The iron–sulphur protein RNase L inhibitor functions in translation termination
Work by the Krebber lab identifies a novel function for the iron-sulfur (Fe/S) containing RNase L inhibitor Rli1 in translation termination. It genetically and physically interacts with the translation termination factors eRF1/Sup45 and eRF3/Sup35. Surprisingly, while the Fe/S cluster is not required for the interaction of Rli1 with eRF1 or its other interacting partner Hcr1 from the initiation complex eIF3, it is required for its activity in translation termination.
Keywords: translation termination, Rli1, eRF1, eRF3, Dbp5
Abstract
The iron–sulphur (Fe–S)-containing RNase L inhibitor (Rli1) is involved in ribosomal subunit maturation, transport of both ribosomal subunits to the cytoplasm, and translation initiation through interaction with the eukaryotic initiation factor 3 (eIF3) complex. Here, we present a new function for Rli1 in translation termination. Through co-immunoprecipitation experiments, we show that Rli1 interacts physically with the translation termination factors eukaryotic release factor 1 (eRF1)/Sup45 and eRF3/Sup35 in Saccharomyces cerevisiae. Genetic interactions were uncovered between a strain depleted for Rli1 and sup35-21 or sup45-2. Furthermore, we show that downregulation of RLI1 expression leads to defects in the recognition of a stop codon, as seen in mutants of other termination factors. By contrast, RLI1 overexpression partly suppresses the read-through defects in sup45-2. Interestingly, we find that although the Fe–S cluster is not required for the interaction of Rli1 with eRF1 or its other interacting partner, Hcr1, from the initiation complex eIF3, it is required for its activity in translation termination; an Fe–S cluster mutant of RLI1 cannot suppress the read-through defects of sup45-2.
Introduction
Translation is well conserved in eukaryotes and can be separated into three distinct steps: initiation, elongation and termination (Kapp & Lorsch, 2004). Initiation involves the assembly of translation-competent ribosomes on messenger RNAs and depends on eukaryotic initiation factors (eIFs) that stimulate ribosome loading. One of the largest initiation factors, eIF3, has six subunits in Saccharomyces cerevisiae (13 in humans), one of which is Hcr1. It stimulates the formation of the 43S pre-initiation complex that binds to the mRNA and scans it to identify the AUG start codon (Hinnebusch, 2006). Interestingly, eIF3 also regulates the dissociation of the ribosome after translation termination (Pisarev et al, 2007).
Termination occurs on synthesis of the polypeptide chain during translation elongation. The process is mediated by recognition of a stop codon through the transfer RNA (tRNA)-mimicking protein eukaryotic release factor 1 (eRF1; Sup45 in S. cerevisiae) and by the subsequent hydrolysis of the ester bond connecting the polypeptide chain and the tRNA, stimulated by the GTPase activity of eRF3 (Sup35 in S. cerevisiae; Jacobson, 2005). Furthermore, the DEAD box RNA helicase Dbp5 has recently been shown to function in translation termination (Gross et al, 2007). It assists eRF1 in stop codon recognition and controls the subsequent eRF1–eRF3 interaction through its dissociation from eRF1 (Gross et al, 2007). The activity of Dbp5 is stimulated by its co-factor, Gle1, and the small molecule inositol hexakisphosphate (Bolger et al, 2008). Interestingly, Gle1 also influences translation initiation, as it interacts genetically and physically with subunits of eIF3, and Gle1 mutants show defects in translation initiation (Bolger et al, 2008).
