Abstract
Magmas, a conserved mammalian protein essential for eukaryotic development, is overexpressed in prostate carcinomas and cells exposed to granulocyte-macrophage colony-stimulating factor (GM-CSF). Reduced Magmas expression resulted in decreased proliferative rates in cultured cells. However, the cellular function of Magmas is still elusive. In this report, we have showed that human Magmas is an ortholog of Saccharomyces cerevisiae Pam16 having similar functions and is critical for protein translocation across mitochondrial inner membrane. Human Magmas shows a complete growth complementation of Δpam16 yeast cells at all temperatures. On the basis of our analysis, we report that Magmas localizes into mitochondria and is peripherally associated with inner mitochondrial membrane in yeast and humans. Magmas forms a stable subcomplex with J-protein Pam18 or DnaJC19 through its C-terminal region and is tethered to TIM23 complex of yeast and humans. Importantly, amino acid alterations in Magmas leads to reduced stability of the subcomplex with Pam18 that results in temperature sensitivity and in vivo protein translocation defects in yeast cells. These observations highlight the central role of Magmas in protein import and mitochondria biogenesis. In humans, absence of a functional DnaJC19 leads to dilated cardiac myophathic syndrome (DCM), a genetic disorder with characteristic features of cardiac myophathy and neurodegeneration. We propose that the mutations resulting in decreased stability of functional Magmas:DnaJC19 subcomplex at human TIM23 channel leads to impaired protein import and cellular respiration in DCM patients. Together, we propose a model showing how Magmas:DnaJC19 subcomplex is associated with TIM23 complex and thus regulates mitochondrial import process.
INTRODUCTION
Mitochondria are essential, complex organelles of eukaryotic organisms required for a variety of metabolic processes including the generation of energy by oxidative phosphorylation (1). Normal mitochondrial function requires 500–2000 different types of proteins depending on the species. However, mitochondrial genome of yeast and human cells encodes only 8 and 13 proteins, respectively (2,3). Thus, the vast majority of proteins that comprise mitochondria are encoded by the nuclear genome and mitochondrial function requires the import and folding of a large number of proteins synthesized on cytosolic ribosomes (4,5).
Owing to distinct compartmentalization into an outer and inner membrane, mitochondria have evolved an efficient system for recognition and transport of precursor proteins across membranes. Import of nuclear encoded proteins into the mitochondrial membranes is a multistep process involving machinery of cytosol, mitochondrial membranes and mitochondrial matrix (6–10). As a first step in the translocation process, the cytosolic facing receptors recognize the mitochondrial targeting sequence of a precursor protein and transfer them to the protein complex of the outer membrane, where Tom40 forms a pore and allows passage of the precursor protein through the membrane (6,8,10). After passage through TOM complex, a major portion of precursor proteins are targeted into mitochondrial matrix; and this process is mediated by presequence translocase (TIM23 complex) of the inner membrane (7–10). The TIM23 complex consists mainly two set of components: (i) channel-forming components which include integral membrane proteins Tim23 and Tim17 that comprise the translocation channel (9,11,12) and (ii) the associated peripheral membrane protein Tim50 (13). The movement of the presequence through the inner membrane requires a membrane potential (14,15), whereas import of the rest of the protein requires translocation channel associated ‘import motor machinery’ (7,9).
The yeast import motor consists of five essential subunits namely, mtHsp70 (Ssc1), Tim44, Pam18, Pam16, Mge1 (8,16–20), and two non-essential subunits, Pam17 and Tam41 (21–23). A critical core component of this machinery is the major mitochondrial 70 kDa heat shock protein (mtHsp70; Ssc1 in yeast), which binds short hydrophobic segments of incoming polypeptide chains (24,25). MtHsp70 is tethered to the import channel via its interaction with an essential peripheral membrane component of the channel, Tim44 (26). This interaction is destabilized upon binding a translocating precursor polypeptide (24,27).
Recently, two additional critical components of the import motor, a J-protein (Pam18) (28–30) and J-like protein (Pam16) (16,17,31) have been identified. Pam18 and Pam16 proteins are highly conserved (16,17). As expected of a J-protein, Pam18 stimulates Ssc1's ATPase activity and stabilizes the interaction with precursor proteins, thus carries out an essential function during the translocation process (28–30). Pam16 regulates Pam18's ATPase stimulating activity by forming a functional heterodimer through its C-terminal domain (31–33). A stable heterodimer is required for the protein translocation and viability of yeast cells (31,34,35). Pam18:Pam16 heterodimer is tethered to the translocon via multiple interactions with other components of the translocation channel and regulates the import motor activity (36). However, the precise mechanism of regulation of the import motor by Pam16 is not clearly understood.
The proteins homologous to yeast Pam16 have been reported in other organisms (37). A homologous deletion mutant in Drosophila was found to be lethal at the first instar larval stage (38). Magmas-like proteins are found essential for the development of murine and Caenorhabditis elegans (39,40). In humans, mitochondria-associated granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling molecule (referred as ‘Magmas’, ortholog of yeast Pam16) was first reported as a protein upregulated in PGMD1 cells cultured in a GM-CSF rich medium and not in cells exposed to Interleukin-3. Reduced Magmas expression in PGMD1 cells under GM-CSF resulted in decreased proliferative rates in a dose-responsive manner (41). During the developmental stages of animals, expression of Magmas was upregulated in muscle, testis, intestinal mucosa and liver (40). Magmas levels were also found to be upregulated in neoplastic prostate in humans, though its expression is restricted only to a subset of tumors (42). On the basis of these observations, it has been proposed that Magmas functions as a ‘signaling molecule’ and perhaps controls anaerobic metabolism, resistance to apoptosis or altered growth sensitivity in mammalian system (41–43).
Although Magmas levels are upregulated in different developmental stages and various pathophysiological conditions including prostate cancer, little functional information about human Magmas is available. On the basis of a weak sequence and predicted structural fold similarity with yPam16's J-like domain, we sought to determine the function of Magmas in humans. We found that Magmas is an ortholog of yeast Pam16 and has similar functions; and it complements the growth of yeast cells deleted for Pam16. Moreover, Magmas interacts with yeast Pam18 as well as human DnaJC19 (ortholog of yeast Pam18) both in vivo and in vitro conditions to form a heterodimeric subcomplex. The residues critical for the association of Magmas with yPam18, DnaJC19 and translocation channel were identified. Our results are consistent with the existence of stable interaction between Magmas and DnaJC19 that plays a crucial role in tethering of DnaJC19 at the translocon and perhaps regulating human import motor activity. We have identified the minimal region of DnaJC19 essential for its association with Magmas. Thus, our study underlines the possible molecular mechanism for the physiological symptoms of dilated cardiomyopathy with ataxia (DCM) syndrome which is associated with a truncated DnaJC19 protein.
RESULTS
Magmas complements growth of Δpam16 yeast strain
The J-like protein Pam16 functions in protein translocation in Saccharomyces cerevisiae and has been conserved throughout eukaryotic evolution (16,17). The sequence analyses of PAM16 revealed its presence as single or multiple copies of genes in all eukaryotes, supporting the idea that the mechanism of protein translocation across mitochondrial inner membrane has been maintained throughout eukaryotic evolution. A multiple protein sequence alignment of predicted Pam16 homologs of different species showed a sequence homology at the C-terminal region across genera (Fig. 1A). In Mus musculus and primates such as Macaca mulatta and Homo sapiens, the Pam16 homologs (also referred as Magmas) showed a high degree of sequence conservation between them. However, the sequence conservation between mammalian Magmas and S. cerevisiae Pam16 was only restricted to the predicted helix III of C-terminal region (Fig. 1A). At the level of protein sequence, Magmas shares ∼41% identity with S. cerevisiae Pam16. Magmas-like proteins are essential in multicellular organisms in various developmental stages (40), but the critical function has not been elucidated. To determine the function in mammalian system, we chose the human protein Magmas which shares weak sequence similarity with yeast Pam16. The primary structure of human Magmas contains 125 amino acid residues and possesses a three-dimensional structural fold similar to J-like domain of yeast Pam16 as reported earlier (32) (Fig. 1B). The J-like domain of Magmas consists of three helices and an exposed loop between tightly packed helix II and helix III with conserved aspartic acid, lysine, serine (DKS) motif (Fig. 1B).
Figure 1.
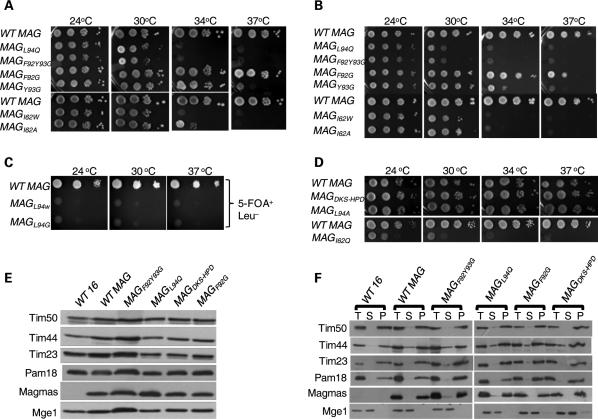
Sequence similarity and growth complementation analysis. (A) Magmas Sequence alignment. Predicted orthologous Magmas sequences from different species; S. cerevisiae (Sc), N. crassa (Nc), C. elegans (Ce), D. melanogaster (Dm), M. musculus (Mm), M. mulata (Mmu) and H. sapiens (Hs) are aligned using ClustalW program. Identical residues are shaded gray and the sequence corresponding to the histidine, proline, aspartic acid (HPD) motif of J-proteins is boxed. (B) Structure of Magmas J-like domain. Magmas J-like domain (47–125 aa) was modeled using yeast Pam16 structure co-ordinates (2GUZ). The N- and C-terminal of the domain is highlighted by N and C, respectively. The helices I, II and III are indicated by HI, HII and HIII, respectively. The amino acid alternations analyzed in this report are highlighted either in stick or ball and stick models. (C and D) In vivo growth phenotype analysis. Diploid Dpam16 cells were transformed with pRS415-MAGMAS construct and subjected to sporulation followed by tetrad dissection analysis. The spores from a single ascus were replica plated on His and Leu drop-out media for selection. Spores carrying the plasmid expressing WTMAGMAS allele (+) or WTPAM16 allele (−) are indicated. To further asses the growth complementation, a serially diluted PAM16ΔHIS/pRS315-WTPAM16 (WT 16) and PAM16ΔHIS/pRS415-WTMAGMAS (WT MAG) cells were spotted on yeast extract peptone dextrose (YPD) (Glu) and YP glycerol (Gly) media followed by incubation at the indicated temperatures for 3 days.
