Abstract
Horn’s parallel analysis (PA) is the method of consensus in the literature on empirical methods for deciding how many components/factors to retain. Different authors have proposed various implementations of PA. Horn’s seminal 1965 article, a 1996 article by Thompson and Daniel, and a 2004 article by Hayton et al., all make assertions about the requisite distributional forms of the random data generated for use in PA. Readily available software is used to test whether the results of PA are sensitive to several distributional prescriptions in the literature regarding the rank, normality, mean, variance, and range of simulated data on a portion of the National Comorbidity Survey Replication (Pennell et al., 2004) by varying the distributions in each PA. The results of PA were found not to vary by distributional assumption. The conclusion is that PA may be reliably performed with the computationally simplest distributional assumptions about the simulated data.
Introduction
Researchers may be motivated to employ principal components analysis (PCA) or factor analysis (FA) in order to facilitate the reduction of multicollinear measures for the sake of analytic dimensionality or as a means of exploring structures underlying multicollinearity of a data set; a critical decision in the process of using PCA or FA is the question of how many components or factors to retain (Preacher & MacCallum, 2003; Velicer & Jackson, 1990). A growing body of review papers and simulation studies has produced a prescriptive consensus that Horn’s (1965) ‘parallel analysis’ (PA) is the best empirical method for component or factor retention in principal components analysis (PCA) or factor analysis (FA) (Cota, Longman, Holden, & Fekken, 1993; Glorfeld, 1995; Hayton, Allen, & Scarpello, 2004; Humphreys & Montanelli, 1976; Lance, Butts, & Michels, 2006; Patil, Singh, Mishra, & Donavan, 2008; Silverstein, 1977; Velicer, Eaton, & Fava, 2000; Zwick & Velicer, 1986). Horn himself subsequently employed PA both in factor analytic research (Hofer, Horn, & Eber, 1997), and in assessment of factor retention criteria (Horn & Engstrom, 1979), though he exhibited a methodological pluralism with respect to factor retention criteria. At least two papers assert that PA applies only to common factor analysis of the common factor model, which is sometimes termed ‘principal factors,’ or ‘principal axes’ (Ford, MacCallum, & Tait, 1986; Velicer et al., 2000). This is unsurprising given the capacity of different FA methods and rotations to alter the sum of eigenvalues by more than an order of magnitude relative to P. From this point forward, FA shall refer to unrotated common factor analysis.
Horn’s Parallel Analysis
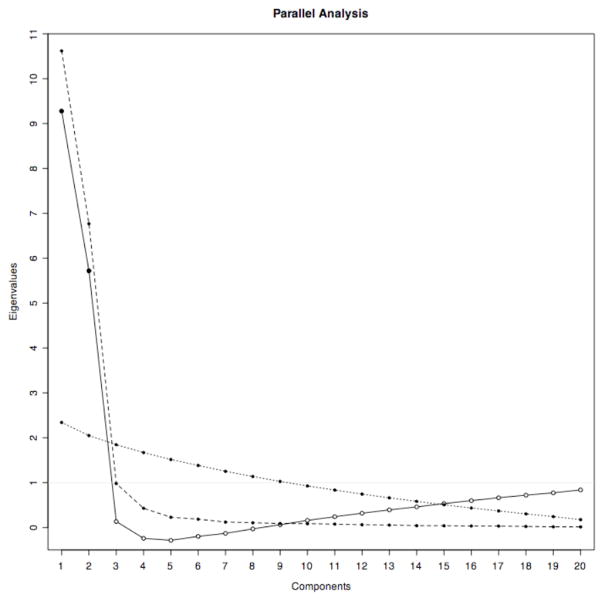
The question of the number of components or factors to retain is critical both for reducing the analytic dimensionality of data, and for producing insight as to structure of latent variables (cf. Velicer & Jackson, 1990). Guttman (1954) formally argued that because in PCA total variance equals the number of variables P, in an infinite population a theoretical lower bound to the number of true components is given by the number of components with eigenvalues greater than one. This insight was later articulated by Kaiser as a retention rule for PCA as the ‘eigenvalue greater than one’ rule (Kaiser, 1960) which has also been called the ‘Kaiser rule’ (cf. Lance et al., 2006), the ‘Kaiser-Guttman’ rule (cf. Jackson, 1993), and the ‘K1’ rule (cf. Hayton et al., 2004). Assessing Kaiser’s prescription, Horn observed that in a finite sample of N observations in P measured variables of uncorrelated data, the eigenvalues from a PCA or FA would be greater than and less than one, due to “sample-error and least squares bias.” Therefore, Horn argued, when making a component-retention decision with respect to observed, presumably correlated data of size N observations by P variables, researchers would want to adjust the eigenvalues of each factor by subtracting the mean sample error from a “reasonably large” number K of uncorrelated N × P data sets, and retaining those components or factors with adjusted eigenvalues greater than one (Horn, 1965). Horn also expressed the PA decision criterion in a mathematically equivalent way, by saying that a researcher would retain those components or factors whose eigenvalues were larger than the mean eigenvalues of the K uncorrelated data sets. Both these formulations are illustrated in Figure 1 which represents PA of a PCA applied to a simulated data set of 50 observations, across 20 variables, with two uncorrelated factors, and %50 total variance.
Figure 1.
Graphical illustration of parallel analysis on a simulated data set of 50 observations, across 20 variables, with two uncorrelated factors, and %50 total variance. The dashed line connects unadjusted eigenvalues of the observed data, the dotted line connects mean eigenvalues of 600 random 50*20 data sets, and the solid line connects adjusted eigenvalues (i.e. subtracting the mean eigenvalues minus one from the observed eigenvalues). The retention criterion is the point at which the adjusted eigenvalues cross the horizontal line at y=1, which is the same point at which the unadjusted eigenvalues cross the line of mean eigenvalues of the random data sets. The solid adjusted eigenvalue markers are those components (or factors, if using factor analysis) that are retained.
Ironically, PA has enjoyed both a substantial affirmation in the methods literature for its performance relative to other retention criteria, while at the same time being one of the least often used methods in actual empirical research (cf. Hayton et al., 2004; Patil et al., 2008; Thompson & Daniel, 1996; Velicer et al., 2000). Methods papers making comparisons between retention decisions in PCA and FA have tended to ratify the idea that PA outperforms all other commonly published component retention methods, particularly the commonly reported Kaiser rule and scree test (Cattell, 1966) methods. Indeed, the panning of the eigenvalue greater than one rule has provoked harsh criticism: “The most disparate results were obtained… with the [K1] criterion…” (Silverstein, 1977, page 398) “Given the apparent functional relation of the number of components retained by Kl to the number of original variables and the repeated reports of the method’s inaccuracy, we cannot recommend the Kl rule for PCA.” (Zwick & Velicer, 1986, page 439) “…the eigenvalues-greater-than-one rule proposed by Kaiser… is the result of a misapplication of the formula for internal consistency reliability.” (Cliff, 1988, page 276) “On average the [K1] rule overestimated the correct number of factors by 66%. This poor performance led to a recommendation against continued use of the [K1] rule.” (Glorfeld, 1995, page 379) “The [K1] rule was extremely inaccurate and was the most variable of all the methods. Continued use of this method is not recommended” (Velicer et al., 2000, page 26). In an article titled “Efficient theory development and factor retention criteria: Abandon the ‘eigenvalue greater than one’ criterion” Patil et al. (2008) wrote on pages 169–170 “With respect to the factor retention criteria, perhaps marketing journals, like some journals in psychology, should recommend strongly the use of PA or minimum average partial and not allow the eigenvalue greater than one rule as the sole criterion. This is essential to avoid proliferation of superfluous constructs and weak theories.” More recent methods include the root mean square error adjustment which evaluates successive maximum likelihood FA models in a progression from zero to some positive number of factors for model fit (Browne & Cudeck, 1992; Steiger & Lind, 1980), and bootstrap methods that account for sampling variability in the estimates of the eigenvalues of the observed data (Lambert, Wildt & Durand, 1990), but have yet to be rigorously evaluated against one another and other retention methods. Recent rerandomization-based hypothesis-test rules (Dray, 2008; Peres-Neto, Jackson & Somers, 2005) appear to promise performance on par with PA.
Despite this consensus in the methods literature, PA remains less used than one might expect. For example, in a Google Scholar search of 867 articles published in Multivariate Behavioral Research mentioning “factor analysis” or “principal components analysis”, only 26 even mention PA as of May 1, 2008. I conjecture that one reason for the lack of widespread adoption of PA among researchers may have been the computational costs in PA due to generating a large number of data sets and performing repeated analyses on them. Both the PCA or FA and the generation of random data sets were computationally quite expensive prior to the advent of cheap and ubiquitous computing. The computational costs of Horn’s PA method (and improvements on it) in the late 20th century encouraged the development of computationally less expensive regression-based models to estimate PA results given only the parameters N and P (Allen & Hubbard, 1986; Keeling, 2000; Lautenschlager, 1989; Longman, Cota, Holden, & Fekken, 1989). However, these techniques have been found to be imprecise approximations to actual PA results, and, moreover, to perform less reliably in making component-retention or factor-retention decisions. (Lautenschlager, 1989; Velicer et al., 2000)
Another possible reason for the lack of the widespread adoption of PA by researchers is the lack of standard implementation in the more commonly used statistical packages including SAS, SPSS, Stata, R, and S-Plus. In fact, several proponents of PA in the literature have published programs or software for performing PA to address just this issue (Hayton et al., 2004; Patil et al., 2008; Thompson & Daniel, 1996).