Here, we have identified the essential iron–sulphur (Fe–S)-containing RNase L inhibitor (Rli1), which belongs to the family of ATP-binding cassette (ABC) proteins, as a new translation termination factor. There are two Fe–S clusters and two ABC domains in Rli1. The crystal structure of archaeal Rli1 shows that the two ABC domains are arranged in a head-to-tail orientation through a hinge domain, suggesting that these domains undergo the tweezer-like power stroke characteristic of ABC enzymes (Karcher et al, 2008). The Rli1 protein requires the mitochondrial and cytosolic Fe–S protein biogenesis machineries for its assembly, and mutations in crucial cysteine residues of Rli1 abolish its association with Fe–S clusters, leading to the loss of cell viability (Kispal et al, 2005; Lill, 2009). The Rli protein associates with polyribosomes (Dong et al, 2004) and with Hcr1, which is proposed to have a dual role in ribosomal RNA processing, as well as in translation initiation. The Rli1 mutants are impaired in precursor rRNA processing and are defective in the export of both ribosomal subunits (Kispal et al, 2005; Yarunin et al, 2005). Furthermore, evidence shows that Rli1 is required for efficient formation and stabilization of 43S and 48S pre-initiation complexes (Dong et al, 2004). The Rli1 protein associates with the components of the eukaryotic translation initiation machinery: eIF2, eIF5 and, in particular, the translation initiation complex eIF3. The Hcr1 protein was enriched visibly in Rli1–TAP (tandem affinity purification; Yarunin et al, 2005). This, together with preliminary yeast two-hybrid experiments (Kispal et al, 2005), suggests that Hcr1 and Rli1 might interact directly, and Hcr1 might link Rli1 to the eIF3 complex and translation initiation.
In human cells, RLI1 was identified originally as an inhibitor of RNase L (Bisbal et al, 1995). The RNase L protein was characterized as a protein that is activated through the interferon system on viral infection (Jacobson, 2005). Recently, an interaction of RNase L with eRF3 was identified, which was then shown to lead to increased translational read-through efficiency at premature termination codons, and to an increased +1 frame-shifting efficiency—which might have an important role in the antiviral response (Jacobson, 2005; Le Roy et al, 2005). Remarkably, Rli1 is highly conserved from yeast to humans, which cannot be explained by a conserved function in viral defence.
In this study, we have identified a new function for the RNase L inhibitor Rli1 as a translation termination factor. We show physical and genetic interactions between Rli1 and both translation termination factors, eRF1 and eRF3. We demonstrate that the second ABC domain of Rli1 is sufficient to mediate the interaction with Hcr1 and eRF1. Furthermore, we find that a functional Fe–S cluster is necessary for the role of Rli1 in stop codon recognition.
Results And Discussion
Two-hybrid interaction of Rli1 and eRF1
To identify new Rli1-interacting proteins, a yeast two-hybrid screen used full-length RLI1 fused to the DNA-binding domain of the transcriptional activator GAL4 and a complementary DNA library fused to the GAL4 activation domain. Approximately five 105 yeast clones were screened and 55 plasmids were identified that gave strong two-hybrid interaction signals with the three reporter genes: ADE2, HIS3 and lacZ (supplementary Fig S1 online). Notably, among these 55 proteins, 32 were identified to encode for HCR1, indicating a favoured interaction between the two proteins. In addition, genes that encode for proteins involved in ribosomal biogenesis (Rpl32, Dbp10, Rps20 and Rps16), glucose metabolism (Pdc1, Std1 and Tdh3) and amino-acid metabolism (His3), as well as in translation termination (Sup45) were identified. Given that ribosomal proteins are known false-positive candidates in yeast two-hybrid screens (Hengen, 1997), and as we do not expect a direct functional role for Rli1 in glucose or amino-acid metabolism, we focused on Hcr1, Sup45 and Dbp10 for further characterization of their interaction with Rli1. Although the interactions of both Sup45 and Hcr1 with Rli1 were verified from both positions (used as a bait or as a prey), an interaction of Rli1 with Dbp10, however, could not be confirmed (data not shown). It will be interesting to characterize the remaining two-hybrid interactions in future experiments and to potentially uncover other important new functions for Rli1. The two-hybrid interaction of Rli1 and Hcr1 is in agreement with earlier results and suggests a direct binding of these proteins (Kispal et al, 2005). By contrast, the two-hybrid interaction detected between Rli1 and Sup45 has not been identified before, and thus both proteins might be new potential interaction partners. Interestingly, a two-hybrid interaction with Sup35 was not identified. This may be due to the potential absence of the gene from the two-hybrid library in the first place, or owing to the possibility that the tagging of the protein prevented an interaction with Rli1.
