Structural basis for the function of DEAH helicases
DEAH helicases catalyse mRNP rearrangements during splicing and ribosome biogenesis. In this paper, Andersen et al. solve the crystal structure of yeast Prp43p in complex with ADP and find homology to DNA helicases and the presence of an OB motif. A β-hairpin from the second RecA domain is wedged between two C-terminal domains and blocks access to the RNA binding site formed by the RecA domains and a C-terminal domain. The structure of Prp43p provides the framework for functional and genetic analysis of all DEAH helicases.
Keywords: helicase, Prp43p, rRNA processing, splicing, X-ray crystallography
Abstract
DEAH helicases participate in pre-messenger RNA splicing and ribosome biogenesis. The structure of yeast Prp43p-ADP reveals the homology of DEAH helicases to DNA helicases and the presence of an oligonucleotide-binding motif. A β-hairpin from the second RecA domain is wedged between two carboxy-terminal domains and blocks access to the occluded RNA binding site formed by the RecA domains and a C-terminal domain. ATP binding and hydrolysis are likely to induce conformational changes in the hairpin that are important for RNA unwinding or ribonucleoprotein remodelling. The structure of Prp43p provides the framework for functional and genetic analysis of all DEAH helicases.
Introduction
RNA helicases use nucleoside triphosphates (NTPs) to unwind double-stranded RNA or remodel protein–RNA complexes (Liu et al, 2008; Hilbert et al, 2009). Prp43p is a DEAH-box helicase involved in the release of the intron lariat during pre-messenger RNA (pre-mRNA) splicing and pre-ribosomal RNA (pre-rRNA) processing in Saccharomyces cerevisiae (Arenas & Abelson, 1997; Combs et al, 2006; Tanaka et al, 2007). Many RNA duplexes that are part of the splicing machinery are short and therefore expected to require proteins for stabilization; such ribonucleoprotein (RNP) complexes might be regulated by remodelling factors (Staley & Guthrie, 1998). Whereas the Prp43p binding site in the spliceosome is still unknown, Prp43p contacts with pre-rRNA have recently been mapped to several different stretches in 18S and 25S rRNA and the helicase was suggested to regulate the association of small nucleolar RNA (snoRNA) with pre-rRNA (Bohnsack et al, 2009). The pre-rRNA binding sites for Prp43p span 14–20 nucleotides, indicating the length of RNA required for Prp43p binding and potential sites of action. Prp43p is an 88 kDa protein that is conserved universally in eukaryotes containing two canonical RecA helicase domains that include eight conserved motifs (I, Ia, Ib, II, III, IV, V and VI; Cordin et al, 2006), followed by a carboxy-terminal region. These features are shared with the other five known DEAH-box proteins in S. cerevisiae: splicing factors Prp2p, Prp16p and Prp22p (Burgess et al, 1990; Chen & Lin, 1990; Company et al, 1991; Arenas & Abelson, 1997), and Dhr1p and Dhr2p, which are involved in ribosome biogenesis (Colley et al, 2000). Prp43p has been reported to interact with Ntr1p, Pfa1p/Sqs1p and Pxr1p/Gno1p (Lebaron et al, 2005; Tsai et al, 2005), all of which contain the conserved G-patch motif (Aravind & Koonin, 1999). The interactions with Ntr1p and Pfa1p/Sqs1p are essential for the biological functions of Prp43p in spliceosomal disassembly and pre-rRNA processing, respectively (Tanaka et al, 2007; Pertschy et al, 2009).
