1016/s0955-0674(02)00334-4. [DOI] [PubMed] [Google Scholar]
- 8.Butler M.G., Hanchett J.M., Thompson T.E. Clinical findings and natural history of Prader–Willi syndrome. In: Butler M.G., Lee P.D.K., Whitman B.Y., editors. Managment of Prader–Willi Syndrome. Springer; 2006. pp. 3–48. [Google Scholar]
- 9.Sahoo T., del Gaudio D., German J.R., Shinawi M., Peters S.U., Person R.E., Garnica A., Cheung S.W., Beaudet A.L. Prader–Willi phenotype caused by paternal deficiency for the HBII-85 C/D box small nucleolar RNA cluster. Nat. Genet. 2008;40:719–721. doi: 10.1038/ng.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kishore S., Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–232. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- 11.Doe C.M., Relkovic D., Garfield A.S., Dalley J.W., Theobald D.E., Humby T., Wilkinson L.S., Isles A.R. Loss of the imprinted snoRNA mbii-52 leads to increased 5htr2c pre-RNA editing and altered 5HT2CR-mediated behaviour. Hum. Mol. Genet. 2009;18:2140–2148. doi: 10.1093/hmg/ddp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakatani J., Tamada K., Hatanaka F., Ise S., Ohta H., Inoue K., Tomonaga S., Watanabe Y., Chung Y.J., Banerjee R., et al. Abnormal behavior in a chromosome-engineered mouse model for human 15q11–13 duplication seen in autism. Cell. 2009;137:1235–1246. doi: 10.1016/j.cell.2009.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiholzer U., Stutz K., Weinmann C., Torresani T., Molinari L., Prader A. Low insulin, IGF-I and IGFBP-3 levels in children with Prader–Labhart–Willi syndrome. Eur. J. Pediatr. 1998;157:890–893. doi: 10.1007/s004310050961. [DOI] [PubMed] [Google Scholar]
- 14.Eiholzer U., Gisin R., Weinmann C., Kriemler S., Steinert H., Torresani T., Zachmann M., Prader A. Treatment with human growth hormone in patients with Prader–Labhart–Willi syndrome reduces body fat and increases muscle mass and physical performance. Eur. J. Pediatr. 1998;157:368–377. doi: 10.1007/s004310050832. [DOI] [PubMed] [Google Scholar]
- 15.Cummings D.E., Clement K., Purnell J.Q., Vaisse C., Foster K.E., Frayo R.S., Schwartz M.W., Basdevant A., Weigle D.S. Elevated plasma ghrelin levels in Prader Willi syndrome. Nat. Med. 2002;8:643–644. doi: 10.1038/nm0702-643. [DOI] [PubMed] [Google Scholar]
- 16.de Lind van Wijngaarden R.F., Otten B.J., Festen D.A., Joosten K.F., de Jong F.H., Sweep F.C., Hokken-Koelega A.C. High prevalence of central adrenal insufficiency in patients with Prader–Willi syndrome. J. Clin. Endocrinol. Metab. 2008;93:1649–1654. doi: 10.1210/jc.2007-2294. [DOI] [PubMed] [Google Scholar]
- 17.Carrel A.L., Lee P.D.K., Mogul H.R. Growth hormone and Prader–Willi syndrome. In: Butler M.G., Lee P.D.K., Whitman B.Y., editors. Managment of Prader–Willi Syndrome. New York: Springer; 2006. pp. 201–241. [Google Scholar]
- 18.Stefan M., Ji H., Simmons R.A., Cummings D.E., Ahima R.S., Friedman M.I., Nicholls R.D. Hormonal and metabolic defects in a Prader–Willi syndrome mouse model with neonatal failure to thrive. Endocrinology. 2005;146:4377–4385. doi: 10.1210/en.2005-0371. [DOI] [PubMed] [Google Scholar]
- 19.Runte M., Huttenhofer A., Gross S., Kiefmann M., Horsthemke B., Buiting K. The IC-SNURF–SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 20.Vitali P., Royo H., Marty V., Bortolin-Cavaille M.L., Cavaille J. Long nuclear-retained non-coding RNAs and allele-specific higher-order chromatin organization at imprinted snoRNA gene arrays. J. Cell. Sci. 2010;123:70–83. doi: 10.1242/jcs.054957. [DOI] [PubMed] [Google Scholar]