Physical interaction of Rli1 with eRF1 and eRF3
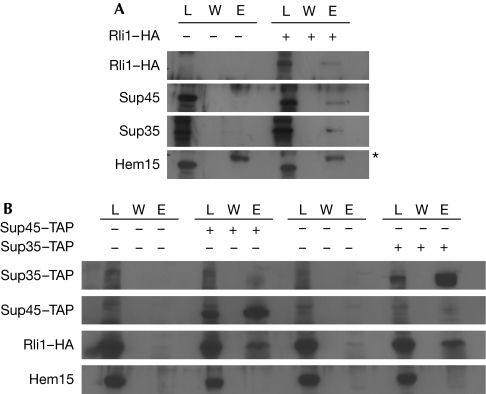
To investigate a potential physical interaction between Rli1 and translation termination factors, we performed interaction studies with eRF1 (Sup45) and eRF3 (Sup35). Wild-type yeast cells were transformed with a plasmid encoding haemagglutinin (HA)-tagged Rli1. Cells were grown to the logarithmic growth phase, before co-immunoprecipitations were performed with Rli1–HA. All samples were treated with RNase to ensure that a direct protein–protein interaction was detected. The eluates were analysed on sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS–PAGE) and the proteins were visualized by immunostaining. As shown in Fig 1A, both translation termination factors interacted with Rli1. To confirm these interactions, we used TAP-tagged Sup45 and Sup35 to co-immunoprecipitate Rli1–HA (Fig 1B). By using both methods, we found Rli1 interacted physically with both translation termination factors.
Figure 1.
Rli1 interacts physically with the translation termination factors eRF1/Sup45 and eRF3/Sup35. (A) Co-immunoprecipitation experiments were performed with HA-tagged Rli1, Sup45 and Sup35 from log-phase yeast cells. Lysates (L), wash fractions (W) and the eluates (E) are shown in western blot analyses. A volume of 10 μl of each lysate and the complete eluates were loaded onto the gel. Each 200 μl eluate including the beads was resuspended in 20 μl of sample buffer and entirely loaded, which represents 20 × the lysate. HA, Sup45, Sup35 and Hem15 (negative control) antibodies were used for detection. The bands visible in the Hem15 eluate fractions are caused by the presence of the cross-reacting antibody chains and are marked by an asterisk. (B) TAP-tagged SUP45 and SUP35 were purified from log-phase yeast cells, carrying a plasmid encoding RLI1–HA. Wild-type cells expressing plasmid-encoded RLI1–HA were the negative control. Western blots of L, W and E are shown. The ratio of lysate to eluate is 1:20. HA and Hem15 (negative control) antibodies were used for detection. HA, haemagglutinin; Rli1, RNase L inhibitor; TAP, tandem affinity purification.
Mapping of the Rli1 interaction domain
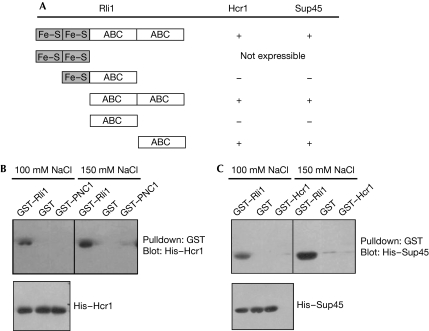
To gain insight into the binding region of Rli1, which mediates the interaction with Sup45 and Hcr1, we performed two-hybrid analyses with truncated versions of the protein. Although rli1 mutants lacking the amino-terminal Fe–S clusters, both Fe–S clusters or the first ABC domain were able to interact with Sup45 and Hcr1, deletion mutants lacking the second carboxy-terminal ATPase domain failed to interact with either Sup45 or Hcr1 (Fig 2A). These results indicate that Fe–S clusters might not be required for the protein interactions. By contrast, we show that the second C-terminal ABC ATPase domain of Rli1 is crucial for the interaction between Rli1 and both Sup45 and Hcr1 in two-hybrid analyses. From these data, we can conclude that the second ABC domain might mediate the interaction of Rli1 with both Hcr1 and Sup45.
