Abstract
Distal arthrogryposis type I (DA1) is a disorder characterized by congenital contractures of the hands and feet for which few genes have been identified. Here we describe a five-generation family with DA1 segregating as an autosomal dominant disorder with complete penetrance. Genome-wide linkage analysis using Affymetrix GeneChip Mapping 10K data from 12 affected members of this family revealed a multipoint LODmax of 3.27 on chromosome 12q. Sequencing of the slow-twitch skeletal muscle myosin binding protein C1 (MYBPC1), located within the linkage interval, revealed a missense mutation (c.706T>C) that segregated with disease in this family and causes a W236R amino acid substitution. A second MYBPC1 missense mutation was identified (c.2566T>C)(Y856H) in another family with DA1, accounting for an MYBPC1 mutation frequency of 13% (two of 15). Skeletal muscle biopsies from affected patients showed type I (slow-twitch) fibers were smaller than type II fibers. Expression of a green fluorescent protein (GFP)-tagged MYBPC1 construct containing WT and DA1 mutations in mouse skeletal muscle revealed robust sarcomeric localization. In contrast, a more diffuse localization was seen when non-fused GFP and MYBPC1 proteins containing corresponding MYBPC3 amino acid substitutions (R326Q, E334K) that cause hypertrophic cardiomyopathy were expressed. These findings reveal that the MYBPC1 is a novel gene responsible for DA1, though the mechanism of disease may differ from how some cardiac MYBPC3 mutations cause hypertrophic cardiomyopathy.
INTRODUCTION
Congenital contractures occur in 0.5–1% of all births (1). Distal limb contractures occur as a spectrum, from isolated contracture of a single limb in talipes equinovarus (also called clubfoot), to severe bilateral contractures of the hands and feet with additional manifestations, including craniofacial anomalies or scoliosis, in some forms of distal arthrogryposis. Of the more than 10 forms of distal arthrogryposis that have been described to date (2), distal arthrogryposis type I (DA1) is the most common, affecting approximately one in 10 000 individuals (3). Patients with DA1 have contractures that are limited to the hands and feet with no additional anomalies. Variable expressivity, including clubfoot, vertical talus, camptodactyly, overriding fingers, ulnar deviation of the fingers, is common in families with autosomal dominant inherited DA1 (3–6).
Mutations in genes encoding the skeletal muscle contractile apparatus have been implicated in distal arthrogryposis. These genes include myosin heavy chain (MYH3 and MYH8), troponin I (TNNI2), troponin T (TNNT3) and tropomyosin (TPM2) (7–11). Much of this research has focused on the most severe forms of distal arthrogryposis, Freeman–Sheldon syndrome (distal arthrogryposis type 2A) and Sheldon–Hall syndrome (distal arthrogryposis type 2B), in which craniofacial anomalies and scoliosis often accompany the distal limb contractures (12). Nearly, all patients with Freeman–Sheldon syndrome and about one-third of all patients with Sheldon–Hall syndrome have mutations in the embryonic myosin heavy chain gene MYH3 (7).
In contrast, the genetic basis of DA1 has been less well established. Mutations in three contractile genes have been identified in patients with DA1, including TPM2 (9), TNNI2 (6) and TNNT3 (13). However, in each case mutations were identified in only a single patient or family, suggesting extensive locus heterogeneity in DA1. Furthermore, mutations in these genes do not exclusively cause a DA1 phenotype, but have also been identified in patients with a wide range of neuromuscular disease, such as cap myopathy (14), nemaline myopathy (15), multiple pterygium syndrome with nemaline myopathy (16) and Freeman–Sheldon syndrome (DA2B) (9).
Here, we provide a clinical description of a large family with autosomal dominant DA1 and identify the first mutations in skeletal muscle slow-twitch myosin binding protein C1 (MYBPC1) in two familial cases of DA1. Because mutations in the cardiac myosin binding protein C3 gene (MYBPC3) are one of the most frequent causes of hypertrophic cardiomyopathy (17,18), this finding suggests that myosin binding protein C is critical for the normal function of both cardiac and skeletal muscle.
RESULTS
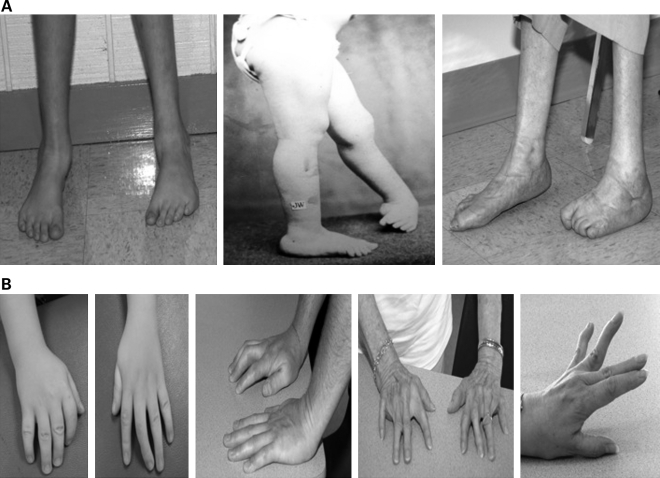
The proband of this large family with DA1 presented with bilateral clubfoot and camptodactyly with ulnar deviation of the fingers (Fig. 1). Facial weakness was absent. He belongs to a multi-generational family (5432) with autosomal dominant inheritance of distal limb contractures with complete penetrance of the lower limb contractures. Nine of 12 affected individuals had congenital talipes equinovarus (clubfoot) and the three other affected individuals had congenital vertical talus (Fig. 1A). The lower limb contractures were bilateral in all cases except one female with left-sided clubfoot and one female with right-sided clubfoot. Surgical treatment of the lower limb contractures occurred in six of 12, whereas the others were treated conservatively. Five of the 12 individuals had hand contractures manifesting as either camptodactyly and ulnar deviation of the fingers (three individuals) or extension contracture of the fourth digit (two individuals) (Fig. 1B). The other seven affected individuals were examined and found to have normal finger flexion creases and no evidence of hand involvement. Detailed neurological examinations were performed on all 12 affected individuals and no facial weakness or weakness of any specific muscle groups was noted with the exception of limitations of finger, toe and foot extensor strength in some individuals. However, the presence of joint contractures limited our ability to fully test these muscle groups. None had hip dysplasia or scoliosis.
Figure 1.