Results And Discussion
The architecture of a DEAH-box helicase
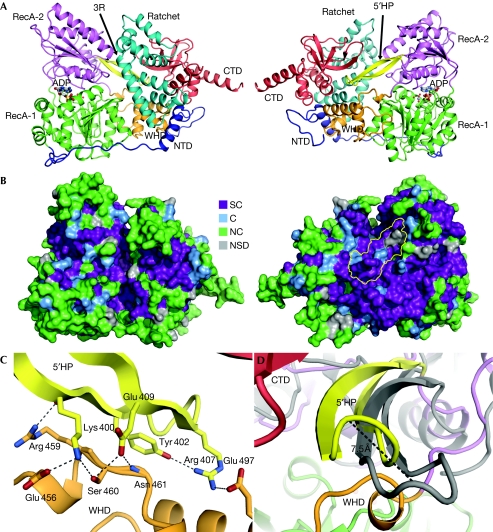
We have determined the crystal structure of yeast Prp43p in complex with ADP at 2.2 Å resolution (Table 1; supplementary Fig S1 online) and traced the entire molecule except for residues 756–767. The molecule is roughly box-shaped with dimensions of 97 × 69 × 65 Å3 and comprises six structural entities (Fig 1; supplementary Fig S2 online). DEAH helicases contain two RecA domains, which in Prp43p-ADP (residues 59–270 and 271–454, respectively) are packed tightly around the nucleotide (Fig 2). As the sequence of Prp43p and other DEAH helicases outside the RecA domains does not show homology to proteins of known structure, we searched for proteins with structural homology by using the DALI server (Holm et al, 2008). In one search with residues 455–634, the best matches were two DNA helicases, Hjm (Research Collaboratory for Structural Bioinformatics (RCSB) entry 2ZJ8, Z-score 8.1, identity 15%) and Hel308 (RCSB entry 2P6R, Z-score 7.3, identity 13%), which are highly related structurally (Buttner et al, 2007; Oyama et al, 2009). This region contains a degenerated winged-helix domain (WHD; residues 455–520) with a weakly defined β-sheet. It is followed by a seven-helical bundle, named the ratchet domain (residues 521–634) owing to its putative function as a ratchet for DNA and RNA (see below; Fig 1A; supplementary Fig S2 online). The second search with residues 635–750 identified structural homology to eIF1A (RCSB entry 2OKQ, Z-score 3.8). We term this region the C-terminal domain (CTD), which includes a five-stranded β-barrel arranged in an oligonucleotide/oligosaccharide-binding (OB) motif (residues 660–712). The CTD is terminated with a long α-helix protruding from the bulk of Prp43p (Fig 1A; supplementary Fig S2 online). The WHD, ratchet domain and CTD make intimate contacts with residues 10–36 of the extended amino-terminal domain (residues 1–58) and the WHD also interacts extensively with RecA-1 (Fig 1A). The interfaces between RecA-2 and the other domains are relatively small and predominantly polar, suggesting that they are dynamic (supplementary Table S1 online).
Table 1. Data collection and refinement statistics.
| Native (SLS PXI) | SeMet (MAX-Lab I911-3) | ||
|---|---|---|---|
| Data collection | |||
| Space group | P3221 | P3221 | |
| Cell dimensions | |||
| a, b, c (Å) | 118.2, 118.2, 253.6 | 118.1, 118.1, 253.8 | |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | |
| Peak | |||
| Wavelength (Å) | 0.980 | 0.9795 | |
| Resolution (Å) | 50–2.2 (2.3–2.2) | 50–2.86 (2.9–2.86) | |
| Rmerge | 0.121 (0.753) | 0.159 (0.733) | |
| I/σI | 16.96 (2.22) | 10.41 (2.22) | |
| Completeness (%) | 99.7 (99.9) | 98.9 (96.9) | |
| Redundancy | 4.2 (4.1) | 5.8 (5.8) | |
| SAD phasing | Refinement | ||
| Resolution (Å) | 30–3.5 | Resolution (Å) | 50–2.2 |
| Number of sites | 34 | Number of reflections | 104407 |
| Figure of merit | 0.293 | Rwork/Rfree | 0.206/0.250 |
| Phasing power | 1.32 | Number of atoms | |
| Cullis R-value | 0.76 | Protein | 12132 |
| Ligand/ion | 56 | ||
| Water | 982 | ||
| B-factors | |||
| Protein | 54.1 | ||
| Ligand/ion | 50.8 | ||
| Water | 50.1 | ||
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.006 | ||
| Bond angles (°) | 1.013 | ||
| One crystal each was used for data collection in both cases. | |||
Figure 1.
The structure of Prp43p. (A) Representation of Prp43p bound to ADP in two orientations differing by a 180° rotation. The extended NTD region is coloured blue, RecA-1 green, RecA-2 magenta, the 5′HP yellow, the WHD orange, the ratchet domain cyan and the CTD red. A long helix in the ratchet domain, containing three conserved arginines potentially involved in RNA interaction, is labelled ‘3R'. (B) Prp43p sequence conservation, based on supplementary Fig S2 online, mapped on the molecular surface in the same orientations as in (A). The 5′HP surface is outlined in yellow in the right panel. (C) Close-up of the interaction between the 5′HP and the WHD. Hydrogen bonds and salt bridges are shown as dashed lines. The orientation is the same as in (A), but the focus is behind the ratchet domain. (D) Predicted movement of the 5′HP between the ADP state (yellow) and a modelled ATP state (grey). The orientation is rotated −120° about the vertical axis with respect to the right panel of (A). C, conserved; CTD, carboxy-terminal domain; NC, non-conserved; NSD, not sufficient data; NTD, amino-terminal domain; SC, strictly conserved; WHD, winged-helix domain.