Figure 2.
Identification of the Rli1 protein interaction domain for Hcr1 and Sup45. (A) A yeast two-hybrid assay with either full-length or the indicated truncations of Rli1 as bait and either Hcr1 or Sup45 as prey. The domain structure of Rli1 is indicated and positive interactions are shown by +, whereas no interaction is shown by −. (B) GST pulldown assays using His-tagged Hcr1 were performed under the indicated salt concentrations with full-length Rli1 (GST–Rli1) or GST and GST–PNC1 as negative controls. The eluate fractions (top) and the input (bottom) are shown. (C) GST pulldown assays using His-tagged Sup45 and GST–Rli1, GST or GST–Hcr1 were performed as described in (B). ABC, ATP-binding cassette; Fe–S, iron–sulphur; GST, glutathione-S-transferase; Rli1, RNase L inhibitor.
Furthermore, we confirmed the interactions between Rli1 and Hcr1 as well as Sup45 by in vitro binding experiments. Recombinant glutathione-S-transferase (GST) fusion was carried out between Rli1 and His-tagged Hcr1 or Sup45. The GST–Rli1 fusion protein was co-expressed in Escherichia coli with either His–Hcr1 or His–Sup45 and purified on glutathione–Sepharose beads. After extensive washing, beads were analysed on SDS–PAGE and proteins were visualized by using immunostaining. Although GST–Rli1 pulled down His–Hcr1 (Fig 2B) and His–Sup45 (Fig 2C), neither GST nor GST–PNC1, used as a negative control, were able to pull down His-tagged Hcr1. Similarly, GST–Rli1, but not GST or GST–Hcr1, interacts with the His-tagged Sup45 protein. Furthermore, binding of Sup45 and Hcr1 to Rli1 was still observed in the presence of a higher salt concentration (150 mM NaCl), indicating a robust interaction of Rli1 with the translation factors. In addition, from these in vitro studies, we conclude that both Rli1 interactions are direct and do not require additional factors.
Genetic interactions of RLI1 with SUP45 and SUP35
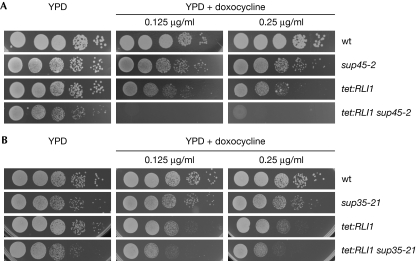
To investigate potential genetic interactions between RLI1 and termination factors, we used a strain in which RLI1 expression can be regulated by the addition of doxocycline (tet:RLI1; Kispal et al, 2005). Doxocycline treatment of tet:RLI1 cells, which completely represses the expression of RLI1, is lethal (supplementary Fig S2 online). However, partial suppression allows growth of the yeast cells, as shown by using an equal number of cells that were spotted in serial dilutions onto full medium (yeast extract–peptone–dextrose) plates and plates containing different concentrations of doxocycline. As shown in Fig 3A, partial repression of RLI1 expression in combination with sup45-2 is synthetically lethal, indicating a strong genetic interaction between RLI1 and SUP45. By contrast, the combination of downregulated RLI1 and sup35-21 shows only a slight growth defect, which might indicate a more significant relationship between Rli1 and Sup45.
Figure 3.
Genetic interactions between RLI1 and translation termination factors eRF1 and eRF3. (A) Serial dilutions of wild type, sup45-2, tet:RLI1 and the double mutant tet:RLI1 sup45-2 were spotted onto full medium plates (YPD) and plates containing the indicated concentrations of doxocycline, which downregulates RLI1 expression. Plates shown are after incubation at 25°C for three days (B) Serial dilutions of wild type, sup35-21, tet:RLI1 and the double mutant tet:RLI1 sup35-21 are shown on YPD and doxocycline plates after incubation at 25°C for three days. eRF, eukaryotic release factor; Rli1, RNase L inhibitor; wt, wild type; YPD, yeast extract–peptone–dextrose.
