Abstract
The Abelson helper integration site 1 (AHI1) gene locus on chromosome 6q23 is among a group of candidate loci for schizophrenia susceptibility that were initially identified by linkage followed by linkage disequilibrium mapping, and subsequent replication of the association in an independent sample. Here, we present results of a replication study of AHI1 locus markers, previously implicated in schizophrenia, in a large European sample (in total 3907 affected and 7429 controls). Furthermore, we perform a meta-analysis of the implicated markers in 4496 affected and 18 920 controls. Both the replication study of new samples and the meta-analysis show evidence for significant overrepresentation of all tested alleles in patients compared with controls (meta-analysis; P = 8.2 × 10−5–1.7 × 10−3, common OR = 1.09–1.11). The region contains two genes, AHI1and C6orf217, and both genes—as well as the neighbouring phosphodiesterase 7B (PDE7B)—may be considered candidates for involvement in the genetic aetiology of schizophrenia.
INTRODUCTION
Schizophrenia is a common mental disorder affecting 0.5–1% of the population and is the seventh most costly medical illness to western societies (1). Psychosis has been ranked the third-most-disabling condition, after quadriplegia and dementia (2), and life expectancy is reduced by approximately 15–20%, due to increased physical health problems and a high suicide rate (3,4). Family, twin and adoption studies show evidence for a strong genetic component in schizophrenia, and the relative contribution of genetic factors has been estimated to be at least 65% (5). Despite this high heritability, only a very limited number of putative susceptibility genes have been replicated to date (6).
Association of AHI1 with schizophrenia was first reported in an inbred Israeli Arab family sample with high incidence of schizophrenia through a genome-wide linkage scan (7), a refined linkage analysis of a linkage peak on 6q (8) and a subsequent fine-mapping study that identified seven markers significantly associating with schizophrenia after correction for multiple testing (9). All seven markers reside within an extended block of high linkage disequilibrium (LD), also harbouring the C6orf217 gene and a predicted micro-RNA gene (MIR548H4). The findings were subsequently replicated in an independent Icelandic case–control sample (10). The 6q region has been linked to schizophrenia in other studies as well (11–13), but possible association with AHI1 markers was not addressed in those samples. Furthermore, other common variants within the AHI1 locus have recently been reported to associate with autism in a candidate gene study (14). This is a notable observation given the recent discoveries of rare genomic microdeletions associated with psychosis on chromosomes 1q21.1, 15q11.2, 15q13.3 (15,16) and 2p16.3 (17,18) that have also been identified in other neurodevelopmental disorders including autism (19,20) and mental retardation (21,22).
Loss-of-function mutations in the AHI1 gene cause Joubert syndrome (OMIM: 213300), an autosomal recessive, cerebellar and cortical neurodevelopmental disorder marked by agenesis of the cerebellar vermis; ataxia; hypotonia; oculomotor apraxia; and various motor, cognitive and behavioural disturbances, including mental retardation (23–26). The AHI1 gene encodes the protein Jouberin which contains seven WD40 repeats, an SH3 domain, potential SH3 binding sites and an N-terminal coiled-coil domain (27). AHI1 is widely expressed in the brain and comparative analysis of the AHI1 locus in primates indicates that the gene has undergone positive selection during the evolution of the human lineage (23). Moreover, expression studies of AHI1 orthologues in mouse (23,24,28) and zebrafish (28) suggest a conserved role of Jouberin in neurodevelopment. The mouse orthologue of Jouberin, Ahi1, binds to huntingtin-associated protein 1 (Hap1) to form a stable protein complex in the brain that is important for maintaining the level of tyrosine kinase receptor B (TrkB), which is critical for neuronal differentiation and brain development (29). Interestingly, the endogenous TrkB ligand in humans—brain derived neurotrophic factor (BDNF)—is a survival factor for parvalbumine-positive interneurons, which have been shown to be specifically altered in a series of post-mortem studies in schizophrenia, and are thought to be involved in the pathobiology of this disorder (30).
Here, we present results of a replication study of seven AHI1 locus markers, previously implicated in schizophrenia (9,10), in a large European sample (in total 3907 affected and 7429 controls). Furthermore, we perform a meta-analysis of the implicated markers in a total sample of 4496 affected patients and 18 920 controls.
RESULTS
A summary of the number of patients and controls in each replication subgroup and the use of surrogate marker alleles (see Materials and Methods) is provided in Table 1 and Supplementary Material, Table S1, respectively, while the allele counts for each marker in each replication subgroup are given in Supplementary Material, Table S2. The distribution of genotypes of all tested markers was consistent with Hardy–Weinberg proportions in all samples after correction for multiple testing (Supplementary Material, Table S3).
Table 1.