Deletion of PAM16 is lethal to yeast cells, indicating that Pam16 has a crucial role in maintaining cell viability (17,31). Therefore, to test whether MAGMAS is a true ortholog of PAM16, we have utilized two genetic approaches for the growth complementation analysis in yeast. First, heterozygous pam16 diploid of PJ53 yeast cells carrying a single chromosomal disrupted copy of PAM16(ΔHIS) was transformed with MAGMAS in pRS415 plasmid (44) and the cells were subjected to sporulation. Tetrads were dissected and replica plated for the selection of MAGMAS positive (HIS+ LEU+) spores. The viability of more than two spores indicates that MAGMAS rescues the growth of Δpam16 strain (Fig. 1C). Secondly, to further confirm the results, we have used plasmid shuffling to rescue the inviability of the Δpam16 strain. A plasmid (pRS415) carrying MAGMAS was transformed into a haploid Δpam16 strain carrying a wild-type copy of PAM16 on a centromeric plasmid having the URA3 gene. The resulting strains were plated on media containing 5-fluoroorotic acid (5-FOA), which selects for cells having lost the URA3-containing plasmid carrying the PAM16 gene. The viable cells were recovered, grown in rich media and subjected to drop test analysis using fermentable and non-fermentable growth media. Magmas showed complete growth complementation of Pam16 deleted yeast cells at all temperatures tested (Fig. 1D), suggesting that it is a true mammalian ortholog of yeast Pam16 carrying out identical function in humans.
Magmas localizes to the mitochondria
In yeast, Pam16 localizes to the mitochondria and is found associated with inner mitochondrial membrane (16,17). To characterize the biological functions of Magmas in human cells, we first set out to establish whether it displayed a similar subcellular localization. To address, we have used in vivo fluorescence imaging and in vitro subcellular fractionation analysis of mitochondria isolated from human cell lines and yeast cells. To determine whether Magmas localizes to the mitochondria in human cells, MAGMAS was cloned in the mammalian expression vector, pEGFP-N3 at the N-terminus and was expressed in HeLa cells. The cells were counter-stained with mitochondrion-specific dye, MitoTracker Red to stain the organelle. Upon confocal imaging analysis of GFP-Magmas fluorescence in HeLa cells, we observed total colocalization with MitoTracker (Fig. 2A), indicating Magmas distribution in the mitochondria. Similar results were obtained in HEK293T cells (data not shown). Our results are consistent with the previous observation in prostate tissue sections using immunogold electron microscopy (41).
Figure 2.
Magmas localization analysis. (A) Magmas localizes to the human mitochondria. Confocal fluorescence images of HeLa cells showing GFP-Magmas fusion protein (green fluorescence) and mitochondria (red) labeled with MitoTracker. (B and C) Subcellular fractionation of Magmas. Purified mitochondria from indicated yeast strains (B) and HEK293T cells (C) were resolved by SDS–PAGE followed by western blotting. The mitochondrial fractions were immunodecorated with respective yeast and human-specific antibodies as indicated. The purity and enrichment of mitochondria obtained from HEK293T cells were analyzed by probing with other organellar-specific antibodies as negative controls. For a comparison, equimolar concentration of total cell lysate and mitochondrial lysate was loaded. (D and E) Mitochondrial membrane association analysis of Magmas. Mitochondria from yeast cells (D) and HEK293T cell lines (E) were sonicated in the presence or absence of high salt (left panel) and carbonate extracted at pH 11.5 (right panel) was subjected to fractionation by ultra centrifugation. The equivalent samples of supernatant (S) and pellet (P) fractions, and unfractionated extract (T) were analyzed by SDS–PAGE and immunoblotted with yeast- and human-specific antibodies as indicated.
We next utilized subcellular fractionation to confirm our GFP fluorescence findings. Analysis of purified mitochondria from HEK293T and yeast cells, followed by immunodecoration with Magmas-specific antibody revealed the presence of a 13.7 kDa band of Magmas within the mitochondrial fraction (Fig. 2B and C). As positive controls, components of the inner membrane translocon such as Tim23, Tim44, Pam18 and Tim17 proteins were probed with specific antibodies against human and yeast, respectively. To ascertain the purity of the mitochondrial preparations obtained from HEK293T cells, immunoblotting was performed with antibodies against other organelle-specific markers such as, cathepsin D as a lysosomal marker, peroxisomal marker protein catalase and superoxide dismutase as a cytosolic/nuclear marker. When compared with control, negligible amounts of these proteins were detected in mitochondrial fraction. Together, these results support our GFP fluorescence analysis and indicate that Magmas localizes to the mitochondrial compartment of human and yeast cells, respectively.
Yeast Pam16 is found associated with the inner mitochondrial membrane compartment. To determine the association of Magmas with the inner mitochondrial membrane of human and yeast, we subjected the mitochondria for hypotonic swelling to generate the mitoplasts. The mitoplasts were gently sonicated in a buffer containing low or high salt followed by separation of soluble and membrane pellet fractions by ultracentrifugation. The samples were resolved on SDS–PAGE and immunodecorated with specific antibodies against respective yeast and human proteins. As expected, the Magmas was found associated with membrane pellet fraction like Tim44, Tim23, Tim17, yPam18 and DnaJC19, whereas soluble matrix protein Mge1 (in yeast) and hTid1 (in human) were released into the supernatant in both high and low salt conditions (Fig. 2D and E). These results are consistent with yeast Pam16's association with the inner membrane of the mitochondria. Upon treatment of human and yeast mitochondria at alkaline pH of 11.5, a considerable amount of Magmas was extracted from yeast, whereas in the case of human mitochondria, a significant amount of Magmas was extracted in the supernatant. However, multi-spanning integral membrane proteins such as Tim23 and Tim17 remained in the membrane fraction (Fig. 2D and E). As an internal control, similar results were obtained for yeast Pam16 (Fig. 2D). In summary, we conclude that like yeast Pam16, human Magmas is a peripheral mitochondrial inner-membrane protein that is exposed to the matrix space and lacks multi-spanning transmembrane segments in the primary structure.
Amino acid substitutions in the J-like domain of Magmas affects in vivo function
On the basis of preliminary genetic analysis and structural investigations, it was evident that Pam16 and yPam18 interact via their helix III to form a stable functional heterodimer (31,32,36). Previously, a temperature sensitive (Ts) mutation in Pam16's J-domain L97W, which lies at the beginning of the helix III region, was isolated. The pam16L97W mutant showed robust in vivo phenotypes including Ts at 34°C and decreased ability to form a stable heterodimer as tested by using mitochondria and in purified system (36). Besides, the helix III of Pam16 homologs are significantly conserved across species including human Magmas (Fig. 1A). On the basis of this evidence, we predicted that the interaction region between Magmas and DnaJC19 or yPam18 is also conserved. As the domain structures of Magmas and Pam16 are well conserved, we created identical mutations in Magmas sequence replacing leucine 94 with either tryptophan or glycine (Fig. 1B). The yeast strains expressing magmas mutants were generated via plasmid shuffling. We observed the yeast cells carrying magmasL94W and magmasL94G mutations (Fig. 1B) were inviable and unable to grow in plates containing 5-FOA (Fig. 3C). By a less drastic amino acid substitution at 94 position into alanine, magmasL94A cells (Fig. 1B) had no obvious growth phenotypes (Fig. 3D), whereas by a glutamine substitution, magmasL94Q (Fig. 1B) cells grew normal as wild-type upto 23°C, grew poorly at 30°C and were inviable at 34°C in rich and minimal media (Fig. 3A and B). Results clearly indicate that the leucine 94 of Magmas is critical for in vivo function. Similarly, we have created additional Magmas Ts mutants by replacing conserved residues at the base of the helix III region. We replaced two aromatic amino acids namely, phenylalanine (F92) and tyrosine (Y93) into glycine residues (Fig. 1B). The single mutant, magmasF92G cells allowed robust growth of Δpam16 cells at all temperatures tested in rich media (Fig. 3A). Although magmasF92G cells formed colonies at 37°C in minimal media, growth was slower when compared with that of wild-type (Fig. 3B). In the other single substitution mutant, magmasY93G cells grew normally upto 34°C and slower at 37°C when plated in rich media, while cells were Ts at 37°C in minimal media (Fig. 3A and B). When we combined both mutations together, as double mutant magmasF92Y93G, a drastic growth phenotype was observed, where the cells were inviable at all temperatures >23°C in both rich and minimal media (Fig. 3A and B). To address the importance of DKS sequence in the loop connecting helix II and helix III (Fig. 1B), we have created a triple mutant by replacing it with the canonical J-protein conserved histidine, proline, aspartic acid (HPD) sequence. The magmasDKS/HPD robustly supported the growth of Δpam16 cells at all temperatures, suggesting that these DKS amino acids are dispensable for in vivo function (Fig. 3D).
Figure 3.