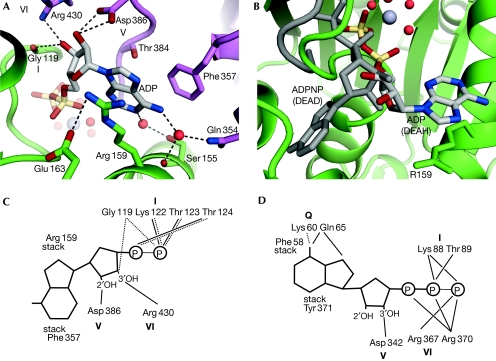
Figure 2.
The ADP binding site. (A) The DEAH-specific binding site for the adenine base between Arg 159 in RecA-1 and Phe357 in RecA-2. Water molecules are shown as red spheres. (B) Comparison of the nucleotide binding sites of a DEAH-box and DEAD-box (grey) helicase (Andersen et al, 2006). (C) Schematic representation of direct interactions between the ADP molecule and Prp43p. Dashed lines represent backbone interactions (eletrostatic or hydrogen bonds) and full lines represent side-chain interactions. The different motifs are shown in bold. (D) The interactions between ADPNP and eIF4AIII (DEAD-box helicase) are shown as in (C). The DEAD-box specific Q motif is responsible for direct recognition of adenine in this class of helicases.
Helicases of the two superfamilies I and II bound to ATP and DNA or RNA adopt a conserved ‘closed' conformation of the two RecA domains (Le Hir & Andersen, 2008), in which single-stranded RNA (ssRNA) or DNA is recognized by motifs Ia and Ib in RecA-1 and motifs IV and V in RecA-2. On ATP hydrolysis and subsequent Pi release (Nielsen et al, 2009), an ‘open' conformation of the helicase is adopted, which can be highly variable (Caruthers et al, 2000; Andersen et al, 2006). To investigate the potential effect of ATP and RNA binding to Prp43p, we first superimposed the RecA-1 domains of eIF4A3-RNA-ADPNP and Prp43p, and then matched the RecA-2 domain of Prp43p onto its counterpart in eIF4A3-RNA-ADPNP. In the resulting model, RecA-2 was rotated approximately 18° relative to Prp43p-ADP (supplementary Fig S3 online). This also allowed us to tentatively model ssRNA bound to the RecA domains (see below).
ADP inhibits binding of RNA to Prp43p
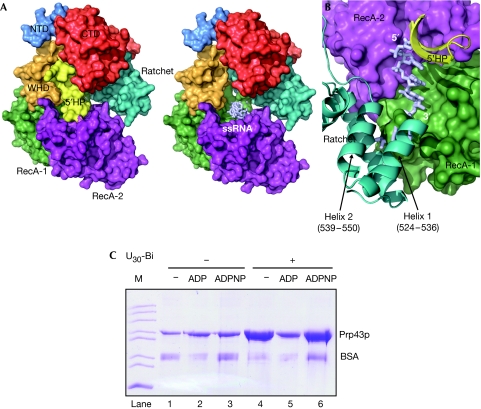
A long, twisted β-hairpin (residues 396–415) located between motifs V and VI of RecA-2 is inserted into a cleft between the WHD and CTD (Figs 1 and 3). We will refer to this hairpin as the 5′HP, as it blocks access to the 5′ end of the putative binding pocket for ssRNA expected to be formed in the ATP state. Pulldown experiments with biotinylated RNA verified in vitro that the presence of ADP inhibits the interaction of Prp43p with ssRNA compared with both Prp43p-ADPNP and apo-Prp43p (Fig 3C). A hairpin of the same length and location is present in the flavivirus NS3 RNA helicase (Luo et al, 2008). Similarly to Prp43p, the NS3 hairpin also closes the 5′ end of the ssRNA binding pocket and interacts with a C-terminal helical region. Other elements bounding this pocket are the conserved helicase motifs Ia, Ib, IV and V in the RecA domains, two loops and the last helix in the Prp43p ratchet domain (Fig 3A,B). This helix (residues 610–630) corresponds to a helix in Hel308, which was suggested to provide a ratchet for the directional movement of ssDNA across the RecA domains (Buttner et al, 2007). An equivalent helix is also present in the RNA helicase Brr2, which is required for unwinding of the extremely stable U4/U6 duplex during splicing (Pena et al, 2009; Zhang et al, 2009). In Prp43p, three conserved arginines in this helix face the 3′-half of this putative RNA binding pocket (Fig 1A; supplementary Fig S2 online), suggesting their involvement in RNA contacts in Prp43p. The 3′ end of this binding pocket is sealed by interactions involving RecA-1, the ratchet domain and the last residues of the WHD (Fig 3B).