Number of patients and controls in different replication samples
| Study | Sample | Patients | Controls |
|---|---|---|---|
| Munich | German | 495 | 1272 |
| SCOPE | Danish | 456 | 995 |
| SCOPE | Norwegian | 264 | 181 |
| SGENE+ | Icelandic | 589a | 11 491 |
| SGENE+ | Finnish General population | 59 | 147 |
| SGENE+ | Finnish Genetic isolate | 123 | 50 |
| SGENE+ | Italian | 84 | 89 |
| SGENE+ | British | 93 | 88 |
| SGENE+ | Scottish | 658 | 661 |
| SGENE+ | German (Bonn) | 483 | 367 |
| SGENE+ | Dutch | 713 | 643 |
| Cardiff | British | 479 | 2936 |
| Total | – | 4496 | 18 920 |
aEssentially the same patients as reported in Ingason et al. (10).
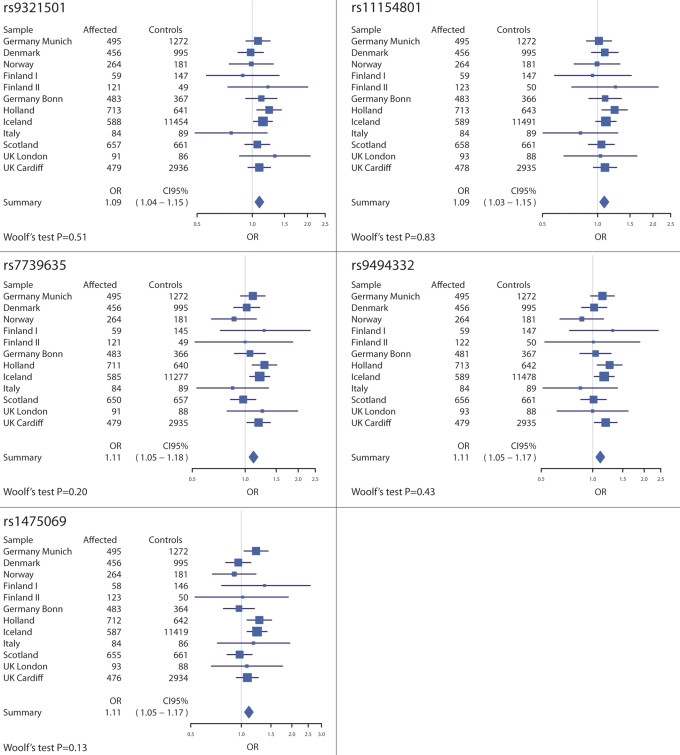
The combined analysis of all new replication samples—Munich, SCOPE, SGENE+ (excluding the previously reported Icelandic replication sample) and Cardiff—showed that all seven alleles were over-represented in affected compared with controls (Table 2). The association remained significant for six alleles using a conservative Bonferroni correction for seven tests, but considering the high LD between many of the markers [rs11154801, rs7750586 and rs9647635 are roughly equivalent; and rs7739635, rs9494332 and rs1475069 are highly inter-correlated (r2 > 0.7)], the number of independent tests is more likely to lie between three and five, in which case all seven alleles remain significant. When we include the Icelandic part of the SGENE+ sample in the analysis, the association is stronger (Table 2) and all seven alleles are significant even with the conservative Bonferroni correction (corrected P = 0.00057–0.012). A Woolf's test of homogeneity of odds ratios across subgroups was non-significant for all markers, i.e. the distribution of odds ratios across subgroups for each marker is compatible with that expected given a common odds ratio. The distribution of odds ratios with 95% confidence intervals along with the results of the Woolf's tests is shown in Figure 1. An exploratory analysis of the SGENE+ samples, using the Illumina HumanHap300 marker set and the SNPTEST imputation program (http://www.stats.ox.ac.uk/∼marchini/software/gwas/snptest.html), found no imputed HapMap marker in the locus among those markers polymorphic in the CEU HapMap population (Phase II) showing significantly stronger association with illness than rs1475069 (data not shown). Similarly, an analysis of all haplotype combinations between the five surrogate markers in the SGENE+ sample, using the NEMO (31) algorithm, found no haplotype with significantly stronger association with illness than rs1475069 (data not shown).
Table 2.