Analysis of mutations affecting Magmas functions. (A–D) In vivo growth phenotypes. Ten-fold serial dilutions of wild-type and mutant cells were spotted onto YPD (A and D) and leucine drop-out media (B) or 5-FOA (C), followed by incubation at the indicated temperatures for 3 days. (E) Expression analysis of TIM23 translocon members in mutants. Equivalent amount mitochondria from wild-type and mutants were separated on SDS–PAGE and subjected to immunoblotting using indicated yeast TIM23 translocon components. (F) Membrane association analysis of Magmas mutants. Sonicated mitochondria from Magmas mutant yeast cells were subjected to ultracentrifugation. The equivalent samples of supernatant (S) and pellet (P), and unfractionated extract (T) were analyzed by SDS–PAGE and immunoblotted with yeast- and human-specific antibodies as indicated.
To aid the identification of residues outside the helix III region of Magmas J-like domain that are important for function, we mutated a highly conserved isoleucine at position 62 into either tryptophan or alanine or glutamine residue (Fig. 1B). The magmasI62W and magmasI62A cells showed normal growth upto 30°C, however no colonies were formed at temperatures >34°C (Fig. 3A and B). Interestingly, the isoleucine to glutamine substituted mutant yeast cells showed severe growth phenotypes as they grew very slowly at 23°C (Fig. 3D). Together, these results highlight the importance of I62 and the contribution of amino acid residues outside helix III for Magmas in vivo function.
To ascertain that the growth phenotypes of mutants are not due to the changes in expression levels, we have analyzed the mitochondria isolated from mutant strains grown at permissive temperatures. Equivalent amounts of mitochondria from the wild-type Magmas and mutants were separated on SDS–PAGE and subjected to immunodecoration with specific antibodies against Magmas and other components of the inner membrane translocon. We observed that the mutation did not alter the steady state levels of Magmas by itself, neither other members of the translocon that we tested (Fig. 3E). Results are in agreement with earlier reported Pam16's conditional mutants (36). To further assess their ability to interact with the inner mitochondrial membrane, we have subjected the mutant mitochondria, magmasL94Q, magmasF92G, magmasF92Y93G and magmasDKS/HPD for fractionation analysis. The membrane pellet and supernatant soluble fraction were separated by ultracentrifugation, followed by SDS–PAGE and immunodecoration with translocon-specific antibodies. Magmas mutants were separated into pellet fraction similar to wild-type and internal control, wt Pam16 (Fig. 3F). In summary, we conclude that the residues spanning the base of helix III region (F92, Y93, L94) and I62 are critical for Magmas in vivo function.
Magmas protein is essential for the import of precursor proteins into mitochondria
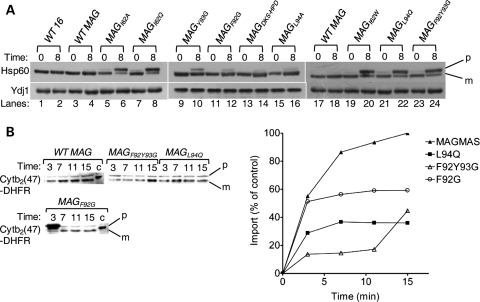
Pam16 is essential for protein translocation across mitochondrial inner membrane (31,45). To demonstrate whether Magmas has a role in mitochondrial protein import in vivo, accumulation of the non-processed precursor form of a nuclear encoded abundant mitochondrial protein, Hsp60, was monitored. This assay has been used in earlier studies to demonstrate the in vivo import process (30). After growing for 8 h at 37°C, wild-type Magmas yeast cells showed undetectable amounts of non-processed Hsp60 precursor similar to that of internal control, wt Pam16 (Fig. 4A, first two lanes). Above result confirms that Magmas completely restores Pam16's functions in yeast and efficiently interacts with the import machinery for the import of precursor proteins into the mitochondria. To further assess the role of Magmas in protein import, we have employed the Ts mutants for the precursor accumulation analysis. When the mutant cells, grown first at 23°C, were shifted to 37°C for 8 h to induce phenotype, they accumulated significant amounts of non-processed Hsp60 precursor form (Fig. 4A, lanes 6, 8, 20, 22, 24); indicative of in vivo import defects associated with the Ts mutants. However, magmasDKS/HPD and magmasL94A yeast cells did not show the accumulation of detectable levels of Hsp60 precursor form supporting their wild-type growth phenotypes (Fig. 4A, lanes 14, 16). The magmasF92G, and magmasY93G yeast cells showed an intermediate levels of precursor accumulation, consistent with the growth phenotypes (Fig. 4A, lanes 10, 12).
Figure 4.
Analysis of protein translocation in Magmas mutants. (A) In vivo precursor accumulation: wild-type and Magmas mutant yeast strains were grown at permissive temperatures in rich media to early log phase. The culture was shifted to 37°C for 8 h. After inducing phenotype, the whole cell lysates were resolved by SDS–PAGE followed by immunoblotting using Hsp60-specific antibodies; (p), precursor form of Hsp60; (m), mature form of Hsp60. (B) In vitro mitochondrial import of Cytb2(47)-DHFR; 100 mg of WT and mutant Magmas (MAGL94Q, MAGF92G and MAGF92GY93G) mitochondria were preincubated in import buffer and reactions were initiated by adding saturated amounts of purified Cytb2(47)-DHFR. Import reaction mixtures were separated on SDS–PAGE and analyzed by immunoblotting using anti-DHFR antibodies. One hundred percentage of the precursor protein offered to the mitochondria was loaded as a control (c). The amount of imported protein (m-Cytb2(47)-DHFR) was quantified using densitometry and plotted against the function of time of import reaction. The amount of m-Cytb2(47)-DHFR in WT mitochondria after the longest import time was set to 100%.
For a detailed kinetic analysis of protein translocation into mitochondria, we have chosen three Ts magmas mutants, namely L94Q, F92G and F92Y93G, to conduct in vitro import experiments using recombinant chemically purified precursor proteins with isolated mitochondria. Mitochondria were isolated from mutant yeast cells grown at permissive temperatures and pre-incubated for 15 min at 37°C to induce the mutant phenotype, and subjected for import reaction using saturating amounts of Cytb2(47)-DHFR precursor protein. When compared with wild-type, magmasL94Q and magmasF92Y93G mitochondria showed substantial reduction in the kinetics of import of Cytb2(47)-DHFR precursor protein, whereas magmasF92G showed an intermediate import defect (Fig. 4B). In summary, the import defects associated with the Magmas conditional mutants are consistent with the growth phenotypes.
Magmas interacts with yPam18 and DnaJC19 to form a stable subcomplex
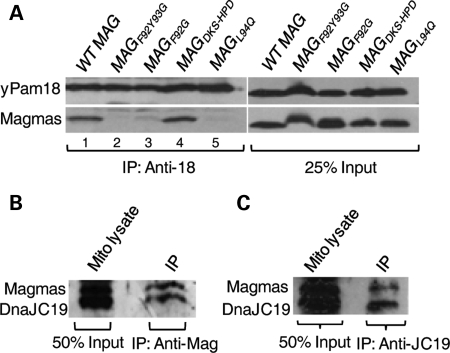
In yeast, previously it has been shown that Pam16 forms a stable heterodimeric subcomplex with yPam18 in the mitochondrial lysates (31,36). The stability of the subcomplex is critical for the translocation process in yeast (36). Therefore, we asked whether Magmas forms a similar subcomplex with yPam18 in yeast mitochondrial lysates. To address, we have purified mitochondria expressing wt Magmas or mutants in Δpam16 yeast strains. Mitochondrial extracts were prepared using 1% Triton X-100 to dissociate channel components, and then subjected for coimmunoprecipitation (CoIP) analysis using cross-linked yPam18 antibody agarose beads. As expected, Magmas co-precipitated with yPam18 from extracts of wild-type mitochondria, indicating that Magmas can form a similar stable subcomplex (Fig. 5A, lane 1). Significantly reduced co-precipitation was observed from magmasF92Y93G, magmasF92G and magmasL94Q extracts, highlighting the importance of these residues for the subcomplex formation in vivo (Fig. 5A, lanes 2, 3, 5). A wild-type level of co-precipitation was observed from magmasDKS/HPD extracts, suggesting that the DKS motif is dispensable for the subcomplex formation with yPam18 in vivo (Fig. 5B, lane 4). These results are consistent with their apparent lack of growth and translocation phenotypes. To demonstrate whether Magmas forms a similar subcomplex in human mitochondria, the mitochondrial lysates were prepared from HEK293T cells and subjected to CoIP analysis using specific antibodies against Magmas and DnaJC19. Regardless of the type of antibodies used, equivalent amounts of co-precipitation of Magmas with DnaJC19 (yPam18 ortholog) supports the existence of a similar subcomplex in humans (Fig. 5B and C).
Figure 5.
Analysis of mutations affecting stability of the Magmas:Pam18/DnaJC19 subcomplex. (A) Co-immunoprecipitation of Magmas:yPam18 subcomplex from yeast mitochondria. Wild-type and mutant Magmas yeast mitochondrial lysate were prepared using 1% Triton X-100. The lysate was subjected to immunoprecipitation using Pam18-specific antibodies, as indicated by bracket, followed by SDS–PAGE and immunoblotting using Pam18- and Magmas-specific antibodies. Twenty-five percentage of soluble material after lysis was used as loading control (25% input). (B and C) Analysis of Magmas:DnaJC19 subcomplex formation in human mitochondria. Human mitochondrial lysate was prepared with 1% Triton X-100 then subjected to immunoprecipitation using Magmas-antibodies (B) or DnaJC19-antibodies (C), followed by SDS–PAGE and immunoblotting using DnaJC19- and Magmas-specific antibodies. Fifty percentage of total soluble material after lysis was used as a loading control (50% input).