Distal limb contractures in affected individuals from family 5432. (A) Photograph of proband with treated bilateral congenital clubfoot and his great aunt with unilateral left-sided clubfoot before and after multiple corrective surgeries. (B) Hand contractures consisting of camptodactyly and ulnar deviation of the fingers in three affected individuals of family 5432. The final photo shows fourth finger extension contractures present in two individuals of this family.
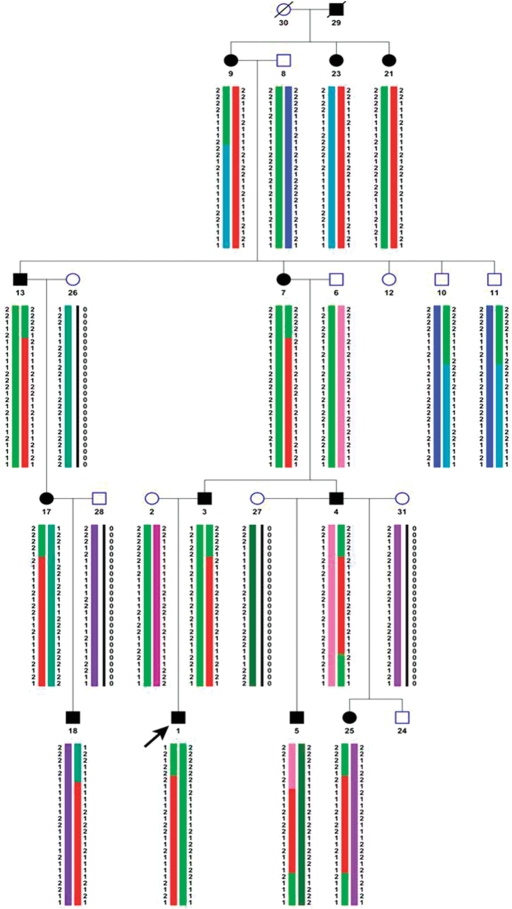
To map the gene causing DA1, a genome-wide linkage scan was performed using data from 12 affected and five unaffected individuals of family 5432. Genotypes for 11 555 SNPs at an average 0.3 cM resolution were determined following hybridization to Affymetrix 10K mapping SNP arrays. Parametric multipoint logarithm of the odds (LOD) score analysis yielded LODmax of 3.27 with markers on chromosome 12q23.2 (Fig. 2). Recombinants delineated the critical region to an interval of 9 cM between rs3869308 and rs1196761, corresponding to a 12 Mb region (chr12: 92 400 000–104 640 000) (Fig. 2). The chromosome 12q23.2 candidate region contains more than 60 genes.
Figure 2.
Haplotype analysis indicating markers on chromosome 12q common to all affected individuals with distal arthrogryposis type I. Common haplotype is shown in red. Arrow indicates proband. + Symbol indicates individuals with hand contractures; VT indicates individuals with vertical talus. All other affected individuals have clubfoot. Haplotypes were created with GENEHUNTER and viewed on Haplopainter (55).
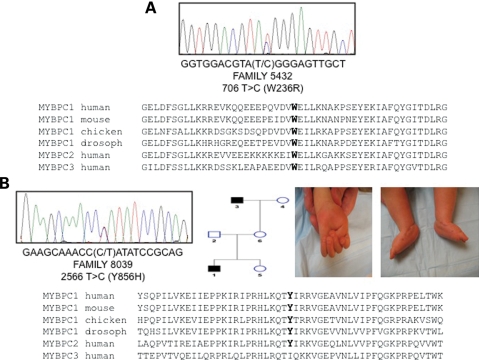
One reasonable candidate within this linkage interval is the skeletal muscle myosin binding protein C1 (MYBPC1) gene. Sequencing of MYBPC1 revealed a missense mutation c.706T>C resulting in the amino acid substitution W236R (Fig. 3A). This mutation was present in all affected family members, and absent in unaffected family members. The W236R amino acid substitution occurs within the MyBP-C unique motif located between the C1 and C2 immunoglobulin domains (Fig. 4). Sequencing of MYBPC1 in 14 additional patients with DA1 identified a missense mutation c.2566T>C in an individual with bilateral vertical talus and hand contractures (Fig. 3B). This mutation results in amino acid substitution Y856H within the C8 repeat (Fig. 4). This mutation was found also in the proband's grandfather who has bilateral clubfoot and no hand contractures. Both mutations occur in regions that are highly conserved and were not identified chromosomes of 400 control individuals of North American Caucasian ancestry.
Figure 3.
MYBPC1 missense mutations in two families with distal arthrogryposis type 1. Chromatograms showing missense mutations in proband of family 5432 (A) and 8039 (B). Both mutations result in amino acid substitutions at highly conserved residues as shown by multiple species alignment of MYBPC1 (skeletal muscle, slow-twitch) and MYBPC2 (skeletal muscle, fast-twitch) and MYBPC3 (cardiac). Also shown in (B) is a pedigree of family 8039 and clinical photographs of hand and foot contractures (congenital vertical talus) occurring in the proband.
Figure 4.
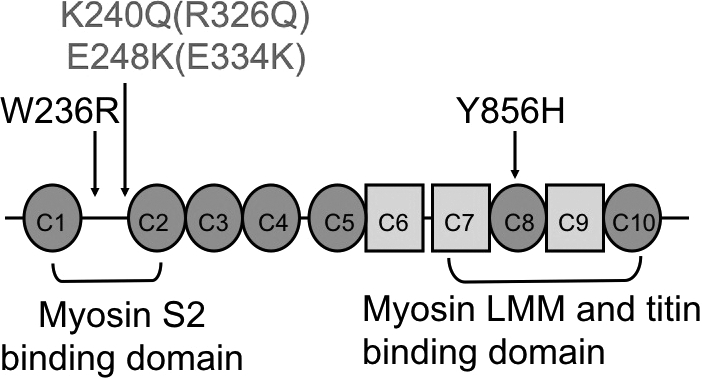
Location of two myosin binding protein C1 (MYBPC1) mutations in two patients with DA1. The W236R mutation is located within the MyBP-C unique motif between the C1 and C2 immunoglobulin domains. This region has been shown to interact with myosin S2 (42). The Y856H mutation is within the C8 repeat, a region known to mediate interactions with LMM portion of myosin (45,46) and titin (47). Two MYBPC3 mutations found in patients with hypertrophic cardiomyopathy, R326Q and E334K, corresponding to MYBPC1 K240Q and E248K, are shown in italics. Immunoglobulin domains are shown as circles and fibronectin type III domains as squares.