Association results from a combined Cochrane–Mantel–Haentzel analysis
| Allelea | New replication samplesb |
All replication samplesc |
||
|---|---|---|---|---|
| P-value | OR (95%CI) | P-value | OR (95%CI) | |
| rs9321501-A | 5.1 × 10−3 | 1.08 (1.02–1.15) | 4.9 × 10−4 | 1.09 (1.04–1.15) |
| rs11154801-C | 6.6 × 10−3 | 1.08 (1.02–1.15) | 1.6 × 10−3 | 1.09 (1.03–1.15) |
| rs7750586-A | 6.6 × 10−3 | 1.08 (1.02–1.15) | 1.7 × 10−3 | 1.09 (1.03–1.15) |
| rs9647635-A | 6.7 × 10−3 | 1.08 (1.02–1.15) | 1.7 × 10−3 | 1.09 (1.03–1.15) |
| rs7739635-C | 2.8 × 10−3 | 1.09 (1.03–1.16) | 8.2 × 10−5 | 1.11 (1.05–1.18) |
| rs9494332-A | 3.0 × 10−3 | 1.09 (1.03–1.17) | 2.4 × 10−4 | 1.11 (1.05–1.17) |
| rs1475069-A | 9.7 × 10−3 | 1.08 (1.01–1.15) | 2.3 × 10−4 | 1.11 (1.05–1.17) |
The details of subsamples, surrogate markers and allele counts are provided in Supplementary Material, Table S2.
aOvertransmitted to affected in Amann-Zalcenstein et al. (9).
bMunich, SCOPE, SGENE+ (excluding Icelandic part) and Cardiff samples.
cAll new samples and the Icelandic part of SGENE+.
Figure 1.
The distribution of odds ratios across subgroups for five of the seven tested alleles at the AHI1 locus, along with the number of affected and control individuals for each subgroup, and the results of Woolf's test of homogeneity of odds ratios. Results for alleles rs7750586-A and rs9647635-A are not included as they are roughly equivalent to those for rs11154801-C.
DISCUSSION
The AHI1 locus was first implicated in genetic schizophrenia susceptibility in a family sample of Israeli Arabs using the classical gene identification approach of positional cloning (7–9). This finding was replicated in an Icelandic case–control study (10). Here, we have provided further support for association in a second replication study, and combined the results with those of the previous replication study (adding additional Icelandic controls) into a powerful meta-analysis of the implicated AHI1 markers including data from 4496 affected and 18 920 controls. The combined results reveal association of all seven tested alleles with increased risk of schizophrenia. The meta-analysis includes the previously reported Icelandic replication sample (10) (as part of the SGENE+ sample), but not the discovery Israeli Arab family sample (9), and should therefore avoid the ‘winner's curse’.
Consistent with the approaches now in use in follow-up studies of genome-wide association data sets (32–34), we have used a one-sided test for analyses that exclude the initial discovery sample. Thus, we specifically test the hypothesis that the alleles that were over-transmitted to affected offspring in the discovery family sample (9) have an increased frequency in affected compared with control individuals in replication samples. The current study originates in the direct genotyping of the previously associated AHI1 markers (or perfect surrogates thereof) in the German Munich sample and the Danish/Norwegian SCOPE sample. Genotype counts for these markers or the closest available surrogate markers (in terms of correlation) were then obtained from genome-wide genotyped samples from collaborating researchers (SGENE+ and Cardiff) and all results combined into a Cochrane–Mantel–Haentzel analysis (as described in Materials and Methods) to estimate the allele frequencies of these markers in affected versus controls across all samples. The researcher responsible for conducting the association analysis was thus blinded to the SGENE+ and Cardiff genotypes of the involved markers prior to contacting researchers from these groups, and remains blinded to other genotypes from these samples. Accordingly, we correct only for seven tests in our interpretation of the significance of the result. This approach is in compliance with the guidelines presented in a recent article on replication studies of genotype–phenotype associations (35).
The use of surrogate markers in a meta-analysis under the assumption of equal odds ratios may be questionable, as it could underestimate the real odds ratios of the original seven markers. However, by the logic of LD mapping, the original seven markers may not necessarily be the true at-risk markers in the locus, but merely surrogates thereof. Thereby, the surrogate markers may provide as good or even better estimate than the original markers themselves, of the risk conferred by the true at-risk variation in the locus. Furthermore, the results of the Woolf's test show that for each marker the distribution of odds ratios across subgroups is compatible with the assumption of a common odds ratio (Fig. 1). In any case, the use of surrogates in this manner is unlikely to inflate the estimate of the true disease risk conferred by genetic variation within the locus.
By applying a Cochrane–Mantel–Haentzel analysis, we compare each sub-sample separately. This is necessary for two reasons; first, because we use data from different populations (allele frequencies of common variants often vary between populations); and second, because samples are genotyped for different surrogate markers that also have subtle differences in allele frequencies. It could seem feasible to pool all the samples into one, since they are all of European origin, but in this case both abovementioned factors would create a bias in the results as the patient-to-control ratios vary substantially between sub-samples.
