Abstract
Amelogenesis imperfecta (AI) describes a broad group of clinically and genetically heterogeneous inherited defects of dental enamel bio-mineralization. Despite identification of a number of genetic mutations underlying AI, the precise causal mechanisms have yet to be determined. Using a multi-disciplinary approach, we describe here a mis-sense mutation in the mouse Amelx gene resulting in a Y → H substitution in the tri-tyrosyl domain of the enamel extracellular matrix protein amelogenin. The enamel in affected animals phenocopies human X-linked AI where similar mutations have been reported. Animals affected by the mutation have severe defects of enamel bio-mineralization associated with absence of full-length amelogenin protein in the developing enamel matrix, loss of ameloblast phenotype, increased ameloblast apoptosis and formation of multi-cellular masses. We present evidence to demonstrate that affected ameloblasts express but fail to secrete full-length amelogenin leading to engorgement of the endoplasmic reticulum/Golgi apparatus. Immunohistochemical analysis revealed accumulations of both amelogenin and ameloblastin in affected cells. Co-transfection of Ambn and mutant Amelx in a eukaryotic cell line also revealed intracellular abnormalities and increased cytotoxicity compared with cells singly transfected with wild-type Amelx, mutant Amelx or Ambn or co-transfected with both wild-type Amelx and Ambn. We hypothesize that intracellular protein–protein interactions mediated via the amelogenin tri-tyrosyl motif are a key mechanistic factor underpinning the molecular pathogenesis in this example of AI. This study therefore successfully links phenotype with underlying genetic lesion in a relevant murine model for human AI.
INTRODUCTION
Amelogenesis imperfecta (AI) is a common group of inherited defects of dental enamel formation that exhibit marked genetic and clinical heterogeneity with at least 14 different sub-types being recognized on the basis of their clinical appearance and mode of inheritance (1–3). Affected individuals show either hypoplastic (thin but seemingly correctly mineralized) or hypomineralized enamel but an overlap of these characteristics is seen in many cases (2). Autosomal dominant, autosomal recessive and X-linked forms of AI are recognized. To date, mutations in the genes encoding amelogenin (AMELX), enamelin (ENAM), enamelysin (MMP20), kallikrein 4 (KLK4) and the FAM83H gene have been shown to underlie non-syndromic forms of AI (4–6). Despite these advances, the underlying molecular pathogenesis of the various forms of AI remains poorly characterized.
Dental enamel is a highly mineralized tissue with ∼85% of its volume occupied by unusually large hydroxyapatite crystals that are organized into prisms (7). Enamel is unique among the mineralized tissues as it is produced by ectodermally derived ameloblasts which pass through a series of discrete differentiation states that correlate with the various stages of enamel formation (5). During the pre-secretory stage, ameloblasts are separated from the adjacent neural crest cell-derived odontoblasts by a basement membrane which is subsequently removed as the ameloblasts enter their secretory phase. During this latter stage, the ameloblasts secrete an eosinophilic, enamel extracellular matrix of which greater than 90% is composed of amelogenin, the remainder comprising non-amelogenin proteins including ameloblastin, enamelin and tuftelin, together with enzymes, including the proteases enamelysin and kallikrein-4 (7). Bio-mineralization of the enamel matrix begins as it is being secreted and there is no non-mineralized ‘pre-enamel’ equivalent to that seen in other mineralized tissues of the skeleton such as bone or dentine. During the secretory phase, extracellular matrix proteins are enzymically processed such that nascent molecules are converted to smaller fragments that form the greater thickness of the matrix (7). As the ameloblasts enter their maturation phase, the organic matrix is degraded completely to allow secondary growth of the hydroxyapatite crystals which ultimately occlude the spaces previously occupied by the enamel extracellular matrix proteins. Ultimately, the ameloblasts atrophy and the cellular layer is lost entirely from the mature erupted tissue (7).
One of the difficulties in elucidating the underlying molecular mechanisms associated with AI in humans is the near-impossibility of obtaining viable, developing (pre-eruptive) teeth. In contrast, the incisor teeth of adult rodents form and erupt continuously throughout life and offer access to all stages of enamel development in a single tooth. The mouse is therefore an excellent model organism to investigate the fundamental events driving dental development and for studying the molecular pathogenesis of AI. In this context, gene targeting experiments have confirmed a central role for amelogenin, enamelysin and ameloblastin in enamel formation. Although ameloblastin may act as a cell adhesion molecule that is essential for maintaining the differentiation state of the ameloblasts (8), both amelogenin and enamelysin are indispensable for the generation of full-thickness enamel of appropriate crystal structure (9–11). Similarly, analyses of N-ethyl-N-nitrosourea (ENU)-induced dominantly inherited mouse mutations have demonstrated that enamelin is required for the initiation of mineral crystal formation during the early stages of enamel formation and the generation of full-thickness enamel (12,13). These studies also reported animals (mouse mutant M100888) with an X-linked enamel phenotype immediately suggesting mutations in the amelogenin gene (Amelx) as this is the only enamel matrix protein currently identified on the X-chromosome. Previously reported human AMELX mutations include those resulting in a total loss of secreted protein (e.g. nonsense mutation in the signal peptide); mutations causing loss of the amelogenin C-terminal, and mutations affecting the amelogenin N-terminal region that includes a lectin-like, tri-tyrosyl domain reported to bind to N-acetyl-d-glucosamine (14).
The amelogenin tri-tyrosyl domain is defined by the sequence PYPSYGYEPMGGW (15; corresponding to P63 → W75 in murine amelogenin) and is highly conserved across the species. A P → T mutation in the same amelogenin domain (underlined above), results in hypomaturation AI in humans (16). In vitro experiments indicated that this mutation not only inhibited normal amelogenin proteolysis (17) but also abrogated lectin-like binding by the tri-tyroysl domain (15). The three tyrosine residues are essential for the lectin-like properties of the amelogenin tri-tyrosyl domain (15). The tri-tyrosyl domain also binds the N-acetyl-d-glucosamine mimicking domain in the N-terminal region of cytokeratin 14 (18) and N-acetyl-d-glucosamine residues on cytokeratin 5 (19). It has therefore been proposed that these cytokeratins may play a role in amelogenin cell trafficking and secretion.
In the current study, we have identified a novel ENU-induced Amelx mutation in M100888 mice which results in a marked dental phenotype and corresponds to similar reported mutations in human AI. Furthermore, we have utilized the continuously erupting incisor teeth of affected animals to identify an apparent impairment of the normal extracellular matrix secretory pathway and so present evidence of a possible underlying mechanism resulting in AI.
RESULTS
Gross morphological analysis of M100888 mutant mice
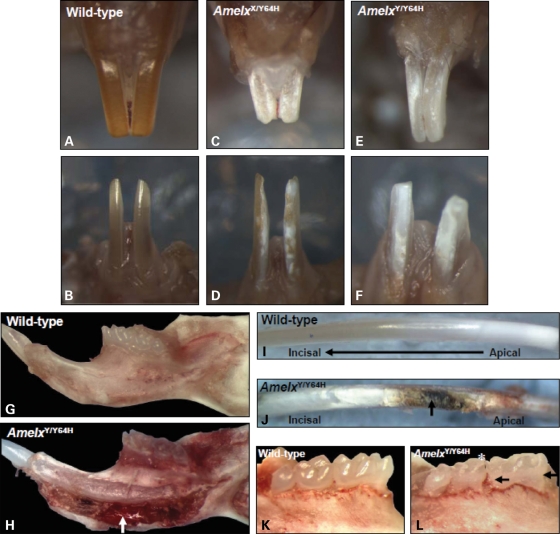
The mouse mutant M100888 was identified during a large-scale screen for dominant abnormal phenotypes of the incisor surface among animals generated from inter-crossing male mice treated with ENU and wild-type (WT) female mice (12). Gross morphological analysis of the dentition of affected mice revealed that the upper and lower incisors were shortened and uneven and presented a chalky white, opaque appearance with roughened/pitted surfaces. In contrast, WT incisors were smooth and opalescent (Fig. 1A–F). Examination of the lower jaw after soft tissue removal indicated that the inferior border of the mandible of affected male mice was enlarged, eroded and discoloured compared with their unaffected counterparts. Subsequent removal of the cortical plate further indicated that the enlarged region of the mandible contained a mass of predominantly soft tissue (Fig. 1G and H). Dissection of the lower incisor teeth indicated that the mineralizing enamel distal to the white opaque zone (where enamel extracellular matrix removal would normally have occurred) was irregular and discoloured with ridges perpendicular to the long axis of the incisor (Fig. 1I and J). Similarly, although the surfaces of the molar teeth of WT mice were smooth and opalescent, those of the affected animals were opaque and roughened with abrasion of the cusps (Fig. 1K and L).
Figure 1.
Morphology of the dentition of Amelx mutant mice. In wild-type (WT) maxillary (A) and mandibular (B) incisor teeth, the enamel appeared smooth, opalescent and orange/brown in colour. (C and D) In heterozygous female (AmelxX/Y64H) mutant mice, the incisor teeth showed patchy regions of roughened, chalky white enamel. (E and F) In hemizygous male (AmelxY/Y64H) mutant mice, the entire enamel surface of the incisor teeth was roughened, opaque and chalky white. Frequently, the incisor teeth of affected mice were shortened with irregular incisal edges (C–F). (G) The inferior border of the WT mandible appeared smooth and unremarkable; (H) in contrast, that of AmelxY/Y64H mutant mice was enlarged, eroded and discoloured containing a large mass of soft tissue (arrow). (I) The surface of dissected WT mandibular incisors was smooth with progressive maturation from white, incompletely mineralized enamel at the apex of the tooth to increasingly pigmented, fully mineralized enamel towards the incisal tip. (J) The surface of mandibular incisors dissected from AmelxY/Y64H mice, at the point where enamel maturation would normally occur, was irregular and discoloured (arrow) with ridges running along the long axis of the tooth. (K) The molar teeth of WT mice were smooth and opalescent whereas (L) those of AmelxY/Y64H mice were opaque and roughened (arrows) with abrasion of the cusps (asterisk).