Figure 3.
RNA binding to Prp43p. (A) Surface representation of Prp43p with the 5′HP present (left) and removed (right) to show its occlusion of a pocket between RecA-1, RecA-2 and the ratchet domains expected to bind ssRNA in the ATP state. An RNA placed by comparison with the exon junction complex (Research Collaboratory for Structural Bioinformatics entry 2HYI) is shown in light-blue sticks. (B) Inside view of the RNA binding area of the two RecA domains with the 5′ end of the binding site obstructed by the 5′HP and the 3′ end bounded by the RecA-1 and ratchet domains (Helix 1 and Helix 2 of the ratchet domain are indicated). (C) SDS–PAGE analysis of streptavidin agarose pulldown using a U30 biotinylated RNA. All reactions contained Prp43p and bovine serum albumin (BSA) to reduce background binding and the indicated nucleotide or U30-biotin. ‘M' denotes marker. CTD, carboxy-terminal domain; NTD, amino-terminal domain; SDS–PAGE, SDS–polyacrylamide gel electrophoresis; ssRNA, single-stranded RNA; WHD, winged-helix domain.
WHDs have been observed to interact with both DNA (Gajiwala & Burley, 2000) and RNA (Alfano et al, 2004), as do OB motifs (Theobald et al, 2003), suggesting that in Prp43p they could have a similar function. The conserved loop connecting β-strands 4 and 5 of the OB motif (residues 701–704) neighbours the 5′HP (supplementary Fig S4A,B online) and is therefore located close to where the 5′ end of ssRNA bound to the Rec-A helicase domains is expected to be, and the equivalent loop in AspRS interacts with the anticodon of transfer RNA (RCSB entry 1ASY). Nearby, in the ratchet domain, helix four and the preceding loop (residues 557–569) contain a highly conserved, positively charged surface patch (supplementary Fig S4B online) and comparison with DNA-bound Hel308 (Buttner et al, 2007) suggests that this region could interact with RNA.
Insertion of the 5′HP between the CTD and WHD is likely to lock the orientation of RecA-2 in the ADP state relative to the rest of the molecule. Both the 5′HP and the residues it contacts are in general highly conserved in Prp43p and other DEAH helicases (Fig 1B,C; supplementary Figs S2 and S5 online). We predict that binding of ATP to Prp43p will induce a conformational change releasing the 5′HP from the RNA binding pocket, with the tip of the β-hairpin moving more than 7 Å (Fig 1D; supplementary Fig S3 online), and will allow binding of ssRNA to the RecA domains. In addition, conformational changes at the interface between RecA-1 and the WHD and ratchet domain are required for opening of the 3′ end of the occluded RNA binding pocket observed in Prp43p-ADP. Similarly, binding of ssRNA to the RecA domains probably causes them to adopt the closed conformation, thereby creating an optimal ATP binding site in agreement with the strong stimulation of Prp43p ATPase activity by ssRNA (Tanaka & Schwer, 2006).