As mentioned in the first paragraph of this section, the Icelandic patients of the SGENE+ sample are essentially the same as from our previous study (10), while the Icelandic SGENE+ control group is much larger than that of the previous study. We therefore decided to use the Icelandic SGENE+ results in the meta-analysis rather than the results of our earlier study. Either way, we feel justified to include the Icelandic sample in the meta-analysis as the previously reported Icelandic study was a replication study and not one of gene discovery. Also, while the association for the seven markers tested here is individually significant in both our meta-analysis of case–control samples and the family-based association study by Amann-Zalcenstein et al. (9), it is of interest to calculate the combined probability of the results of both studies. By applying Fisher's combined probability test, we estimate the two-sided probability for no association given the results of both studies to range from 2 × 10−8 for rs7739635 and rs1475069 to 1 × 10−5 for rs7750586 (with 4 × 10−8, 3 × 10−7, 1 × 10−6 and 2 × 10−6 for rs11154801, rs9494332, rs9647635 and rs9521501, respectively).
The biology of the AHI1 gene makes it a plausible candidate for involvement in schizophrenia aetiology. As mentioned earlier, it is widely expressed in brain and appears to participate in cellular processes important for early development of the brain. The association seen by Alvarez Retuerto et al. (14) between AHI1 markers and autism is also interesting, as there is known phenotypic and perhaps also aetiological overlap between autism and schizophrenia. The most significantly overtransmitted haplotype of Alvarez Retuerto et al. (14) is tagged by the C allele of rs17707754. The imputation results of the SGENE+ data set using SNPTEST suggest that this allele is over-represented in cases (P = 0.003, common OR = 1.72), but the relative statistical information is low (29%) due to poor tagging information for rs17707754 in the HumanHap300 marker set, and therefore the result has to be interpreted with caution.
The LD structure, arrangement of known genes, and markers typed in this study are shown in Figure 2. The genes include—in addition to AHI1—C6orf217 and PDE7B as well as the predicted micro-RNA gene MIR548H4. C6orf217 is expressed in brain, but its function remains unknown. Phosphodiesterase 7B is highly expressed in brain and thought to be involved in the control of cAMP-mediated neural activity and cAMP metabolism in the brain (36), and in the striatum, PDE7B transcription is activated by dopamine receptor D1 signalling mediated through the cAMP/cAMP-dependent protein kinase/cAMP-response element binding protein pathway (37). Although the LD block containing the markers tested here does not extend into PDE7B itself, regulatory elements affecting its transcription may be embedded in the sequence upstream of the gene, and thus be in LD with the markers tested here (Fig. 1). It should though be emphasized that the existence of such elements is entirely speculative at present.
Figure 2.
View of markers, genes and LD structure at the AHI1 locus using the UCSC Genome Browser (http://genome.ucsc.edu). The markers genotyped in different sub-samples to assess association of markers from the original family association study (9) are shown at top, followed by a track showing genes from the RefSeq database. Linkage disequilibrium (r2) in the HapMap CEU population (http://www.hapmap.org) is shown at bottom. Genomic coordinates are according to NCBI genome build 36.
We observe low odds ratios for the associated markers (1.09–1.11); this is not unexpected as it has been widely established through genome-wide association studies (GWAS) of common disease, that more often than not, the risk conferred by truly associating variants is indeed modest. For example, the marker rs1344706 was recently identified as associating with schizophrenia and bipolar disorder following a three-phased GWAS, with a common odds ratio of 1.12 derived from the combined samples (38). This association was subsequently replicated with similar odds ratios in two studies (39,40). Similar odds ratios are also seen for genome-wide significantly associating markers that have recently been identified in three large genome-wide studies in schizophrenia (41–43). It is believed that genetic variants associated with such subtle disease risk, interact with other susceptibility variants, as well as environmental factors in precipitating disease onset. Bearing this in mind, the recent discovery of a linkage region on chromosome 10 interacting with the linkage region of 6q23 (44) in the same Israeli Arab families wherein AHI1 association to schizophrenia was first described (9), may provide a hint for further studies seeking genetic variants interacting with AHI1 variants to increase schizophrenia risk.
In conclusion, we have found convincing evidence for association of common variation in the 6q23 region around AHI1, C6orf217 and PDE7B with schizophrenia in a large European case–control sample. This evidence further consolidates the locus as one of the few established genetic loci harbouring common schizophrenia risk variants.
MATERIALS AND METHODS
Samples and genotyping
Munich
The Munich sample included 495 schizophrenia patients and 1272 healthy controls from Munich, Germany. Six out of seven markers from the previous studies were tested in this sample; the assay for rs7739635 failed and a surrogate marker, rs12196952 (r2 = 0.85 in HapMap CEU trios), was typed instead. The Munich sample was genotyped at the Genetics Research Centre GmbH in Munich, Germany. One nanogram of DNA was assayed using the iPLEX assay on the MassARRAY MALDI-TOF mass spectrometer (SEQUENOM, Hamburg, Germany). DNA concentration was adjusted using the PicoGreen quantitation reagent (Invitrogen, Karlsruhe, Germany).