Asserted Distributional Requirements of Parallel Analysis
Among some PA proponents there is a history of asserting the sensitivity of PA to the distributional form of the simulated data used to conduct it. In his 1965 paper, Horn wrote that the data in the simulated data sets needed to be normally distributed, with no explicit indication of the mean or variance of simulated data. Moreover, he asserted of PA that “if the distribution is not normal, the expectations outlined here need to be modified” (Horn, 1965, page 179). Thompson and Daniel (1996, page 200) asserted that the simulated data in PA “requires a raw matrix of the same ‘rank’ as the actual raw data matrix. For example, if one had 1-to-5 Likert-scale data for 103 subjects on 19 variables, a 103-by-19 raw data matrix consisting of 1s, 2s, 3s, 4s, or 5s would be generated.” And more recently, Hayton et al. (2004) wrote a review of component and factor retention decisions that justified the use of PA, and published SPSS code for conducting it. However, in doing so, the authors made the following claim (page 198) about the distributional assumptions of the data simulated in PA:
In addition to these changes, it is also important to ensure that the values taken by the random data are consistent with those in the comparison data set. The purpose of line 7 is to ensure that the random variables are normally distributed within the parameters of the real data. Therefore, line 7 must be edited to reflect the maximum and midpoint values of the scales being analyzed. For example, if the measure being analyzed is a 7-point Likert-type scale, then the values 5 and 3 in line 7 must be edited to 7 and 4, respectively. Lines 14 and 15 ensure that the random data assumes only values found in the comparison data and so must be edited to reflect the scale minimum and maximum, respectively.
This claim was described operationally in the accompanying code on page 201:
-
7)
COMPUTE V = RND (NORMAL (5/6) + 3).
-
8)
COMMENT This line relates to the response levels
-
9)
COMMENT 5 represents the maximum response value for the scale.
-
10)
COMMENT Change 5 to whatever the appropriate value may be.
-
11)
COMMENT 3 represents the middle response value.
-
12)
COMMENT Change 3 to whatever the actual middle response value may be.
-
13)
COMMENT (e.g., 3 is the midpoint for a 1 to 5 Likert scale).
-
14)
IF (V LT 1)V = 1.
-
15)
IF(V GT 5)V = 5.
The three notable features of the claim by Hayton et al. (2004), are (1) that the simulated data must be normally distributed, (2) that they have the same minimum and maximum values as the observed data, and (3) that the mid-point of the range of observed data must serve as the mean of the simulated data. This is a stronger injunction than Horn’s and that made by Thompson and Daniel.
This claim is interesting since Hayton et al. (2004) employed in their article a Monte Carlo sampled greater-than-median percentile estimate of sample bias published by Glorfeld (1995). However, Glorfeld included a detailed section in his article in which he found his extension of PA insensitive to distributional assumptions (Glorfeld, 1995, pages 383–386) using normally-, uniformly-, and G-distributed simulations, and simulations mixing all three of these distributions.
In order to assess whether PA is sensitive to the distributional forms of simulated data, and in particular to attend to the distributional prescriptions by Hayton et al. (2004), I conducted parallel PCAs and maximum likelihood FAs on nine combinations of number of observations and number of variables using ten different distributional assumptions. The results of PA could only be affected by the distribution of randomly generated data through the estimates of random eigenvalues due to “sample-error and least squares bias.” Therefore, I examine differences in these estimates, rather than interpreting adjusted eigenvalues.
In varying the distribution of random data, I emphasized varying the parameters of the normal distribution to carefully attend to the claims by Hayton et al. (2004), attended to the implication that the simulated data required the same univariate distribution as the observed data by employing two resampling methods (rerandomization, and bootstrap), and explored the sensitivity of PA to distributions that varied the skewness and kurtosis of the simulated data while maintaining the same mean and variance as the observed data. I also included binomially distributed data with extremely different distributional forms than the observed data.
I conducted a simulation experiment to test whether PA in PCA and FA is sensitive to the specific distribution of the data used in the randomly generated data sets used in it. I varied the distributional properties of the random data among parallel analyses in PCA and FA on simulated data sets as described below.
Method
Distributional Characteristics of Random Data for Parallel Analysis
I contrasted ten different distribution methods (A–J) for the simulation portion of PA as described below.
Method A
Each variable was drawn from a uniform distribution with mean of zero, and variance of one to comply with Horn’s (1965) assertion.
Method B
As described by Hayton et al. (2004), each variable was drawn from a normal distribution with mean equal to the mid-point of the observed data, variance nominally equal to the observed variance, and which was bounded within the observed minimum and maximum by recoding values outside this range to boundary values. The variance was nominal, because the recoding of extreme values of simulated data tended to shrink its variance.
Method C
Each variable was drawn from a normal distribution with a random mean and variance.
Method D
Each separate simulated variable was drawn from a rerandomized sample of the univariate distribution of a separate variable in the observed data (i.e. resampling without replacement) to provide simulated data in which each variable has precisely the same distribution as the corresponding observed variable.
Method E
Each separate simulated variable was drawn from a bootstrap sample of the univariate distribution of each separate variable in the observed data with replacement to provide simulated data in which each variable estimates the population distribution producing the univariate distribution of the corresponding observed variable.
Method F
Each variable was drawn from a Beta distribution with α = 2 and β = .5, scaled to the observed variance and centered on the observed mean, in order to produce data distributed with large skewness (−1.43) and large positive kurtosis (1.56).
Method G
Each variable was drawn from a Beta distribution with α = 1 and β = 2.86, scaled to the observed variance and centered on the observed mean, in order to produce data distributed with moderate skewness (0.83) and no kurtosis (<0.01).
Method H
Each variable was drawn from a Laplace distribution with a mean of zero and a scale parameter of one, scaled to the observed variance and centered on the observed mean, in order to produce data distributed with minimal skewness (mean skewness > 0.01 for 5000 iterations) and large positive kurtosis (mean kurtosis is 5.7 for 5000 iterations).
Method I
Each variable was drawn from a binomial distribution, with each variable having a randomly assigned probability of success drawn from a uniform distribution bounded by 10/N to 1–10/N.
Method J
Each variable was drawn from a uniform distribution with minimum of zero and maximum of one. This is the least computationally intensive method of generating random variables.
Data Simulated With Pre-Specified Component/Factor Structure
In order to rigorously assess whether PA is sensitive to the distributional form of random data, nine data sets were produced with low, medium and high values of the number of observations N (75, 300, 1200) and low, medium and high values of the number of variables P (10, 25, 50). Other characteristics such as the uniqueness of each variable, the number of components or factors, and factor correlations were not considered in these simulations, because they affect neither the construction of random data sets, nor the estimates of bias in classical PA in any way. Having variables with differing distributions (particularly those that may look like the Likert-type data appearing in scale items) also offers an opportunity to evaluate the performance of, for example, the two resampling methods in producing unbiased eigenvalue estimates. Each data set D was produced as described by Equation 1.
| (1) |
Where:
L is the component/factor loading matrix, and is defined as a P by F matrix of random values distributed uniform from −4 to 4.
F is a random integer between 1 and P for each data set.
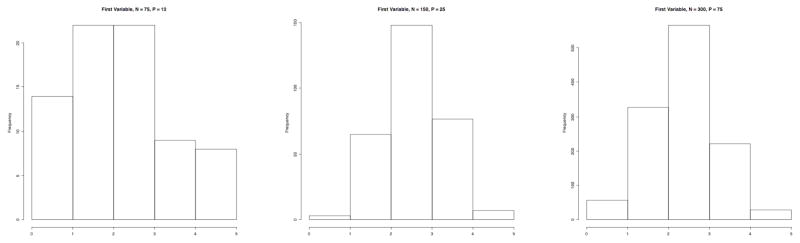
C is the component/factor matrix, and was defined as an N by F matrix of random variables. The values of each factor was distributed Beta with α distributed uniform with minimum of two and maximum of four, and β =0.8. (Equation 2). The linear combination of these factors produced variables that also had varying distributions. (see Figure 2)
Figure 2.
Histograms showing the distributions of the first variable from three of nine simulated data sets. All variables have five values (the integers from 1 to 5), and variable distributions based on different parameterizations of the Beta distribution plus an amount of uniform noise.
| (2) |
U is the uniqueness matrix of a simulation, and describes how much random error contributes to each variable relative to the component/factor contribution. U is defined as an N by P matrix of random values distributed uniformly with mean of zero and variance of one.
Data Analysis of Simulation
All distribution methods were applied to parallel analyses to estimate means and posterior 95% quantile intervals of the eigenvalues of 5,000 random data sets each. The 97.5th centile allowed an assessment of the sensitivity of Glorfeld’s PA variant to different random data distributions. Each analysis was also replicated using only 50 iterations to assess whether any sensitivity to distributional form or parameterization depends on the number of iterations used in PA. Each PA was conducted for both a PCA and an unrotated common FA on each of the nine simulated data sets. Multiple ordinary least squares (OLS) regressions were conducted for all eigenvalues corresponding to each data set, upon effect coded variables for each random data distribution for parallel analyses with 5,000 and with 50 iterations. This is indicated in Equation 3, where p indexes the first through tenth eigenvalues, r indicates the average is over the random data sets, and is distributed standard normal. Multiple comparisons control procedures were used because there were 2550 comparisons made for each of the four analyses (three data sets times ten mean eigenvalues times ten methods, plus three data sets times 25 mean eigenvalues times ten methods, plus three data sets times 50 mean eigenvalues times ten methods). Strong control of the familywise error rate is a valid procedure to account for multiple comparisons when an analysis produces a decision of one best method among a number of competing methods, (Benjamini & Hochberg, 1995), and therefore the Holm adjustment was employed as a control procedure (Holm, 1979). However, multiple comparisons procedures with weak control of the familywise error rate provide more power, so a procedure (Benjamini & Yekutieli, 2001) controlling the false discovery rate was also used to allow patterns in simulation method to emerge.
| (3) |
All analyses were carried out using a version of paran, (freely available for Stata by typing “net describe paran, from(http://www.doyenne.com/stata)” within Stata, and for R at http://cran.r-project.org/web/packages/paran/index.html and on the many CRAN mirrors). A modified version of paran for R was used to accommodate the different data generating methods described above. This modified file, and a file used to generated the simulated data presented here are available at: http://doyenne.com/Software/files/PAInsensitivityFiles.zip.