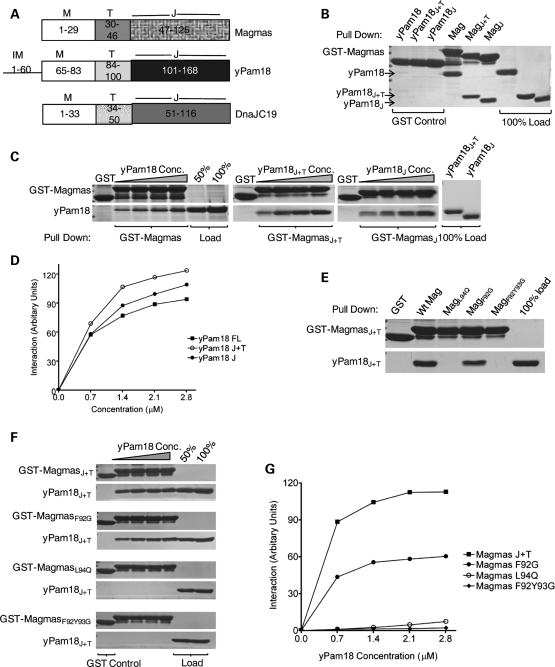
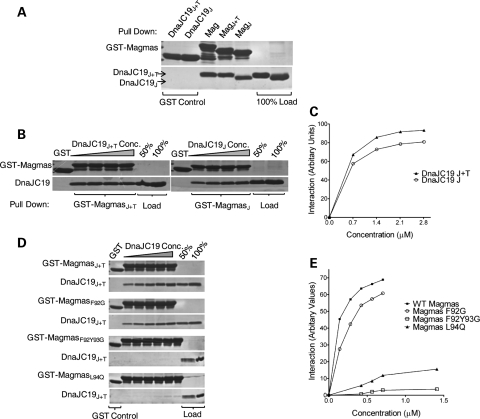
To further confirm our in vivo findings, we assessed whether Magmas is capable of forming a subcomplex with yPam18 in vitro by glutathione S-transferase (GST) pull-down analysis using purified proteins. Consistent with results obtained in mitochondrial lysates, we found that upon mixing equimolar amounts of full-length Magmas and yPam18, >90% of yPam18 was pulled down using full-length GST-Magmas. In order to investigate the molecular basis of DCM syndrome, we employed in vitro pull-down assay and generated N-terminal truncations of DnaJC19 to determine the region essential for its interaction with Magmas. Similar truncations were also generated for yPam18 based on previous reports that Pam16 interacts with yPam18 through their C-terminal-related J-domains. Therefore, to determine the minimal region of Magmas required for the interaction with yPam18 and DnaJC19, we mixed J + T and J-domain fragments (Fig. 6A) of yPam18 and DnaJC19 together with Magmas and performed GST pull-down analysis. As with full-length protein, a similar pull down was observed for these smaller fragments containing the J + T and J-domains (Figs 6B and 7A). To determine the relative affinities of these fragments, we have incubated GST-bound Magmas (full-length, J + T and J-like domain) with increasing concentrations of full-length, J + T and J-domains of either yPam18 or DnaJC19 followed by pull-down analysis. By quantitative analysis, we observed that the fragments of Magmas interacted with yPam18 and DnaJC19 fragments with nearly similar affinities when compared with full-length proteins (Figs 6C, D and 7B, C). These findings clearly indicate that the subcomplex formation is mediated through their related J-domains.
Figure 6.
In vitro interaction analysis of Magmas with yPam18. (A) Domain organization of Magmas, Pam18 and DnaJC19. On the basis of sequence alignment, the amino acids corresponding to the predicted regions are indicated; intermembrane space (IM), membrane association (M), predicted mitochondria targeting (T) and J-domains/J-like domains (J). (B–D) Magmas interacts with the J-domain of yPam18. Immobilized GST-Magmas, GST-MagmasJ+T and GST-MagmasJ were incubated with equimolar concentration (B) or increasing concentrations (C) of his6-tagged full-length Pam18, Pam18J+T and Pam18J, respectively. The bound proteins were washed and analyzed by SDS–PAGE followed by Coomassie dye staining, and quantified by densitometry (D). GST alone was used as a negative control, and 100% input of Pam18, Pam18J+T, PAM18J used as a loading control. (E–G) Magmas mutants are defective in forming subcomplex with yPam18. Equimolar concentration (E) or increasing concentrations (F) of yPam18 was incubated with GST-Magmas, GST-MagmasL94Q, GST-MagmasF92G and GST-MagmasF92Y93G beads for 30 min for complex formation. The beads were washed and separated on SDS–PAGE gel, followed by Coomassie dye staining and quantified by densitometric analysis (G).
Figure 7.
In vitro interaction analysis of Magmas with DnaJC19. (A–C) Magmas interact with the J-domain of DnaJC19 protein. The interaction analysis between GST-Magmas and DnaJC19 was carried out essentially as described in Figure 6B–D. (D and E) Magmas mutants are defective in forming subcomplex with DnaJC19. GST-Magmas, GST-MagmasL94Q, GST-MagmasF92G and GST-MagmasF92Y93G were incubated with increasing concentrations of DnaJC19 for 30 min to form the subcomplex. The samples were analyzed as mentioned in Figure 6F and G.
We next evaluated whether the amino acid alterations in Magmas J-like domain affects its binding to yPam18. To address, we have utilized Ts mutants of Magmas which were found defective in forming a stable subcomplex with yPam18 in mitochondrial lysates. Similar amino acid substitutions were carried out in MagmasJ+T and subjected to GST pull-down analysis using yPam18J+T protein. When compared with wild-type, in magmasL94Q and magmasF92Y93G undetectable levels of yPam18 binding, whereas in the case of magmasF92G, significant reduced interaction was observed (Fig. 6E). These results are in agreement with our earlier findings of mitochondrial lysates, where Ts mutants fail to form a stable subcomplex. To determine the relative affinities for the binding, we have incubated GST bound Magmas mutants with increasing concentrations of yPam18J+T and subjected to GST pull-down analysis. A 2-fold reduction in the binding affinity was observed for magmasF92G, whereas a significant reduction in the affinities for Pam18 binding was observed in magmasL94Q and magmasF92Y93G mutants (Fig. 6F and G). To test whether Ts mutants have a similar interaction defects with human Pam18 ortholog DnaJC19, we have conducted a similar GST pull-down analysis in vitro. As expected, a similar reduction in the relative affinities for DnaJC19 binding was observed for the mutants (Fig. 7D and E). Together, these results suggest that Magmas interacts with yPam18 and DnaJC19 through its J-like domain and forms a stable subcomplex in vivo and in vitro condition. The stability of the subcomplex is critical for the protein translocation and cell viability.
Magmas is a part of presequence translocase of the inner mitochondrial membrane
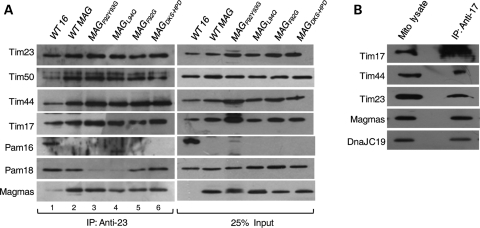
The translocon of the inner mitochondrial membrane is composed of two components—the TIM23 core channel and the import motor (8). Previously, it has been shown that yeast Pam16 is a part of import motor and precisely tethers yPam18 to the translocon so that it efficiently stimulates ATPase activity of mtHsp70 (36,45). To demonstrate whether Magmas forms a part of the translocon machinery and is involved in the interaction with yPam18 of TIM23 complex, mitochondria were isolated from yeast strains expressing wild-type Magmas and Ts mutants. Purified mitochondria were lysed with digitonin and the supernatant was subjected to CoIP analysis using cross-linked Tim23 antibody agarose beads. As expected from earlier studies, both yPam18 and Pam16/Magmas were equally co-precipitated from wt Magmas and internal control wt Pam16 extracts along with other components of the translocon including Tim17, Tim50 and Tim44 (Fig. 8A, lanes 1 and 2). However, the levels of yPam18 co-precipitated from magmasL94Q and magmasF92Y93G lysates were significantly reduced (Fig. 8A, lanes 3 and 4). A 2-fold reduction was observed in the case of magmasF92G mitochondrial lysate, whereas magmasDKS/HPD showed wild-type levels of yPam18 association with the translocon (Fig. 8A, lanes 5 and 6). Interestingly, in all the mutants, the levels of Magmas association were roughly similar to that of wild-type mitochondria (Fig. 8A). These results are in agreement with earlier reported observations made for pam16L97W (36). Together, we conclude that a mutational defect in Magmas C-terminal region impairs the association of yPam18 with the translocon.
Figure 8.
Magmas association with TIM23 translocase. (A) Magmas interaction with yeast Tim23-core complex. Equivalent amounts of mitochondria from yeast cells expressing wt Magmas or mutants were solubilized by incubating in buffer containing 1% digitonin. The soluble supernatant was subjected to co-immunoprecipitation using Tim23 specific antibodies. Samples were resolved on SDS–PAGE and immunodecorated with antibodies specific to Magmas-, yPam16-, yPam18-, yTim17-, yTim23-, yTim44- and yTim50. (B) Magmas association with human Tim23-core complex. Mitochondria obtained from HEK293T cells were lysed in 1% digitonin, and the supernatant was subjected to immunoprecipitation using anti-Tim17 antibodies. Samples were analyzed on SDS–PAGE, followed by immunoblotting with antibodies against Magmas-, DnaJC19-, hTim17-, hTim23- and hTim44. Twenty-five percentage of the total soluble supernatant after lysis was used as loading control (25% input).