SCOPE
The SCOPE sample included 456 and 264 patients affected with schizophrenia or related psychoses, and 995 and 181 healthy controls from Denmark and Norway, respectively. Four out of the seven markers from previous studies were tested in the SCOPE sample; markers rs11154801 and rs9647635 were not typed due to very high LD with rs7750586 (r2 = 0.96 and 1, respectively, in HapMap CEU trios); the surrogate markers rs9494335 and rs9399158 were used for markers rs9494332 and rs1475069, respectively [as in Ingason et al. (10), r2 = 1 in HapMap CEU trios in both cases]; and the assay for rs7739635 failed. The SCOPE sample was genotyped at deCODE Genetics using the Centaurus platform (Nanogen Inc., San Diego, CA, USA).
SGENE+
The SGENE+ sample studied here includes 2802 patients affected with schizophrenia or related psychoses and 13 536 control individuals from Iceland, Scotland, Germany, UK, Italy, Finland and the Netherlands. The Icelandic patients of the SGENE+ sample are essentially the same as reported in the previous replication study (10), but the Icelandic SGENE+ control group is much larger than the one previously reported. Therefore, while we omit the Icelandic samples altogether from the analysis of new samples, we use the results from the Icelandic SGENE+ sample (rather than those previously reported) in the meta-analysis of all replication samples. The core SGENE sample was genotyped on the HumanHap300 BeadArrayTM at deCODE genetics. The additional SGENE+ samples from Aberdeen were genotyped, respectively, on the HumanHap550v2 BeadArrayTM (Illumina, San Diego, CA, USA), at Duke University in collaboration with GlaxoSmithKline. The SGENE+ samples from Germany and the Netherlands were genotyped on the HumanHap550v3 BeadArrayTM (Illumina, San Diego, CA, USA) at Bonn University and UCLA, respectively. In the SGENE+ sample only subjects with at least 98% genotype yield (on the entire micro-array) were used in the association analysis.
Cardiff
The Cardiff sample included 479 schizophrenia patients and 2936 controls from the UK. The patient sample was genotyped on the Affymetrix GeneChip 500K Mapping Array as part of the Wellcome Trust Case-Control Consortium pipeline (though it was not part of that study), and the control genotypes are those reported by the WTCCC (45).
Ethical approval was obtained from the local Ethics Committees. All participants gave written informed consent. A summary of the sample sizes is given in Table 1, while a more detailed sample description is provided in Supplementary Material online.
Association analysis
We performed a combined analysis using a one-sided exact Cochrane–Mantel–Haentzel test for enrichment of the over-transmitted alleles in patients. In this analysis, allele frequencies in patients and controls are compared separately for each sub-sample and a common odds ratio derived for all sub-samples (i.e. it is assumed that odds ratios calculated for each sub-sample are distributed around a common ‘true’ odds ratio). We assumed that odds ratios of surrogate alleles are the same as those of the alleles of the original study (9). Markers rs12196952 and rs9494335 served as surrogates for rs7739635 in the Munich and SCOPE samples, respectively, and rs7750586 for rs1154801 and rs9647635 in the SCOPE sample only. In the SGENE+ samples, surrogate markers on Illumina micro-arrays were used for the five markers not present on the arrays. In the Cardiff sample, surrogate markers on the Affymetrix GeneChip 500K Mapping Array were used for the six markers not present on the array. A summary of the surrogate alleles used in the current study to assess association of the markers implicated from previous studies showing marker correlations derived from the HapMap CEU sample (available at http://www.hapmap.org) is provided in Supplementary Material, Table S1. A Woolf's test was performed to estimate homogeneity of odds ratios across subgroups in the Cochrane–Mantel–Haentzel meta-analysis. Because all the tested markers are in LD, most missing genotypes could be imputed from genotypes at other marker loci through a likelihood approach implemented in the program NEMO (31). Inclusion of this partial information allowed findings for each marker to be better comparable, as they were based on the same set of individuals. Although P-values in the meta-analysis are one-sided, the 95% confidence intervals for common odds ratios are derived from a two-sided alternative hypothesis.