The functional importance of the 5′HP is supported by several observations. First, there is a strong asymmetry in the distribution of conserved surface residues in Prp43p, and the 5′HP is located at the most conserved surface (Fig 1B). Second, the 5′HP interacts functionally with the N-terminal region of Ntr1 (Tanaka et al, 2007), which significantly stimulates the helicase activity of Prp43p (Tanaka & Schwer, 2006; Pertschy et al, 2009). A similar hairpin in the same location in Hel308 is probably responsible for DNA unwinding (Buttner et al, 2007), suggesting a potential role of the 5′HP in RNA unwinding and/or RNP remodelling catalysed by DEAH helicases. Third, a mutation in the 5′HP of Prp43p (Ser 414) contributes to accumulation of 20S pre-rRNA during ribosome biogenesis (Pertschy et al, 2009; supplementary Table S2 and Fig S6 online). Hence, the 5′-HP might have at least two functions; in the ADP state it probably has an autoinhibitory role, preventing binding of ssRNA to Prp43p, and in the ATP state it possibly participates in strand separation or remodelling of protein–RNA interactions.
A novel binding site for the ADP molecule
An ADP molecule is sandwiched between the two RecA domains (Fig 2). Compared with DEAD box and other helicases, the adenine base is stacked at a novel binding site between Phe 359 and Arg 159, which are conserved in all DEAH helicases (Fig 2; supplementary Figs S2 and S5 online). Asp 386 from motif V and Arg 430 from motif VI interact with the ribose, and the conserved Ser 155 and Gln 354 coordinate water molecules to form hydrogen bonds with the adenine, but the base is not specifically recognized as seen in the ATP-specific DEAD-box proteins (Fig 2C,D; supplementary Fig S7 online). In agreement with a non-specific nucleotide binding site, DEAH helicases Prp43p (Tanaka & Schwer, 2006), Prp2p (Kim et al, 1992), Prp16p (Schwer & Guthrie, 1992) and Prp22p (Tanaka & Schwer, 2005) have all been shown to hydrolyse various NTPs and dNTPs. The ADP phosphates are coordinated by interactions with Gly 119, Lys 122 and Thr 123 from motif I and the adjacent Thr 124 involving both backbone and side-chain interactions (supplementary Fig S7 online). The Mg2+ ion is coordinated by four water molecules, a β-phosphate oxygen and Thr 123. Three of the water molecules coordinating the Mg2+ ion are kept in place by Thr 381 and Ser 382 from motif V (supplementary Fig S7 online). Motifs I, II, V and VI interact through contacts between Thr 123–Asp 215, Glu 216–Thr 381 and Arg 430–Gly 119, while Glu 216 and His 218 in motif II form a salt bridge (supplementary Fig S7 online). In the ATP state of DEAH helicases, we expect no significant changes in the adenine binding pocket, whereas the triphosphate moiety probably binds similarly to other superfamily I and II helicases surrounded by motifs I, II and VI (Le Hir & Andersen, 2008).
In summary, our structure defines the structural building blocks present in Prp43p, and based on sequence comparisons, these are also present in the other DEAH helicases. These findings might also be extended to DExH RNA helicases, such as DHX9, that are involved in transcription (Fuller-Pace, 2006). It also reveals an unexpected structural homology of these DEAH helicases to the Hel308 and Hjm DNA helicases and establishes the presence of motifs involved in RNA binding in other proteins, thereby providing straightforward suggestions for regions that are potentially involved in intermolecular interactions. Finally, our structure provides the rationale for the well-established functional importance of the 5′HP in Prp43p shown by genetic and functional studies of both pre-mRNA splicing and ribosome biogenesis (Tanaka et al, 2007; Pertschy et al, 2009). Together with the recent identification of regions in pre-rRNA and snoRNA that can be crosslinked to Prp43p (Bohnsack et al, 2009), our structure paves the way for a rational approach to obtain a detailed molecular understanding of the function of Prp43p. However, several important questions about the mode of action of Prp43p remain, including (i) whether Prp43p is a processive or non-processive helicase; (ii) whether it unwinds double-stranded RNA and/or remodels messenger RNP (mRNP); (iii) and how binding partners recruit and localize Prp43p to a specific place for unwinding and mRNP remodelling (Guenther & Jankowsky, 2009).
Methods
Methods for cloning, expression and purification of Prp43p can be found in the supplementary information online.
RNA pulldown. High Capacity Streptavidin Agarose Resin (Thermo Scientific, Waltham, MA, USA) and 3′ biotinylated pU30 (Dharmacon, Lafayette, CO, USA) were used for RNA pulldown of Prp43p. For each reaction, 20 μl of streptavidin agarose resin was washed three times with 500 μl of binding buffer (20 mM Tris–HCl, pH 7.6, 200 mM NaCl, 5 mM MgCl2, 0.5 mM dithiothreitol and 10% glycerol), then mixed together with 0.1 nmol of biotinylated polyU 10 μg Prp43p and, when necessary, 1 mM of ADP or ADPNP in the binding buffer to a final volume of 100 μl. The reaction mixtures were incubated at 4°C overnight. After thorough washing of the resin, Prp43p was eluted by resuspending the resin in 20 μl 2.5 × SDS–polyacrylamide gel electrophoresis loading buffer and heated for 5 min at 100°C. The bound Prp43p was visualized by Coomassie staining.