Mutation analysis in M100888 mutant mice
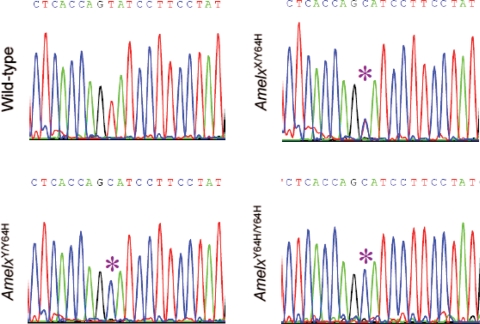
As the mutation underlying the phenotype observed in M100888 mutant mice had been mapped to the X chromosome (http://www.gsc.riken.jp/Mouse/), direct sequencing of all the coding exons and the associated splice junctions of the Amelx gene was performed. The only difference found between the WT and affected mice was a T to C transition at nucleotide 249 of the Amelx coding sequence which resulted in the mis-sense mutation Y64H based on the ENSEMBL sequence ENSMUST00000112119/P63277-4 (Fig. 2). This mutation affects the first tyrosine residue (Y64) within the conserved tri-tyrosyl motif (PYPSYGYEPMGGW) of amelogenin which is positioned towards the C-terminus of the tyrosine-rich amelogenin peptide (TRAP) domain. Although this mutation was present in all affected male (n = 72) and female (n = 54) mice analysed, it was not detected in their WT littermates (n = 160) nor in a panel of eight inbred mouse strains. Affected males and affected female mice that were homozygous for the mutation were more severely affected than heterozygous females.
Figure 2.
Identification of the Amelx Y64H mutation. Partial sequence chromatograms of Amelx depicting the WT sequence, and the T → C transition (asterisks) that leads to the Y64H mutation in amelogenin in heterozygous female (AmelxX/Y64H), hemizygous male (AmelxY/Y64H) and homozygous female (AmelxY64C/Y64H) mutant mice.
Enamel thickness, mineral density and ultrastructural appearance in M100888 mutant mice
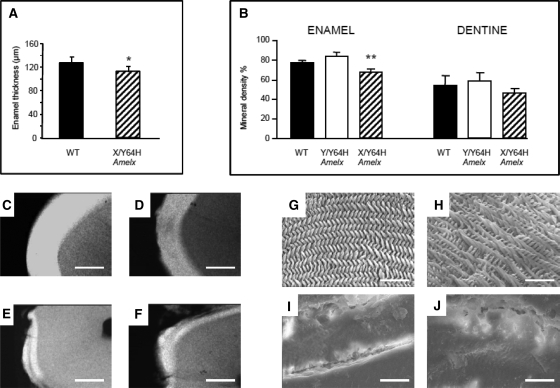
Enamel thickness was significantly reduced in AmelxX/Y64H heterozygous females compared with their WT littermates (Fig. 3A). Measurement of enamel thickness for affected males was impossible to carry out in a meaningful way as large areas of enamel were missing entirely from the teeth. Quantitative microradiography of transverse sections through WT and affected teeth demonstrated a significantly reduced enamel mineral content in AmelxX/Y64H females (Fig. 3–F; P < 0.001). Enamel mineral density in affected males was not significantly different to that of WT animals but this finding only related to the small amount of enamel that was available for measurement. There were no significant differences in mineral levels in the dentine of the teeth of affected animals compared with WT tissue (Fig. 3B).
Figure 3.
Enamel phenotype of Amelx mutant mice. (A) Enamel thickness in heterozygous female (AmelxX/Y64H) mutant mice was significantly reduced compared with WT (P < 0.05). Enamel thickness in hemizygous males could not be reliably measured due to large areas of enamel being missing from the teeth and thus no data are presented. (B) Enamel mineral density in AmelxX/Y64H females was also significantly reduced compared with WT littermates (P < 0.001). Mineral density in hemizygous males was, however, not significantly different from WT. There were no significant differences in mineral levels in the dentine of the teeth of affected animals compared with WT tissue. (C–F) Representative transverse sections of mature enamel prepared for quantitative microradiography clearly show qualitative differences in enamel from WT (C), heterozygous females (AmelxX/Y64H) (D) and hemizygous males (AmelxY/Y64H) (E, F). Scale bars = 100 µm. (G–J) Scanning electron micrographs of WT enamel (G) exhibited decussating prism architecture characteristic of rodent enamel. Enamel in AmelxX/Y64H females also showed decussating prisms (H) but they were less highly ordered and possessed a more open structure compared with WT. Enamel from AmelxY/Y64H affected males could only be imaged where remnants of the tissue were present and revealed severely dysplastic non-prismatic ‘enamel’ with a smooth glass-like appearance (I, J). Scale bars = 20 µm.
The tissue architecture and ultrastructural appearance of mature incisor enamel in mice carrying the Y64H mutation were compared with those of WT animals using scanning electron microscopy (Fig. 3G–J). WT enamel exhibited the herring bone pattern of decussating prisms characteristic of rodent enamel (Fig. 3G). Although the enamel in AmelxX/Y64H mice also showed prismatic structure, this was less highly ordered than that of WT tissue, with greater inter-prismatic spacing (Fig. 3H). Enamel from AmelxY/Y64H mice could only be imaged where remnants of the tissue remained. Scanning electron microscopy revealed severely dysplastic non-prismatic ‘enamel’ with a smooth glass-like appearance and no evidence of clear prismatic structure (Fig. 3I and J).
Histological appearance of the enamel organ and developing enamel in M100888 mutant mice
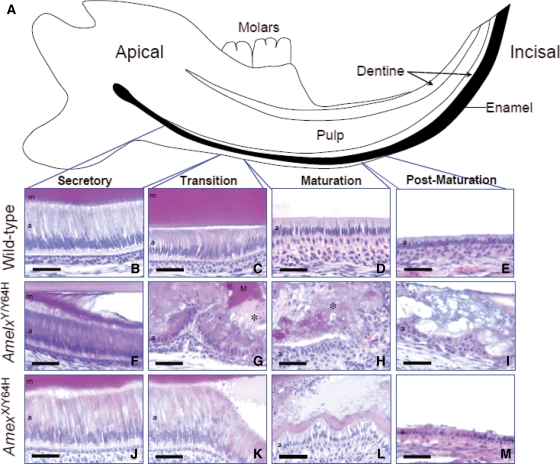
To characterize further the dental phenotype of mice affected by the Y64H mutation, we performed histological analysis of mandibular incisors and enamel organs from 2- to 3-month old WT, affected male (AmelY/Y64H), heterozygous female (AmelxX/Y64H) and homozygous female (AmelxY64H/Y64H) mice (Fig. 4). In WT lower incisors, the normal cellular components of the enamel organ were clearly evident. Towards the apex of the developing incisor, an epithelial sheet was observed which differentiated longitudinally towards the incisal tip to form the tall ameloblasts with distinctive Tomes’ processes that characterize the secretory stage of amelogenesis (20,21). The eosinophilic, enamel extracellular matrix was also clearly visible (Fig. 4B). Following the secretory stage, the ameloblasts decreased in height and increased in width as they progressed through the transitional/early maturation stage of amelogenesis during which maximum enzyme-mediated matrix degradation occurs (Fig. 4C). Further towards the incisal tip, this specialized epithelium entered the true maturation phase where the ameloblasts shortened further and lost their Tomes’ process and secretory characteristics (Fig. 4D). Following maturation, the ameloblasts became cuboidal as they entered the post-maturation phase (Fig. 4E). These cells progressed forwards until they reached the gingival margin where the tooth erupts from the mandible.
Figure 4.
Histological analysis of the lower incisor teeth. (A) Diagrammatic representation of the mouse lower incisor tooth within the mandible. As indicated in black, enamel production commences at the apical region of the tooth and proceeds towards the incisal edge. (B–E) The secretory, transition, maturation and post-maturation stages of amelogenesis showed characteristically normal morphology in the WT lower incisor tooth. (F) The secretory ameloblasts of hemizygous male (AmelxY/Y64H) mutant mice differentiated normally but soon lost contact with the newly secreted enamel matrix and became abnormally eosinophilic. (G and H) In the transition and maturation zones, the ameloblasts of AmelxY/Y64H mice adopted a highly abnormal morphology and formed irregular, multi-cellular masses. In these regions, large quantities of amorphous material containing cellular debris (asterisks) were apparent. (I) The multi-cellular masses frequently persisted through the post-maturation zone to the gingival margin. (J) Although the secretory ameloblasts of heterozygous female (AmelxX/Y64H) mutant mice appeared largely normal, a proportion exhibited increased eosinophilia. (K and L) In the transition and maturation zones, contact of the ameloblasts with the enamel matrix was occasionally lost leading to the formation of blister-like structures. (M) In contrast, the appearance of the post-maturation ameloblasts resembled that of WT mice. m, enamel matrix; a, ameloblasts. Scale bars, 50 µm.
Ameloblasts of AmelxY/Y64H and AmelxY64H/Y64H mice appeared to differentiate normally, acquiring typical secretory characteristics and initially elaborating an eosinophilic, extracellular matrix; however, soon after commencement of enamel matrix production the ameloblasts appeared to lose contact with their sub-adjacent matrix and their cytoplasm became atypically eosinophilic (Fig. 4F). At this point, a matrix-free space was observed between the newly formed enamel and the ameloblasts. The departure from the phenotypic norm became more pronounced as amelogenesis proceeded. In what would normally be regarded as the transition and maturation zones, the cells became highly disorganized with the ameloblasts losing their characteristic columnar morphology and forming a multi-cellular mass coincident with that observed macroscopically associated with erosion of the inferior border of the mandible (Fig. 4G and H). Surprisingly, in the post-maturation zone, the ameloblasts regained a semblance of their normal cuboidal morphology (Fig. 4I). Heterozygous AmelxX/Y64H mice exhibited a milder phenotype with occasional small ‘blister-like’ structures of variable size apparent from the late secretory stage onwards (Fig. 4J–M). Within these ‘blister-like’ structures, aberrant matrix was present with areas appearing less eosinophilic than the original enamel matrix (Fig. 4J–M). Analysis of molar teeth derived from mutant mice at post-natal day 7 and their WT littermates revealed a similar histological appearance for AmelxY/Y64H molars to that noted for the incisor teeth with eosinophilic ameloblasts that had lost their characteristic morphology and were detached from the enamel matrix forming multi-cellular masses (Supplementary Material, Fig. S1).
Immunofluorescence analysis of the enamel organ and developing enamel in M100888 mutant mice
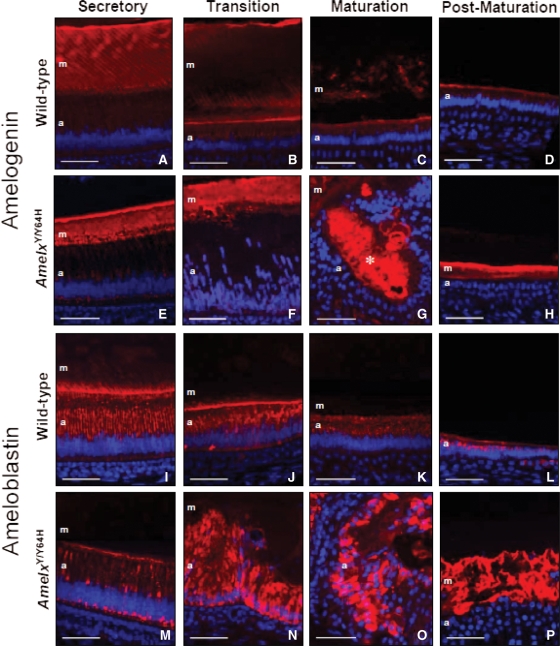
To determine the effects of the Amelx Y64H mutation on the distribution of the major enamel extracellular proteins, we performed immunofluorescence studies using antibodies raised against amelogenin and ameloblastin, which represent >95% of all developing enamel matrix proteins (Fig. 5 and Supplementary Material, Fig. S2).