SUPPLEMENTARY MATERIAL
FUNDING
Funding for the project was provided by the Wellcome Trust under award 076113. Part of the genotyping of the Munich sample was done at the Genetics Research Centre (GmbH) which is a joint venture between GlaxoSmithKline Germany and the Department of Psychiatry, Ludwig-Maximilians-University. This work was funded by EU grants LSHM-CT-2006-037761 (Project SGENE) and PIAP-GA-2008-218251 (Project PsychGene). The Cardiff research was funded by grants from the MRC and the Wellcome Trust. The UCLA-Utrecht research was funded by NIH grant R01 MH078075.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the participating subjects, their relatives and staff at the recruitment centres. We also thank David Goldstein and colleagues, and Hreinn Stefansson and colleagues for permission to use the genotype data from the Munich and Aberdeen samples typed at Duke University, and the Icelandic SGENE sample typed at deCODE Genetics, respectively, and Stacy Steinberg at deCODE Genetics for performing imputation and haplotype analyses of the SGENE+ data. This study makes use of data generated by the Wellcome Trust Case Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. GROUP Investigators include: Department of Psychiatry, Rudolf Magnus Institute of Neuroscience, University Medical Centre Utrecht, Utrecht, The Netherlands (René S. Kahn, Wiepke Cahn); Department of Psychiatry, Academic Medical Centre University of Amsterdam, Amsterdam, The Netherlands (Don Linszen, Lieuwe de Haan); Maastricht University Medical Centre, EURON South Limburg Mental Health Research and Teaching Network, Maastricht, The Netherlands (Jim van Os, Lydia Krabbendam, Inez Myin-Germeys); University Medical Centre Groningen, Department of Psychiatry, University of Groningen, The Netherlands (Durk Wiersma, Richard Bruggeman).
Conflict of Interest statement. None declared.
REFERENCES
- 1.Freedman R. Schizophrenia. N. Engl. J. Med. 2003;349:1738–1749. doi: 10.1056/NEJMra035458. [DOI] [PubMed] [Google Scholar]
- 2.Ustün T.B., Rehm J., Chatterji S., Saxena S., Trotter R., Room R., Bickenbach J. Multiple-informant ranking of the disabling effects of different health conditions in 14 countries. WHO/NIH Joint Project CAR Study Group. Lancet. 1999;354:111–115. doi: 10.1016/s0140-6736(98)07507-2. [DOI] [PubMed] [Google Scholar]
- 3.Harris E.C., Barraclough B. Excess mortality of mental disorder. Br. J. Psychiatry. 1998;173:11–53. doi: 10.1192/bjp.173.1.11. [DOI] [PubMed] [Google Scholar]
- 4.Newman S.C., Bland R.C. Mortality in a cohort of patients with schizophrenia: a record linkage study. Can. J. Psychiatry. 1991;36:239–245. doi: 10.1177/070674379103600401. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein P., Yip B.H., Björk C., Pawitan Y., Cannon T.D., Sullivan P.F., Hultman C.M. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet. 2009;373:234–239. doi: 10.1016/S0140-6736(09)60072-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rujescu D., Collier D.A. Dissecting the many genetic faces of schizophrenia. Epidemiol. Psichiatr. Soc. 2009;18:91–95. [PubMed] [Google Scholar]
- 7.Lerer B., Segman R.H., Hamdan A., Kanyas K., Karni O., Kohn Y., Korner M., Lanktree M., Kaadan M., Turetsky N., et al. Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol. Psychiatry. 2003;8:488–498. doi: 10.1038/sj.mp.4001322. [DOI] [PubMed] [Google Scholar]
- 8.Levi A., Kohn Y., Kanyas K., Amann D., Pae C.U., Hamdan A., Segman R.H., Avidan N., Karni O., Korner M., et al. Fine mapping of a schizophrenia susceptibility locus at chromosome 6q23: increased evidence for linkage and reduced linkage interval. Eur. J. Hum. Genet. 2005;13:763–771. doi: 10.1038/sj.ejhg.5201406. [DOI] [PubMed] [Google Scholar]
- 9.Amann-Zalcenstein D., Avidan N., Kanyas K., Ebstein R.P., Kohn Y., Hamdan A., Ben-Asher E., Karni O., Mujaheed M., Segman R.H., et al. AHI1, a pivotal neurodevelopmental gene, and C6orf217 are associated with susceptibility to schizophrenia. Eur. J. Hum. Genet. 2006;14:1111–1119. doi: 10.1038/sj.ejhg.5201675. [DOI] [PubMed] [Google Scholar]
- 10.Ingason A., Sigmundsson T., Steinberg S., Sigurdsson E., Haraldsson M., Magnusdottir B.B., Frigge M.L., Kong A., Gulcher J., Thorsteinsdottir U., et al. Support for involvement of the AHI1 locus in schizophrenia. Eur. J. Hum. Genet. 2007;15:988–991. doi: 10.1038/sj.ejhg.5201848. [DOI] [PubMed] [Google Scholar]
- 11.Cao Q., Martinez M., Zhang J., Sanders A.R., Badner J.A., Cravchik A., Markey C.J., Beshah E., Guroff J.J., Maxwell M.E., et al. Suggestive evidence for a schizophrenia susceptibility locus on chromosome 6q and a confirmation in an independent series of pedigrees. Genomics. 1997;43:1–8. doi: 10.1006/geno.1997.4815. [DOI] [PubMed] [Google Scholar]