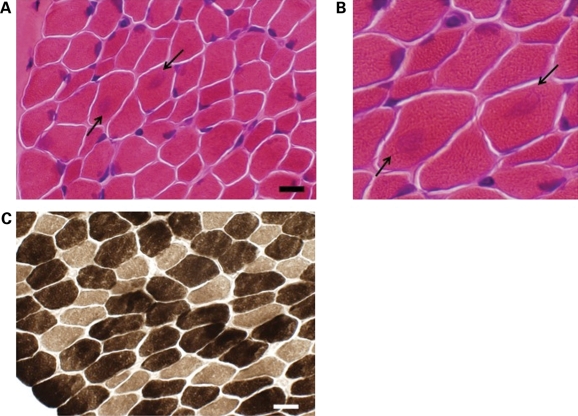
Routine ATPase stains demonstrated type I fiber smallness from abductor hallucis muscle of both probands with MYBPC1 W236R (Fig. 5) and Y856H mutations (data not shown). Muscle biopsies were obtained at 4 and 2 years of age, respectively. For the biopsy shown in Figure 5 (W236R), type 1 fibers were 24% smaller than type 2 fibers (14.0 versus 18.4 µm), meeting the criteria for type 1 fiber atrophy (greater than 12% difference in size) (19). In the biopsy of the second patient (Y856H), the type 1 fibers were 33% smaller than type 2 fibers (6.2 versus 9.3 µm). Although there was an increase in the number of type 2 fibers in the biopsy shown in Figure 5 (W236R) (66% type 1 fibers compared with 34% type 1 fibers), this did not quite meet the definition of >2:1 ratio required for fiber type predominance (20,21). There was no fiber predominance in the biopsy of the second patient (Y856H).
Figure 5.
Histochemistry of muscle biopsy of the proband with MYBPC1 W236R mutation showing small type I fibers. (A) Hematoxylin and eosin staining demonstrating variability in fiber size as well as the presence of centrally localized basophilia in some larger fibers (arrows). Scale bar equals 18 µm. (B) Higher magnification of hematoxylin and eosin stained image shown in (A). (C) ATPase 9.4 staining highlighting smaller type I fibers (lightly stained). Scale bar equals 19 µm.
The biopsy from the patient with MYBPC1 W236R mutation showed focal regions of basophilic staining centrally on hematoxylin and eosin staining in larger fibers (Fig. 5A). These regions were not appreciated on other histochemical stains such as Gomori trichrome, nicotinamide adenine dinucleotide, ATPase or succinic dehydrogenase. Additionally, they did not stain with an MYBPC1 antibody, dystrophy-related stains (dystrophin, α-dystroglycan, α/β/γ/δ sarcoglycan, α2-laminin, lamp-2, caveolin-3) or with antibodies to other proteins known to aggregate including desmin, VCP, ubiquitin, SMI-31 or TDP-43 (22).
Skeletal muscle MYBPC1 protein expression did not differ between patients with the W236R and Y856H mutations and controls on western blot (data not shown). However, we were unable to distinguish between wild-type and mutant MYBPC1 protein and therefore the relative abundance of each could not be determined. Both muscle biopsies were evaluated with routine electron microscopy and no abnormalities were noted (data not shown).
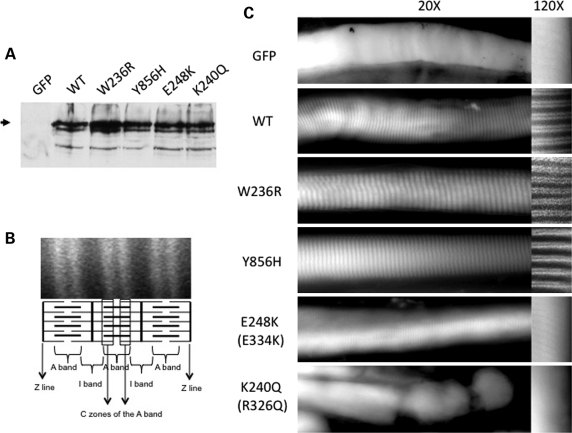
As MYBPC1 protein expression levels appeared to be unaffected by DA1 mutations, we elected to evaluate their localization in mature skeletal muscle. To do this, we generated MYBPC1 expression constructs with a C-terminal fused green fluorescent protein (GFP) tag consisting of WT MYBPC1, DA1 mutants W236R and Y856H, and the homologous mutations to the human MYBPC3 R326Q and E334K that are associated with hypertrophic cardiomyopathy (K240Q and E248K in MYBPC1, respectively). When transiently transfected into cultured cells, all constructs expressed a similar size MYBPC1 fusion protein migrating at 160 kDa that was immunoreactive with both GFP and MYBPC1 antibodies (Fig. 6A and data not shown). To determine the localization of the mutant proteins in vivo, each expression construct as well as a control unfused GFP control vector was electroporated into mouse epitrochlearis muscle. Following 5 days of expression, WT MYBPC1-GFP localized to the sarcomeric C-zone of the A band, resulting in characteristic transverse stripes occurring as a doublet pattern when single muscle fibers were evaluated for GFP fluorescence (Fig. 6B and C) as expected for MYBPC1 expression (23). This pattern was similar to that seen with MYBPC1 W236R and Y856H mutants, suggesting that DA1 mutations did not alter their localization. In contrast, when the MYBPC1 K240Q and E248K missense mutants corresponding to MYBPC3 R326Q and E334K and mutations were expressed there was reduced or absent localization to the sarcomere. This diffuse staining pattern was similar to that seen following electroporation of an unfused GFP control vector.
Figure 6.