Figure 5.
Amelogenin and ameloblastin distribution in Amelx mutant mice. (A–H) Amelogenin distribution. (A) Strong amelogenin immunolabelling of the enamel matrix, arranged as closely packed parallel lines, was observed in the secretory zone of WT mice. (B) Enamel matrix degradation commences in the transition zone and this was reflected in the WT enamel matrix where the intensity of immunostaining was markedly reduced. (C) Little amelogenin immunoreactivity was detected in the maturation zone where the process of enamel matrix degradation is completed. (D) No amelogenin immunoreactivity was observed in the post-maturation enamel matrix. (E) In the secretory zone of hemizygous male (AmelxY/Y64H) mutant mice strong amelogenin immunolabelling of the enamel matrix was observed; however, the pattern of parallel lines observed in WT mice was lost. Additionally, the secretory ameloblasts of AmelxY/Y64H mice showed increased immunostaining in the cytoplasm. (F and G) Where the ameloblasts became disorganized in AmelxY/Y64H mice, strong amelogenin immunostaining was detected both in the enamel matrix and within the multi-cellular masses (asterisk). (H) Amelogenin immunoreactivity was often detected in the post-maturation zone where enamel matrix is normally absent. (I–P) Ameloblastin distribution. (I) In the secretory zone of WT mice, strong ameloblastin immunostaining of the enamel matrix was detected in the vicinity of the ameloblast Tomes’ processes. Strong ameloblastin immunoreactivity was also observed in the supranuclear compartment of the secretory ameloblasts. (J–L) Ameloblastin expression in the cytoplasm of WT ameloblasts was detected throughout the subsequent stages of amelogenesis. (M) In hemizygous male (AmelxY/Y64H) mutant mice, the secretory ameloblasts were immunoreactive for ameloblastin; however, the regular pattern of supra-nuclear immunoreactivity observed in WT mice was replaced by patchy aggregates of ameloblastin immunostaining. Immunolabelling of the enamel matrix was very weak in AmelxY/Y64H mice. (N and O) Where the ameloblasts became disorganized in those regions corresponding to the transition and maturation zones of WT mice, intracellular ameloblastin immunostaining became more intense in AmelxY/Y64H mice. (P) Extracellular ameloblastin immunoreactivity was frequently observed in the post-maturation stage of enamel formation in AmelxY/Y64H mice. m, enamel matrix; a, ameloblasts. Scale bars, 50 µm.
Amelogenin distribution
In the secretory zone of WT incisors, strong amelogenin immunolabelling of the enamel extracellular matrix was detected as closely packed parallel lines corresponding to the prismatic pattern characteristic of the mature tissue. Staining was particularly strong at the junction between the enamel and dentine (Fig. 5A). In the transition zone, where matrix degradation occurs, this staining pattern was largely lost (Fig. 5B), whereas in the maturation and post-maturation zones, amelogenin staining of the enamel was lost entirely (Fig. 5C and D). In contrast, although strong amelogenin immunolabelling was detected in the secretory enamel of AmelxY/Y64H and AmelxY64H/Y64H mice, no obvious staining pattern was present other than within a limited region in the vicinity of the Tomes’ processes of the ameloblasts (Fig. 5E). Where the ameloblasts were severely disorganized, strong amelogenin staining was observed both within the enamel matrix and within the multi-cellular masses (Fig. 5F and G). Surprisingly, extracellular amelogenin staining persisted into the post-maturation enamel of the incisors of AmelxY/Y64H and AmelxY64H/Y64H mice; no organic matrix was detectable in the corresponding tissue of WT mice (Fig. 5H). Amelogenin staining of the enamel matrix in the secretory zone of AmelxX/Y64H incisors showed a similar, albeit less well-defined, pattern to that observed in WT mice; however, amelogenin immunostaining with no obvious structural organization was also detected in the blister-like structures present in transition and maturation zones (Supplementary Material, Fig. S2).
Ameloblastin distribution
In WT mice, ameloblastin immunostaining was observed within secretory ameloblasts, predominantly localizing within the supra-nuclear region. Less intense ameloblastin immunostaining was also present towards the apical region (Fig. 5I). Newly formed enamel matrix, close to the Tomes’ processes of the ameloblasts also stained intensely (Fig. 5I). Ameloblastin continued to be detected within the ameloblasts as maturation proceeded, with relatively little staining present in the underlying enamel matrix (Fig. 5J–L). In AmelxY/Y64H and AmelxY64H/Y64H mice, ameloblastin staining within the ameloblasts appeared to accumulate throughout the secretory zone, with strong staining towards the proximal pole of the cells. In contrast, ameloblastin staining was very weak in the newly secreted enamel matrix (Fig. 5M). In those regions where the ameloblasts were severely disorganized, strong ameloblastin staining was observed both within the multi-cellular masses and within the enamel matrix (Fig. 5N–P). AmelxX/Y64H mice displayed a staining pattern that was intermediate between that seen in WT mice and that observed in AmelxY/Y64H and AmelxY64H /Y64H mice (Supplementary Material, Fig. S2).
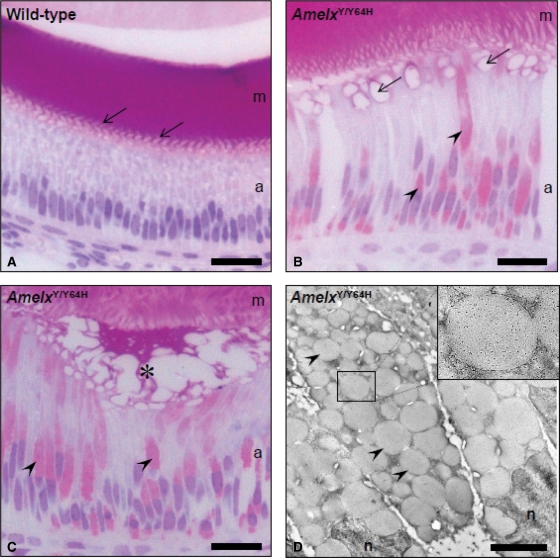
Resin histology and immuno-electron microscopy of the enamel organ and developing enamel of M100888 mutant mice
The increased eosinophilia and amelogenin immunofluorescence intensity within the secretory ameloblasts of AmelxY/Y64H mice prompted us to explore their morphology in greater detail. Haematoxylin and eosin-stained, semi-thin resin sections of the secretory zone of WT mice showed very little eosinophilia in the ameloblasts with a regular ‘herring-bone’ pattern corresponding to the insertion of the Tomes’ processes into the strongly stained enamel matrix (Fig. 6A). Examination of similar sections from AmelxY/Y64H mice revealed strong eosinophilia within many of the ameloblasts (Fig. 6B and C). These eosinophilic structures were visible throughout the cell and appeared to be vesicular in nature. At the interface of the ameloblasts and enamel matrix the regular pattern of Tomes’ process insertions was absent but showed a disorganized vesicular pattern (Fig. 6B) or contact of the ameloblasts with the enamel matrix was often lost leading to the formation of out-pocketings containing eosinophilic material (Fig. 6C). Immuno-electron microscopy using an antibody against amelogenin indicated that the affected secretory ameloblasts were engorged with amelogenin-containing vacuoles (Fig. 6D).
Figure 6.
Acrylic resin histology and immunoelectron microscopy. (A) Semi-thin, transverse sections of the secretory zone of WT mice revealed pale-staining ameloblasts and an intensely eosinophilic, enamel matrix. The interdigitations of the ameloblast Tomes’ processes were clearly evident in the region where new enamel matrix was forming (arrows). (B and C) In hemizygous male (AmelxY/Y64H) mutant mice, many of the ameloblasts contained large numbers of eosinophilic vesicles (arrowheads). Although the enamel matrix was intensely eosinophilic as observed in WT mice, it was highly disorganized (asterisk) and contact with the ameloblasts was lost. (C) Where new enamel matrix was being secreted, the interdigitating pattern seen in WT mice was replaced by large vacuolar structures (arrows). (D) Immunoelectron microscopy of AmelxY/Y64H secretory ameloblasts revealed the cytoplasm to be engorged by vacuoles of up to 2 µm diameter which were present in such large numbers that they led to the ameloblast nuclei becoming distorted. These vacuoles were strongly immunoreactive (black dots) for amelogenin (inset). m, enamel matrix; a, ameloblasts; n, nuclei. Scale bars in A–C, 25 µm and in D, 2 µm.
Further characterization of the enamel organ in M100888 mutant mice
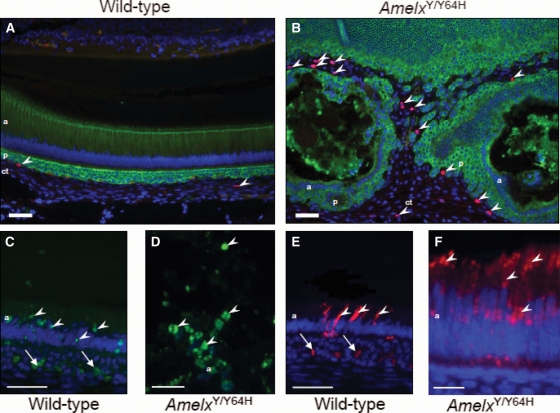
To characterize further the disorganized, multi-cellular masses observed in the incisor teeth of AmelxY/Y64H and AmelxY64H/Y64H mice, immunodetection of keratins and vimentin was carried out. Additionally, immunodetection of keratins was performed in conjunction with bromodeoxyuridine (5-bromo-2-deoxyuridine) (BrdU) and terminal deoxynucleotidyl transferase dUTP nick end labelling (TUNEL)/caspase-3 staining to determine cell proliferation and apoptosis, respectively. In WT mice, cells of ectodermal origin (ameloblasts, stratum intermedium and papillary layer) were immunoreactive for a pan-keratin antibody although those cells derived from the mesenchyme (fibroblasts, vascular endothelia) stained positively for vimentin (Supplementary Material, Fig. S3A and B). Immunohistochemistry using both antibodies in AmelxY/Y64H and AmelxY64H/Y64H mice revealed both pan-keratin and vimentin-positive cells within the abnormal multi-cellular masses; these appeared to be localized within distinct cellular cohorts consistent with their developmental origins (Supplementary Material, Fig. S3C–E). In WT mice, although occasional BrdU-positive cells were observed within the mesenchymally derived tissue compartment defined above, they were not detected in the enamel organ (Fig. 7A). This was also the case for AmelxY/Y64H and AmelxY64H/Y64H mice (Fig. 7B). At the point where the multi-cellular masses begin to form within the enamel organ of the AmelxY/Y64 H mice, cellular debris was observed suggesting that many of the ameloblasts were dying. Apoptotic ameloblast cell death is usual in the enamel organ of WT mice; some 25% are lost during the transition stage and a further 25% are lost during the maturation stage of amelogenesis (22). To investigate whether the increased cell death observed in AmelxY/Y64H mice was apoptotic, we performed TUNEL assays and immunolabelling for activated caspase-3 (Fig. 7C–F). Examination of ameloblasts within the transition zone of WT mice revealed occasional TUNEL-positive nuclei and cells labelled strongly for activated caspase-3 (Fig. 7C and E). In AmelxY/Y64H mice, much of the cellular debris was either TUNEL- or activated caspase-3-positive (Fig. 7D and F). Furthermore, the ameloblasts immediately proximal to the area of observable cell death were frequently immunoreactive for activated caspase-3.