To demonstrate existence of a similar system in human mitochondria, immunoprecipitation of the translocon was carried out with digitonin-lysed mitochondria isolated from HEK293T cells using α-Tim17-conjugated beads. Pull down of Magmas along with other translocon components such as hTim44, hTim23, DnaJC19 provides direct evidence for the association of Magmas with the human translocon (Fig. 8B). Therefore, we propose that the subunits' organization of inner mitochondrial translocon is well conserved during evolution.
Magmas inhibits ATPase stimulatory activity of Pam18 and DnaJC19
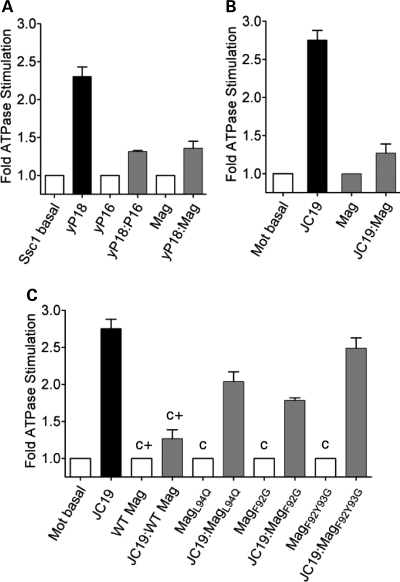
Pam18 is a J-protein and critical component of the import motor tethered at the translocon to stimulate Ssc1's ATPase activity (30). Pam16, a J-like protein negatively regulates the import motor activity (Ssc1's ATPase activity) by forming heterodimeric complex with yPam18 through their related J-domains (34). However, the physiological significance of such negative regulation is not clearly understood. To test whether Magmas regulates yPam18's ATPase stimulatory activity, we have incubated preformed radioactive Ssc1–ATP complex with different molar ratio of purified proteins; yPam18 alone, Magmas alone and yPam18:Magmas complex. As an internal control, Pam16 alone and Pam16:yPam18 complex were used to monitor the stimulation of Ssc1's ATPase activity under single turnover conditions. As expected, Pam16:yPam18 heterodimer was 40–50% as active as yPam18 alone in stimulating Ssc1's ATPase activity over different ratios of heterodimer concentrations (Fig. 9A). Fifty to 60% ATPase stimulating activity was retained by Magmas:yPam18 complex when compared with yPam18 alone over a range of complex concentration (Fig. 9A), consistent with the previous observations (33). A comparable amount of inhibition by Pam16 and Magmas indicates that both function in similar mechanism in regulating the import motor activity.
Figure 9.
Inhibition of mtHsp70's ATPase activity. (A) Effect of yPam18:Magmas subcomplex: Ssc1:ATP(a-32P) complex was incubated with 2-fold excess of either yPam18 (yP18) alone, Magmas alone or yPam18:Magmas subcomplex and the rate of hydrolysis was measured as a function of time at 23°C. The experiment was repeated using yPam16 (P16) as a positive control. (B) Effect of DnaJC19:Magmas subcomplex: 2-fold excess of DnaJC19 (JC19) alone, Magmas alone or DnaJC19:Magmas subcomplex was incubated with the human Mortalin:ATP(a-32P) complex and the rate of ATP hydrolysis was monitored at identical conditions as mentioned earlier. The basal rate of ATP hydrolysis by Mortalin alone was set to 1. The rate of conversion to ADP was measured and represented as fold stimulation over the basal rate. (C) Effect of DnaJC19:Magmas mutant subcomplex: Human Mortalin:ATP(α-32P) complex was incubated with 2-fold excess of DnaJC19 alone, Magmas mutants alone (negative control; c) or DnaJC19:Magmas mutant subcomplexes as indicated. Magmas alone and Magmas:DnaJC19 subcomplex were used as a positive controls (c+); as mentioned in B. The assay was carried out and represented as described above. The assays were carried out in duplicates and the standard deviation was represented as standard error bars.
To confirm whether Magmas regulates the human import motor activity in a similar mechanism, we have purified human Hsp70 (Mortalin) and human Pam18 ortholog DnaJC19 and subjected to ATPase stimulation analysis under single turnover conditions. By pre-incubating different molar ratios of Magmas:DnaJC19 complex and DnaJC19 alone, we have observed upto 70% reduction in ATPase stimulating activity of human Mortalin by Magmas:DnaJC19 complex (Fig. 9B). To further support the CoIP analysis, all Magmas mutants were found defective in inhibiting DnaJC19 ATPase stimulation (Fig. 9C). A total recapitulation of results obtained from yeast and human system thus provide further evidences for the mechanism of regulation of human import motor by Magmas during the translocation process.
DISCUSSION
Protein transport is a highly regulated process that depends on the critical functioning of inner mitochondrial ‘import motor’ components (8,9). The proteins related to the yeast import motor are conserved in mammalian mitochondria including humans (46). Depending upon the metabolic state of the cell, the expression of human import motor components is highly regulated, thus controlling the import process. The mechanism of regulation of the import process in humans is largely unknown; however, altered regulation leads to severe mitochondrial disorders including neuromuscular diseases and malignancy (47–49). Mammalian Magmas proteins are ubiquitously expressed; and in humans, it was identified as GM-CSF specific signaling molecule which gets overexpressed in neoplastic prostate (41,42). Although it has been assumed that Magmas proteins are predicted homologs of yeast Pam16 (16,17), the primary function and mechanism of regulation in mammalian system was still elusive. The data presented here on human Magmas provides first experimental evidences to show that the mammalian Magmas proteins are orthologs of yeast Pam16 having similar functions and are essential part of mammalian import motor. The primary structure of human Magmas is related to yeast Pam16 and share a common domain organization. It consists of predicted N-terminal membrane association domain (TM), middle targeting region (T) and C-terminal J-like domain (J). However, it lacks C-terminal extension region of Pam16 which is functionally dispensable in yeast (32).
Human Magmas is an ortholog of yeast Pam16 in several aspects and performs the similar function. Several biochemical and genetic evidences are presented here to support this idea. First, it shows a complete growth support of yeast cells deleted for essential PAM16 gene at all conditions. These observations highlight a possible similar essential functional role for Magmas in humans. Second, it localizes into mitochondria when expressed in Δpam16 cells as well as in human HeLa and HEK293T cells when expressed as GFP fusion proteins. These observations were further supported by enrichment of the protein levels in purified mitochondria from yeast and human cell lines, respectively. Third, in both yeast and human, Magmas is tightly associated with the inner mitochondrial membrane. Similar to Pam16, Magmas was fractionated into inner membrane pellet fraction and resistant to high salt extraction. However, majority of Magmas was extracted from pellet fraction at alkaline pH, indicating that it is a peripherally associated with inner mitochondrial membrane protein. Fourth, like Pam16, Magmas is associated with mitochondrial TIM23 complex in Δpam16 yeast cells as well as in humans. Fifth, Magmas regulates the ATPase stimulation activity of yPam18 as well as DnaJC19 in humans by forming a stable subcomplex through their C-terminal regions, thus regulating Ssc1 and human mtHsp70 (Mortalin) activity during the import process. Sixth, like Pam16, Magmas plays an essential role in import of precursor proteins into the mitochondrial matrix. Previously, it has been shown that downregulation of Pam16 level leads to the accumulation of precursor form of Hsp60 in yeast cells suggesting its essential role in protein import (16,17). Similarly, Δpam16 yeast cells expressing wt Magmas did not show the accumulation of precursor form of Hsp60 when exposed to non-permissive temperature, indicating that Magmas can complement the import function of Pam16. In addition to that, Magmas also supported a wild-type level of protein import of model preprotein, Cytb2(47)-DHFR into the Δpam16 mitochondria.
Our results demonstrate that Magmas protein interacts with yPam18 and DnaJC19 via their J-like and J-domains to form a stable subcomplex. The J-like and J-domain of Magmas and yPam18/DnaJC19, respectively, are competent to form a subcomplex with similar affinities and a stable Magmas:yPam18 and Magmas:DnaJC19 subcomplex was co-immunoprecipitated in yeast and human mitochondrial lysates. Formation of a similar subcomplex between Pam16 and Pam18 was reported earlier in yeast. Hence, our findings clearly broaden the idea of evolutionary conserved existence of such subcomplex from yeast to human systems.
A stable interaction between J-like domain of Magmas and J-domain of yPam18/DnaJC19 is required for in vivo function. A single amino acid alteration in helix I and helix III region of the J-like domain of Magmas compromises growth of yeast cells and is associated with an unstable Magmas/yPam18 subcomplex. The mutant proteins also showed a dramatic decrease in affinity for their interaction with yPam18 and human Pam18 ortholog DnaJC19 proteins; envisioning a similar defect in subcomplex formation in human mitochondria. The corresponding in vivo and in organellar protein translocation defect associated with the Magmas mutants is consistent with the idea that the protein import and growth defects of cells expressing the mutant proteins are caused by instability of the subcomplex.
Our findings support the hypothesis of Magmas playing a critical role in positioning of J-protein, Pam18/DnaJC19 to the translocon and perhaps regulating the import motor activity in mammalian system. In yeast, it was demonstrated that Pam16 plays a crucial role in tethering Pam18 to the translocon possibly by interacting through Tim44 with its N-terminal domain (36). Although the Ts mutants of Pam16 showed a compromised heterodimer subcomplex formation, a significantly reduced interaction of Pam18 at the translocon was observed, but Pam16 association remained unaffected (36). Supporting this data, Magmas Ts mutants also showed a similar reduced co-precipitation of yPam18, but not of Magmas with the translocon when Magmas:yPam18 subcomplex formation was compromised. Owing to compromised heterodimer formation, the Magmas mutants were found to be defective in regulating ATPase activity of human import motor. This is consistent with an important role for Magmas in tethering yPam18/DnaJC19 to the translocon. On the basis of our analysis, we conclude that both Magmas and Pam16 share a common function across species and evolved precisely to alter Pam18's level at the translocon by forming subcomplex through their C-terminal regions, thus regulating the mitochondrial activity.