Crystallization and structure determination. Needle-shaped crystals with maximum dimensions of 500 × 20 × 20 μm3 of the Prp43p–ADP complex were grown in hanging drops by vapour diffusion at 4°C against reservoirs containing 100 mM Mes-NaOH, pH 6.75, 14% PEG8000, 300 mM NaOAc and 100 mM BaCl2. The crystals were transferred to a cryoprotection buffer (100 mM Mes-NaOH, pH 6.75, 200 mM NaCl, 15% PEG8000, 20% glycerol) and flash frozen in liquid nitrogen. Native data were recorded at 100 K at the PXI beamline of the Swiss Light Source (Villigen, Switzerland), and selenomethionine derivative data were collected at the I911-3 beamline of MAX-lab (Lund, Sweden). Diffraction data were processed and scaled by using the XDS package (Kabsch, 2001). SHELXD (Sheldrick, 2008) was used to identify 34 of 36 possible selenium sites from the anomalous data with a correlation coefficient of 0.54 for the resolution range 50–4 Å. Phasing and initial model building was done using the AutoSol and Resolve procedures as implemented in PHENIX (Adams et al, 2002). Iteratively improved solvent flattened and averaged maps starting from combined phases (experimental phases and current model phases) were obtained with CNS (Brunger et al, 1998) until the model was refined to an Rfree of 35%. Subsequently, all rebuilding was done in averaged 2mFo−DFc maps. Manual rebuilding was done in O ( Jones et al, 1991) and COOT (Emsley & Cowtan, 2004), after which the model was refined with PHENIX using tight non-crystallographic symmetry restraints on the two copies of Prp43p-ADP present in the asymmetric unit. Refinement without non-crystallographic symmetry restraints resulted in an increase in Rfree and was therefore abandoned. In the final model, 96.28% of the residues fell into the favoured regions of the Ramachandran plot and there were no outliers. Structure analysis was done with MolProbity (Davis et al, 2004), DynDom (Hayward & Berendsen, 1998) and PISA (Krissinel & Henrick, 2007), and figures with PyMOL (DeLano, 2002) or ALINE (Bond & Schuttelkopf, 2009).
Accession codes. Coordinates and structure factors are deposited at the RCSB protein data bank as entry 3KX2.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We are grateful to the staff at MAX-lab, European Synchrotron Radiation Facility and Swiss Light Source for their help with data collection, Lars Sottrup-Jensen for amino acid analysis, and Mickaél Blaise, Laure Yatime and Christian B.F. Andersen for their help with data processing. K.H.N. was supported by the Alfred Benzon Foundation. G.R.A. was supported by the Danish Science Research Council (FNU), the Danish National Research Foundation and a Hallas-Møller stipend from the Novo-Nordisk Foundation.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58: 1948–1954 [DOI] [PubMed] [Google Scholar]
- Alfano C, Sanfelice D, Babon J, Kelly G, Jacks A, Curry S, Conte MR (2004) Structural analysis of cooperative RNA binding by the La motif and central RRM domain of human La protein. Nat Struct Mol Biol 11: 323–329 [DOI] [PubMed] [Google Scholar]
- Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira C, Pedersen JS, Seraphin B, Le Hir H, Andersen GR (2006) Structure of the exon junction core complex with a trapped DEAD-Box ATPase bound to RNA. Science 313: 1968–1972 [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV (1999) G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci 24: 342–344 [DOI] [PubMed] [Google Scholar]
- Arenas JE, Abelson JN (1997) Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc Natl Acad Sci USA 94: 11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D (2009) Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell 36: 583–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Schuttelkopf AW (2009) ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr 65: 510–512 [DOI] [PubMed] [Google Scholar]
- Brunger AT et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Burgess S, Couto JR, Guthrie C (1990) A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60: 705–717 [DOI] [PubMed] [Google Scholar]
- Buttner K, Nehring S, Hopfner KP (2007) Structural basis for DNA duplex separation by a superfamily 2 helicase. Nat Struct Mol Biol 14: 647–652 [DOI] [PubMed] [Google Scholar]
- Caruthers JM, Johnson ER, McKay DB (2000) Crystal structure of yeast initiation factor 4A, a DEAD-box RNA helicase. Proc Natl Acad Sci USA 97: 13080–13085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JH, Lin RJ (1990) The yeast PRP2 protein, a putative RNA-dependent ATPase, shares extensive sequence homology with two other pre-mRNA splicing factors. Nucleic Acids Res 18: 6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley A, Beggs JD, Tollervey D, Lafontaine DL (2000) Dhr1p, a putative DEAH-box RNA helicase, is associated with the box C+D snoRNP U3. Mol Cell Biol 20: 7238–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combs DJ, Nagel RJ, Ares M Jr, Stevens SW (2006) Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol 26: 523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J (1991) Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493 [DOI] [PubMed] [Google Scholar]