Rli1 is required for proper stop codon recognition
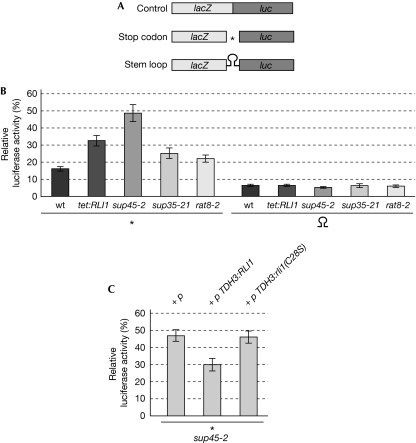
Physical and genetic interactions between Rli1 and the termination factors eRF1 and eRF3 indicate a new function for Rli1 in translation termination. To test this, we investigated whether the downregulation of Rli1 would lead to defects in the recognition of a stop codon by using an established dual reporter assay that detects read-through efficiency (Bhattacharya et al, 2000). The assay is based on compared expression of β-galactosidase and luciferase open reading frames, separated by a stop codon (Fig 4A). As a control, we used a reporter with a stem loop. This assay enabled us to compare the frequency of translational read-through in different strain backgrounds, such as the rat8-2 strain that has known mRNA export defects (Cole & Scarcelli, 2006). All measured luciferase activity was, therefore, calculated in relation to the β-galactosidase activity (Fig 4B,C and supplementary Fig S4B online). Similar to the rat8-2 mutant strain, the downregulation of RLI1 results in an approximately 50% reduced translation rate as compared with wild type (supplementary Fig S3A online), which in case of RLI1 is due to defects in translation initiation (Dong et al, 2004). However, the dual reporter construct enabled us to analyse the true defects in translation termination by calculating the ratio of luciferase activity with regard to the β-galactosidase activity (supplementary Fig S3B online). In agreement with previous results, we observed a basal read-through activity of about 15–17% in wild-type cells (Bhattacharya et al, 2000). Although RLI1 repression did not influence read-through activities in the presence of the stem loop, Rli1 was required for the efficient recognition of the termination codon, similarly to the other termination factors: eRF1, eRF3 and Dbp5 (Fig 4B).
Figure 4.
Rli1 is required for efficient stop codon recognition. (A) Scheme of the reporter constructs used to determine the read-through activity. (B) Read-through activities are shown for wild type, a strain that downregulates the RLI1 expression (tet:RLI1), sup45-2, sup35-21 and rat8-2, encoding the defective DEAD-box RNA helicase Dbp5. All strains carrying either of the reporter constructs were grown to log phase and incubated at 37°C for 15 min before cell lysis. β-Galactosidase and luciferase activities were measured and their ratios were used to calculate the relative molar luciferase expression. (C) Suppression of the termination read-through defects of sup45-2 by increased Rli1 level (TDH3:RLI1), but not an Fe–S cluster mutant (TDH3:rli1(C28S)), is shown. All strains carrying the reporter construct and the indicated plasmids were treated as described in (B). The results of at least 10 independent experiments are shown. The asterisks represent the termination codon UAG. Rli1, RNase L inhibitor; wt, wild type.
In addition, we observed that the defects of sup45-2 in termination codon recognition were partly suppressed by overexpression of RLI1 using the strong TDH3 promoter. Yeast cells containing the reporter construct and either an empty vector (p), plasmids encoding wild-type RLI1 (TDH3:RLI1) or a mutant defective in the Fe–S cluster (TDH3:rli1(C28S)) were grown to the log phase, before the termination read-through activity was determined. As shown in Fig 4C, overexpression of wild-type RLI1, but not the Fe–S cluster mutant, was able to partly suppress the termination read-through defects of sup45-2. The functionality of the TDH3:RLI1 plasmid is shown in supplementary Fig S2 online.