MYBPC1 W236R and Y856H mutant proteins localize to the sarcomeric C-zone similar to wild-type protein. (A) Immunoblot of transiently transfected U20S cells expressing WT, DA1 mutants W236R or Y856H or the homologous MYBCP3 mutations E248K or K240Q using a GFP antibody. Note that all proteins express an intact full-length protein. The location of full-length MYBPC1 protein is shown with an arrowhead. (B) Fluorescent image of mouse epitrochlearis muscle fibers after electroporation with GFP WT MYBPC1-GFP showing localization of the protein to the C zones of the A band. Schematic illustration shows three sarcomeres with thin (actin) and thick (myosin) filaments oriented horizontally and Z lines oriented vertically. (C) Fluorescent images of mouse epitrochlearis muscle fibers after electroporation with GFP-tagged constructs. The unfused GFP construct, containing only green fluorescent protein, shows diffuse cytoplasmic staining of single skeletal muscle fibers. High-powered images (at right) show the characteristic doublet pattern of MYBPC1 expression in the C-zone of the sarcomere with the expression of wild-type (WT) MYBPC1, W236R and Y856H mutant GFP-tagged proteins. However, expression of two MYBPC1–GFP constructs (K240Q and E248K) containing the corresponding MYBPC3 mutations (R326Q and E334K) identified in hypertrophic cardiomyopathy patients show poor or absent sarcomeric localization and instead show diffuse cytoplasmic staining.
DISCUSSION
In this report, we have identified the first human mutations in the slow skeletal myosin binding protein C1 (MYBPC1) gene, thus providing additional evidence that distal arthrogryposis is a disease of the skeletal muscle sarcomere. Although MYBPC1 mutations have not previously been reported, mutations in the cardiac myosin binding protein C3 (MYBPC3) gene represent one of the most common causes of familial hypertrophic cardiomyopathy (17,18). The three known myosin binding protein C genes (MYBPC1, MYBPC2 and MYBPC3) are each encoded on a different chromosome and show differential expression in striated muscle. Although the function of myosin binding protein C (MyBP-C) has not been fully elucidated, it appears to stabilize sarcomere structure through its interactions with myosin and titin and may modulate muscle contraction by phosphorylation-dependent interactions with myosin-S2 (reviewed in 24). Now that MYBPC1 mutations have been identified in DA1, MyBP-C joins a group of sarcomeric proteins, including myosin heavy chain, troponin T, troponin I, tropomyosin and actin, which can cause either cardiac or skeletal muscle phenotypes, depending on whether the mutation occurs in the gene encoding either the cardiac or skeletal muscle isoform (25,26).
Like the DA1 families described here with MYBPC1 mutations, MYBPC3 mutations often result in a less severe phenotype with decreased penetrance and a more benign course compared with other gene mutations (27). Intrafamilial phenotypic variability, particularly in the penetrance of hand contractures, was present in both families with MYBPC1 mutations and is considerable in DA1 (4,6,28), suggesting an important role for additional genetic or environmental modifiers. Given the increasingly large number of genes responsible for distal arthrogryposis, it is likely that additive effects of multiple gene mutations or common variants contributes to this variability, as has been shown in hypertrophic cardiomyopathy where as many as 5% of affected individuals harbor multiple gene mutations (18,29).
Although multiple hypotheses have been proposed regarding the pathogenesis of distal arthrogryposis, a leading hypothesis is that the contractures occur in response to in utero lack of joint movement. The congenital nature of the DA1 contractures resulting from MYBPC1 mutations may be explained by the early embryonic expression of this gene. Although not restricted to embryonic development, MYBPC1 is the only MYBPC gene expressed in skeletal muscle prior to birth at which time the fast skeletal MYBPC2 gene begins to be expressed (30,31). A similar pattern is seen with the slow troponin I gene being expressed prior to the subsequent switch to fast troponin I gene in fast-twitch skeletal muscle (32). Therefore, mutations affecting the only MYBPC gene expressed during skeletal muscle development, namely MYBPC1, are likely to negatively impact early muscle development and fetal movement resulting in congenital contracture. The prior identification of embryonic or fetally expressed gene mutations in congenital contracture syndromes provides significant support for this theory. Specific examples include the embryonic myosin heavy chain (MYH3) causing distal arthrogryposis type 2 (7), perinatal myosin heavy chain (MYH8) causing trismus pseudocamptodactyly syndrome (11) and acetylcholine receptor embryonal subunits (CHRNG) causing multiple pterygium syndrome (33).
On the basis of the prior identification of mutations in genes encoding fast-twitch contractile proteins in distal arthrogryposis syndromes, it was previously proposed that DA syndromes result from abnormalities of fast-twitch myofibers (9). However, the gene responsible for DA1 in these two families described here is the slow-twitch skeletal muscle MYBPC1. In mouse, the slow skeletal MYBPC1 mRNA is the sole MYBPC gene expressed in the soleus (a muscle with a high proportion of slow-twitch fibers), whereas it is coexpressed with the fast skeletal MYBPC2 in mouse extensor digitorum longus (31). A similar pattern is revealed in adult human muscle where the slow skeletal MYBPC1 is expressed solely in type 1 fibers (slow-twitch), but fast skeletal MYBPC2 and slow skeletal MYBPC1 are coexpressed in type 2 fibers (30 and unpublished data). Although these expression patterns may therefore explain the type 1 fiber atrophy present histologically in muscle biopsies of two individuals with the MYBPC1 Y856H and W236R mutations, they do not explain the enigmatic restriction of the contractures to the distal musculature.
Most sarcomeric gene mutations causing distal arthrogryposis and hypertrophic cardiomyopathy are missense mutations causing gain of function or dominant-negative effects on function, although haploinsufficiency has been proposed as a mechanism of disease resulting from MYBPC3 mutations. Evidence to support this comes from the fact that nearly two-thirds of the cardiac MYBPC3 mutations identified in individuals with hypertrophic cardiomyopathy are truncation mutations (17,18,34) as well as the lack of evidence for truncated protein expression in patient myocardial biopsies (35,36). Nonsense mutations in MYBPC1 were not identified in this study and MYBPC1 knockout mice have not been reported, thus it is not known if MYBPC1 haploinsufficiency results in disease. Few studies have examined the functional effects of MYBPC3 missense mutations that are most relevant to the MYBPC1 missense mutations described here. One study reported a slight reduction in cardiac MYBPC3 protein expression in myocardial biopsies from two patients with MYBPC3 missense mutations (35). Another study reported reduced expression and more rapid degradation of the MYBPC3 E334K protein expressed in COS-7 cells (37). In the mutants studied here, abnormalities in the sarcomeric localization of the MYBPC3 missense mutants R326Q and E334K were noted in comparison with the MYBPC1 mutants that appeared to localize correctly. This suggests that MYBPC1 and MYBPC3 missense mutations may result in disease through different mechanisms, including failure to localize to the sarcomere.