Data Analysis of Empirical Data
PA using the ten distributional models described above was also applied to coded responses from the National Comorbidity Survey Replication (NCS-R) portion of the Collaborative Psychiatric Epidemiology Surveys 2001–2003 (Pennell et al., 2004). This analysis employed 51 variables from questions loosely tapping the Axis I and Axis II conceptual domains of depression, irritability, anxiety, explosive anger, positive affect, and psychosis (i.e. variables NSD1–NSD5J, excluding variables NSD3A and NSD3B). Analyses were performed using 5,000 iterations for both PCA and common FA, and both classical PA and PA with a Monte Carlo extension at the 97.5th centile.
Results
Results for Simulation Data Analysis
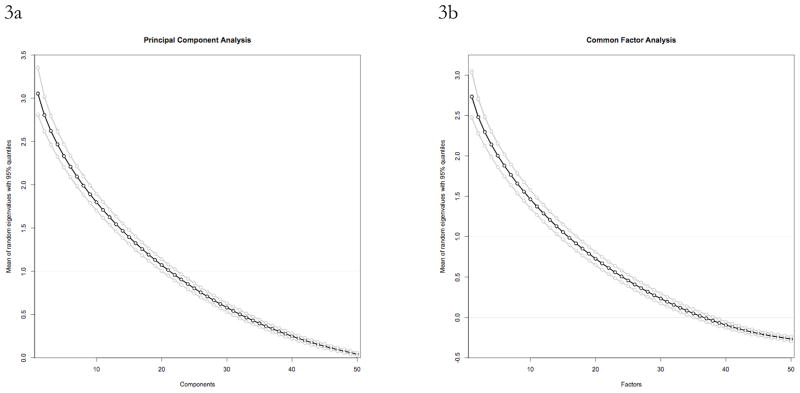
The first, third and fifth mean eigenvalues with 95% quantile intervals are presented in Table 1 (PCA) and Table 2 (FA). For each of the nine data sets analyzed the estimated mean eigenvalues from the random data do not vary noticeably with the distributional form of the simulated data. This same pattern is true for the quantile estimates, including the 97.5th centile estimate which is an implementation of Glorfeld’s (1995) Monte Carlo improvement to PA. This pattern holds for low, medium and high numbers of observations, and for low, medium and high numbers of variables. The means and standard deviations of the mean and quantile estimates across all distribution methods for each data set are presented for PA with PCA in Table 3a (5000 iterations) and Table 3b (50 iterations), and presented for PA with common FA in Table 4a (5000 iterations) and Table 4b (50 iterations). The extremely low standard deviations quantitatively characterize the between distribution differences in eigenvalue estimates of random data sets, even for the analyses with only 50 iterations, and are evidence of the absence of an effect of the distributional form of simulated data in PA. None of the distribution methods consistently overestimate or underestimate the mean or quantile of the random data eigenvalues. These results are represented visually in Figures 3a and 3b, which show essentially undifferentiable random eigenvalue estimates across all distribution methods for low N and high P for both PCA and FA with 5,000 iterations. These results suggest that mean and centile estimates of Horn’s “sample-error and least squares bias” (1965, page 180) are insensitive to distributional assumptions. High variances for 50-iteration analyses relative to the 5,000 iteration analyses suggest that Horn’s “sufficiently large” K may require many iterations.
Table 1.
Mean Estimates for First, Third, and Fifth Random Eigenvalues From Parallel Analyses Using Principal Component Analysis with No Rotation
| Data Set | Distribution method | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | P | A | B | C | D | E | F | G | H | I | J |
| First eigenvalue | |||||||||||
| l | l | 1.620 (1.438,1.853) | 1.621 (1.44,1.863) | 1.619 (1.44,1.846) | 1.623 (1.445,1.844) | 1.619 (1.437,1.844) | 1.618 (1.436,1.846) | 1.626 (1.443,1.861) | 1.622 (1.438,1.86) | 1.621 (1.443,1.85) | 1.621 (1.443,1.851) |
| m | l | 1.295 (1.214,1.396) | 1.296 (1.215,1.399) | 1.296 (1.214,1.395) | 1.296 (1.215,1.398) | 1.297 (1.214,1.4) | 1.295 (1.211,1.393) | 1.296 (1.212,1.399) | 1.296 (1.213,1.401) | 1.295 (1.213,1.397) | 1.296 (1.214,1.399) |
| h | l | 1.144 (1.105,1.191) | 1.144 (1.105,1.193) | 1.143 (1.104,1.189) | 1.144 (1.105,1.191) | 1.144 (1.106,1.192) | 1.143 (1.104,1.189) | 1.144 (1.105,1.192) | 1.144 (1.104,1.19) | 1.144 (1.104,1.191) | 1.144 (1.106,1.191) |
| l | m | 2.244 (2.034,2.508) | 2.248 (2.035,2.515) | 2.248 (2.037,2.524) | 2.245 (2.037,2.514) | 2.246 (2.033,2.505) | 2.249 (2.034,2.511) | 2.246 (2.037,2.499) | 2.246 (2.028,2.521) | 2.244 (2.031,2.505) | 2.249 (2.031,2.518) |
| m | m | 1.568 (1.479,1.676) | 1.566 (1.476,1.676) | 1.566 (1.479,1.67) | 1.568 (1.481,1.674) | 1.567 (1.478,1.674) | 1.567 (1.478,1.674) | 1.568 (1.477,1.677) | 1.568 (1.477,1.674) | 1.566 (1.477,1.675) | 1.568 (1.48,1.676) |
| h | m | 1.269 (1.23,1.316) | 1.269 (1.23,1.315) | 1.269 (1.23,1.316) | 1.269 (1.229,1.317) | 1.271 (1.229,1.318) | 1.270 (1.231,1.319) | 1.269 (1.23,1.318) | 1.269 (1.23,1.317) | 1.270 (1.23,1.319) | 1.269 (1.23,1.317) |
| l | h | 3.052 (2.807,3.349) | 3.053 (2.81,3.351) | 3.050 (2.811,3.354) | 3.057 (2.808,3.358) | 3.054 (2.807,3.351) | 3.055 (2.813,3.354) | 3.053 (2.805,3.347) | 3.057 (2.803,3.357) | 3.056 (2.806,3.36) | 3.054 (2.818,3.353) |
| m | h | 1.894 (1.796,2.008) | 1.895 (1.798,2.009) | 1.895 (1.8,2.004) | 1.893 (1.799,2.005) | 1.893 (1.801,2.006) | 1.893 (1.8,2.004) | 1.895 (1.801,2.008) | 1.895 (1.798,2.008) | 1.894 (1.8,2.008) | 1.893 (1.801,2.004) |
| h | h | 1.414 (1.374,1.462) | 1.413 (1.374,1.461) | 1.414 (1.373,1.46) | 1.414 (1.375,1.463) | 1.414 (1.374,1.462) | 1.413 (1.374,1.46) | 1.413 (1.374,1.46) | 1.413 (1.374,1.459) | 1.414 (1.374,1.461) | 1.413 (1.373,1.461) |
| Third eigenvalue | |||||||||||
| l | l | 1.264 (1.155,1.385) | 1.262 (1.154,1.382) | 1.264 (1.156,1.388) | 1.263 (1.151,1.387) | 1.264 (1.153,1.384) | 1.264 (1.155,1.383) | 1.264 (1.151,1.386) | 1.262 (1.151,1.382) | 1.263 (1.151,1.381) | 1.263 (1.151,1.384) |
| m | l | 1.135 (1.081,1.195) | 1.134 (1.081,1.194) | 1.134 (1.082,1.193) | 1.134 (1.079,1.193) | 1.135 (1.082,1.192) | 1.134 (1.083,1.19) | 1.135 (1.081,1.194) | 1.134 (1.081,1.196) | 1.134 (1.081,1.193) | 1.134 (1.081,1.192) |
| h | l | 1.068 (1.043,1.096) | 1.068 (1.043,1.097) | 1.068 (1.042,1.098) | 1.068 (1.042,1.096) | 1.068 (1.041,1.096) | 1.068 (1.043,1.096) | 1.067 (1.041,1.095) | 1.068 (1.042,1.097) | 1.068 (1.042,1.097) | 1.068 (1.042,1.097) |
| l | m | 1.857 (1.723,2.009) | 1.859 (1.724,2.008) | 1.857 (1.725,2.003) | 1.859 (1.723,2.007) | 1.860 (1.72,2.008) | 1.860 (1.723,2.011) | 1.858 (1.725,2.007) | 1.856 (1.715,2.009) | 1.858 (1.723,2.009) | 1.859 (1.714,2.01) |
| m | m | 1.408 (1.348,1.474) | 1.408 (1.348,1.472) | 1.409 (1.35,1.474) | 1.409 (1.351,1.473) | 1.408 (1.349,1.473) | 1.408 (1.35,1.474) | 1.409 (1.348,1.474) | 1.409 (1.35,1.476) | 1.409 (1.349,1.475) | 1.409 (1.351,1.476) |