Figure 7.
Analysis of cell proliferation and apoptosis. (A) Pan-cytokeratin (green) and BrdU (red) dual immunolabelling of the enamel organ of WT mice showing occasional BrdU-positive nuclei (arrowheads), confined to the underlying connective tissue compartment. (B) In the multi-cellular masses observed in hemizygous male (AmelxY/Y64H) mutant mice, BrdU-positive nuclei remained confined to the connective tissue element (arrowheads). (C) TUNEL-positive nuclei (arrowheads) were observed in the transition zone of WT mice. (D) In hemizygous male (AmelxY/Y64H) mutant mice, numerous TUNEL-positive nuclei (arrowheads) were observed in the amorphous material associated with the multi-cellular masses. (E) Immunolabelling for activated caspase 3 in WT mice revealed strong immunoreactivity in transition stage ameloblasts (arrowheads). (F) The ameloblasts immediately proximal to the area where multi-cellular masses form in AmelxY/Y64H mice were strongly immunoreactive for activated caspase 3 (arrowheads). The arrows in (C) and (E) indicate autofluorescence from erythrocytes. a, ameloblasts; p, stratum intermedium/papillary layer complex; ct, connective tissue. Scale bars in A–C and E, 50 µm and in D and F, 25 µm.
Effect of the amelogenin Y64H mutation on the pattern of the developing enamel matrix proteome
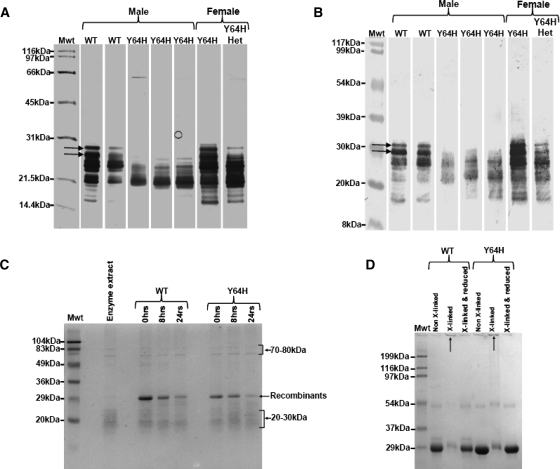
SDS PAGE and subsequent Western analysis with anti-amelogenin antibodies (Fig. 8B) revealed that the developing enamel matrix proteome was atypical for both AmelxX/Y64H and AmelxY/Y64H animals. WT controls showed the characteristic pattern of rodent amelogenin proteins, with components migrating at apparent molecular weights of between 5 and 31 kDa. The larger amelogenins (26–31 kDa) correspond to a spectrum of nascent molecules generated by alternative splicing (Fig. 8A and B). Most of the lower molecular weight molecules are the result of extracellular enzymatic processing of these nascent amelogenins. Matrix proteins extracted from AmelxY/Y64H mice appeared to be devoid of the higher molecular weight nascent amelogenins and exhibited a different spectrum of processing products (Fig. 8A). Matrix proteins extracted from AmelxX/Y64H mice showed a protein profile between that of WT and affected male mice (Fig. 8A and B).
Figure 8.
Biochemical investigations relating to the Y64H developing enamel proteome. (A) SDS–PAGE of secretory stage developing enamel removed from WT and Y64H mouse mandibular incisors showing apparent lack of full-length amelogenin proteins in the mutant animals (arrowed). Male and female WT animals had clear evidence of higher molecular weight amelogenins migrating up to ∼30 kDa. Y64H affected males had no proteins corresponding to this molecular weight range. Female mice, heterozygous with respect to the Y64H mutation did have higher molecular weight amelogenin present but at reduced amounts compared with WT. (B) Western blot of secretory stage developing enamel removed from WT and Y64H mouse mandibular incisors confirming the apparent lack of full-length amelogenin proteins (arrowed) in the mutant animals observed by SDS–PAGE (A). (C) SDS–PAGE of WT and Y64H recombinant amelogenins before and after incubation with secretory enamel protease extracts. After 24 h, Coomassie Blue staining revealed clear evidence of proteolytic degradation for both recombinant proteins, with progressive loss of intact protein (SDS Mr = 29 kDa). Proteins migrating at 70–80 kDa and around 20 kDa were also present in the MMP20 extracts and can be ignored with regard to the degradation of the recombinants. (D) SDS–PAGE of WT and Y64H recombinant amelogenins subjected to near-neighbour molecular cross-linking. Both WT and Y64H recombinant amelogenins were cross-linked to form supra-molecular complexes that failed to penetrate the SDS–PAGE stacking gel. On reduction of the cross-links, the amelogenins forming these complexes were returned to their monomeric state migrating at around 29 kDa.
To determine whether the observed lack of high molecular weight amelogenins in the developing enamel of AmelxY/Y64H animals was due to differential proteolytic processing of the mutant protein, recombinant WT and Y64H mutant amelogenins were expressed, isolated and purified. Western blotting coupled with mass spectroscopy of the prepared recombinant proteins confirmed their identities. SDS–PAGE showed that both WT and Y64H mutant amelogenins migrating at 29 kDa were similarly degraded over 24 h in the presence of secretory stage enamel extract enriched with MMP-20 (‘enamelysin’; Fig. 8C).
Effect of the amelogenin Y64H mutation on amelogenin–amelogenin interactions
Amelogenin forms self-assembled structures (‘nanospheres’) in vivo and in vitro (23,24). We used a near-neighbour, cross-linking strategy to determine any gross effects of the Y64H mutation on amelogenin–amelogenin interactions and found that no effects were discernible between Y64H and WT recombinant amelogenin behaviour. Both WT and Y64H recombinant amelogenins were cross-linked to form supra-molecular complexes that failed to penetrate the SDS–PAGE stacking gel (Fig. 8D). On reduction of the cross-links, the amelogenins forming these complexes were returned to their monomeric state migrating at around 29 kDa (Fig. 8D).
Effect of the Y64H mutation on amelogenin mRNA expression
In situ hybridization
To investigate whether the Y64H mutation affected amelogenin expression, we performed in situ hybridization using an amelogenin riboprobe corresponding to the full-length (excluding exon 4) transcript. In WT lower incisors, amelogenin mRNA was observed within the secretory ameloblasts and persisted into the early maturation stage ameloblasts after which its expression ceased abruptly (Supplementary Material, Fig. S4A). In AmelxY/Y64H mice, amelogenin mRNA expression showed a similar pattern extending to a region analogous to that of early maturation seen in WT mice (Supplementary Material, Fig. S4B).
RT–PCR
Our results using in situ hybridization suggested that the lack of high molecular weight amelogenin in the developing enamel matrix of Y64H affected animals was not due to lack of amelogenin expression in the enamel organ. However, to determine whether full-length transcripts of amelogenin were present, RT–PCR was also carried out. The results showed no differences in transcript length between animals containing the Y64H mutation and their WT littermates (Supplementary Material, Fig. S4C). Transcripts corresponding to the predicted size of the major, full-length amelogenin transcript were clearly seen in the AmelxY/Y64H mice [including a transcript corresponding to alternatively spliced transcripts containing exon 4 (25)].
Effect of Y64H amelogenin on epithelial cell survival
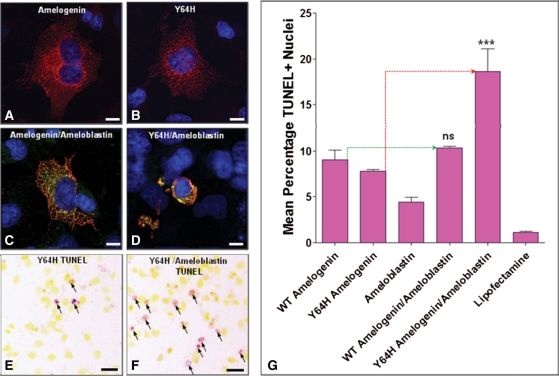
To investigate potential effects of the Y64H mutation on cell survival, we transfected COS-7 cells with expression vectors containing WT and Y64H amelogenin cDNAs (Fig. 9). Additionally, in light of the retention of ameloblastin in the ameloblasts of M100888 mutant mice (Fig. 5M–O), we also co-transfected an ameloblastin expression vector into COS-7 cells. Both WT and Y64H amelogenin were expressed in transfected cells, which were generally viable (Fig. 9A and B). Similarly, ameloblastin expression and co-expression of WT amelogenin and ameloblastin did not lead to a marked reduction in cell viability (Fig. 9C). In contrast, large numbers of cells with condensed nuclei were observed following co-transfection with ameloblastin and Y64H amelogenin, suggesting that cell survival was compromised (Fig. 9D). In light of these observations, we determined the incidence of apoptosis in transfected cells using the TUNEL assay (Fig. 9E and F). Comparison of cells singly transfected with WT amelogenin and those co-transfected with WT amelogenin and ameloblastin revealed no statistically significant difference in the mean percentage of TUNEL-positive cell nuclei (Fig. 9G). In contrast, comparison of cells singly transfected with Y64H amelogenin and those co-transfected with Y64H amelogenin and ameloblastin revealed a statistically significant (P < 0.0001) increase in the mean percentage of TUNEL-positive nuclei (Fig. 9G).
Figure 9.