Assessing the additional functional effects of MYBPC1 missense mutations is complicated by the large number of proteins with which MYBPC1 interacts, including titin (38), myosin [both at S2 and at light meromyosin (LMM) protein] (39), actin (40) and four and a half LIM domain 1 (FHL1) (41). The DA1 mutant W236R occurs within the MyBP-C unique motif located between the C1 and C2 immunoglobulin domains. One function of this MyBP-C unique motif is to regulate muscle contraction through a phosphorylation-dependent interaction with the S2 region of myosin (42). Although these phosphorylation-regulated interactions have been well studied in heart (43,44), their significance in skeletal muscle is less clear since these consensus phosphorylation sites are poorly conserved in MYBPC1 or MYBPC2. The highest affinity interaction of MyBP-C with myosin takes place between the LMM portion of myosin and the MyBP-C C-terminal C7–C10 domains (45,46) near the location of the DA1 Y856H mutation. These same MyBP-C domains participate in the interactions with titin (47). Because the MYBPC1 W236R and Y856H mutations identified in DA1 patients appear to localize to the sarcomere correctly in an in vivo expression system, we predict that these mutations may have more subtle effects on the regulatory aspects of this protein and could reduce the ability of muscle fibers to contract properly. However, we also cannot rule out the possibility that some of the basophilic stained material in the patient muscle biopsy (W236R) may represent an aggregation of misfolded protein, though it was not recognized by antibodies to MYBPC1 or other antigens typically present in protein aggregates. Further studies will be needed to better understand the mechanism by which these mutations cause human disease.
Given that the majority of individuals with DA1 do not have mutations in slow skeletal MYBPC1, DA1 is likely more genetically heterogeneous than some types of distal arthrogryposis such as Freeman–Sheldon syndrome (DA2B) where nearly all patients have MYH3 mutations (7). Recently, the feasibility of the whole exome sequencing approach for gene discovery was demonstrated using a small number of unrelated distal arthrogryposis patient DNA samples (48). Whole genome sequencing projects are likely to play an important role in identifying additional genes responsible for distal arthrogryposis and the genetic modifiers that apply to individual patients.
MATERIALS AND METHODS
The study protocol was approved by the Institutional Review Board, and all subjects and/or their parents gave informed consent. Individuals were considered affected, if one or more major clinical manifestations (2) were present since the family history was positive. Major diagnostic criteria of the upper limbs include ulnar deviation of the fingers, camptodactyly, hypoplastic and/or absent flexion creases, or overriding fingers at birth. Major diagnostic criteria for the lower limbs include talipes equinovarus (clubfoot), calcaneovalgus deformity, vertical talus and metatarsus varus. Control DNA consisted of 400 Caucasian individuals negative for congenital limb anomalies. DNA was isolated from peripheral blood or spit samples (Oragene, DNA Genotek, Inc., Ottawa, ON Canada). Muscle biopsies were processed in the Washington University Neuromuscular Laboratory. Cryostat sections of rapidly frozen muscle were processed for muscle histochemistry in standard fashion (49). Abnormalities in fiber sizes and fiber type proportions were based on measurements and counts of 100 representative fibers from each biopsy.
Linkage analysis
Genome-wide linkage analysis was performed with Affymetrix Mapping 10K XbaI array data from 12 affected and five unaffected individuals from family 5432. The Affymetrix 10K SNP mapping array genotyping was performed by the NIH Neuroscience Microarray Consortium. All linkage programs were accessed through the EasyLinkage package (50,51). PedCheck was used for the detection of Mendelian errors (52) and non-Mendelian errors were identified with Merlin (53). Multipoint linkage analysis was performed with GENEHUNTER (54) using sets of 100 markers and repeated with sets of 60 markers for verification. Linkage was performed assuming autosomal dominant inheritance with 95% penetrance, a disease allele frequency of 0.01%, and no phenocopies. Caucasian allele frequencies and marker locations were obtained from Affymetrix. Haplotypes were created with GENEHUNTER and viewed on Haplopainter (55). LOD score graphs were displayed using EasyLinkage software (50,51).
Sequencing of candidate genes
Primers were designed to amplify all coding exons of MYBPC1 using Primer3 and amplicons were sequenced with an ABI 3730 Sequencer using the Big Dye Terminator Cycle Sequencing Ready Reaction sequencing kit (PE Biosystems). Sequence was analyzed with Sequencher v4.2 software (Gene Codes Corporation, Ann Arbor, MI, USA). MYBPC1 sequence annotation corresponds to NM_002465 coding sequence. The frequency of the MYBPC1 mutations was determined in control patient samples at the Washington University Division of Human Genetics Sequenom Genotyping Core. The CLUSTALW program was used for multiple sequence alignment (http://align.genome.jp). Prediction of functional effects of polymorphisms/variants was performed using PolyPhen (http://genetics.bwh.harvard.edu/pph/index.html) (56).
Plasmid construction
The DNA construct encoding MYBPC1 isoform 4 (AM392734) was obtained from the ORFeome Collaboration (www.orfeomecollaboration.org) as a Gateway (Invitrogen) entry vector (pENTR201) and transferred to a C-terminus GFP destination vector (pcDNA6.2/GFP-DEST). Both MYBPC1 missense mutations were created by engineering single nucleotide substitutions into the MYBPC1-GFP plasmid with the Quick Change Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). The MYBPC3 R326Q and E334K mutations (18,37) were made at the corresponding conserved residues K240Q and E248K of MYBPC1-GFP plasmid sequence. All mutations were verified by sequencing. Plasmid DNA was prepared for each of the five constructs using the Qiagen MaxiPrep kit.
Western blot
Skeletal muscle lysates were obtained from biopsy tissue that was flash frozen in liquid nitrogen within 30 min. Briefly, tissue was homogenized in lysis buffer [20 mm Tris pH 8.0, 100 mm NaCl, 1% Triton X-100, 1 mm PMSF, 10 mm benzamidine and 1 mm protease inhibitor cocktail (Sigma)], and centrifuged at 12 000g for 10 min. In the case of tissue culture cells, a 60 mm dish of human osteosarcoma cells (U20S) were transfected with 4 μg plasmid DNA using lipofectamine 2000 per company recommendations (Invitrogen). At 48 h after transfection, cells were rinsed in 1× PBS, pelleted and lysed in RIPA buffer (25 mm Tris•HCl pH 7.6, 150 mm NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS) and briefly sonicated. Supernatant protein samples (25 μg) were electrophoresed on SDS–PAGE 9% gel and transferred to PVDF membrane (Millipore, Bedford, MA, USA). Immunoblotting was performed with an anti-MYBPC1 antibody (Abcam, UK) or with GFP antibody (Sigma) both at 1:1000 dilution. Immune complexes were visualized with the ECL Plus kit (Amersham Biosciences, Piscataway, NJ, USA).