| h | m | 1.199 (1.172,1.23) | 1.199 (1.172,1.23) | 1.199 (1.172,1.229) | 1.199 (1.172,1.229) | 1.200 (1.172,1.23) | 1.199 (1.172,1.23) | 1.199 (1.173,1.227) | 1.199 (1.171,1.23) | 1.199 (1.171,1.229) | 1.199 (1.171,1.23) |
| l | h | 2.622 (2.467,2.791) | 2.624 (2.468,2.799) | 2.622 (2.464,2.802) | 2.622 (2.464,2.793) | 2.623 (2.467,2.801) | 2.623 (2.46,2.8) | 2.620 (2.461,2.793) | 2.619 (2.457,2.801) | 2.623 (2.466,2.796) | 2.622 (2.462,2.795) |
| m | h | 1.733 (1.668,1.806) | 1.733 (1.667,1.803) | 1.733 (1.67,1.802) | 1.732 (1.668,1.8) | 1.733 (1.669,1.805) | 1.733 (1.667,1.802) | 1.733 (1.67,1.804) | 1.733 (1.667,1.804) | 1.733 (1.667,1.805) | 1.732 (1.669,1.801) |
| h | h | 1.346 (1.318,1.377) | 1.346 (1.318,1.376) | 1.346 (1.318,1.377) | 1.347 (1.317,1.376) | 1.347 (1.318,1.378) | 1.346 (1.318,1.379) | 1.347 (1.318,1.376) | 1.346 (1.318,1.376) | 1.346 (1.318,1.376) | 1.346 (1.318,1.376) |
| Fifth eigenvalue | |||||||||||
| l | l | 1.021 (0.926,1.118) | 1.022 (0.926,1.118) | 1.021 (0.928,1.117) | 1.020 (0.927,1.115) | 1.021 (0.925,1.118) | 1.022 (0.927,1.115) | 1.020 (0.925,1.112) | 1.021 (0.93,1.115) | 1.021 (0.925,1.114) | 1.022 (0.93,1.116) |
| m | l | 1.018 (0.972,1.066) | 1.019 (0.972,1.066) | 1.018 (0.972,1.063) | 1.017 (0.971,1.064) | 1.018 (0.973,1.066) | 1.019 (0.973,1.065) | 1.019 (0.973,1.066) | 1.019 (0.974,1.065) | 1.019 (0.972,1.065) | 1.019 (0.974,1.064) |
| h | l | 1.012 (0.99,1.036) | 1.011 (0.988,1.035) | 1.011 (0.988,1.035) | 1.011 (0.989,1.034) | 1.011 (0.989,1.034) | 1.011 (0.988,1.034) | 1.011 (0.988,1.034) | 1.011 (0.988,1.035) | 1.011 (0.988,1.034) | 1.011 (0.988,1.035) |
| l | m | 1.598 (1.487,1.715) | 1.598 (1.485,1.714) | 1.598 (1.487,1.71) | 1.599 (1.489,1.714) | 1.600 (1.492,1.714) | 1.599 (1.49,1.712) | 1.598 (1.492,1.713) | 1.597 (1.486,1.716) | 1.600 (1.493,1.715) | 1.598 (1.489,1.712) |
| m | m | 1.298 (1.25,1.352) | 1.299 (1.249,1.351) | 1.299 (1.249,1.352) | 1.299 (1.25,1.352) | 1.299 (1.25,1.35) | 1.298 (1.251,1.349) | 1.299 (1.249,1.351) | 1.299 (1.25,1.351) | 1.299 (1.25,1.351) | 1.299 (1.249,1.352) |
| h | m | 1.149 (1.125,1.174) | 1.149 (1.126,1.173) | 1.149 (1.127,1.174) | 1.149 (1.126,1.174) | 1.149 (1.126,1.174) | 1.149 (1.125,1.174) | 1.149 (1.125,1.173) | 1.149 (1.126,1.175) | 1.149 (1.126,1.173) | 1.149 (1.125,1.173) |
| l | h | 2.331 (2.204,2.469) | 2.333 (2.201,2.472) | 2.332 (2.203,2.474) | 2.332 (2.204,2.467) | 2.330 (2.201,2.47) | 2.330 (2.199,2.47) | 2.331 (2.202,2.47) | 2.329 (2.199,2.468) | 2.333 (2.206,2.475) | 2.330 (2.201,2.463) |
| m | h | 1.621 (1.568,1.679) | 1.622 (1.568,1.679) | 1.622 (1.568,1.679) | 1.622 (1.569,1.681) | 1.621 (1.568,1.678) | 1.622 (1.57,1.679) | 1.622 (1.569,1.679) | 1.621 (1.569,1.677) | 1.621 (1.568,1.679) | 1.622 (1.568,1.68) |
| h | h | 1.299 (1.275,1.324) | 1.299 (1.275,1.323) | 1.299 (1.275,1.324) | 1.299 (1.276,1.325) | 1.299 (1.275,1.324) | 1.299 (1.274,1.324) | 1.299 (1.275,1.325) | 1.299 (1.275,1.324) | 1.299 (1.275,1.324) | 1.299 (1.275,1.324) |
Analyses using 5,000 iterations each with 95% quantile intervals of nine data sets using ten different distributions. Simulated data sets vary in terms of low (l), medium (m), or high (h) numbers of observations (N) and numbers of variables (P). The upper quantile is an implementation of Glorfeld’s (1995) estimate.
Table 2.
Mean Estimates for First, Third, and Fifth Random Eigenvalues From Parallel Analyses Using Common Factor Analysis with No Rotation
| Data Set | Distribution method | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | P | A | B | C | D | E | F | G | H | I | J |
| First eigenvalue | |||||||||||
| l | l | 0.770 (0.547, 1.040) | 0.771 (0.551, 1.036) | 0.774 (0.544, 1.038) | 0.770 (0.547, 1.033) | 0.772 (0.55, 1.043) | 0.773 (0.550, 1.052) | 0.770 (0.549, 1.039) | 0.780 (0.545, 1.064) | 0.769 (0.546, 1.040) | 0.769 (0.546, 1.051) |
| m | l | 0.334 (0.240, 0.449) | 0.333 (0.239, 0.449) | 0.333 (0.238, 0.448) | 0.334 (0.240, 0.448) | 0.334 (0.243, 0.448) | 0.334 (0.240, 0.450) | 0.334 (0.240, 0.450) | 0.334 (0.240, 0.447) | 0.334 (0.240, 0.455) | 0.334 (0.240, 0.447) |
| h | l | 0.154 (0.112, 0.205) | 0.154 (0.112, 0.204) | 0.154 (0.111, 0.205) | 0.153 (0.110, 0.205) | 0.154 (0.112, 0.205) | 0.154 (0.112, 0.207) | 0.153 (0.113, 0.202) | 0.153 (0.110, 0.204) | 0.153 (0.112, 0.204) | 0.154 (0.113, 0.204) |
| l | m | 1.603 (1.368, 1.892) | 1.606 (1.372, 1.879) | 1.599 (1.361, 1.889) | 1.603 (1.364, 1.883) | 1.604 (1.364, 1.887) | 1.601 (1.363, 1.893) | 1.603 (1.367, 1.892) | 1.611 (1.365, 1.912) | 1.604 (1.367, 1.900) | 1.606 (1.373, 1.893) |
| m | m | 0.658 (0.559, 0.775) | 0.658 (0.562, 0.776) | 0.658 (0.558, 0.774) | 0.658 (0.561, 0.780) | 0.658 (0.561, 0.776) | 0.658 (0.564, 0.776) | 0.658 (0.561, 0.776) | 0.659 (0.559, 0.777) | 0.659 (0.560, 0.774) | 0.658 (0.559, 0.776) |
| h | m | 0.293 (0.250, 0.344) | 0.293 (0.250, 0.344) | 0.292 (0.249, 0.342) | 0.293 (0.250, 0.342) | 0.293 (0.250, 0.343) | 0.292 (0.249, 0.343) | 0.293 (0.251, 0.343) | 0.293 (0.250, 0.343) | 0.292 (0.250, 0.341) | 0.293 (0.250, 0.343) |
| l | h | 2.733 (2.473, 3.040) | 2.733 (2.483, 3.033) | 2.729 (2.479, 3.030) | 2.729 (2.476, 3.037) | 2.732 (2.474, 3.030) | 2.733 (2.469, 3.051) | 2.734 (2.478, 3.040) | 2.741 (2.480, 3.060) | 2.729 (2.476, 3.041) | 2.732 (2.476, 3.035) |
| m | h | 1.070 (0.969, 1.188) | 1.069 (0.967, 1.189) | 1.071 (0.968, 1.189) | 1.071 (0.969, 1.190) | 1.069 (0.971, 1.185) | 1.070 (0.971, 1.186) | 1.069 (0.970, 1.188) | 1.072 (0.970, 1.193) | 1.070 (0.970, 1.187) | 1.068 (0.967, 1.184) |
| h | h | 0.458 (0.416, 0.506) | 0.458 (0.416, 0.506) | 0.458 (0.416, 0.508) | 0.458 (0.417, 0.506) | 0.459 (0.418, 0.507) | 0.458 (0.417, 0.506) | 0.459 (0.416, 0.507) | 0.459 (0.416, 0.508) | 0.459 (0.416, 0.509) | 0.459 (0.418, 0.508) |
| Third eigenvalue | |||||||||||
| l | l | 0.381 (0.242, 0.535) | 0.382 (0.242, 0.539) | 0.383 (0.240, 0.541) | 0.381 (0.243, 0.536) | 0.383 (0.240, 0.547) | 0.384 (0.246, 0.546) | 0.383 (0.240, 0.539) | 0.380 (0.238, 0.547) | 0.383 (0.242, 0.545) | 0.381 (0.243, 0.536) |
| m | l | 0.164 (0.102, 0.233) | 0.163 (0.105, 0.232) | 0.163 (0.105, 0.231) | 0.163 (0.102, 0.234) | 0.164 (0.102, 0.231) | 0.164 (0.104, 0.232) | 0.164 (0.103, 0.230) | 0.164 (0.104, 0.232) | 0.164 (0.102, 0.232) | 0.163 (0.103, 0.235) |