Effect of the Y64H mutant amelogenin on cell survival. (A) COS-7 cell transfected with the WT amelogenin construct. The cell shows strong amelogenin expression (red fluorescence) and appears large and viable. (B) A similar cell transfected with Y64H amelogenin construct showing strong amelogenin expression (red fluorescence) is also typically large and viable. (C) A cell co-transfected with WT amelogenin and ameloblastin constructs shows strong immunolabelling for both proteins (orange–yellow fluorescence) and also appears large and viable. (D) Cells co-transfected with Y64H mutant amelogenin and ameloblastin constructs (orange–yellow fluorescence) were typically small with a condensed nucleus (blue) consistent with apoptosis. (E and F) TUNEL analysis of cells transfected with (E), mutant amelogenin alone and (F), mutant amelogenin and ameloblastin constructs. To improve discrimination of apoptotic cell nuclei, the images of the cells subjected to TUNEL analysis were pseudo-coloured such that TUNEL-positive nuclei appear red (arrows) and normal nuclei appear yellow. Scale bars represent 10 µm. (G) Results of TUNEL assays on four replicate experiments of each transfection/co-transfection. One-way ANOVA followed by a Bonferroni multiple comparison test showed that the large increase in mean percentage of TUNEL-positive nuclei detected in the Y64H mutant amelogenin/ameloblastin co-transfections was statistically significant (P < 0.0001) compared with cells transfected with Y64H mutant amelogenin alone. No statistically significant difference in the incidence of apoptosis could be shown between cells transfected with WT amelogenin alone and those co-transfected with WT amelogenin and ameloblastin.
DISCUSSION
Human X-linked AI is associated predominantly (although not exclusively) with mutations in the AMELX gene which encodes the enamel extracellular matrix protein amelogenin and some 15 mutations have been described to date (4). In general, mutations predicted to cause a total loss of amelogenin function, for example signal peptide mutations, or mutations causing loss of the amelogenin C-terminus, result in enamel hypoplasias, although mineralization defects may also occur (14). In contrast, although only a small number of mis-sense mutations have been described, those which affect the N-terminal region, especially those causing changes in the tyrosine-rich (TRAP) region of amelogenin, result in a predominantly hypomineralization/hypomaturation AI phenotype. In these cases, mature enamel may retain amelogenin and be discoloured. Understanding the relationship between phenotype and underlying genetic lesion is made more complex by the extensive alternative splicing of AMELX such that a point mutation could potentially affect several different amelogenin proteins but have no effect on others.
Despite the progress that has been made in determining the genetic basis of X-linked AI, the underlying molecular pathology remains poorly characterized. This is due to the paucity of available teeth from affected individuals suitable for biochemical and histological examination and the difficulty of obtaining developing human dental tissues. Mouse models are therefore an essential resource for furthering our understanding of the role of amelogenin in enamel bio-mineralization. Analysis of amelogenin-null mice has demonstrated that although ameloblast differentiation is normal, the resulting enamel is hypoplastic, appearing as an abnormally thin layer that lacks the highly organized prismatic structure characteristic of the mature tissue, suggesting that amelogenin is essential for the control of enamel bio-mineralization, structure and thickness (9,26). In this respect, the amelogenin-null mouse combines the various human phenotypes into a single model (9).
In the current article, we report the novel mis-sense mutation Y64H in amelogenin as underlying the AI phenotype observed in mice obtained as part of a large-scale ENU mutagenesis programme (http://www.gsc.riken.jp/Mouse/). The Y64H mutation occurs within the conserved tri-tyrosyl motif (PYPSYGYEPMGGW) of amelogenin which binds to N-acetyl-d-glucosamine and is positioned towards the C-terminus of the TRAP region (15). In humans, a P → T mutation, which also lies in this domain, underlies a unique but consistent AI phenotype in three families (14,16,27). Our results show that the murine Y64H mutation results in enamel that is hypomineralized in the case of AmelxX/Y64H mice or severely hypoplastic in the case of AmelxY/Y64H and AmelxY64H/Y64H animals indicating that the M100888 mutant mice phenocopy AI patients presenting with a P → T mutation in the tri-tyrosyl motif of amelogenin.
Large areas of enamel were missing from the teeth of Y64H homozygotes (both AmelxY/Y64H males and AmelxY64H/Y64H females), though the mineral levels in the enamel that remained were within the normal range; however, the ultrastructural appearance was severely dysplastic. It is unclear whether the apparent loss of enamel from these teeth results from post-eruptive breakdown or whether this is a primary developmental defect. Our preliminary investigations using nano-CT scanning of unerupted teeth suggests the latter (data not shown). The physical properties of dental enamel are closely associated with its structure and prismatic organization. Previous studies have shown that nascent amelogenin localizes to prism cores in the newly secreted enamel (28) suggesting a role in mineral organization. Using SDS–PAGE and Western analysis, we failed to demonstrate the presence of full-length nascent amelogenin within the secretory stage matrix of AmelxY/Y64H teeth. The lower molecular weight amelogenin components detected in these animals are likely to reflect proteins arising from differentially spliced Amelx transcripts, the most abundant of which is thought to be the leucine-rich amelogenin peptide (LRAP). LRAP would not contain the Y64H mutation due to splicing out of the 5′ end of exon 6 containing this mutation.
The lack of full-length amelogenin in the developing enamel matrix of affected animals suggested therefore that the observed bio-mineralization defects could be due to (i) failure to express full-length amelogenin containing the Y64H mutation; (ii) impaired secretion of Y64H amelogenin or (iii) an unusually high extracellular degradation rate for amelogenins carrying this mutation. Both in situ hybridization and RT–PCR indicated that full-length amelogenin was expressed by ameloblasts in AmelxY/Y64H mice, eliminating failure to express this transcript as a possible pathogenic mechanism. Our preliminary degradation studies using recombinant WT and Y64H mutant amelogenins incubated with MMP20 extracts showed no gross differences in degradation rate or pattern of resulting degradation products at the time intervals used. This is in contrast to previous reports that showed amelogenin hydrolysis to be compromised as a result of a P → T mutation in this same domain (17). Our data tend to eliminate the possibility that the Y64H mutation increases the susceptibility of Y64H amelogenin to normal post-secretory processing and enhanced sensitivity to extracellular proteolysis does not appear to be a mechanism underpinning the observed pathology. We therefore hypothesize that the paucity of full-length Y64H amelogenin in the Y64H mutant enamel matrix results from a failure to successfully traffic and secrete amelogenin into the enamel matrix. Our immunohistochemical data indicate that both amelogenin and ameloblastin accumulate within the ameloblasts of affected mice and that these accumulations become increasingly marked as amelogenesis proceeds. Transmission electron microscopy further confirmed this finding, clearly showing ameloblasts with abnormal cell morphology and engorged endoplasmic reticulum/Golgi apparatus heavily immunolabelled with an anti-amelogenin antibody, indicating intracellular retention of Y64H amelogenin.
In light of the impaired secretion of amelogenin and ameloblastin in M100888 mutant mice, we analysed the effects of WT amelogenin, Y64H mutant amelogenin and ameloblastin expression, both singly and in combination, in a transfected eukaryotic cell line of epithelial origin. This approach is relevant since amelogenin has been shown to interact directly through its tri-tyrosine motif with native and recombinant ameloblastin (29). In addition, ameloblastin and amelogenin share a common secretory pathway and co-localize within identical secretory granules within ameloblasts (30) where they may interact. Co-expression of Y64H mutant amelogenin and ameloblastin in our transfected epithelial cell lines resulted in a marked decrease in cell survival compared with when the three proteins were transfected in isolation or when WT amelogenin and ameloblastin were co-transfected. This observation suggests that a combination of Y64H mutant amelogenin and ameloblastin is a pathological factor in these transfected cells that might also be present in vivo in the M100888 mutants. The precise mechanism by which Y64H amelogenin in combination with ameloblastin leads to decreased cell survival in our transfection experiments is unclear but it could be due to Y64H amelogenin–ameloblastin interactions resulting in complexes that can be no longer trafficked appropriately prior to secretion due to conformational anomalies. The accumulating protein could then lead to ER stress and cell death.
Mice expressing a truncated ameloblastin (lacking exons 5 and 6), share similar histopathological features to the Y64H amelogenin mutant mice reported here (8,31). The ameloblasts of these mice lose contact with their meagre enamel matrix during the secretory stage of amelogenesis and become disorganized, forming multi-cellular masses often enclosing isolated accretions of matrix-like material. This raises the possibility that the M100888 mutant phenotype may be due in part to disturbances involving ameloblastin secretion and function that normally rely on amelogenin–ameloblastin interactions—these interactions being compromised in the presence of Y64H mutant amelogenin.
What is certain is that biological interplay between amelogenin and ameloblastin appears to be necessary to sustain ameloblasts during the secretory stage of amelogenesis. In mice null for both amelogenin and ameloblastin the deleterious effect on enamel formation was greater than in mice null for amelogenin alone or null for ameloblastin alone suggesting a synergistic role for these proteins in amelogenesis (32).
Taken together, these data suggest that the ameloblasts in M100888 mutant mice endure a secretory defect which impairs not only the secretion of amelogenin but also that of ameloblastin. The impairment of ameloblastin secretion may be responsible for the altered behaviour of the ameloblasts in the Y64H amelogenin mutant mouse acting similarly to the loss-of-function mutation in the Ambn mutant mouse. It is also plausible that the retention of both proteins within ameloblasts would induce cellular stresses which culminate in the extensive apoptosis we observed in the Y64H mutant mouse; a phenomenon that is clearly modelled by our transfection studies.
However, when considering amelogenin–ameloblastin interactions and their potential role in amelogenesis in vivo, the relative abundances of the two proteins must be considered. The intra-cellular concentrations of amelogenin and ameloblastin are unknown but there is a huge excess of amelogenin over ameloblastin in the extracellular matrix. It is therefore conceivable that the synthesis of both proteins differs commensurately and that most amelogenin remains un-associated with the much scarcer ameloblastin in the intracellular secretory pathway. Abnormal amelogenin–ameloblastin interactions may therefore be only one part of the reason for impairment of secretion in Y64H mutant mice. Other protein–protein interactions may also be affected, including amelogenin–cytokeratin interactions. The tri-tyrosyl motif has been shown to bind to a conserved N-acetyl-d-glucosamine mimicking sequence present in cytokeratin 14 (33) and N-acetyl-d-glucosamine residues on cytokeratin 5 (19). Amelogenin and cytokeratin 14 co-localize at the apical regions of ameloblasts and dissociate at the Tomes’ processes prior to secretion leading to suggestions of a possible role for cytokeratin 14 in chaperoning amelogenin during amelogenesis (19,33). Failure to bind cytokeratin 14 was demonstrated when the human P → T mutation was introduced into the amelogenin tri-tyrosyl motif or when all three tyrosine residues were replaced by phenylalanine (33). It is therefore possible that the Y64H mutation also affects binding between amelogenin and cytokeratin 14 which is necessary for amelogenin secretion. Whether either or both of these possible mechanisms is driving phenotype in the Y64H mutant mice, we suggest that failure to secrete matrix proteins would lead to cell stress and disruption of normal ameloblast organization and behaviour.