Overexpression of RLI1 in wild-type cells is toxic and leads to translation initiation defects (Dong et al, 2004). The interaction domain of Rli1 with both Hcr1 and Sup45 was determined to the second ABC domain of the protein. Therefore, one would expect that overexpression of this domain in wild-type cells might disturb those interactions and inhibit cellular growth. Indeed, expression of the second ABC domain from the strong GAL1 promoter leads to inhibition of growth (supplementary Fig S4A online), but not to termination read-through defects (supplementary Fig S4B online). There could be several reasons for this: expression of the ABC domain might result in an unnatural folding of this domain; alternatively, it might remain associated with Hcr1 in the initiation complex; or it might associate with other proteins with which the full-length protein has no contact.
Taken together, our results indicate that Rli1 is required for translation termination as the protein interacts with both eRF1 and eRF3 in vivo (Fig 1). The Rli1 protein might regulate the termination process through eRF1, as a strong genetic interaction was detected specifically with a mutant of eRF1 (sup45-2; Fig 3). Consistently, we find that the physical interaction between Rli1 and eRF1 is direct, as no additional factor is required for the interaction in vitro (Fig 2C). Furthermore, although the repression of RLI1 expression leads to defects in the recognition of a stop codon, its overexpression partly suppresses the read-through defects of an eRF1 mutant (sup45-2; Fig 4C). Interestingly, although we show that the Fe–S cluster is not required for the interaction of Rli1 with eRF1 or Hcr1 (Fig 2), it is required for its activity in translation termination, as an Fe–S cluster mutant of Rli1 is not able to suppress the read-through defects of sup45-2 (Fig 4C). We mapped the protein interaction domain of Rli1 to its second C-terminal ATPase domain (Fig 2). Interestingly, Rli1 requires the second ABC domain for its interaction with the termination factor eRF1 and the initiation factor Hcr1. The Hcr1 protein has recently been shown to have a role in ribosomal subunit dissociation and recycling, together with eRF1 (Pisarev et al, 2007). Thus, it is tempting to speculate that Rli1 might also be involved in ribosome recycling. However, as the disassembly of post-termination complexes was successfully performed with purified eIF3/Hcr1 and eRF1 (Pisarev et al, 2007), a potential role of Rli1 in this process is limited. Future experiments are needed to investigate the potential functions for Rli1 beyond translation termination.
Methods
Strains and plasmids. All yeast strains used in this study are listed in supplementary Table S1 online, and the plasmids are listed in supplementary Table S2 online. To analyse several proteins in the same S. cerevisiae strain, we used wild-type cells with genomically tagged genes and plasmids encoding functionally tagged versions of the analysed proteins. Strains were cultivated in standard medium at 25°C. Details on strain constructions and cloning strategies can be found in the supplementary information online. All growth tests are shown in 10-fold serial dilution starting with approximately 105 cells per drop.
Yeast two-hybrid library screen. Full-length RLI1 was fused to the DNA-binding domain of the transcriptional activator GAL4 and used as bait for a two-hybrid screen of a yeast cDNA library fused to the GAL4 activation domain. A detailed description of the procedure is described in the supplementary information online.
Co-immunoprecipitations. The experiment was performed as described in the study by Gross et al (2007). Co-immunoprecipitations were performed using protein G Sepharose beads (Amersham Biosciences, Freiburg, Germany) conjugated to HA-specific antibodies or IgG Sepharose beads (Amersham Biosciences) in the presence or absence of 200 μg/ml RNase. Purification of the TAP-tagged proteins was performed under similar conditions. A detailed description of the procedure is described in the supplementary information online.
GST pulldown analysis. Rosetta II (DE3) cells were transformed with pGEX6P3-Rli1 and with either pET30a-Sup45 or pET28b-Hcr1. After cell lysis and removal of cell debris, the supernatant was incubated with GST Sepharose beads. After washing, beads were analysed on SDS–PAGE followed by western blotting and immunostaining. For a comprehensive protocol, see supplementary information online.