| h | l | 0.075 (0.047, 0.107) | 0.075 (0.047, 0.106) | 0.075 (0.047, 0.107) | 0.075 (0.047, 0.106) | 0.075 (0.047, 0.107) | 0.075 (0.047, 0.106) | 0.075 (0.047, 0.107) | 0.075 (0.047, 0.106) | 0.075 (0.048, 0.106) | 0.075 (0.047, 0.106) |
| l | m | 1.204 (1.035, 1.382) | 1.201 (1.038, 1.378) | 1.199 (1.038, 1.381) | 1.201 (1.045, 1.378) | 1.203 (1.040, 1.379) | 1.199 (1.039, 1.374) | 1.200 (1.041, 1.368) | 1.201 (1.038, 1.381) | 1.198 (1.040, 1.369) | 1.201 (1.039, 1.374) |
| m | m | 0.494 (0.424, 0.567) | 0.495 (0.425, 0.568) | 0.495 (0.427, 0.568) | 0.495 (0.427, 0.570) | 0.495 (0.425, 0.571) | 0.495 (0.427, 0.569) | 0.494 (0.425, 0.567) | 0.494 (0.426, 0.569) | 0.495 (0.426, 0.571) | 0.494 (0.426, 0.568) |
| h | m | 0.220 (0.191, 0.254) | 0.221 (0.190, 0.253) | 0.221 (0.191, 0.254) | 0.221 (0.190, 0.252) | 0.221 (0.191, 0.253) | 0.221 (0.191, 0.253) | 0.221 (0.191, 0.253) | 0.221 (0.190, 0.255) | 0.220 (0.191, 0.254) | 0.221 (0.191, 0.253) |
| l | h | 2.295 (2.121, 2.476) | 2.295 (2.128, 2.481) | 2.295 (2.123, 2.484) | 2.295 (2.123, 2.477) | 2.297 (2.126, 2.483) | 2.294 (2.125, 2.478) | 2.297 (2.125, 2.484) | 2.299 (2.122, 2.497) | 2.295 (2.129, 2.483) | 2.295 (2.122, 2.487) |
| m | h | 0.905 (0.833, 0.982) | 0.905 (0.833, 0.985) | 0.905 (0.834, 0.981) | 0.905 (0.835, 0.982) | 0.905 (0.835, 0.983) | 0.905 (0.834, 0.983) | 0.905 (0.834, 0.981) | 0.905 (0.833, 0.983) | 0.905 (0.835, 0.982) | 0.904 (0.833, 0.982) |
| h | h | 0.389 (0.360, 0.421) | 0.390 (0.360, 0.421) | 0.390 (0.359, 0.422) | 0.389 (0.360, 0.421) | 0.389 (0.359, 0.422) | 0.390 (0.359, 0.421) | 0.390 (0.360, 0.423) | 0.390 (0.359, 0.423) | 0.390 (0.359, 0.423) | 0.390 (0.360, 0.423) |
| Fifth eigenvalue | |||||||||||
| l | l | 0.123 (0.023, 0.239) | 0.122 (0.026, 0.239) | 0.123 (0.023, 0.244) | 0.124 (0.023, 0.246) | 0.123 (0.022, 0.241) | 0.122 (0.020, 0.241) | 0.123 (0.024, 0.238) | 0.123 (0.023, 0.235) | 0.124 (0.024, 0.242) | 0.122 (0.020, 0.240) |
| m | l | 0.043 (−0.003, 0.095) | 0.043 (−0.004, 0.097) | 0.043 (−0.005, 0.095) | 0.044 (−0.003, 0.095) | 0.043 (−0.004, 0.095) | 0.044 (−0.003, 0.095) | 0.043 (−0.004, 0.098) | 0.043 (−0.003, 0.093) | 0.043 (−0.003, 0.095) | 0.043 (−0.003, 0.095) |
| h | l | 0.017 (−0.005, 0.042) | 0.017 (−0.005, 0.042) | 0.018 (−0.005, 0.042) | 0.017 (−0.006, 0.042) | 0.018 (−0.006, 0.042) | 0.018 (−0.005, 0.042) | 0.017 (−0.006, 0.042) | 0.017 (−0.005, 0.042) | 0.017 (−0.006, 0.042) | 0.018 (−0.005, 0.043) |
| l | m | 0.930 (0.794, 1.077) | 0.931 (0.798, 1.074) | 0.932 (0.795, 1.072) | 0.929 (0.792, 1.070) | 0.931 (0.793, 1.074) | 0.931 (0.799, 1.076) | 0.931 (0.796, 1.070) | 0.928 (0.793, 1.072) | 0.929 (0.799, 1.072) | 0.929 (0.794, 1.071) |
| m | m | 0.381 (0.323, 0.441) | 0.381 (0.324, 0.442) | 0.381 (0.322, 0.439) | 0.380 (0.322, 0.441) | 0.381 (0.324, 0.441) | 0.381 (0.323, 0.442) | 0.380 (0.323, 0.440) | 0.380 (0.323, 0.441) | 0.381 (0.325, 0.441) | 0.380 (0.322, 0.441) |
| h | m | 0.169 (0.144, 0.197) | 0.170 (0.145, 0.196) | 0.170 (0.145, 0.196) | 0.169 (0.144, 0.196) | 0.169 (0.144, 0.196) | 0.169 (0.144, 0.196) | 0.169 (0.144, 0.195) | 0.170 (0.144, 0.196) | 0.169 (0.145, 0.195) | 0.169 (0.144, 0.196) |
| l | h | 2.002 (1.860, 2.155) | 2.004 (1.860, 2.156) | 2.004 (1.862, 2.150) | 2.004 (1.864, 2.152) | 2.003 (1.861, 2.157) | 2.004 (1.861, 2.154) | 2.002 (1.860, 2.146) | 2.003 (1.860, 2.156) | 2.004 (1.864, 2.150) | 2.003 (1.861, 2.155) |
| m | h | 0.791 (0.731, 0.854) | 0.793 (0.733, 0.854) | 0.792 (0.731, 0.854) | 0.791 (0.730, 0.855) | 0.792 (0.731, 0.856) | 0.791 (0.729, 0.856) | 0.792 (0.730, 0.855) | 0.792 (0.730, 0.857) | 0.791 (0.732, 0.854) | 0.791 (0.731, 0.852) |
| h | h | 0.341 (0.316, 0.368) | 0.341 (0.316, 0.368) | 0.341 (0.315, 0.369) | 0.341 (0.316, 0.368) | 0.341 (0.315, 0.369) | 0.342 (0.315, 0.369) | 0.341 (0.315, 0.368) | 0.341 (0.315, 0.368) | 0.341 (0.316, 0.369) | 0.341 (0.315, 0.368) |
Analyses using 5,000 iterations each with 95% quantile intervals of nine data sets using ten different distributions. Simulated data sets vary in terms of low (l), medium (m), or high (h) numbers of observations (N) and numbers of variables (P). The upper quantile is an implementation of Glorfeld’s (1995) estimate.
Table 3a.
Means and Standard Deviations of Eigenvalue Estimates Across the Parallel Analyses Using Principal Component Analysis with No Rotation
| Data Set | Mean (SD)
|
|||
|---|---|---|---|---|
| N | P | Mean | 2.5th centile | 97.5th centile |
| First eigenvalue | ||||
| l | l | 1.620 (0.002) | 1.438 (0.003) | 1.853 (0.007) |
| m | l | 1.295 (0.001) | 1.214 (0.001) | 1.396 (0.002) |
| h | l | 1.144 (<0.001) | 1.105 (0.001) | 1.191 (0.001) |
| l | m | 2.244 (0.002) | 2.034 (0.003) | 2.508 (0.008) |
| m | m | 1.568 (0.001) | 1.479 (0.001) | 1.676 (0.002) |
| h | m | 1.269 (<0.001) | 1.230 (<0.001) | 1.316 (0.001) |
| l | h | 3.052 (0.002) | 2.807 (0.004) | 3.349 (0.004) |
| m | h | 1.894 (0.001) | 1.796 (0.002) | 2.008 (0.002) |
| h | h | 1.414 (<0.001) | 1.374 (0.001) | 1.462 (0.001) |
| Third eigenvalue | ||||
| l | l | 1.264 (0.001) | 1.155 (0.002) | 1.385 (0.002) |
| m | l | 1.135 (<0.001) | 1.081 (0.001) | 1.195 (0.002) |
| h | l | 1.068 (<0.001) | 1.043 (0.001) | 1.096 (0.001) |
| l | m | 1.857 (0.001) | 1.723 (0.004) | 2.009 (0.002) |
| m | m | 1.408 (<0.001) | 1.348 (0.001) | 1.474 (0.001) |
| h | m | 1.199 (<0.001) | 1.172 (0.001) | 1.230 (0.001) |
| l | h | 2.622 (0.002) | 2.467 (0.003) | 2.791 (0.004) |
| m | h | 1.733 (<0.001) | 1.668 (0.001) | 1.806 (0.002) |
| h | h | 1.346 (<0.001) | 1.318 (<0.001) | 1.377 (0.001) |
| Fifth eigenvalue | ||||
| l | l | 1.021 (0.001) | 0.926 (0.002) | 1.118 (0.002) |
| m | l | 1.018 (<0.001) | 0.972 (0.001) | 1.066 (0.001) |
| h | l | 1.012 (<0.001) | 0.990 (0.001) | 1.036 (0.001) |
| l | m | 1.598 (0.001) | 1.487 (0.003) | 1.715 (0.002) |
| m | m | 1.298 (<0.001) | 1.250 (0.001) | 1.352 (0.001) |
| h | m | 1.149 (<0.001) | 1.125 (0.001) | 1.174 (0.001) |
| l | h | 2.331 (0.001) | 2.204 (0.002) | 2.469 (0.003) |
| m | h | 1.621 (<0.001) | 1.568 (0.001) | 1.679 (0.001) |
| h | h | 1.299 (<0.001) | 1.275 (<0.001) | 1.324 (0.001) |
Analyses using 5,000 iterations each, and ten different distributional methods.