In this study, we have shown here that abnormal retention of matrix components within the ameloblasts is coincident with loss of ameloblast organization and appearance of multi-cellular masses, frequently associated with tissue necrosis, in the Y64H animals. Analysis of cell proliferation showed that increased cell proliferation was a feature of the surrounding connective tissue rather than the ameloblasts themselves. Investigations of cell death revealed a pronounced loss of ameloblasts associated with apoptosis at around the point where the multi-cellular masses began to form. This massive loss of ameloblasts was seen histologically as a region of necrosis. The increased proliferation within the surrounding connective tissue may therefore be a pathological reaction to this necrosis leading to the observed connective tissue invasion and fibrosis of the persisting enamel matrix. Such fibrosis is a well-known occurrence in many chronic, necrotizing pathologies (34–37). The presence of multi-cellular masses in the jaws of the M100888 mice reported here has also been observed in transgenic mice expressing the P → T mutation in the human amelogenin tri-tyrosyl motif (38), further paralleling the Y64H mutation with that found in humans.
We predict that the Y64H amelogenin mutation results in a failure to traffic and secrete protein along the usual pathways resulting in the observed accumulation of amelogenin and ameloblastin in the ameloblast endoplasmic reticulum. This could be due to one of two different mechanisms: the presence of the Y64H mutation in the amelogenin tri-tyrosyl domain could preclude normal chaperoning and intracellular trafficking. Our preliminary results using recombinant Y64H and WT amelogenins show that amelogenin–amelogenin near neighbour interactions are similar. It is therefore possible that other protein–protein binding, including ameloblastin–amelogenin binding and/or cytokeratin–amelogenin binding, is necessary to prevent intracellular amelogenin self-assembly which in turn would lead to secretory failure. Other examples of this in man include the premature oligomerization of mutated forms of myelin proteolipid protein in the endoplasmic reticulum, shown to cause secretory failure and resulting in dysmyelinating disease (39).
In summary, we describe here a mis-sense mutation in the murine Amelx tri-tyrosyl domain of the enamel extracellular matrix protein amelogenin that phenocopies human X-linked AI where similar mutations have been reported in the same sequence. Animals affected by the mutation have severe enamel bio-mineralization defects associated with absence of full-length amelogenin protein in the developing enamel matrix, loss of ameloblast phenotype, increased ameloblast apoptosis and formation of multi-cellular masses. We present evidence to demonstrate that affected ameloblasts express but fail to secrete amelogenin leading to ER/Golgi engorgement and hypothesize that this is a key mechanistic factor underpinning the pathology. This study therefore successfully links phenotype with underlying genetic lesion in a relevant murine model for human AI.
MATERIALS AND METHODS
Source of material and gene sequencing
The mutant mouse line M100888 was obtained from RIKEN GSC (http://www.gsc.riken.jp/Mouse/) and maintained on a DBA/2J genetic background. To search for the causative mutation, we designed oligonucleotide primer sequences based on the genomic sequence of Amelx (ENSMUSG00000031354) and sequenced each exon and the associated splice junctions using dye primer chemistry. The sequences obtained were compared with those generated from a panel of inbred strains (A/J, AKR/J, BALB/c, C57BL/6J, CAST/Ei, CBA/J, DBA/2J and SPRET/Ei). All experiments were performed in accordance with the Animals (Scientific Procedures) Act, UK, 1986.
Preparation of teeth for microradiography and scanning electron microscopy
Lower incisors were dissected from 2- to 3-month-old, WT, hemizygous (AmelxY/Y64H) mutant male and heterozygous mutant females (AmelxX/Y64H) mice (n = 3 in each category). Adherent soft tissue was removed and the incisors freeze-dried and embedded in methyl methacrylate. The embedded teeth were sectioned transverse to the long axis of the tooth using an Accutom-5 cutter with a peripheral diamond cutting disc and minimal water cooling to minimize loss of organic components within the enamel. Sections were taken at points corresponding to the end of secretion, maturation and fully mature enamel and polished plano-parallel with 1200 grade carborundum paper to a thickness of ∼100 µm (final thickness being determined by micrometer measurement) then acid-etched for 15 s with 35% phosphoric acid to remove the smear layer and thoroughly washed in distilled water (pH 7.0).
Quantitative determination of enamel mineral density
Mineral density was determined across the full thickness of enamel within the sections using transverse microradiography (TMR) as described previously (40). Briefly, sections were contact microradiographed for 6 min on Kodak High Resolution plate, type 1A, by Ni-filtered Cu–K radiation at 20 kV, 4 mA. An aluminium step wedge was included to aid quantitation. The microradiographs were developed in Kodak HRP developer and analysed by TMRW software (version 20.0.27.16, 2000; Inspektor Research Systems, The Netherlands). The enhanced images of the microradiographs were analysed under standard conditions of light intensity and magnification and were processed, along with data from measurement of the aluminium step wedge, by the TMR program. Mineral density levels were compared, where possible, using one way ANOVA.
Ultrastructural appearance of enamel
Following microradiography, the sections were mounted on aluminium stubs and sputter-coated with gold. Microstructural analysis was undertaken using a Jeol 35 SEM fitted with the Deben ‘Genie’ upgrade (Deben Engineering, UK).
Histological and immunofluorescence analysis of the enamel organ and developing enamel
Mandibles from 2- to 3-month-old hemizygous mutant male, heterozygous mutant female, homozygous mutant female and WT male mice (n = 4 in each category) were dissected following cervical dislocation and fixed in neutral buffered formalin at room temperature for 48 h. Following fixation, the mandibles were decalcified in 5% EDTA, dehydrated through a graded ethanol series, cleared in chloroform, embedded as hemi-mandibles in paraffin wax, sectioned and stained with haematoxylin and eosin. Sections were examined using a DMRB microscope (Leica) with Spot™ digital camera and associated software (RTKE/SE Diagnostic Instruments Inc.). Immunofluorescence analysis was performed on mandibles prepared as above using antibodies raised against activated caspase 3 (Abcam), amelogenin (FL191, Santa Cruz Biotechnology) and ameloblastin (C17, Santa Cruz Biotechnology). Primary antibodies were detected using biotinylated secondary antibodies (Vector laboratories) followed by Cy-3-conjugated streptavidin (Sigma) and mounted in fluorescence mountant containing DAPI (Vector laboratories). The TUNEL assay was performed according to the manufacturer's instructions (Roche Diagnostics). Tissue sections were examined as above.
High-resolution histology and immuno-electron microscopy
Hemi-mandibles dissected from WT and hemizygous mutant male mice (n = 3) were fixed in 4% paraformaldehyde in 0.1 m cacodylate buffer (pH 7.4) overnight at 4°C, rinsed in PBS and decalcified as described above. Following decalcification, samples were re-fixed with 4% paraformaldehyde in 0.1 m cacodylate buffer (pH 7.4) overnight. The next morning, samples were dehydrated through a graded ethanol series and embedded as transverse segments corresponding to the secretory and maturation regions of the tooth in acrylic resin (LR White) and polymerized at 40°C for 24 h. Semi-thin sections were prepared using a glass knife, mounted on Superfrost Plus glass slides (Thermo Shandon), stained with haematoxylin and eosin and examined as above.
Ultrathin sections were cut with a Diatome diamond knife on a Reichert-Jung Ultracut microtome and were mounted on formvar/carbon-coated copper grids. For immunolabelling, grids were incubated with blocking solution (1% bovine serum albumin, 0.15 m glycine, 0.1% Tween-20, 0.1 m phosphate buffer, pH 7.4) for 15 min, then with primary antibody to amelogenin (FL191, Santa Cruz Biotechnology) for 1 h, then with secondary antibodies conjugated with 10 nm gold particles for 30 min and washed extensively with distilled water. Sections were contrasted with 1% uranyl acetate for 10 min and observed using an FEI Tecnai 12 Biotwin electron microscope (FEI company, The Netherlands) at an accelerating voltage of 100 kV.
Determination of amelogenin expression by in situ hybridization
Amelogenin cDNA was cloned into pBluescript II SK (Stratagene) and the resulting construct used to synthesize a digoxygenin-labelled, amelogenin antisense riboprobe using T7 polymerase (Promega) and digoxygenin-conjugated dUTP (Roche). In situ hybridization was performed as described previously (41,42), however, riboprobe binding was detected immunohistochemically using an alkaline phosphatase-conjugated, anti-digoxygenin antibody (Roche) followed by histochemical demonstration of alkaline phosphatase activity using BM purple (Roche) as the chromogenic substate.
Determination of ameloblast cell proliferation using BrdU labelling
Cell proliferation was detected by intra-peritoneal injection of BrdU labelling reagent (Amersham Biosciences) at 100 µg/g body weight, into 2–3-month-old, WT and hemizygous mutant male mice (n = 4). The mice were killed by cervical dislocation 2 h later and their mandibles processed for histological analysis as described above. Sections were immunolabelled using an antibody against BrdU (Abcam) following retrieval in 10 mm sodium citrate (pH 6.0) and detected using a biotinylated secondary antibody (Vector laboratories) and Cy3-conjugated streptavidin (Sigma). A pan-cytokeratin antibody (Neomarkers) was used to detect epithelial elements in the same sections and detected with a fluorescent secondary antibody (AlexaFluor 488, Molecular Probes).
SDS–PAGE analysis and western blotting
WT, hemizygous AmelxY/Y64H mutant male mice and Amelx X/Y64H heterozygous female mice were killed by cervical dislocation and the mandibles removed. The contralateral incisors were dissected from the mandibular bone and the enamel organs wiped away with damp paper tissue. After ∼1 min, the white opaque zone indicative of the maturation stage became apparent. Contiguous samples of secretory, early maturation and late maturation stage enamel were microdissected from the dentine. In this way, results obtained reflect protein per volume of enamel. All samples were stored at −80°C.
Enamel samples were dissolved in 20 µl of 10% acetic acid. Once dissolved, 180 µl of SDS sample loading buffer were added and the samples neutralized by addition of 7.5 µl of 5 m NaOH. Samples were loaded at 10 µl per lane and subjected to gel electrophoresis (Bio-Rad Protean III) using 15% Tris/glycine gels run at a constant 200 V. Resolved proteins were detected by silver staining. Alternatively, separated proteins were transferred from the gels to nitrocellulose sheets and probed using monospecific, polyclonal antibodies to amelogenin (Santa Cruz FL-191). Nitrocellulose membranes were blocked overnight at room temperature in TBS containing 5% casein. Membranes were washed briefly in TBS and incubated for 2 h in rabbit polyclonal anti-full-length amelogenin antibodies. Immuno-reactivity was visualized using an amplified alkaline phosphatase detection system (Bio-Rad, Immun-Blot amplified AP assay kit) in accordance with the manufacturer's instructions.