- 12.Martinez M., Goldin L.R., Cao Q., Zhang J., Sanders A.R., Nancarrow D.J., Taylor J.M., Levinson D.F., Kirby A., Crowe R.R., et al. Follow-up study on a susceptibility locus for schizophrenia on chromosome 6q. Am. J. Med. Genet. 1999;88:337–343. [PubMed] [Google Scholar]
- 13.Levinson D.F., Holmans P., Straub R.E., Owen M.J., Wildenauer D.B., Gejman P.V., Pulver A.E., Laurent C., Kendler K.S., Walsh D., et al. Multicenter linkage study of schizophrenia candidate regions on chromosomes 5q, 6q, 10p, and 13q: schizophrenia linkage collaborative group III. Am. J. Hum. Genet. 2000;67:652–663. doi: 10.1086/303041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez Retuerto A.I., Cantor R.M., Gleeson J.G., Ustaszewska A., Schackwitz W.S., Pennacchio L.A., Geschwind D.H. Association of common variants in the Joubert syndrome gene (AHI1) with autism. Hum. Mol. Genet. 2008;17:3887–3896. doi: 10.1093/hmg/ddn291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stefansson H., Rujescu D., Cichon S., Pietiläinen O.P., Ingason A., Steinberg S., Fossdal R., Sigurdsson E., Sigmundsson T., Buizer-Voskamp J.E., et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirov G., Gumus D., Chen W., Norton N., Georgieva L., Sari M., O'Donovan M.C., Erdogan F., Owen M.J., Ropers H.H., Ullmann R. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum. Mol. Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 18.Rujescu D., Ingason A., Cichon S., Pietiläinen O.P., Barnes M.R., Toulopoulou T., Picchioni M., Vassos E., Ettinger U., Bramon E., et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum. Mol. Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A., Green T., et al. Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 20.Kim H.G., Kishikawa S., Higgins A.W., Seong I.S., Donovan D.J., Shen Y., Lally E., Weiss L.A., Najm J., Kutsche K., et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am. J. Hum. Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp A.J., Mefford H.C., Li K., Baker C., Skinner C., Stevenson R.E., Schroer R.J., Novara F., De Gregori M., Ciccone R., et al. A recurrent 15q13.3 microdeletion syndrome associated with mental retardation and seizures. Nat. Genet. 2008;40:322–328. doi: 10.1038/ng.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mefford H.C., Sharp A.J., Baker C., Itsara A., Jiang Z., Buysse K., Huang S., Maloney V.K., Crolla J.A., Baralle D., et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N. Engl. J. Med. 2008;359:1685–1699. doi: 10.1056/NEJMoa0805384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferland R.J., Eyaid W., Collura R.V., Tully L.D., Hill R.S., Al-Nouri D., Al-Rumayyan A., Topcu M., Gascon G., Bodell A., et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat. Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 24.Dixon-Salazar T., Silhavy J.L., Marsh S.E., Louie C.M., Scott L.C., Gururaj A., Al-Gazali L., Al-Tawari A.A., Kayserili H., Sztriha L., Gleeson J.G. Mutations in the AHI1 gene, encoding Jouberin, cause Joubert syndrome with cortical polymicrogyria. Am. J. Hum. Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisi M.A., Doherty D., Eckert M.L., Shaw D.W., Ozyurek H., Aysun S., Giray O., Al Swaid A., Al Shahwan S., Dohayan N., et al. AHI1 mutations cause both retinal dystrophy and renal cystic disease in Joubert syndrome. J. Med. Genet. 2006;43:334–339. doi: 10.1136/jmg.2005.036608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valente E.M., Brancati F., Silhavy J.L., Castori M., Marsh S.E., Barrano G., Bertini E., Boltshauser E., Zaki M.S., Abdel-Aleem A., et al. AHI1 gene mutations cause specific forms of Joubert syndrome-related disorders. Ann. Neurol. 2006;59:527–534. doi: 10.1002/ana.20749. [DOI] [PubMed] [Google Scholar]
- 27.Jiang X., Hanna Z., Kaouass M., Girard L., Jolicoeur P. Ahi-1, a novel gene encoding a modular protein with WD40-repeat and SH3 domains, is targeted by the Ahi-1 and Mis-2 provirus integrations. J. Virol. 2002;76:9046–9059. doi: 10.1128/JVI.76.18.9046-9059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doering J.E., Kane K., Hsiao Y.C., Yao C., Shi B., Slowik A.D., Dhagat B., Scott D.D., Ault J.G., Page-McCaw P.S., Ferland R.J. Species differences in the expression of Ahi1, a protein implicated in the neurodevelopmental disorder Joubert syndrome, with preferential accumulation to stigmoid bodies. J. Comp. Neurol. 