Table 3b.
Means and Standard Deviations of Eigenvalue Estimates Across the Parallel Analyses Using Principal Component Analysis with No Rotation
| Data Set | Mean (SD)
|
|||
|---|---|---|---|---|
| N | P | Mean | 2.5th centile | 97.5th centile |
| First eigenvalue | ||||
| l | l | 1.602 (0.016) | 1.408 (0.023) | 1.788 (0.043) |
| m | l | 1.295 (0.010) | 1.207 (0.013) | 1.391 (0.022) |
| h | l | 1.143 (0.002) | 1.112 (0.005) | 1.183 (0.007) |
| l | m | 2.231 (0.017) | 2.044 (0.017) | 2.396 (0.056) |
| m | m | 1.575 (0.006) | 1.473 (0.010) | 1.670 (0.011) |
| h | m | 1.268 (0.003) | 1.237 (0.004) | 1.303 (0.008) |
| l | h | 3.068 (0.028) | 2.843 (0.031) | 3.349 (0.070) |
| m | h | 1.888 (0.006) | 1.797 (0.016) | 1.984 (0.017) |
| h | h | 1.414 (0.003) | 1.376 (0.005) | 1.458 (0.007) |
| Third eigenvalue | ||||
| l | l | 1.252 (0.007) | 1.168 (0.006) | 1.341 (0.029) |
| m | l | 1.137 (0.004) | 1.088 (0.008) | 1.194 (0.012) |
| h | l | 1.069 (0.001) | 1.050 (0.006) | 1.088 (0.005) |
| l | m | 1.860 (0.008) | 1.744 (0.015) | 2.038 (0.032) |
| m | m | 1.413 (0.004) | 1.349 (0.006) | 1.476 (0.009) |
| h | m | 1.195 (0.003) | 1.172 (0.002) | 1.221 (0.007) |
| l | h | 2.621 (0.010) | 2.486 (0.025) | 2.814 (0.023) |
| m | h | 1.730 (0.005) | 1.678 (0.007) | 1.790 (0.014) |
| h | h | 1.344 (0.002) | 1.320 (0.002) | 1.375 (0.007) |
| Fifth eigenvalue | ||||
| l | l | 1.023 (0.007) | 0.935 (0.017) | 1.123 (0.015) |
| m | l | 1.020 (0.004) | 0.968 (0.010) | 1.065 (0.005) |
| h | l | 1.011 (0.001) | 0.992 (0.004) | 1.032 (0.003) |
| l | m | 1.591 (0.012) | 1.500 (0.024) | 1.694 (0.010) |
| m | m | 1.303 (0.004) | 1.255 (0.006) | 1.350 (0.011) |
| h | m | 1.149 (0.002) | 1.127 (0.005) | 1.173 (0.006) |
| l | h | 2.337 (0.011) | 2.209 (0.016) | 2.453 (0.014) |
| m | h | 1.622 (0.003) | 1.585 (0.009) | 1.682 (0.008) |
| h | h | 1.299 (0.001) | 1.282 (0.005) | 1.319 (0.004) |
Analyses using 50 iterations each, and ten different distributional methods.
Table 4a.
Means and Standard Deviations of Eigenvalue Estimates Across the Parallel Analyses Using Common Factor Analysis with No Rotation
| Data Set | Mean (SD)
|
|||
|---|---|---|---|---|
| N | P | Mean | 2.5th centile | 97.5th centile |
| First eigenvalue | ||||
| l | l | 0.770 (0.003) | 0.547 (0.002) | 1.040 (0.009) |
| m | l | 0.334 (0.001) | 0.240 (0.001) | 0.449 (0.002) |
| h | l | 0.154 (<0.001) | 0.112 (0.001) | 0.205 (0.001) |
| l | m | 1.603 (0.003) | 1.368 (0.004) | 1.892 (0.009) |
| m | m | 0.658 (<0.001) | 0.559 (0.002) | 0.775 (0.002) |
| h | m | 0.293 (<0.001) | 0.250 (0.001) | 0.344 (0.001) |
| l | h | 2.733 (0.003) | 2.473 (0.004) | 3.040 (0.009) |
| m | h | 1.070 (0.001) | 0.969 (0.001) | 1.188 (0.003) |
| h | h | 0.458 (<0.001) | 0.416 (0.001) | 0.506 (0.001) |
| Third eigenvalue | ||||
| l | l | 0.381 (0.001) | 0.242 (0.002) | 0.535 (0.005) |
| m | l | 0.164 (<0.001) | 0.102 (0.001) | 0.233 (0.002) |
| h | l | 0.075 (<0.001) | 0.047 (<0.001) | 0.107 (<0.001) |
| l | m | 1.204 (0.002) | 1.035 (0.002) | 1.382 (0.005) |
| m | m | 0.494 (0.001) | 0.424 (0.001) | 0.567 (0.002) |
| h | m | 0.220 (<0.001) | 0.191 (<0.001) | 0.254 (0.001) |
| l | h | 2.295 (0.001) | 2.121 (0.003) | 2.476 (0.006) |
| m | h | 0.905 (<0.001) | 0.833 (0.001) | 0.982 (0.001) |
| h | h | 0.389 (<0.001) | 0.360 (<0.001) | 0.421 (0.001) |
| Fifth eigenvalue | ||||
| l | l | 0.123 (0.001) | 0.023 (0.002) | 0.239 (0.003) |
| m | l | 0.043 (<0.001) | −0.003 (0.001) | 0.095 (0.001) |
| h | l | 0.017 (<0.001) | −0.005 (<0.001) | 0.042 (<0.001) |
| l | m | 0.930 (0.001) | 0.794 (0.003) | 1.077 (0.002) |
| m | m | 0.381 (<0.001) | 0.323 (0.001) | 0.441 (0.001) |
| h | m | 0.169 (<0.001) | 0.144 (<0.001) | 0.197 (0.001) |
| l | h | 2.002 (0.001) | 1.860 (0.002) | 2.155 (0.004) |
| m | h | 0.791 (0.001) | 0.731 (0.001) | 0.854 (0.002) |
| h | h | 0.341 (<0.001) | 0.316 (0.001) | 0.368 (<0.001) |
Analyses using 5,000 iterations each, and ten different distributional methods.
Table 4b.
Means and standard deviations of eigenvalue estimates across the parallel analyses using FA with ten distributional methods with only 50 iterations.
| Data Set | Mean (SD)
|
|||
|---|---|---|---|---|
| N | P | Mean | 2.5th centile | 97.5th centile |
| First eigenvalue | ||||
| l | l | 0.757 (0.016) | 0.589 (0.021) | 1.007 (0.047) |
| m | l | 0.330 (0.009) | 0.245 (0.011) | 0.412 (0.019) |
| h | l | 0.151 (0.002) | 0.120 (0.004) | 0.182 (0.010) |
| l | m | 1.604 (0.019) | 1.397 (0.038) | 1.869 (0.036) |
| m | m | 0.658 (0.006) | 0.592 (0.013) | 0.774 (0.010) |
| h | m | 0.297 (0.003) | 0.263 (0.004) | 0.345 (0.010) |
| l | h | 2.734 (0.020) | 2.467 (0.033) | 3.006 (0.055) |
| m | h | 1.070 (0.006) | 0.980 (0.015) | 1.216 (0.020) |
| h | h | 0.460 (0.004) | 0.425 (0.006) | 0.500 (0.008) |
| Third eigenvalue | ||||
| l | l | 0.375 (0.009) | 0.238 (0.015) | 0.463 (0.024) |
| m | l | 0.170 (0.005) | 0.091 (0.008) | 0.246 (0.011) |
| h | l | 0.079 (0.002) | 0.052 (0.005) | 0.105 (0.005) |
| l | m | 1.209 (0.012) | 1.054 (0.027) | 1.381 (0.035) |
| m | m | 0.494 (0.005) | 0.425 (0.011) | 0.569 (0.011) |
| h | m | 0.223 (0.002) | 0.192 (0.003) | 0.250 (0.004) |
| l | h | 2.295 (0.011) | 2.081 (0.032) | 2.477 (0.025) |
| m | h | 0.913 (0.006) | 0.835 (0.014) | 0.998 (0.020) |
| h | h | 0.390 (0.003) | 0.365 (0.007) | 0.420 (0.007) |
| Fifth eigenvalue | ||||
| l | l | 0.117 (0.009) | 0.028 (0.013) | 0.203 (0.022) |
| m | l | 0.042 (0.003) | −0.005 (0.005) | 0.094 (0.010) |
| h | l | 0.018 (0.001) | −0.008 (0.004) | 0.034 (0.005) |
| l | m | 0.928 (0.008) | 0.833 (0.019) | 1.048 (0.030) |
| m | m | 0.377 (0.005) | 0.335 (0.009) | 0.433 (0.010) |
| h | m | 0.174 (0.003) | 0.153 (0.004) | 0.197 (0.005) |
| l | h | 2.011 (0.014) | 1.850 (0.030) | 2.134 (0.013) |
| m | h | 0.793 (0.004) | 0.734 (0.011) | 0.850 (0.009) |
| h | h | 0.338 (0.002) | 0.320 (0.003) | 0.359 (0.007) |
Analyses using 50 iterations each, and ten different distributional methods.