Expression of recombinant proteins
Full-length WT and Y64H mutant amelogenin proteins were expressed using a pET28 expression vector (Novagen, Merck Chemicals Ltd) modified with a HRV 3C protease site in Rosetta DE3 Escherichia Coli cells (Novagen). The cells were harvested by centrifugation and lyzed using a lysis buffer containing 100 mm NaH2PO4, 10 mm Tris and 6 M guanidine-HCl (pH 8.0). Expressed proteins were purified using nickel affinity chromatography (GE Healthcare HiTrap Nickel column) in the presence of 8 m urea. The histidine tag was removed by incubating the protein with HRV 3C protease (A.G. Scientific, Inc.) overnight in 50 mm Tris buffer (pH 8.0) at a concentration of 25 µg/ml. Final purification was achieved by preparative SDS–PAGE (Bio-Rad Model 491 Prepcell) using an 8 cm 12% polyacrylamide gel run at a constant power of 12 W. Fractions were collected for 3.33 min each at a flow rate of 220 µl/min. Fractions containing purified recombinant protein were identified by standard SDS–PAGE. Relevant fractions were desalted against 125 mm formic acid using a Hi Prep 26/10 desalting column (GE Healthcare) and the proteins lyophilized prior to use. Protein identity was confirmed by western blotting using anti-amelogenin antibodies as described above. Mass spectroscopy of WT and Y64H recombinant proteins gave molecular masses of 20 590 and 20 564 kDa, respectively (predicted masses = 20 591 and 20 565 kDa, respectively).
Determination of amelogenin–amelogenin interactions using molecular cross-linking
To compare amelogenin–amelogenin interactions, a near neighbour cross-linking analysis was performed using a modification of methods described previously (24). Briefly, dithiobis(succinimidylpropionate) (DSP) (Thermo Scientific) was dissolved in DMSO at 100 mm and added to 100 µl of synthetic enamel fluid (43) containing recombinant WT or Y64H amelogenin at 2 mg/ml to give a final DSP concentration of 5 mm. The mixture was incubated at room temperature for 10 min before quenching the reaction by the addition of 20 µl of 500 mm Tris (pH 6.8). Samples were diluted 3:1 (vol/vol) with either reducing or non-reducing 4× concentrated SDS–PAGE loading buffer and the molecular size of cross-linked amelogenins determined by loading 5 µl on 12% SDS gels containing 6m urea run at a constant 200 V. Resolved proteins were detected by Coomassie Blue staining.
In vitro degradation of recombinant WT and Y64H amelogenins by secretory stage developing enamel proteases
Secretory enamel protease extract (containing principally MMP20 enzyme) was obtained from the developing enamel of rat mandibular incisors. Male Wistar rats were sacrificed by cervical dislocation and the lower incisors dissected free of the jaws. The adhering enamel organs were wiped away using damp paper tissue and the teeth allowed to air dry until the white opaque zone was visible. A scalpel was used to dissect the developing enamel lying apical to the white opaque zone clear of the underlying dentine. Secretory stage enamel from two contra-lateral teeth was then extracted in 100 µl of 50 mm Tris (pH 7.4) containing 10 mm CaCl2 by grinding with a fine glass rod. The extracts were centrifuged at 20 000g for 10 min and the supernatant containing extracted MMP20 removed. WT and Y64H recombinant amelogenins were prepared in 50 mm Tris (pH 8.0) at a concentration of 1 µg/ml and mixed in equal volumes with the MMP20 extract. A 10 µl aliquot of the mixture was removed immediately, diluted with 10 µl of SDS–PAGE sample loading buffer containing a cocktail of protease inhibitors and stored at −80°C until required. The remaining sample was incubated at 37°C and further aliquots were removed after 8 and 24 h. Samples were analysed by loading 10 µl per lane on 15% SDS–PAGE mini gels and stained with Coomassie Blue.
Determination of transcript size using RT–PCR
Lower incisors dissected from WT and hemizygous mutant male mice mandibles (n = 3) were immediately placed into RNAlater™ solution (Applied Biosystems) for RNA preservation in accordance with the manufacturer's instructions. RNA was extracted from whole mice teeth using the Ribopure™ kit (Applied Biosystems) according to the manufacturer's instructions. RNA was evaluated using spectrophotometry and gel electrophoresis and subjected to DNase treatment to remove contaminating DNA using the Turbo DNA-free™ kit (Applied Biosystems). Reverse transcription was performed using the Transcriptor First Strand cDNA synthesis kit (Roche). The resulting cDNA was used in a PCR reaction (Phire hot start DNA polymerase Finnzymes—New England Biolabs) with primers designed to amplify a 536 bp product of the gene (5′-atgcccctaccacctcatc-3′ and 5′-acttcttcccgcttggtctt-3′) along with other potential PCR products corresponding to alternatively spliced amelogenin mRNAs.
Site-directed mutagenesis and plasmid construction
The Y64H mutation was introduced into a rat amelogenin cDNA (44) using the ‘Quick Change’ site-directed mutagenesis kit (Stratagene) and the primers 5′-GCATGATAAGGCAGCCGCATCCTTCCTATGGTTAC-3′ and 5′-GTAACCATAGGAAGGATGCGGCTGCCTTATCATGC-3′. This strategy yielded a mutant amelogenin cDNA of identical amino acid sequence to that of the mouse. Subsequently, cDNAs encoding WT and mutant amelogenin were sub-cloned into the expression vector pSG5-HA. A full-length ameloblastin cDNA (IMAGE clone 30504437; Geneservice, Cambridge, UK) was sub-cloned into the expression vector pSG5-FLAG. All constructs were verified by sequence analysis.
Transfection experiments
COS-7 cells were cultured on coverslips in DMEM containing 10% fetal bovine serum at 37°C and 5% CO2. Once cells had reached 90% confluence, they were transfected with 8 µg of each construct using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen) and incubated as above for 18 h. Transfections omitting the DNA constructs were set up as negative controls. Subsequently, cells were fixed in ice cold 4% paraformaldehyde and autofluorescence was quenched by treating cells first with 50 mm ammonium chloride in PBS and then in 0.1 m glycine. The cells were then made permeable using 0.1% Triton X-100 in PBS and amelogenin and ameloblastin expression detected using the appropriate fluorescently labelled antibody (see above). Immunolabelled cells were examined on a Delta Vision RT (Applied Precision) restoration microscope using a 100x/1.4 Uplan Apo objective and the Sedat filter set (Chroma 89000 v2). Images were collected using a Coolsnap HQ (Photometrics) camera with a Z optical spacing of 0.2 µm. Raw images were then deconvoluted using the Softworx software and displayed as maximum intensity projections.
In parallel experiments, COS-7 cells were transfected as above and cell death was detected by TUNEL assay using the ‘In situ cell death’ kit (Roche) according to the manufacturer's instructions. Ten contiguous images of each sample were collected using a ×20 objective lens and the percentage of TUNEL-positive cells in each sample determined. Results were analysed by 1-way ANOVA followed by a Bonferroni multiple comparison test using Graphpad Prism® v5.0 software (Graphpad, San Diego, CA, USA). Results were taken to be statistically significant at the P < 0.05 level.
SUPPLEMENTARY MATERIAL
FUNDING
M.J.D. is supported by the NIHR Manchester Biomedical Research Centre. This work was supported by a Wellcome Trust Programme Grant (grant number 075945).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Sarah Myers for expert technical assistance and the electron microscopic service in the Faculty of Life Sciences, University of Manchester, for their help with the transmission electron microscopy. We are grateful to Dr W.A. Bonass for supplying the rat cDNA used in this work.
Conflict of Interests statement. None declared.