In vivo electroporation and microscopic evaluation
Mice were anesthetized using pentobarbital (50 mg/kg body weight). The skin of the upper arms was shaved, and 50 µg in 50 µl of the expression construct was injected into the region of the epitrochlearis muscle by using a 0.5 ml syringe fitted with a 29-gauge needle. Two-needle array electrodes (BTX, San Diego, CA, USA) were inserted into the muscle immediately after DNA delivery for electroporation. The distance between the electrodes was 5 mm, and the array was inserted longitudinally relative to the muscle fibers. In vivo electroporation parameters were the following: voltage, 100 V; pulse length, 50 ms; no. of pulse, six pulses; pulse interval, 200 ms; desired field strength, 200 V/cm, given by a BTX ECM830 Electro Square Porator. After 5 days of recovery, epitrochlearis muscles were removed from anesthetized mice. The epitrochlearis is a small and flat muscle that lies along the medial surface of the upper forelimb and is sufficiently thin to allow microscopic evaluation of intact fibers. The muscles were then fixed in 3% paraformaldehyde on glass cover. After fixation, the tissues were rinsed with PBS and distilled water and examined for GFP using a Nikon Eclipse 80i microscope with an apochromat 40× objective. Image processing was performed using 2D deconvolution and NIS elements software. Images are representative of at least three independent injections.
FUNDING
This work was supported by grants from the National Institutes of Health [K12 HD001459]; the Children's Discovery Institute; March of Dimes Basil O'Connor Starter Scholar Research Award; St Louis Children's Hospital Foundation; Pediatric Orthopaedic Society of North America; Shriners Hospital and the Muscular Dystrophy Association.
Supplementary Material
ACKNOWLEDGEMENTS
We kindly thank the National Institutes of Health Neuroscience Microarray Consortium for performing the Affymetrix microarray analyses.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Bamshad M., Van Heest A.E., Pleasure D. Arthrogryposis: a review and update. J. Bone. Joint. Surg. Am. 2009;91:40–46. doi: 10.2106/JBJS.I.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bamshad M., Jorde L.B., Carey J.C. A revised and extended classification of the distal arthrogryposes. Am. J. Med. Genet. 1996;65:277–281. doi: 10.1002/(SICI)1096-8628(19961111)65:4<277::AID-AJMG6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 3.Hall J.G. Genetic aspects of arthrogryposis. Clin. Orthop. Relat. Res. 1985;194:44–53. [PubMed] [Google Scholar]
- 4.Klemp P., Hall J.G. Dominant distal arthrogryposis in a Maori family with marked variability of expression. Am. J. Med. Genet. 1995;55:414–419. doi: 10.1002/ajmg.1320550406. [DOI] [PubMed] [Google Scholar]
- 5.Bamshad M., Bohnsack J.F., Jorde L.B., Carey J.C. Distal arthrogryposis type 1: clinical analysis of a large kindred. Am. J. Med. Genet. 1996;65:282–285. doi: 10.1002/(SICI)1096-8628(19961111)65:4<282::AID-AJMG7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 6.Jiang M., Zhao X., Han W., Bian C., Li X., Wang G., Ao Y., Li Y., Yi D., Zhe Y., et al. A novel deletion in TNNI2 causes distal arthrogryposis in a large Chinese family with marked variability of expression. Hum. Genet. 2006;120:238–242. doi: 10.1007/s00439-006-0183-4. [DOI] [PubMed] [Google Scholar]
- 7.Toydemir R.M., Rutherford A., Whitby F.G., Jorde L.B., Carey J.C., Bamshad M.J. Mutations in embryonic myosin heavy chain (MYH3) cause Freeman–Sheldon syndrome and Sheldon–Hall syndrome. Nat. Genet. 2006;38:561–565. doi: 10.1038/ng1775. [DOI] [PubMed] [Google Scholar]
- 8.Sung S.S., Brassington A.M., Krakowiak P.A., Carey J.C., Jorde L.B., Bamshad M. Mutations in TNNT3 cause multiple congenital contractures: a second locus for distal arthrogryposis type 2B. Am. J. Hum. Genet. 2003;73:212–214. doi: 10.1086/376418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sung S.S., Brassington A.M., Grannatt K., Rutherford A., Whitby F.G., Krakowiak P.A., Jorde L.B., Carey J.C., Bamshad M. Mutations in genes encoding fast-twitch contractile proteins cause distal arthrogryposis syndromes. Am. J. Hum. Genet. 2003;72:681–690. doi: 10.1086/368294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tajsharghi H., Kimber E., Holmgren D., Tulinius M., Oldfors A. Distal arthrogryposis and muscle weakness associated with a beta-tropomyosin mutation. Neurology. 2007;68:772–775. doi: 10.1212/01.wnl.0000256339.40667.fb. [DOI] [PubMed] [Google Scholar]
- 11.Veugelers M., Bressan M., McDermott D.A., Weremowicz S., Morton C.C., Mabry C.C., Lefaivre J.F., Zunamon A., Destree A., Chaudron J.M., et al. Mutation of perinatal myosin heavy chain associated with a Carney complex variant. N. Engl. J. Med. 2004;351:460–469. doi: 10.1056/NEJMoa040584. [DOI] [PubMed] [Google Scholar]
- 12.Stevenson D.A., Carey J.C., Palumbos J., Rutherford A., Dolcourt J., Bamshad M.J. Clinical characteristics and natural history of Freeman–Sheldon syndrome. Pediatrics. 2006;117:754–762. doi: 10.1542/peds.2005-1219. [DOI] [PubMed] [Google Scholar]