Although we have assigned a primary function for Magmas, understanding the mechanisms of regulation of Magmas as well as DnaJC19 is of utmost importance for the mitochondrial physiology. There are numerous connections between altered Magmas levels and DnaJC19's function with mitochondrial pathophysiology: (i) Magmas levels is highly upregulated in prostate carcinoma and in response to GM-CSF treatment (41,42). (ii) Truncated DnaJC19 (devoid of its J-domain) protein causes a severe genetic disorder called DCM syndrome characterized by respiratory, cardiac and neurological symptoms (47). DCM syndrome is an autosomal recessive disorder prevalent in Canadian and ancestral European population and an IVS3-1G→C mutation was identified upon genotyping the disease haplotypes (47). The mutation causes abnormal splicing that result in the truncation of exon 4 followed by a stop codon. Translation of the Δexon 4 transcript produces a protein having a truncated J-domain (47). On the basis of our studies, we propose the molecular etiology underlying the disorder. As a functional J or J-like domain is essential for the formation of a stable heterodimer, we predict that the truncated protein is defective in forming a functional Magmas:DnaJC19 subcomplex and hence, the interaction of DnaJC19 with human TIM23 complex gets compromised. This leads to the deregulation of import motor activity causing improper protein import thereby, affecting mitochondria biogenesis and cellular respiration. The consequent impairment of energy metabolism in the cell especially in mitochondria-rich cardiac and neuronal tissues results in symptoms such as cardiomyopathy, peripheral neurodegeneration and ataxia. However, the relevance of upregulated Magmas levels other than protein import function is not well established, underlines the importance of future investigation.
MATERIALS AND METHODS
Yeast strains, plasmids construction and genetic analysis
MAGMAS and DNAJC19 gene were amplified from cDNA library of HeLa cells (Stratagene). MAGMAS was cloned in pRS415 yeast expression vector under TEF promoter. Yeast strain deleted for PAM16 has been previously described (31). The haploid PAM16ΔHIS/pRS415-WTMAGMAS yeast strain was obtained by transforming pRS415-WTMAGMAS construct into the diploid Δpam16 cells and subjected to sporulation in potassium acetate plates for 5 days followed by tetrad dissection analysis. The viable spores were grown at 30°C in yeast extract peptone dextrose (YPD) media for 2 days and replica plated on drop-out media (His+, Leu+) for selection. In a similar way, the haploid PAM16ΔHIS/pRS316-WTPAM16 yeast strain was generated and selected on (His+, Ura+) drop-out media.
To obtain temperature sensitive (Ts) mutants, a series of point mutations in pRS415-MAGMAS were created through site-directed mutagenesis using the QuikChange protocol (Stratagene). Δpam16 carrying pRS316-WTPAM16 was transformed with pRS415-MAGMAS mutants and incubated at 30°C on leucine omission plates. Transformants were streaked onto 5-FOA plates (US Biologicals) to select for candidates that could grow at permissible temperatures but not at non-permissible temperatures. The Ts mutants were rescued on rich media and subjected to drop test analysis on YPD, leucine omission plates and YPG to confirm the growth phenotypes. All in vivo experiments were carried out in the W303 genetic background in derivatives of PJ53 (Genotype: trp1-1/trp1-1 ura3-1/ura3-1 leu2-3,112/leu2-3/112 his3-11,15/his3-11,15 ade2-1/ade2-1 can1-100/can1-100 GAL2+/GAL2+ met2-Δ1/met2-Δ1 lys2-Δ2/lys2-Δ2).
GST-fusion full-length, J + T and J-domain constructs of MAGMAS were generated by introducing the EcoRI–XhoI site at the C-terminal end of GST in pGEX-KG vector (50). Hexahistidine-tagged full-length, J + T and J-domain constructs of MAGMAS and DNAJC19 were generated by cloning them into pET3a and pET21d vector (Novagen), respectively.
Cell culture and in vivo imaging
HeLa and HEK293T cells were cultured in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Gibco) and 1% penicillin–streptomycin (Sigma). The cells were incubated at 37°C in 5% CO2. The adherent cultures of HEK293T cells were grown to ∼90% confluency. The cells were trypsinized and centrifuged at 600g for 5 min. The cell pellet was washed with ice-cold PBS and subjected to mitochondria isolation. For in vivo imaging analysis, the gene encoding Magmas was cloned at the 5′-terminus of GFP in mammalian plasmid expression vector pEGFP-N3 (Clontech). One microgram of transgene was transfected into HeLa and HEK293T cell lines using Lipofectamine 2000 (Invitrogen) as described (51). The cells were cultured overnight and were stained with 100 ng/ml of MitoTracker Red 580 (Invitrogen) in complete medium at 37°C in a CO2 incubator for 45 min. Cells were washed twice with PBS before analysis without fixation. Images were acquired using Zeiss LSM 510 Meta and 63× NA 1.4 objective lens. The images were processed using LSM software.
Fractionation of mitochondria
Yeast or human mitochondria, corresponding to 0.5 mg/ml of total mitochondrial protein was subjected to fractionation by hypotonic swelling in sonication buffer [30 mm HEPES–KOH pH 7.6, 150 mm KCl, 10 mm magnesium acetate and 1 mm phenylmethyl sulfonyl fluoride (PMSF)] for 10 min followed by sonication (three times, 15 s) at 30% duty per cycle. For fractionation experiments in high salt conditions, 500 mm NaCl was included in the sonication buffer. Extraction of peripheral membrane proteins was performed by incubating the mitochondria in 0.1 m Na2CO3 pH 11.5 for 20 min. The samples were then separated into membrane and soluble fractions by ultracentrifugation at 100 000 g (TLA 120.2 rotor) for 1 h at 4°C in Optima™ TLX table-top ultracentrifuge (Beckman-Coulter). The supernatant fraction was precipitated using 12.5% trichloroacetic acid and was subsequently washed in ice-cold acetone. Fractionated samples were separated on SDS–PAGE and subjected to immunoblot analysis using polyclonal antibodies specific for Magmas, yPam18, yTim23, yTim44, yTim50 and yMge1. In the case of mitochondria isolated from HEK293T cells, the mitochondrial fractions were analyzed using polyclonal antibodies specific to Magmas, DnaJC19, hTim17 and hTid1 and monoclonal antibodies specific for hTim23 and hTim44.
Mitochondrial CoIP analysis
The interaction between Magmas and yPam18 or DnaJC19 was analyzed under conditions where they are separated from the translocon in both yeast and human mitochondrial lysates. Lysates from 2 mg of wild-type or mutant yeast mitochondria were prepared in lysis buffer (20 mm MOPS–KOH, pH 7.4, 250 mm sucrose, 80 mm KCl, 5 mm EDTA and 1 mm PMSF) in the presence of 1% Triton X-100 (US Biochemicals) on ice for 30 min by gentle vortexing. Lysates were centrifuged for 15 min at 16 000g at 4°C. Twenty microliter (bed volume) of saturated Pam18 cross-linked antibody beads were incubated with the supernatants for 1 h at 4°C. The beads were washed four times with mitochondrial lysis buffer containing 1% Triton X-100. Samples were separated by SDS–PAGE, followed by immunoblot analysis using antibodies against Magmas and yPam18. In the case of HEK293T mitochondrial lysates, soluble supernatants were incubated with 20 µl Magmas- or DnaJC19-specific antibody and subjected to CoIP analysis as described earlier. The samples were resolved in SDS–PAGE and subjected to western analysis. The blot was probed with DnaJC19- and Magmas-specific antibodies separately and sequentially, followed by secondary antibody decoration.
For analysis of the interaction of Magmas with the yeast or human translocon, 2 mg of wild-type or mutant mitochondria were gently lysed and solubilized in 1 ml of lysis buffer (25 mm Tris, pH 7.5, 10% glycerol, 80 mm KCl, 5 mm EDTA and 1 mm PMSF) in the presence of 1% digitonin (Calbiochem) at 4°C for 50 min using nutator. Lysates were centrifuged for 30 min at 16 000g for 30 min at 4°C; 15 µl (bed volume) of saturated cross-linked Tim23 (for yeast) and Tim17 (for human) antibody beads were incubated with supernatants of the lysates for 2 h at 4°C. The beads were washed four times with the corresponding mitochondrial lysis buffer containing 0.1% digitonin. Samples were resolved on SDS–PAGE, followed by immunoblot analysis using antibodies against yPam16, Magmas, yPam18, yTim17, yTim50 and yTim23. In the case of human mitochondrial CoIP, the immunoblotting was done against human-specific antibodies such as Magmas, DnaJC19, hTim23, hTim44 and hTim17.
Protein purification
For purification of J + T and J-domains Magmas, a six histidine tag was introduced at the N-terminus of the respective constructs. Overexpression was carried out in Escherichia coli (C41) (52) by allowing growth at 24°C to an A600 of 0.5, followed by induction using 0.5 mm IPTG for 5 h. Protein was purified by standard affinity chromatography using Ni-NTA agarose (GE Healthcare). One liter cell pellet was resuspended in 10 ml of buffer A (20 mm HEPES pH 6.8, 40 mm imidazole, 0.5 m NaCl and 5% glycerol) containing 0.2 mg/ml lysozyme and protease inhibitor cocktail followed by incubation at 4°C for 1 h. The sample was gently lysed with 0.2% DOC followed by DNase I (10 µg/ml, 10 mm MgCl2) treatment for 15 min at 4°C. The cell lysate was clarified by centrifuging at 28 000g for 30 min at 4°C. The soluble supernatant was incubated with 500 µl bed volume of Ni-NTA agarose for 2 h at 4°C. Unbound proteins were removed by washing three times with 15 ml of buffer A. The resin was washed with 15 ml of buffer B (20 mm HEPES pH 6.8, 80 mm imidazole, 1 m NaCl and 5% glycerol) at 4°C. To remove bacterial DnaK contamination, the resin was washed with 15 ml of buffer C (20 mm HEPES pH 6.8, 80 mm imidazole, 0.5 m NaCl, 0.1 mm ATP, 10 mm MgCl2 and 5% glycerol) at 4°C. Bound proteins were eluted with buffer D (20 mm HEPES pH 6.8, 250 mm imidazole, 250 mm NaCl and 5% glycerol) and dialyzed against buffer appropriate for use in particular experiments.