- 21.Kishore S., Stamm S. Regulation of alternative splicing by snoRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006;LXXI:329–334. doi: 10.1101/sqb.2006.71.024. [DOI] [PubMed] [Google Scholar]
- 22.Cavaille J., Bachellerie J.P. SnoRNA-guided ribose methylation of rRNA: structural features of the guide RNA duplex influencing the extent of the reaction. Nucleic Acids Res. 1998;26:1576–1587. doi: 10.1093/nar/26.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rehmsmeier M., Steffen P., Hochsmann M., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kishore S., Khanna A., Stamm S. Rapid generation of splicing reporters with pSpliceExpress. Gene. 2008;427:104–110. doi: 10.1016/j.gene.2008.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ender C., Krek A., Friedlander M.R., Beitzinger M., Weinmann L., Chen W., Pfeffer S., Rajewsky N., Meister G. A human snoRNA with microRNA-like functions. Mol. Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 26.Saraiya A.A., Wang C.C. snoRNA, a novel precursor of microRNA in Giardia lamblia. PLoS Pathog. 2008;4:e1000224. doi: 10.1371/journal.ppat.1000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott M.S., Avolio F., Ono M., Lamond A.I., Barton G.J. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput. Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutzinger R., Feederle R., Mrazek J., Schiefermeier N., Balwierz P.J., Zavolan M., Polacek N., Delecluse H.J., Huttenhofer A. Expression and processing of a small nucleolar RNA from the Epstein-Barr virus genome. PLoS Pathog. 2009;5:e1000547. doi: 10.1371/journal.ppat.1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cavaille J., Buiting K., Kiefmann M., Lalande M., Brannan C.I., Horsthemke B., Bachellerie J.P., Brosius J., Huttenhofer A. Identification of brain-specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc. Natl Acad. Sci. USA. 2000;97:14311–14316. doi: 10.1073/pnas.250426397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 31.Lee K.A., Bindereif A., Green M.R. A small-scale procedure for preparation of nuclear extracts that support efficient transcription and pre-mRNA splicing. Gene Anal. Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., Darnell J.C., Darnell R.B. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stamm S., Zhu J., Nakai K., Stoilov P., Stoss O., Zhang M.Q. An alternative-exon database and its statistical analysis. DNA Cell Biol. 2000;19:739–756. doi: 10.1089/104454900750058107. [DOI] [PubMed] [Google Scholar]
- 35.Vitali P., Basyuk E., Le Meur E., Bertrand E., Muscatelli F., Cavaille J., Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J. Cell Biol. 2005;169:745–753. doi: 10.1083/jcb.200411129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding F., Prints Y., Dhar M.S., Johnson D.K., Garnacho-Montero C., Nicholls R.D., Francke U. Lack of Pwcr1/MBII-85 snoRNA is critical for neonatal lethality in Prader–Willi syndrome mouse models. Mamm. Genome. 2005;16:424–431. doi: 10.1007/s00335-005-2460-2. [DOI] [PubMed] [Google Scholar]
- 37.Kanber D., Giltay J., Wieczorek D., Zogel C., Hochstenbach R., Caliebe A., Kuechler A., Horsthemke B., Buiting K. A paternal deletion of MKRN3, MAGEL2 and NDN does not result in Prader–Willi syndrome. Eur. J. Hum. Genet. 2009;17:582–590. doi: 10.1038/ejhg.2008.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stoss O., Stoilov P., Hartmann A.M., Nayler O., Stamm S. The in vivo minigene approach to analyze tissue-specific splicing. Brain Res. Protoc. 1999;4:383–394. doi: 10.1016/s1385-299x(99)00043-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.