- 13.Gurnett C.A., Alaee F., Desruisseau D., Boehm S., Dobbs M.B. Skeletal muscle contractile gene (TNNT3, MYH3, TPM2) mutations not found in vertical talus or clubfoot. Clin. Orthop. Relat. Res. 2009;467:1195–1200. doi: 10.1007/s11999-008-0694-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehtokari V.L., Ceuterick-de Groote C., de Jonghe P., Marttila M., Laing N.G., Pelin K., Wallgren-Pettersson C. Cap disease caused by heterozygous deletion of the beta-tropomyosin gene TPM2. Neuromuscul. Disord. 2007;17:433–442. doi: 10.1016/j.nmd.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 15.Donner K., Ollikainen M., Ridanpaa M., Christen H.J., Goebel H.H., de Visser M., Pelin K., Wallgren-Pettersson C. Mutations in the beta-tropomyosin (TPM2) gene—a rare cause of nemaline myopathy. Neuromuscul. Disord. 2002;12:151–158. doi: 10.1016/s0960-8966(01)00252-8. [DOI] [PubMed] [Google Scholar]
- 16.Monnier N., Lunardi J., Marty I., Mezin P., Labarre-Vila A., Dieterich K., Jouk P.S. Absence of beta-tropomyosin is a new cause of Escobar syndrome associated with nemaline myopathy. Neuromuscul. Disord. 2009;19:118–123. doi: 10.1016/j.nmd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 17.Watkins H., Conner D., Thierfelder L., Jarcho J.A., MacRae C., McKenna W.J., Maron B.J., Seidman J.G., Seidman C.E. Mutations in the cardiac myosin binding protein-C gene on chromosome 11 cause familial hypertrophic cardiomyopathy. Nat. Genet. 1995;11:434–437. doi: 10.1038/ng1295-434. [DOI] [PubMed] [Google Scholar]
- 18.Richard P., Charron P., Carrier L., Ledeuil C., Cheav T., Pichereau C., Benaiche A., Isnard R., Dubourg O., Burban M., et al. Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- 19.Dubowitz V. Muscle Biopsy. A Practical Approach. 2nd edn. London: Elsevier; 1985. [Google Scholar]
- 20.Brooke M.H., Engel W.K. The histographic analysis of human muscle biopsies with regard to fiber types. 4. Children's biopsies. Neurology. 1969;19:591–605. doi: 10.1212/wnl.19.6.591. [DOI] [PubMed] [Google Scholar]
- 21.Sirca A., Erzen I., Pecak F. Histochemistry of abductor hallucis muscle in children with idiopathic clubfoot and in controls. J. Pediatr. Orthop. 1990;10:477–482. [PubMed] [Google Scholar]
- 22.Temiz P., Weihl C.C., Pestronk A. Inflammatory myopathies with mitochondrial pathology and protein aggregates. J. Neurol. Sci. 2009;278:25–29. doi: 10.1016/j.jns.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 23.Craig R., Offer G. The location of C-protein in rabbit skeletal muscle. Proc. R. Soc. Lond. B. Biol. Sci. 1976;192:451–461. doi: 10.1098/rspb.1976.0023. [DOI] [PubMed] [Google Scholar]
- 24.Oakley C.E., Chamoun J., Brown L.J., Hambly B.D. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int. J. Biochem. Cell Biol. 2007;39:2161–2166. doi: 10.1016/j.biocel.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 25.Laing N.G., Nowak K.J. When contractile proteins go bad: the sarcomere and skeletal muscle disease. Bioessays. 2005;27:809–822. doi: 10.1002/bies.20269. [DOI] [PubMed] [Google Scholar]
- 26.Morimoto S. Sarcomeric proteins and inherited cardiomyopathies. Cardiovasc. Res. 2008;77:659–666. doi: 10.1093/cvr/cvm084. [DOI] [PubMed] [Google Scholar]
- 27.Niimura H., Bachinski L.L., Sangwatanaroj S., Watkins H., Chudley A.E., McKenna W., Kristinsson A., Roberts R., Sole M., Maron B.J., et al. Mutations in the gene for cardiac myosin-binding protein C and late-onset familial hypertrophic cardiomyopathy. N. Engl. J. Med. 1998;338:1248–1257. doi: 10.1056/NEJM199804303381802. [DOI] [PubMed] [Google Scholar]
- 28.Bamshad M., Watkins W.S., Zenger R.K., Bohnsack J.F., Carey J.C., Otterud B., Krakowiak P.A., Robertson M., Jorde L.B. A gene for distal arthrogryposis type I maps to the pericentromeric region of chromosome 9. Am. J. Hum. Genet. 1994;55:1153–1158. [PMC free article] [PubMed] [Google Scholar]
- 29.Ingles J., Doolan A., Chiu C., Seidman J., Seidman C., Semsarian C. Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J. Med. Genet. 2005;42:e59. doi: 10.1136/jmg.2005.033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gautel M., Furst D.O., Cocco A., Schiaffino S. Isoform transitions of the myosin binding protein C family in developing human and mouse muscles: lack of isoform transcomplementation in cardiac muscle. Circ. Res. 1998;82:124–129. doi: 10.1161/01.res.82.1.124. [DOI] [PubMed] [Google Scholar]
- 31.Kurasawa M., Sato N., Matsuda A., Koshida S., Totsuka T., Obinata T. Differential expression of C-protein isoforms in developing and degenerating mouse striated muscles. Muscle Nerve. 1999;22:196–207. doi: 10.1002/(sici)1097-4598(199902)22:2<196::aid-mus7>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 32.Sutherland C.J., Esser K.A., Elsom V.L., Gordon M.L., Hardeman E.C. Identification of a program of contractile protein gene expression initiated upon skeletal muscle differentiation. Dev. Dyn. 1993;196:25–36. doi: 10.1002/aja.1001960104. [DOI] [PubMed] [Google Scholar]
- 33.Morgan N.V., Brueton L.A., Cox P., Greally M.T., Tolmie J., Pasha S., Aligianis I.A., van Bokhoven H., Marton T., Al-Gazali L., et al. Mutations in the embryonal subunit of the acetylcholine receptor (CHRNG) cause lethal and Escobar variants of multiple pterygium syndrome. Am. J. Hum. Genet. 2006;79:390–395. doi: 10.1086/506256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andersen P.S., Havndrup O., Bundgaard H., Larsen L.A., Vuust J., Pedersen A.K., Kjeldsen K., Christiansen M. Genetic and phenotypic characterization of mutations in myosin-binding protein C (MYBPC3) in 81 families with familial hypertrophic cardiomyopathy: total or partial haploinsufficiency. Eur. J. Hum. Genet. 2004;12:673–677. doi: 10.1038/sj.ejhg.5201190. [DOI] [PubMed] [Google Scholar]