Full-length yPam18, DnaJC19J+T and DnaJC19J proteins were purified from insoluble fraction. The pellet fraction was solubilized by incubating with 10 ml of buffer E (buffer A + 1% Triton X-100, 500 mm KCl and 3 m urea) for 2 h at 4°C. The soluble fraction was further clarified by centrifuging at 28 000g for 45 min at 4°C. The supernatant was incubated with Ni-NTA agarose beads for 2 h at 4°C. The protein bound beads were washed two times with 15 ml buffer E, followed by 15 ml of buffer F (buffer A + 0.5% Triton X-100, 250 mm KCl and 2 m urea) at 4°C. The resin was further washed with 15 ml of buffer G (buffer A + 0.5% Triton X-100, 250 mm KCl and 1 m urea) and two times with buffer H (buffer A + 0.5% Triton X-100 and 250 mm KCl). The bound proteins were eluted with buffer I (buffer A + 0.5% Triton X-100, 150 mm KCl and 300 mm imidazole) and dialyzed against buffer appropriate for use in defined experiments.
A hexahistidine tag was introduced at the N-termini of wild-type yeast Pam18, Pam18J+T, Pam18J and C-terminus of the wild-type Pam16 proteins for purification from E. coli. The protein purification is done essentially according to the procedure as described (31,36). The full-length GST-Magmas, GST-MagmasJ+T, GST-MagmasJ and mutant proteins were purified according to the published protocols with minor modifications (50). The buffer components have been changed to 20 mm Tris, pH 7.5 instead of phosphate buffer in GST-fusion protein purification protocols. The Ssc1His (24) and human Mortalin (53) were purified from yeast as described.
In vitro GST pull-down analysis
Equimolar concentration of purified full-length GST-Magmas, GST-MagmasJ+T and GST-MagmasJ were incubated with 10 µl bed volume of glutathione-agarose beads (GE Healthcare) in 150 µl GST-buffer (50 mm Tris pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.2% Triton X-100 and 1 mm PMSF). Unbound proteins were removed by washing the beads two times with GST buffer. The samples were blocked with 0.1% BSA for 20 min at 23°C followed by washing two times with GST-buffer. The beads were resuspended in 200 µl GST binding buffer and incubated with equimolar or increasing concentration of either full-length, J + T, J-domains of yPam18 or DnaJC19 for half an hour at 23°C. The GST beads were washed three times with GST-buffer and resolved on SDS–PAGE followed by Coomassie dye staining. A similar protocol has been employed for the GST pull-down analysis of purified GST-MagmasL94Q, GST-MagmasF92G and GST-MagmasF92Y93G mutants by incubating with equimolar or increasing concentrations of yPam18J+T and DnaJC19J+T proteins.
Miscellaneous
Yeast mitochondria were purified according to the published protocols (25,54) and resuspended to 20 mg protein ml−1 concentration in SEM buffer (250 mm sucrose, 1 mm EDTA and 10 mm MOPS–KOH, pH 7.2). The human mitochondria were isolated using Cell Mitochondria Isolation Kit™ (Sigma-Aldrich) as per manufacturer's instructions. The mutant mitochondria were pre-incubated at 37°C prior to any kind of analysis. For expression analysis, 50 µg of yeast or 100 µg human mitochondria was subjected to SDS–PAGE, followed by immunodecoration with specific antibodies as indicated in the figures. The affinity purification of antibodies (24) and ATPase assays (30) and in vitro import assays (55) were carried out as described. For generation of anti-Magmas, anti-DnaJC19 and anti-hTim17 anti-sera rabbits were injected with cleaved GST fragments of Magmas (30–125 aa), DnaJC19 (34–116 aa) and KLH-conjugated peptide (CPKDGTPAPGYPSYQ) corresponding to the C-terminal region of hTim17, respectively (Imgenex Biotech). Anti-yTim23 and anti-yTim50 antibodies were generated by using N-terminal region of yTim23 (1–98 aa) and C-terminal region of yTim50 (133–476 aa), respectively, as antigens. For the generation of anti-DHFR antibodies, the rabbits were injected with the recombinant protein. Antibodies against yHsp60 (30), yTim44 (20), yMge1 (18), yPam18 (31), yPam16 (31) were gifted by Prof. Elizabeth A. Craig's laboratory, University of Wisconsin-Madison. Antibodies specific to human proteins, namely hTim23, hTim44, SOD and cathepsin D were obtained from BD Biosciences. Anti-catalase antibody was obtained from Calbiochem. Immunoblot analysis was carried out by using the ECL system (Amersham Pharmacia) according to the manufacturer's instructions. The reagents used for the experiments were obtained from Sigma-Aldrich unless specified.
FUNDING
This work was supported by The Wellcome Trust International Senior Research Fellowship in Biomedical Science WT081643MA (to P.D.S) and Council of Scientific and Industrial Research Fellowship (to D.S).
ACKNOWLEDGEMENTS
We thank Dr Elizabeth A. Craig, Dr William Walter and Dr Brenda Schilke for yeast strains and yeast specific antibodies, Dr Apurva Sarin and Dr Manjula Nagala for the confocal images and Dr Ganesh Nagaraju for helpful comments on the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu. Rev. Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 2.Hoogenraad N.J., Ryan M.T. Translocation of proteins into mitochondria. IUBMB Life. 2001;51:345–350. doi: 10.1080/152165401753366096. [DOI] [PubMed] [Google Scholar]
- 3.Pfanner N., Geissler A. Versatility of the mitochondrial protein import machinery. Nat. Rev. Mol. Cell Biol. 2001;2:339–349. doi: 10.1038/35073006. [DOI] [PubMed] [Google Scholar]
- 4.Neupert W., Hartl F.U., Craig E.A., Pfanner N. How do polypeptides cross the mitochondrial membranes? Cell. 1990;63:447–450. doi: 10.1016/0092-8674(90)90437-j. [DOI] [PubMed] [Google Scholar]
- 5.Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep. 2008;9:42–49. doi: 10.1038/sj.embor.7401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamenskii P.A., Vinogradova E.N., Krasheninnikov I.A., Tarasov I.A. [Directed import of macromolecules into mitochondria] Mol. Biol. (Mosk) 2007;41:216–233. [PubMed] [Google Scholar]
- 7.Neupert W., Brunner M. The protein import motor of mitochondria. Nat. Rev. Mol. Cell Biol. 2002;3:555–565. doi: 10.1038/nrm878. [DOI] [PubMed] [Google Scholar]
- 8.Neupert W., Herrmann J.M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- 9.Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- 10.Rehling P., Wiedemann N., Pfanner N., Truscott K.N. The mitochondrial import machinery for preproteins. Crit. Rev. Biochem. Mol. Biol. 2001;36:291–336. doi: 10.1080/20014091074200. [DOI] [PubMed] [Google Scholar]
- 11.Alder N.N., Jensen R.E., Johnson A.E. Fluorescence mapping of mitochondrial TIM23 complex reveals a water-facing, substrate-interacting helix surface. Cell. 2008;134:439–450. doi: 10.1016/j.cell.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Jensen R.E., Dunn C.D. Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim. Biophys. Acta. 2002;1592:25–34. doi: 10.1016/s0167-4889(02)00261-6. [DOI] [PubMed] [Google Scholar]
- 13.Geissler A., Chacinska A., Truscott K.N., Wiedemann N., Brandner K., Sickmann A., Meyer H.E., Meisinger C., Pfanner N., Rehling P. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–518. doi: 10.1016/s0092-8674(02)01073-5. [DOI] [PubMed] [Google Scholar]
- 14.Martin J., Mahlke K., Pfanner N. Role of an energized inner membrane in mitochondrial protein import. Delta psi drives the movement of presequences. J. Biol. Chem. 1991;266:18051–18057. [PubMed] [Google Scholar]
- 15.Ungermann C., Guiard B., Neupert W., Cyr D.M. The delta psi- and Hsp70/MIM44-dependent reaction cycle driving early steps of protein import into mitochondria. EMBO J. 1996;15:735–744. [PMC free article] [PubMed] [Google Scholar]
- 16.Frazier A.E., Dudek J., Guiard B., Voos W., Li Y., Lind M., Meisinger C., Geissler A., Sickmann A., Meyer H.E., et al. Pam16 has an essential role in the mitochondrial protein import motor. Nat. Struct. Mol. Biol. 2004;11:226–233. doi: 10.1038/nsmb735. [DOI] [PubMed] [Google Scholar]
- 17.Kozany C., Mokranjac D., Sichting M., Neupert W., Hell K. The J domain-related cochaperone Tim16 is a constituent of the mitochondrial TIM23 preprotein translocase. Nat. Struct. Mol. Biol. 2004;11:234–241. doi: 10.1038/nsmb734. [DOI] [PubMed] [Google Scholar]
- 18.Voos W., Gambill B.D., Laloraya S., Ang D., Craig E.A., Pfanner N. Mitochondrial GrpE is present in a complex with hsp70 and preproteins in transit across membranes. Mol. Cell. Biol. 1994;14:6627–6634. doi: 10.1128/mcb.14.10.6627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhattacharyya T., Karnezis A.N., Murphy S.P., Hoang T., Freeman B.C., Phillips B., Morimoto R.I. Cloning and subcellular localization of human mitochondrial hsp70. J. Biol. Chem. 1995;270:1705–1710. doi: 10.1074/jbc.270.4.1705. [DOI] [PubMed] [Google Scholar]