Figure 3.
Figures 3a and 3b. Plot connecting the means (black) and 95% quantiles (grey) of 5000 random eigenvalues for simulated data sets with 75 observations and 50 variables for parallel analyses conducted with ten different random data distributions for principal components analysis (3a) and factor analysis (3b). The near perfect overlap of the means and quantiles across the entire range of factors with such a small sample size illustrates the absolute or virtual insensitivity of parallel analysis to the distributional form of simulated data.
As shown in Table 5, only the Laplace distribution (high kurtosis, zero skewness), was ever a significant predictor in the multiple OLS regressions using the Holm procedure to account for multiple comparisons; specifically in 6 out of 255 tests for the PA using common FA with 5000 iterations. When accounting for multiple comparisons using the false discovery rate procedure, a few methods were found to be significant for each of the four parallel analyses. None of the distribution methods were found to be significant predictors across all four analyses, although method H, appears to be slightly more likely to predict in parallel analyses using 5000 iterations. The absence of a clear pattern in predicting eigenvalues, and the very small number of significant predictions out of the total number of tests (<1% in all eight cases) supports the idea that the distribution of random data in PA is unrelated to the distribution of eigenvalues.
Table 5.
Number of Distributional Methods Found to be Significant Predictors of Eigenvalues in Parallel Analysis
| PCA (K = 5000) | PCA (K = 50) | FA (K = 5000) | FA (K = 50) | |
|---|---|---|---|---|
| FWER | 0 (intercepts only) | 0 (intercepts only) | 6 (H: 6) | 0 (intercepts only) |
| FDR | 6 (E: 1, G: 1, H: 4) | 4 (D: 2, I: 2) | 11 (D: 2, H: 9) | 2 (D: 1, H: 1) |
Procedures correcting for multiple comparisons used the family-wise error rate (FWER, see Holm, 1979) and false discovery rate (FDR, see Benjamini & Yekutieli, 2001) in 255 eigenvalues, in four parallel analyses of nine data sets. Specific methods indicated in parentheses. Adjusted intercept terms were always significant for both procedures and always with smaller adjusted p values than any predictor.
Results for Empirical Data Analysis
The results for the NCS-R data (Table 6) are unambiguously insensitive to distributional assumptions. The adjusted eigenvalues for classical PA, and PA with Glorfeld’s (1995) Monte Carlo extension vary not at all, or only by one thousandth in the reported first ten components and common factors. It is notable that PCA and the FA give disparate results for the NCS-R data. PCA (both with and without Monte Carlo) retains as true seven components (adjusted eigenvalues greater than one), while common FA (both with and without Monte Carlo) retains as true 15 common factors (adjusted eigenvalues greater than zero).
Table 6.
Adjusted Eigenvalues for the First Ten Components and Common Factors from the National Comorbidity Study Replicate Data Using Ten Different Distributions in the Simulated Data
| Component | Distribution method
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | J | |
| 1 | 15.201 (15.165) | 15.201 (15.164) | 15.201 (15.164) | 15.201 (15.166) | 15.201 (15.166) | 15.201 (15.165) | 15.201 (15.165) | 15.201 (15.165) | 15.201 (15.163) | 15.201 (15.166) |
| 2 | 2.693 (2.665) | 2.693 (2.667) | 2.693 (2.666) | 2.693 (2.666) | 2.693 (2.666) | 2.693 (2.665) | 2.693 (2.666) | 2.693 (2.666) | 2.693 (2.667) | 2.693 (2.667) |
| 3 | 2.140 (2.117) | 2.140 (2.116) | 2.140 (2.116) | 2.140 (2.116) | 2.139 (2.116) | 2.140 (2.116) | 2.140 (2.117) | 2.140 (2.116) | 2.140 (2.117) | 2.140 (2.117) |
| 4 | 1.675 (1.654) | 1.675 (1.653) | 1.674 (1.653) | 1.675 (1.653) | 1.675 (1.654) | 1.675 (1.655) | 1.675 (1.654) | 1.675 (1.653) | 1.675 (1.654) | 1.675 (1.654) |
| 5 | 1.465 (1.445) | 1.466 (1.446) | 1.465 (1.447) | 1.465 (1.446) | 1.465 (1.447) | 1.466 (1.446) | 1.465 (1.446) | 1.465 (1.446) | 1.465 (1.446) | 1.465 (1.446) |
| 6 | 1.163 (1.144) | 1.163 (1.145) | 1.162 (1.144) | 1.162 (1.145) | 1.163 (1.003) | 1.162 (1.145) | 1.163 (1.145) | 1.163 (1.145) | 1.163 (1.145) | 1.163 (1.145) |
| 7 | 1.020 (1.003) | 1.020 (1.003) | 1.020 (1.002) | 1.020 (1.002) | 1.020 (0.886) | 1.020 (1.003) | 1.020 (1.002) | 1.020 (1.003) | 1.020 (1.003) | 1.020 (1.002) |
| 8 | 0.902 (0.885) | 0.903 (0.887) | 0.903 (0.886) | 0.902 (0.886) | 0.902 (0.746) | 0.903 (0.886) | 0.902 (0.886) | 0.902 (0.886) | 0.903 (0.886) | 0.902 (0.886) |
| 9 | 0.761 (0.745) | 0.762 (0.746) | 0.761 (0.746) | 0.761 (0.745) | 0.761 (0.746) | 0.762 (0.746) | 0.761 (0.746) | 0.761 (0.745) | 0.761 (0.746) | 0.761 (0.746) |
| 10 | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) | 0.731 (0.716) |
| Factor | ||||||||||
| 1 | 14.666 (14.629) | 14.666 (14.629) | 14.666 (14.629) | 14.666 (14.629) | 14.666 (14.629) | 14.666 (14.628) | 14.666 (14.628) | 14.666 (14.629) | 14.666 (14.629) | 14.666 (14.630) |
| 2 | 2.092 (2.063) | 2.092 (2.064) | 2.092 (2.063) | 2.092 (2.064) | 2.092 (2.063) | 2.092 (2.064) | 2.092 (2.063) | 2.092 (2.064) | 2.092 (2.064) | 2.092 (2.064) |
| 3 | 1.655 (1.630) | 1.655 (1.630) | 1.655 (1.630) | 1.655 (1.630) | 1.655 (1.631) | 1.655 (1.630) | 1.655 (1.630) | 1.654 (1.630) | 1.655 (1.630) | 1.655 (1.630) |
| 4 | 1.042 (1.020) | 1.042 (1.020) | 1.042 (1.019) | 1.042 (1.020) | 1.042 (1.020) | 1.042 (1.019) | 1.042 (1.019) | 1.041 (1.019) | 1.041 (1.019) | 1.042 (1.019) |
| 5 | 0.897 (0.876) | 0.898 (0.876) | 0.898 (0.877) | 0.898 (0.877) | 0.897 (0.877) | 0.897 (0.877) | 0.897 (0.876) | 0.897 (0.877) | 0.897 (0.876) | 0.897 (0.877) |
| 6 | 0.609 (0.589) | 0.609 (0.590) | 0.609 (0.590) | 0.609 (0.589) | 0.609 (0.590) | 0.609 (0.589) | 0.609 (0.589) | 0.609 (0.590) | 0.609 (0.590) | 0.609 (0.589) |
| 7 | 0.472 (0.453) | 0.471 (0.454) | 0.471 (0.453) | 0.471 (0.453) | 0.471 (0.453) | 0.471 (0.452) | 0.471 (0.453) | 0.472 (0.453) | 0.471 (0.453) | 0.471 (0.452) |
| 8 | 0.298 (0.280) | 0.297 (0.280) | 0.297 (0.280) | 0.297 (0.279) | 0.297 (0.280) | 0.297 (0.280) | 0.298 (0.280) | 0.298 (0.280) | 0.297 (0.280) | 0.298 (0.280) |
| 9 | 0.195 (0.177) | 0.195 (0.178) | 0.194 (0.178) | 0.195 (0.178) | 0.195 (0.177) | 0.195 (0.178) | 0.195 (0.177) | 0.195 (0.177) | 0.195 (0.178) | 0.195 (0.177) |
| 10 | 0.137 (0.121) | 0.137 (0.120) | 0.137 (0.121) | 0.137 (0.121) | 0.137 (0.121) | 0.137 (0.121) | 0.137 (0.121) | 0.137 (0.121) | 0.137 (0.121) | 0.137 (0.121) |
There were 1918 complete observations on 51 variables. Adjusted eigenvalues are reported for classical parallel analysis and in parentheses for parallel analysis with Glorfeld’s (1995) Monte Carlo estimate using the 97.5th centile.
Conclusion
PA appears to be either absolutely or virtually insensitive to both the distributional form, and parameterization of uncorrelated variables in the simulated data sets. Both mean and centile estimates were stable across varied distributional assumptions in ten different data generating methods for PA, including Horn’s (1965) original method. For both PCA and common FA, a small number of iterations was associated with larger variances than for 5,000 iterations. The results with an empirical data set were more striking with almost no variance in the adjusted eigenvalues for either PCA or FA, for PA with and without Monte Carlo.
Strengths and Limitations
Because the distribution of random data used in Horn’s PA can only affect the analysis though estimates based on random eigenvalues, this simulation study did not consider variation or heterogeneity in the number of true components or factors, uniqueness, correlations among factors, or patterns in the true loading structure. The finding of simulated data insensitivity holds for changes in any of these qualities.
I conclude that component-retention decision guided by ‘classical’ PA and the large-sample Monte Carlo improvement upon it, are unaffected by the distributional form of the random data in the analysis. There appears to be no reason to use anything other than a simple data-generation scheme when conducting PA. Given the computational costs of the more complicated distributions for random data generation (e.g. rerandomization takes longer to generate than uniformly distributed numbers), and the insensitivity of PA to distributional form, there appears to be no reason to use anything other than the simplest distributional methods such as uniform(0,1) or standard normal distributions.