- 35.Marston S., Copeland O., Jacques A., Livesey K., Tsang V., McKenna W.J., Jalilzadeh S., Carballo S., Redwood C., Watkins H. Evidence from human myectomy samples that MYBPC3 mutations cause hypertrophic cardiomyopathy through haploinsufficiency. Circ. Res. 2009;105:219–222. doi: 10.1161/CIRCRESAHA.109.202440. [DOI] [PubMed] [Google Scholar]
- 36.Moolman J.A., Reith S., Uhl K., Bailey S., Gautel M., Jeschke B., Fischer C., Ochs J., McKenna W.J., Klues H., et al. A newly created splice donor site in exon 25 of the MyBP-C gene is responsible for inherited hypertrophic cardiomyopathy with incomplete disease penetrance. Circulation. 2000;101:1396–1402. doi: 10.1161/01.cir.101.12.1396. [DOI] [PubMed] [Google Scholar]
- 37.Bahrudin U., Morisaki H., Morisaki T., Ninomiya H., Higaki K., Nanba E., Igawa O., Takashima S., Mizuta E., Miake J., et al. Ubiquitin–proteasome system impairment caused by a missense cardiac myosin-binding protein C mutation and associated with cardiac dysfunction in hypertrophic cardiomyopathy. J. Mol. Biol. 2008;384:896–907. doi: 10.1016/j.jmb.2008.09.070. [DOI] [PubMed] [Google Scholar]
- 38.Furst D.O., Vinkemeier U., Weber K. Mammalian skeletal muscle C-protein: purification from bovine muscle, binding to titin and the characterization of a full-length human cDNA. J. Cell Sci. 1992;102:769–778. doi: 10.1242/jcs.102.4.769. [DOI] [PubMed] [Google Scholar]
- 39.Moos C., Offer G., Starr R., Bennett P. Interaction of C-protein with myosin, myosin rod and light meromyosin. J. Mol. Biol. 1975;97:1–9. doi: 10.1016/s0022-2836(75)80017-9. [DOI] [PubMed] [Google Scholar]
- 40.Moos C., Mason C.M., Besterman J.M., Feng I.N., Dubin J.H. The binding of skeletal muscle C-protein to F-actin, and its relation to the interaction of actin with myosin subfragment-1. J. Mol. Biol. 1978;124:571–586. doi: 10.1016/0022-2836(78)90172-9. [DOI] [PubMed] [Google Scholar]
- 41.McGrath M.J., Cottle D.L., Nguyen M.A., Dyson J.M., Coghill I.D., Robinson P.A., Holdsworth M., Cowling B.S., Hardeman E.C., Mitchell C.A., et al. Four and a half LIM protein 1 binds myosin-binding protein C and regulates myosin filament formation and sarcomere assembly. J. Biol. Chem. 2006;281:7666–7683. doi: 10.1074/jbc.M512552200. [DOI] [PubMed] [Google Scholar]
- 42.Gruen M., Gautel M. Mutations in beta-myosin S2 that cause familial hypertrophic cardiomyopathy (FHC) abolish the interaction with the regulatory domain of myosin-binding protein-C. J. Mol. Biol. 1999;286:933–949. doi: 10.1006/jmbi.1998.2522. [DOI] [PubMed] [Google Scholar]
- 43.Sadayappan S., Gulick J., Osinska H., Martin L.A., Hahn H.S., Dorn G.W., II, Klevitsky R., Seidman C.E., Seidman J.G., Robbins J. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ. Res. 2005;97:1156–1163. doi: 10.1161/01.RES.0000190605.79013.4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadayappan S., Gulick J., Klevitsky R., Lorenz J.N., Sargent M., Molkentin J.D., Robbins J. Cardiac myosin binding protein-C phosphorylation in a {beta}-myosin heavy chain background. Circulation. 2009;119:1253–1262. doi: 10.1161/CIRCULATIONAHA.108.798983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gilbert R., Kelly M.G., Mikawa T., Fischman D.A. The carboxyl terminus of myosin binding protein C (MyBP-C, C-protein) specifies incorporation into the A-band of striated muscle. J. Cell Sci. 1996;109:101–111. doi: 10.1242/jcs.109.1.101. [DOI] [PubMed] [Google Scholar]
- 46.Okagaki T., Weber F.E., Fischman D.A., Vaughan K.T., Mikawa T., Reinach F.C. The major myosin-binding domain of skeletal muscle MyBP-C (C protein) resides in the COOH-terminal, immunoglobulin C2 motif. J. Cell. Biol. 1993;123:619–626. doi: 10.1083/jcb.123.3.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freiburg A., Gautel M. A molecular map of the interactions between titin and myosin-binding protein C. Implications for sarcomeric assembly in familial hypertrophic cardiomyopathy. Eur. J. Biochem. 1996;235:317–323. doi: 10.1111/j.1432-1033.1996.00317.x. [DOI] [PubMed] [Google Scholar]
- 48.Ng S.B., Turner E.H., Robertson P.D., Flygare S.D., Bigham A.W., Lee C., Shaffer T., Wong M., Bhattacharjee A., Eichler E.E., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mozaffar T., Pestronk A. Myopathy with anti-Jo-1 antibodies: pathology in perimysium and neighbouring muscle fibres. J. Neurol. Neurosurg. Psychiatry. 2000;68:472–478. doi: 10.1136/jnnp.68.4.472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lindner T.H., Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–407. doi: 10.1093/bioinformatics/bti009. [DOI] [PubMed] [Google Scholar]
- 51.Hoffmann K., Lindner T.H. easyLINKAGE-Plus—automated linkage analyses using large-scale SNP data. Bioinformatics. 2005;21:3565–3567. doi: 10.1093/bioinformatics/bti571. [DOI] [PubMed] [Google Scholar]
- 52.O'Connell J.R., Weeks D.E. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 54.Kruglyak L., Daly M.J., Reeve-Daly M.P., Lander E.S. Parametric and nonparametric linkage analysis: a unified multipoint approach. Am. J. Hum. Genet. 1996;58:1347–1363. [PMC free article] [PubMed] [Google Scholar]
- 55.Thiele H., Nurnberg P. HaploPainter: a tool for drawing pedigrees with complex haplotypes. Bioinformatics. 2005;21:1730–1732. doi: 10.1093/bioinformatics/bth488. [DOI] [PubMed] [Google Scholar]
- 56.Sunyaev S., Ramensky V., Koch I., Lathe W., III, Kondrashov A.S., Bork P. Prediction of deleterious human alleles. Hum. Mol. Genet. 2001;10:591–597. doi: 10.1093/hmg/10.6.591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.