- 20.Blom J., Kubrich M., Rassow J., Voos W., Dekker P.J., Maarse A.C., Meijer M., Pfanner N. The essential yeast protein MIM44 (encoded by MPI1) is involved in an early step of preprotein translocation across the mitochondrial inner membrane. Mol. Cell. Biol. 1993;13:7364–7371. doi: 10.1128/mcb.13.12.7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamura Y., Harada Y., Yamano K., Watanabe K., Ishikawa D., Ohshima C., Nishikawa S., Yamamoto H., Endo T. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. J. Cell Biol. 2006;174:631–637. doi: 10.1083/jcb.200603087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Laan M., Chacinska A., Lind M., Perschil I., Sickmann A., Meyer H.E., Guiard B., Meisinger C., Pfanner N., Rehling P. Pam17 is required for architecture and translocation activity of the mitochondrial protein import motor. Mol. Cell. Biol. 2005;25:7449–7458. doi: 10.1128/MCB.25.17.7449-7458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallas M.R., Dienhart M.K., Stuart R.A., Long R.M. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Mol. Biol. Cell. 2006;17:4051–4062. doi: 10.1091/mbc.E06-04-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., D'Silva P., Walter W., Marszalek J., Craig E.A. Regulated cycling of mitochondrial Hsp70 at the protein import channel. Science. 2003;300:139–141. doi: 10.1126/science.1083379. [DOI] [PubMed] [Google Scholar]
- 25.Gambill B.D., Voos W., Kang P.J., Miao B., Langer T., Craig E.A., Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J. Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moro F., Okamoto K., Donzeau M., Neupert W., Brunner M. Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J. Biol. Chem. 2002;277:6874–6880. doi: 10.1074/jbc.M107935200. [DOI] [PubMed] [Google Scholar]
- 27.Schiller D., Cheng Y.C., Liu Q., Walter W., Craig E.A. Residues of Tim44 involved in both association with the translocon of the inner mitochondrial membrane and regulation of mitochondrial Hsp70 tethering. Mol. Cell. Biol. 2008;28:4424–4433. doi: 10.1128/MCB.00007-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokranjac D., Sichting M., Neupert W., Hell K. Tim14, a novel key component of the import motor of the TIM23 protein translocase of mitochondria. EMBO J. 2003;22:4945–4956. doi: 10.1093/emboj/cdg485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Truscott K.N., Voos W., Frazier A.E., Lind M., Li Y., Geissler A., Dudek J., Muller H., Sickmann A., Meyer H.E., et al. A J-protein is an essential subunit of the presequence translocase-associated protein import motor of mitochondria. J. Cell Biol. 2003;163:707–713. doi: 10.1083/jcb.200308004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.D'Silva P.D., Schilke B., Walter W., Andrew A., Craig E.A. J protein cochaperone of the mitochondrial inner membrane required for protein import into the mitochondrial matrix. Proc. Natl Acad. Sci. USA. 2003;100:13839–13844. doi: 10.1073/pnas.1936150100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D'Silva P.R., Schilke B., Walter W., Craig E.A. Role of Pam16's degenerate J domain in protein import across the mitochondrial inner membrane. Proc. Natl Acad. Sci. USA. 2005;102:12419–12424. doi: 10.1073/pnas.0505969102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mokranjac D., Bourenkov G., Hell K., Neupert W., Groll M. Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J. 2006;25:4675–4685. doi: 10.1038/sj.emboj.7601334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elsner S., Simian D., Iosefson O., Marom M., Azem A. The mitochondrial protein translocation motor: structural conservation between the human and yeast Tim14/Pam18-Tim16/Pam16 co-chaperones. Int. J. Mol. Sci. 2009;10:2041–2053. doi: 10.3390/ijms10052041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y., Dudek J., Guiard B., Pfanner N., Rehling P., Voos W. The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J. Biol. Chem. 2004;279:38047–38054. doi: 10.1074/jbc.M404319200. [DOI] [PubMed] [Google Scholar]
- 35.Iosefson O., Levy R., Marom M., Slutsky-Leiderman O., Azem A. The Pam18/Tim14-Pam16/Tim16 complex of the mitochondrial translocation motor: the formation of a stable complex from marginally stable proteins. Protein Sci. 2007;16:316–322. doi: 10.1110/ps.062459607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.D'Silva P.R., Schilke B., Hayashi M., Craig E.A. Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol. Biol. Cell. 2008;19:424–432. doi: 10.1091/mbc.E07-08-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peng J., Huang C.H., Short M.K., Jubinsky P.T. Magmas gene structure and evolution. In Silico Biol. 2005;5:251–263. [PubMed] [Google Scholar]
- 38.Becker S., Gehrsitz A., Bork P., Buchner S., Buchner E. The black-pearl gene of Drosophila defines a novel conserved protein family and is required for larval growth and survival. Gene. 2001;262:15–22. doi: 10.1016/s0378-1119(00)00548-5. [DOI] [PubMed] [Google Scholar]
- 39.Gonczy P., Echeverri C., Oegema K., Coulson A., Jones S.J., Copley R.R., Duperon J., Oegema J., Brehm M., Cassin E., et al. Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature. 2000;408:331–336. doi: 10.1038/35042526. [DOI] [PubMed] [Google Scholar]
- 40.Jubinsky P.T., Short M.K., Mutema G., Witte D.P. Developmental expression of Magmas in murine tissues and its co-expression with the GM-CSF receptor. J. Histochem. Cytochem. 2003;51:585–596. doi: 10.1177/002215540305100504. [DOI] [PubMed] [Google Scholar]
- 41.Jubinsky P.T., Messer A., Bender J., Morris R.E., Ciraolo G.M., Witte D.P., Hawley R.G., Short M.K. Identification and characterization of Magmas, a novel mitochondria-associated protein involved in granulocyte-macrophage colony-stimulating factor signal transduction. Exp. Hematol. 2001;29:1392–1402. doi: 10.1016/s0301-472x(01)00749-4. [DOI] [PubMed] [Google Scholar]
- 42.Jubinsky P.T., Short M.K., Mutema G., Morris R.E., Ciraolo G.M., Li M. Magmas expression in neoplastic human prostate. J. Mol. Histol. 2005;36:69–75. doi: 10.1007/s10735-004-3840-8. [DOI] [PubMed] [Google Scholar]
- 43.Kawano T., Sugawara S., Hosono M., Tatsuta T., Nitta K. Alteration of gene expression induced by Silurus asotus lectin in Burkitt's lymphoma cells. Biol. Pharm. Bull. 2008;31:998–1002. doi: 10.1248/bpb.31.998. [DOI] [PubMed] [Google Scholar]
- 44.Sikorski R.S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mokranjac D., Berg A., Adam A., Neupert W., Hell K. Association of the Tim14.Tim16 subcomplex with the TIM23 translocase is crucial for function of the mitochondrial protein import motor. J. Biol. Chem. 2007;282:18037–18045. doi: 10.1074/jbc.M701895200. [DOI] [PubMed] [Google Scholar]
- 46.Bauer M.F., Gempel K., Reichert A.S., Rappold G.A., Lichtner P., Gerbitz K.D., Neupert W., Brunner M., Hofmann S. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J. Mol. Biol. 1999;289:69–82. doi: 10.1006/jmbi.1999.2751. [DOI] [PubMed] [Google Scholar]
- 47.Davey K.M., Parboosingh J.S., McLeod D.R., Chan A., Casey R., Ferreira P., Snyder F.F., Bridge P.J., Bernier F.P. Mutation of DNAJC19, a human homologue of yeast inner mitochondrial membrane co-chaperones, causes DCMA syndrome, a novel autosomal recessive Barth syndrome-like condition. J. Med. Genet. 2006;43:385–393. doi: 10.1136/jmg.2005.036657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wadhwa R., Takano S., Kaur K., Deocaris C.C., Pereira-Smith O.M., Reddel R.R., Kaul S.C. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int. J. Cancer. 2006;118:2973–2980. doi: 10.1002/ijc.21773. [DOI] [PubMed] [Google Scholar]
- 49.Yi X., Luk J.M., Lee N.P., Peng J., Leng X., Guan X.Y., Lau G.K., Beretta L., Fan S.T. Association of mortalin (HSPA9) with liver cancer metastasis and prediction for early tumor recurrence. Mol. Cell Proteomics. 2008;7:315–325. doi: 10.1074/mcp.M700116-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Guan K.L., Dixon J.E. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 51.Dalby B., Cates S., Harris A., Ohki E.C., Tilkins M.L., Price P.J., Ciccarone V.C. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004;33:95–103. doi: 10.1016/j.ymeth.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 52.Miroux B., Walker J.E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 53.Zhai P., Stanworth C., Liu S., Silberg J.J. The human escort protein Hep binds to the ATPase domain of mitochondrial hsp70 and regulates ATP hydrolysis. J. Biol. Chem. 2008;283:26098–26106. doi: 10.1074/jbc.M803475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Voisine C., Craig E.A., Zufall N., von Ahsen O., Pfanner N., Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell. 1999;97:565–574. doi: 10.1016/s0092-8674(00)80768-0. [DOI] [PubMed] [Google Scholar]
- 55.Bihlmaier K., Bien M., Herrmann J.M. In vitro import of proteins into isolated mitochondria. Methods Mol. Biol. 2008;457:85–94. doi: 10.1007/978-1-59745-261-8_6. [DOI] [PubMed] [Google Scholar]