Postscript
A reply to the current article was written by Dr. James Hayton, and is published immediately following the current article. This postscript contains a short reply by the author to the comments from Dr. Hayton.
Hayton makes several important points in response to my article. We disagree somewhat as regards the historical assertion of a requisite distributional form of the random data employed in PA, but are largely in agreement as to the implications of the insensitivity to distributional form which I report, and strongly agree with one another as regards the need for editors of research journals to push for better standards in acceptance of submitted research.
It is indeed the case that prior scholars have asserted the necessity of specific distributional forms for the simulation data in PA. In Horn’s seminal article, this is a subtle point couched in the behavior of correlation matrices motivating the entire article, and is debatably an assertion of necessity. Subsequent authors have been more blunt. As I quoted them in this article, Thompson and Daniels were explicit in linking “rank” to the distributional properties of the simulation—in their case giving an example of how to simulate a discrete variable. Hayton et al, clearly and unambiguously describe simulation using the observed number of observations and variables in the last paragraph on page 197 of their article. So, in the first paragraph of the next page, it is difficult to read their stressed importance of ensuring “that the values taken by the random data are consistent with those in the comparison set,” (emphasis added) as anything other than a distributional requisite. This is also evident in the comments of their SPSS code regarding the distributional particulars of the simulation, such as mid-points, maximum, etc.. In any event, in this article and Hayton’s response to it the question of a distributional requirement has been explicitly raised, and the importance of its answer considered. In point of historical fact, rather than invoking a straw man, my study was motivated by the concrete programming choices and challenges I faced in writing software which cleaved to published assertions about how to perform PA.
Hayton and I agree that PA appears to be insensitive to distributional form of its simulated data as long as they are independently and identically distributed. We also agree that this gives researchers the advantage of dispensing with both computationally costly rerandomization methods, and with related issues of inferring the distribution of the process generating the observed data. As a direct consequence parallel analysis software can implement random data simulation in the computationally cheapest manner, and researchers can confidently direct their concerns to issues other than the distributional forms of observed data.
PCA and FA are major tools in scale and theory development in many scientific disciplines. Dimensionality, and the question of retention are critical in application, yet, as Hayton eloquently argues, best practice is impeded by researcher and editor complicity in the sanction of the K1 rule. I wonder if funders’ methodology reviewers are also complicit. Certainly, Hayton is correct to draw attention to the complicity of the designers of the default behavior of major statistical computing packages. Until some definitive improvement comes along, PA can serve as a standard by which empirical component or factor retention decisions are made. As there are a growing number of fast free software tools available for any researcher to employ, the bar ought to be raised.
Acknowledgments
This article was prepared while the author was funded by National Institutes of Health postdoctoral training grant (CA-113710).
References
- Allen SJ, Hubbard R. Regression equations for the latent roots of random data correlation matrices with unities on the diagonal. Multivariate Behavioral Research. 1986;21:393–96. doi: 10.1207/s15327906mbr2103_7. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. The Control of the False Discovery Rate in Multiple Testing under Dependency. The Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- Browne MW, Cudeck R. Alternative Ways of Assessing Model Fit. Sociological Methods & Research. 1992;21:230. [Google Scholar]
- Cattell RB. The scree test for the number of factors. Multivariate Behavioral Research. 1966;1:245–276. doi: 10.1207/s15327906mbr0102_10. [DOI] [PubMed] [Google Scholar]
- Cliff N. The eigenvalues-greater-than-one rule and the reliability of components. Psychological bulletin. 1988;103:276–279. [Google Scholar]
- Cota AA, Longman RS, Holden RR, Fekken GC. Comparing Different Methods for Implementing Parallel Analysis: A Practical Index of Accuracy. Educational and Psychological Measurement. 1993;53:865–876. [Google Scholar]
- Dray S. On the number of principal components: A test of dimensionality based on measurements of similarity between matrices. Computational Statistics and Data Analysis. 2008;52:2228–2237. [Google Scholar]
- Ford JK, MacCallum RC, Tait M. The application of exploratory factor analysis in applied psychology: A critical review and analysis. Personnel Psychology. 1986;39:291–314. [Google Scholar]
- Glorfeld LW. An Improvement on Horn’s Parallel Analysis Methodology for Selecting the Correct Number of Factors to Retain. Educational and Psychological Measurement. 1995;55:377–393. [Google Scholar]
- Guttman L. Some necessary conditions for common-factor analysis. Psychometrika. 1954;19:149–161. [Google Scholar]
- Hayton JC, Allen DG, Scarpello V. Factor Retention Decisions in Exploratory Factor Analysis: a Tutorial on Parallel Analysis. Organizational Research Methods. 2004;7:191–205. [Google Scholar]
- Hofer SM, Horn JL, Eber HW. A robust five-factor structure of the 16PF: Strong evidence from independent rotation and confirmatory factorial invariance procedures. Personality and Individual Differences. 1997;23:247–269. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scandinavian Journal of Statistics. 1979;6:1979. [Google Scholar]
- Horn JL. A rationale and test for the number of factors in factor analysis. Psychometrika. 1965;30:179–185. doi: 10.1007/BF02289447. [DOI] [PubMed] [Google Scholar]
- Horn JL, Engstrom R. Cattell’s Scree Test In Relation To Bartlett’s Chi-Square Test And Other Observations On The Number Of Factors Problem. Multivariate Behavioral Research. 1979;14:283–300. doi: 10.1207/s15327906mbr1403_1. [DOI] [PubMed] [Google Scholar]
- Humphreys LG, Montanelli RG. Latent roots of random data correlation matrices with squared multiple correlations on the diagonal: A monte carlo study. Psychometrika. 1976;41:341–348. [Google Scholar]
- Jackson DA. Stopping Rules in Principal Components Analysis: A Comparison of Heuristical and Statistical Approaches. Ecology. 1993;74:2204–2214. [Google Scholar]
- Kaiser H. The Application of Electronic Computers to Factor Analysis. Educational and Psychological Measurement. 1960;20:141–151. [Google Scholar]
- Keeling K. A Regression Equation for Determining the Dimensionality of Data. Multivariate Behavioral Research. 2000;35:457–468. doi: 10.1207/S15327906MBR3504_02. [DOI] [PubMed] [Google Scholar]
- Lambert ZV, Wildt AR, Durand RM. Assessing Sampling Variation Relative to Number-of-Factors Criteria. Educational and Psychological Measurement. 1990;50:33. [Google Scholar]
- Lance CE, Butts MM, Michels LC. The Sources of Four Commonly Reported Cutoff Criteria: What Did They Really Say? Organizational Research Methods. 2006;9:202–220. [Google Scholar]
- Lautenschlager GJ. A comparison of alternatives to conducting Monte Carlo analyses for determining parallel analysis criteria. Multivariate Behavioral Research. 1989;24:365–395. doi: 10.1207/s15327906mbr2403_6. [DOI] [PubMed] [Google Scholar]
- Longman RS, Cota AA, Holden RR, Fekken GC. A regression equation for the parallel analysis criterion in principal components analysis: Mean and 95th percentile eigenvalues. Multivariate Behavioral Research. 1989;24:59–69. doi: 10.1207/s15327906mbr2401_4. [DOI] [PubMed] [Google Scholar]
- Patil V, Singh S, Mishra S, Todd Donavan D. Efficient theory development and factor retention criteria: Abandon the ‘eigenvalue greater than one’ criterion. Journal of Business Research. 2008;61:162–170. [Google Scholar]
- Pennell B, Bowers A, Carr D, Chardoul S, Cheung G, Dinkelmann K, Gebler N, Hansen S, Pennell S, Torres M. The development and implementation of the National Comorbidity Survey Replication, the National Survey of American Life, and the National Latino and Asian American Survey. International Journal of Methods in Psychiatric Research. 2004;13:241–69. doi: 10.1002/mpr.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres-Neto P, Jackson D, Somers K. How many principal components? stopping rules for determining the number of non-trivial axes revisited. Computational Statistics and Data Analysis. 2005;49:974–997. [Google Scholar]
- Preacher KJ, MacCallum RC. Repairing Tom Swift’s Electric Factor Analysis Machine. Understanding Statistics. 2003;2:13–43. [Google Scholar]
- Silverstein AB. Comparison of two criteria for determining the number of factors. Psychological Reports. 1977;41:387–390. [Google Scholar]
- Steiger JH, Lind JC. Handout at the annual meeting of the Psychometric Society. Iowa City, IA: 1980. May, Statistically based tests for the number of common factors. [Google Scholar]
- Thompson B, Daniel LG. Factor Analytic Evidence for the Construct Validity of Scores: A Historical Overview and Some Guidelines. Educational and Psychological Measurement. 1996;56:197–208. [Google Scholar]
- Velicer WF, Eaton CA, Fava JL. Construct explication through factor or component analysis: A review and evaluation of alternative procedures for determining the number of factors or components. In: Goffen RD, Helms E, editors. Problems and Solutions in Human Assessment – Honoring Douglas N Jackson at Seventy. Norwell, MA: Springer; 2000. pp. 41–71. [Google Scholar]
- Velicer WF, Jackson DN. Component Analysis versus Common Factor Analysis: Some Issues in Selecting an Appropriate Procedure. Multivariate Behavioral Research. 1990;25:1–28. doi: 10.1207/s15327906mbr2501_1. [DOI] [PubMed] [Google Scholar]
- Zwick WR, Velicer WF. A comparison of five rules for determining the number of factors to retain. Psychological Bulletin. 1986;99:432–442. [Google Scholar]