2008;511:238–256. doi: 10.1002/cne.21824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheng G., Xu X., Lin Y.F., Wang C.E., Rong J., Cheng D., Peng J., Jiang X., Li S.H., Li X.J. Huntingtin-associated protein 1 interacts with Ahi1 to regulate cerebellar and brainstem development in mice. J. Clin. Invest. 2008;118:2785–2795. doi: 10.1172/JCI35339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lisman J.E., Coyle J.T., Green R.W., Javitt D.C., Benes F.M., Heckers S., Grace A.A. Circuit-based framework for understanding neurotransmitter and risk gene interactions in schizophrenia. Trends Neurosci. 2008;31:234–242. doi: 10.1016/j.tins.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amundadottir L.T., Sulem P., Gudmundsson J., Helgason A., Baker A., Agnarsson B.A., Sigurdsson A., Benediktsdottir K.R., Cazier J.B., Sainz J., et al. A common variant associated with prostate cancer in European and African populations. Nat. Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 32.Lasky-Su J., Lyon H.N., Emilsson V., Heid I.M., Molony C., Raby B.A., Lazarus R., Klanderman B., Soto-Quiros M.E., Avila L., et al. On the replication of genetic associations: timing can be everything! Am. J. Hum. Genet. 2008;82:849–858. doi: 10.1016/j.ajhg.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller T.D., Hinney A., Scherag A., Nguyen T.T., Schreiner F., Schäfer H., Hebebrand J., Roth C.L., Reinehr T. ‘Fat mass and obesity associated' gene (FTO): no significant association of variant rs9939609 with weight loss in a lifestyle intervention and lipid metabolism markers in German obese children and adolescents. BMC Med. Genet. 2008;9:85. doi: 10.1186/1471-2350-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herder C., Rathmann W., Strassburger K., Finner H., Grallert H., Huth C., Meisinger C., Gieger C., Martin S., Giani G., et al. Variants of the PPARG, IGF2BP2, CDKAL1, HHEX, and TCF7L2 genes confer risk of type 2 diabetes independently of BMI in the German KORA studies. Horm. Metab. Res. 2008;40:722–726. doi: 10.1055/s-2008-1078730. [DOI] [PubMed] [Google Scholar]
- 35.NCI-NHGRI Working Group on Replication in Association Studies. Replicating genotype–phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 36.Sasaki T., Kotera J., Yuasa K., Omori K. Identification of human PDE7B, a cAMP-specific phosphodiesterase. Biochem. Biophys. Res. Commun. 2000;271:575–583. doi: 10.1006/bbrc.2000.2661. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki T., Kotera J., Omori K. Transcriptional activation of phosphodiesterase 7B1 by dopamine D1 receptor stimulation through the cyclic AMP/cyclic AMP-dependent protein kinase/cyclic AMP-response element binding protein pathway in primary striatal neurons. J. Neurochem. 2004;89:474–483. doi: 10.1111/j.1471-4159.2004.02354.x. [DOI] [PubMed] [Google Scholar]
- 38.O'Donovan M.C., Craddock N., Norton N., Williams H., Peirce T., Moskvina V., Nikolov I., Hamshere M., Carroll L., Georgieva L., et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat. Genet. 2008;40:1053–1055. doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- 39.Riley B., Thiselton D., Maher B.S., Bigdeli T., Wormley B., McMichael G.O., Fanous A.H., Vladimirov V., O'Neill F.A., Walsh D., Kendler K.S. Replication of association between schizophrenia and ZNF804A in the Irish Case-Control Study of Schizophrenia sample. Mol. Psychiatry. 2010;15:29–37. doi: 10.1038/mp.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steinberg S., Mors O., Børglum A.D., Gustafsson O., Werge T., Mortensen P.B., Andreassen O.A., Sigurdsson E., Thorgeirsson T.E., Böttcher Y., et al. Expanding the range of ZNF804A variants conferring risk of psychosis. Mol. Psychiatry. 2010 doi: 10.1038/mp.2009.149. Epub ahead of print January 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefansson H., Ophoff R.A., Steinberg S., Andreassen O.A., Cichon S., Rujescu D., Werge T., Pietiläinen O.P., Mors O., Mortensen P.B., et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.The International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi J., Levinson D.F., Duan J., Sanders A.R., Zheng Y., Pe'er I., Dudbridge F., Holmans P.A., Whittemore A.S., Mowry B.J., et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alkelai A., Kohn Y., Olender T., Sarner-Kanyas K., Rigbi A., Hamdan A., Ben-Asher E., Lancet D., Lerer B. Evidence for an interaction of schizophrenia susceptibility loci on chromosome 6q23.3 and 10q24.33-q26.13 in Arab Israeli families. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:914–925. doi: 10.1002/ajmg.b.30918. [DOI] [PubMed] [Google Scholar]
- 45.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.