Dual reporter β-galactosidase luciferase assay. The reporter assays were carried out as described previously (Bidou et al, 2000). Different transformants of strains carrying the dual reporter vector and the indicated plasmids were grown to an absorbance of (A600)=0.6–0.8 and lysed. The temperature-sensitive mutants were transferred to 37°C for 30 min before lysis. β-Galactosidase and luciferase activities were assayed from the same cell culture, as described previously (Bidou et al, 2000). Cells were lysed using acid-washed glass beads and vigorous vortexing for 20 s and 4.5 m/s using FastPrep-24 (MP Biomedicals, Illkirch, France). The re-coding efficiency, expressed in percentage, was determined by dividing the luciferase/β-galactosidase ratios of each reporter in relation to the in-frame control construct (also see Bidou et al, 2000).
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank D.M. Bedwell, R. Lill and U. Mühlenhoff, for providing strains and/or plasmids and/or antibodies, and B. Boll and L. Schreiber for providing plasmids and experimental assistance.
Footnotes
The authors declare that they have no conflict of interest.
References
- Bhattacharya A, Czaplinski K, Trifillis P, He F, Jacobson A, Peltz SW (2000) Characterization of the biochemical properties of the human Upf1 gene product that is involved in nonsense-mediated mRNA decay. RNA 6: 1226–1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidou L, Stahl G, Hatin I, Namy O, Rousset JP, Farabaugh PJ (2000) Nonsense-mediated decay mutants do not affect programmed -1 frameshifting. RNA 6: 952–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T (1995) Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J Biol Chem 270: 13308–13317 [DOI] [PubMed] [Google Scholar]
- Bolger TA, Folkmann AW, Tran EJ, Wente SR (2008) The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell 134: 624–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CN, Scarcelli JJ (2006) Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol 18: 299–306 [DOI] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG (2004) The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem 279: 42157–42168 [DOI] [PubMed] [Google Scholar]
- Gross T, Siepmann A, Sturm D, Windgassen M, Scarcelli JJ, Seedorf M, Cole CN, Krebber H (2007) The DEAD-box RNA helicase Dbp5 functions in translation termination. Science 315: 646–649 [DOI] [PubMed] [Google Scholar]
- Hengen PN (1997) False positives from the yeast two-hybrid system. Trends Biochem Sci 22: 33–34 [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG (2006) eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci 31: 553–562 [DOI] [PubMed] [Google Scholar]
- Jacobson A (2005) The end justifies the means. Nat Struct Mol Biol 12: 474–475 [DOI] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR (2004) The molecular mechanics of eukaryotic translation. Annu Rev Biochem 73: 657–704 [DOI] [PubMed] [Google Scholar]
- Karcher A, Schele A, Hopfner KP (2008) X-ray structure of the complete ABC enzyme ABCE1 from Pyrococcus abyssi. J Biol Chem 283: 7962–7971 [DOI] [PubMed] [Google Scholar]
- Kispal G, Sipos K, Lange H, Fekete Z, Bedekovics T, Janaky T, Bassler J, Aguilar Netz DJ, Balk J, Rotte C, Lill R (2005) Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein Rli1p and mitochondria. EMBO J 24: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roy F, Salehzada T, Bisbal C, Dougherty JP, Peltz SW (2005) A newly discovered function for RNase L in regulating translation termination. Nat Struct Mol Biol 12: 505–512 [DOI] [PubMed] [Google Scholar]
- Lill R (2009) Function and biogenesis of iron–sulphur proteins. Nature 460: 831–838 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV (2007) Recycling of eukaryotic post-termination ribosomal complexes. Cell 131: 286–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarunin A, Panse VG, Petfalski E, Dez C, Tollervey D, Hurt EC (2005) Functional link between ribosome formation and biogenesis of iron–sulfur proteins. EMBO J 24: 580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.