- Cordin O, Banroques J, Tanner NK, Linder P (2006) The DEAD-box protein family of RNA helicases. Gene 367: 17–37 [DOI] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC (2004) MolProbity: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res 32: W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL User's Manual. Palo Alto, CA, USA: San Carlos DeLano Scientific [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV (2006) DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34: 4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajiwala KS, Burley SK (2000) Winged helix proteins. Curr Opin Struct Biol 10: 110–116 [DOI] [PubMed] [Google Scholar]
- Guenther UP, Jankowsky E (2009) Helicase multitasking in ribosome assembly. Mol Cell 36: 537–538 [DOI] [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ (1998) Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins 30: 144–154 [PubMed] [Google Scholar]
- Hilbert M, Karow AR, Klostermeier D (2009) The mechanism of ATP-dependent RNA unwinding by DEAD box proteins. Biol Chem 390: 1237–1250 [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Cowan S, Zou J-Y, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kabsch W (2001) XDS. In International Tables for Crystallography: Vol F, Crystallography of Biological Macromolecules, Rossman MG, Arnold E (eds), pp 730–733. Dordrecht, The Netherlands: Kluwer Academic [Google Scholar]
- Kim SH, Smith J, Claude A, Lin RJ (1992) The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J 11: 2319–2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Andersen GR (2008) Structural insights into the exon junction complex. Curr Opin Struct Biol 18: 112–119 [DOI] [PubMed] [Google Scholar]
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y (2005) The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol 25: 9269–9282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Putnam A, Jankowsky E (2008) ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc Natl Acad Sci USA 105: 20209–20214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D et al. (2008) Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J 27: 3209–3219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen KH, Chamieh H, Andersen CB, Fredslund F, Hamborg K, Le Hir H, Andersen GR (2009) Mechanism of ATP turnover inhibition in the EJC. RNA 15: 67–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Oka H, Mayanagi K, Shirai T, Matoba K, Fujikane R, Ishino Y, Morikawa K (2009) Atomic structures and functional implications of the archaeal RecQ-like helicase Hjm. BMC Struct Biol 9: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena V, Jovin SM, Fabrizio P, Orlowski J, Bujnicki JM, Luhrmann R, Wahl MC (2009) Common design principles in the spliceosomal RNA helicase Brr2 and in the Hel308 DNA helicase. Mol Cell 35: 454–466 [DOI] [PubMed] [Google Scholar]
- Pertschy B, Schneider C, Gnadig M, Schafer T, Tollervey D, Hurt E (2009) RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 284: 35079–35091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Guthrie C (1992) A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 11: 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64: 112–122 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C (1998) Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Schwer B (2005) Characterization of the NTPase, RNA-binding, and RNA helicase activities of the DEAH-box splicing factor Prp22. Biochemistry 44: 9795–9803 [DOI] [PubMed] [Google Scholar]
- Tanaka N, Schwer B (2006) Mutations in PRP43 that uncouple RNA-dependent NTPase activity and pre-mRNA splicing function. Biochemistry 45: 6510–6521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Aronova A, Schwer B (2007) Ntr1 activates the Prp43 helicase to trigger release of lariat-intron from the spliceosome. Genes Dev 21: 2312–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theobald DL, Mitton-Fry RM, Wuttke DS (2003) Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol Struct 32: 115–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai RT, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC (2005) Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 19: 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R (2009) Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol 16: 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.