REFERENCES
- 1.Backman B., Holm A.K. Amelogenesis imperfecta: prevalence and incidence in a northern Swedish county. CommunITY Dent. Oral Epidemiol. 1986;14:43–47. doi: 10.1111/j.1600-0528.1986.tb01493.x. [DOI] [PubMed] [Google Scholar]
- 2.Backman B. Amelogenesis imperfecta—clinical manifestations in 51 families in a northern Swedish county. Scand. J. Dent. Res. 1988;96:505–516. doi: 10.1111/j.1600-0722.1988.tb01590.x. [DOI] [PubMed] [Google Scholar]
- 3.Witkop C.J., Jr Amelogenesis imperfecta, dentinogenesis imperfecta and dentin dysplasia revisited: problems in classification. J. Oral Pathol. 1988;17:547–553. doi: 10.1111/j.1600-0714.1988.tb01332.x. [DOI] [PubMed] [Google Scholar]
- 4.Stephanopoulos G., Garefalaki M.E., Lyroudia K. Genes and related proteins involved in amelogenesis imperfecta. J. Dent. Res. 2005;84:1117–1126. doi: 10.1177/154405910508401206. [DOI] [PubMed] [Google Scholar]
- 5.Hu J.C., Chun Y.H., Al Hazzazzi T., Simmer J.P. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186:78–85. doi: 10.1159/000102683. [DOI] [PubMed] [Google Scholar]
- 6.Kim J.W., Lee S.K., Lee Z.H., Park J.C., Lee K.E., Lee M.H., Park J.T., Seo B.M., Hu J.C., Simmer J.P. FAM83H mutations in families with autosomal-dominant hypocalcified amelogenesis imperfecta. Am. J. Hum. Genet. 2008;82:489–494. doi: 10.1016/j.ajhg.2007.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith C.E. Cellular and chemical events during enamel maturation. Crit. Rev. Oral Biol. Med. 1998;9:128–161. doi: 10.1177/10454411980090020101. [DOI] [PubMed] [Google Scholar]
- 8.Fukumoto S., Kiba T., Hall B., Iehara N., Nakamura T., Longenecker G., Krebsbach P.H., Nanci A., Kulkarni A.B., Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J. Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson C.W., Yuan Z.A., Hall B., Longenecker G., Chen E., Thyagarajan T., Sreenath T., Wright J.T., Decker S., Piddington R., et al. Amelogenin-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2001;276:31871–31875. doi: 10.1074/jbc.M104624200. [DOI] [PubMed] [Google Scholar]
- 10.Caterina J.J., Skobe Z., Shi J., Ding Y., Simmer J.P., Birkedal-Hansen H., Bartlett J.D. Enamelysin (matrix metalloproteinase 20)-deficient mice display an amelogenesis imperfecta phenotype. J. Biol. Chem. 2002;277:49598–49604. doi: 10.1074/jbc.M209100200. [DOI] [PubMed] [Google Scholar]
- 11.Bartlett J.D., Skobe Z., Lee D.H., Wright J.T., Li Y., Kulkarni A.B., Gibson C.W. A developmental comparison of matrix metalloproteinase-20 and amelogenin null mouse enamel. Eur. J. Oral Sci. 2006;114(Suppl. 1):18–23. doi: 10.1111/j.1600-0722.2006.00292.x. discussion 39–41, 379. [DOI] [PubMed] [Google Scholar]
- 12.Masuya H., Shimizu K., Sezutsu H., Sakuraba Y., Nagano J., Shimizu A., Fujimoto N., Kawai A., Miura I., Kaneda H., et al. Enamelin (Enam) is essential for amelogenesis: ENU-induced mouse mutants as models for different clinical subtypes of human amelogenesis imperfecta (AI) Hum. Mol. Genet. 2005;14:575–583. doi: 10.1093/hmg/ddi054. [DOI] [PubMed] [Google Scholar]
- 13.Seedorf H., Klaften M., Eke F., Fuchs H., Seedorf U., Hrabe de Angelis M. A mutation in the enamelin gene in a mouse model. J. Dent. Res. 2007;86:764–768. doi: 10.1177/154405910708600815. [DOI] [PubMed] [Google Scholar]
- 14.Wright J.T., Hart P.S., Aldred M.J., Seow K., Crawford P.J., Hong S.P., Gibson C.W., Hart T.C. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect. Tissue Res. 2003;44(Suppl. 1):72–78. [PubMed] [Google Scholar]
- 15.Ravindranath R.M., Moradian-Oldak J., Fincham A.G. Tyrosyl motif in amelogenins binds N-acetyl-d-glucosamine. J. Biol. Chem. 1999;274:2464–2471. doi: 10.1074/jbc.274.4.2464. [DOI] [PubMed] [Google Scholar]
- 16.Collier P.M., Sauk J.J., Rosenbloom S.J., Yuan Z.A., Gibson C.W. An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch. Oral Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- 17.Li W., Gibson C.W., Abrams W.R., Andrews D.W., DenBesten P.K. Reduced hydrolysis of amelogenin may result in X-linked amelogenesis imperfecta. Matrix Biol. 2001;19:755–760. doi: 10.1016/s0945-053x(00)00121-9. [DOI] [PubMed] [Google Scholar]
- 18.Ravindranath R.M., Tam W.Y., Bringas P., Jr, Santos V., Fincham A.G. Amelogenin–cytokeratin 14 interaction in ameloblasts during enamel formation. J. Biol. Chem. 2001;276:36586–36597. doi: 10.1074/jbc.M104656200. [DOI] [PubMed] [Google Scholar]
- 19.Ravindranath R.M., Basilrose R.M., Sr, Ravindranath N.H., Vaitheesvaran B. Amelogenin interacts with cytokeratin-5 in ameloblasts during enamel growth. J. Biol. Chem. 2003;278:20293–20302. doi: 10.1074/jbc.M211184200. [DOI] [PubMed] [Google Scholar]
- 20.Hay M.F. The development in vivo and in vitro of the lower incisor and molars of the mouse. Arch. Oral Biol. 1961;3:86–109. doi: 10.1016/0003-9969(61)90142-x. [DOI] [PubMed] [Google Scholar]
- 21.Smith C.E., Warshawsky H. Histological and 3 dimensional organization of odontogenic organ in lower incisor of 100 gram rats. Am. J. Anat. 1975;142:403–429. doi: 10.1002/aja.1001420402. [DOI] [PubMed] [Google Scholar]
- 22.Smith C.E., Warshawsky H. Quantitative-analysis of cell turnover in enamel organ of rat incisor—evidence for ameloblast death immediately after enamel matrix secretion. Anat. Rec. 1977;187:63–97. doi: 10.1002/ar.1091870106. [DOI] [PubMed] [Google Scholar]
- 23.Fincham A.G., Moradianoldak J., Diekwisch T.G.H., Lyaruu D.M., Wright J.T., Bringas P., Slavkin H.C. Evidence for amelogenin nanospheres as functional components of secretory-stage enamel matrix. J. Struct. Biol. 1995;115:50–59. doi: 10.1006/jsbi.1995.1029. [DOI] [PubMed] [Google Scholar]
- 24.Brookes S.J., Kirkham J., Lyngstadaas S.P., Shore R.C., Wood S.R., Robinson C. Spatially related amelogenin interactions in developing rat enamel as revealed by molecular cross-linking studies. Arch. Oral Biol. 2000;45:937–943. doi: 10.1016/s0003-9969(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 25.Hu C.C., Ryu O.H., Qian Q., Zhang C.H., Simmer J.P. Cloning, characterization, and heterologous expression of exon-4-containing amelogenin mRNAs. J. Dent. Res. 1997;76:641–647. doi: 10.1177/00220345970760020401. [DOI] [PubMed] [Google Scholar]
- 26.Li Y., Suggs C., Wright J.T., Yuan Z.A., Aragon M., Fong H., Simmons D., Daly B., Golub E.E., Harrison G., et al. Partial rescue of the amelogenin null dental enamel phenotype. J. Biol. Chem. 2008;283:15056–15062. doi: 10.1074/jbc.M707992200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hart S., Hart T., Gibson C., Wright J.T. Mutational analysis of X-linked amelogenesis imperfecta in multiple families. Arch. Oral Biol. 2000;45:79–86. doi: 10.1016/s0003-9969(99)00106-5. [DOI] [PubMed] [Google Scholar]
- 28.Uchida T., Tanabe T., Fukae M., Shimizu M., Yamada M., Miake K., Kobayashi S. Immunochemical and immunohistochemical studies, using antisera against porcine 25 kDa amelogenin, 89 kDa enamelin and the 13–17 kDa nonamelogenins, on immature enamel of the pig and rat. Histochemistry. 1991;96:129–138. doi: 10.1007/BF00315983. [DOI] [PubMed] [Google Scholar]
- 29.Ravindranath H.H., Chen L.S., Zeichner-David M., Ishima R., Ravindranath R.M. Interaction between the enamel matrix proteins amelogenin and ameloblastin. Biochem. Biophys. Res. Commun. 2004;323:1075–1083. doi: 10.1016/j.bbrc.2004.08.207. [DOI] [PubMed] [Google Scholar]
- 30.Zalzal S.F., Smith C.E., Nanci A. Ameloblastin and amelogenin share a common secretory pathway and are co-secreted during enamel formation. Matrix Biol. 2008;27:352–359. doi: 10.1016/j.matbio.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Wazen R.M., Moffatt P., Zalzal S.F., Yamada Y., Nanci A. A mouse model expressing a truncated form of ameloblastin exhibits dental and junctional epithelium defects. Matrix Biol. 2009;28:292–303. doi: 10.1016/j.matbio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatakeyama J., Fukumoto S., Nakamura T., Haruyama N., Suzuki S., Hatakeyama Y., Shum L., Gibson C.W., Yamada Y., Kulkarni A.B. Synergistic roles of amelogenin and ameloblastin. J. Dent. Res. 2009;88:318–322. doi: 10.1177/0022034509334749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravindranath R.M., Tam W.Y., Nguyen P., Fincham A.G. The enamel protein amelogenin binds to the N-acetyl-D-glucosamine-mimicking peptide motif of cytokeratins. J. Biol. Chem. 2000;275:39654–39661. doi: 10.1074/jbc.M006471200. [DOI] [PubMed] [Google Scholar]
- 34.Johnson M.A., Barron M.J. Muscle biopsy analysis. In: Lane R.J.M., editor. Handbook of Muscle Disease. New York: Marcel Dekker; 1996. pp. 61–81. [Google Scholar]
- 35.Contempre B., de Escobar G.M., Denef J.F., Dumont J.E., Many M.C. Thiocyanate induces cell necrosis and fibrosis in selenium- and iodine-deficient rat thyroids: a potential experimental model for myxedematous endemic cretinism in central Africa. Endocrinology. 2004;145:994–1002. doi: 10.1210/en.2003-0886. [DOI] [PubMed] [Google Scholar]
- 36.Ammann R., Jacobs P. Decubitus ulcers—how to measure the risks. Krankenpfl Soins Infirm. 1996;89:16–20. [PubMed] [Google Scholar]
- 37.Ammann R.W., Heitz P.U., Kloppel G. Course of alcoholic chronic pancreatitis: a prospective clinicomorphological long-term study. Gastroenterology. 1996;111:224–231. doi: 10.1053/gast.1996.v111.pm8698203. [DOI] [PubMed] [Google Scholar]
- 38.Gibson C.W., Yuan Z.A., Li Y., Daly B., Suggs C., Aragon M.A., Alawi F., Kulkarni A.B., Wright J.T. Transgenic mice that express normal and mutated amelogenins. J. Dent. Res. 2007;86:331–335. doi: 10.1177/154405910708600406. [DOI] [PubMed] [Google Scholar]
- 39.Swanton E., Holland A., High S., Woodman P. Disease-associated mutations cause premature oligomerization of myelin proteolipid protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 2005;102:4342–4347. doi: 10.1073/pnas.0407287102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barron M.J., Brookes S.J., Draper C.E., Garrod D., Kirkham J., Shore R.C., Dixon M.J. The cell adhesion molecule nectin-1 is critical for normal enamel formation in mice. Hum. Mol. Genet. 2008;17:3509–3520. doi: 10.1093/hmg/ddn243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knight A.S., Schutte B.C., Jiang R., Dixon M.J. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev. Dyn. 2006;235:1441–1447. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- 42.Wilkinson D.G., Bailes J.A., McMahon A.P. Expression of the protooncogene int-1 is restricted to specific neural cells in the developing mouse embryo. Cell. 1987;50:79–88. doi: 10.1016/0092-8674(87)90664-7. [DOI] [PubMed] [Google Scholar]
- 43.Brookes S.J., Lyngstadaas S.P., Robinson C., Shore R.C., Wood S.R., Kirkham J. Enamelin compartmentalization in developing porcine enamel. Connect. Tissue Res. 2002;43:477–481. doi: 10.1080/03008200290000862. [DOI] [PubMed] [Google Scholar]
- 44.Bonass W.A., Robinson P.A., Kirkham J., Shore R.C., Robinson C. Molecular cloning and DNA sequence of rat amelogenin and a comparative analysis of mammalian amelogenin protein sequence divergence. Biochem. Biophys. Res. Commun. 1994;198:755–763. doi: 10.1006/bbrc.1994.1109. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.