TBCCD1, a new centrosomal protein, is required for centrosome and Golgi apparatus positioning
In this report, Soares and colleagues describe the first characterization of human TBCCD1, a protein related to tubulin cofactor C, as a new player in the centrosome-nucleus association. TBCCD1 localizes to the centrosome and the spindle midzone, as well as to the midbody and basal bodies of primary and motile cilia. TBCCD1 depletion in RPE-1 cells results in centrosome/microtubule aster misplacement, Golgi disorganization, defects in cell migration and in the ability to form primary cilia.
Keywords: cell migration, centrosome, Golgi apparatus, primary cilia, TBCCD1
Abstract
In animal cells the centrosome is positioned at the cell centre in close association with the nucleus. The mechanisms responsible for this are not completely understood. Here, we report the first characterization of human TBCC-domain containing 1 (TBCCD1), a protein related to tubulin cofactor C. TBCCD1 localizes at the centrosome and at the spindle midzone, midbody and basal bodies of primary and motile cilia. Knockdown of TBCCD1 in RPE-1 cells caused the dissociation of the centrosome from the nucleus and disorganization of the Golgi apparatus. TBCCD1-depleted cells are larger, less efficient in primary cilia assembly and their migration is slower in wound-healing assays. However, the major microtubule-nucleating activity of the centrosome is not affected by TBCCD1 silencing. We propose that TBCCD1 is a key regulator of centrosome positioning and consequently of internal cell organization.
Introduction
The centrosome, the major microtubule (MT)-organizing centre in animal cells, is a key organelle comprising a pair of centrioles surrounded by the pericentriolar matrix. By nucleating and organizing the spatial distribution of MTs in interphase, the centrosome has been implicated in organelle positioning—for example, Golgi apparatus (GA)—cell migration, adhesion and polarity, and in mitosis it assists spindle pole formation (reviewed in Lüders & Stearns, 2007). The ability to nucleate the ciliary axoneme, becoming a basal body, constitutes another main role of the centrosome. Usually, the centrosome is maintained at the cell centre, closely associated with the nucleus. This has been shown to be essential, for example, in the early development of Caenorhabditis elegans (Malone et al, 2003). The centrosome is repositioned to peripheral sites during cell-state transitions such as wound healing, cell migration and cell growth and differentiation (Yvon et al, 2002; de Anda et al, 2005).
The geometrical constraints imposed by the substratum have a crucial role in centrosome positioning and the internal organization of the cell (Pouthas et al, 2008), but probably do not provide the sole answer to this question. The MTs and forces exerted on them by actomyosin and dynein are also crucial (Wittmann & Waterman-Storer, 2001; Burakov et al, 2003). A variety of data support the existence of a physical link between the centrosome and the nuclear envelope and implicate proteins such as Zyg-12 and Emerin (Malone et al, 2003; Salpingidou et al, 2007). This interaction seems to be regulated by the p160ROCK Rho-associated kinase and the coordinated activity of Polo/Greatwall (Gwl) mitotic kinases (Chevrier et al, 2002; Archambault et al, 2007).
A dynamic nucleus–centrosome connection also seems important for directed cell migration—for example, in neurons (Higginbotham & Gleeson, 2007)—and several studies reported centrosome reorientation towards the leading edge in migrating cells (Yvon et al, 2002). Nevertheless, this is not true for all cell types and is also substratum-dependent, leaving the relevance of this reorientation for cell migration still open to debate (Yvon et al, 2002).
In recent years, tubulin cofactors (TBC) A–E, which participate in the tubulin-folding pathway, have emerged as proteins with crucial roles, not always associated directly to their expected role in the pathway, but still related to the cytoskeleton. For example, TBCD is a centrosomal protein required for γ-tubulin ring complex recruitment and mitotic spindle organization (Cunningham & Kahn, 2008). These data and the fact that specific TBC domains were identified in the functionally related proteins RP2 and E-like (Bartolini et al, 2002, 2005), suggest that they could have evolved to deal with specific requirements of MT assembly and dynamics, possibly in specific cell types. This prompted us to search the human genome database for other proteins with TBC-specific domains. During this process we found TBCC-domain containing 1 (TBCCD1), which is conserved through the phylogenetic tree (Stephan et al, 2007). TBCCD1 is related to TBCC, which, together with TBCD, acts as a β-tubulin GTPase-activating protein (GAP; Fontalba et al, 1993; Tian et al, 1999). TBCCD1 is also related to RP2, which overlaps functionally with TBCC (Bartolini et al, 2002), seeming to participate in tubulin quality control at the basal body of the Trypanosoma flagellum (Stephan et al, 2007).
Here, we report the first characterization of human TBCCD1 and show that it is required for centrosome and GA perinuclear positioning in RPE-1 cells. We also show that TBCCD1-silenced cells are less efficient in primary cilia assembly and affected in cell migration.
Results And Discussion
TBCCD1 localizes at the centrosome and basal bodies
In addition to the TBCC domain, TBCC, RP2 and TBCCD1 possess a CARP domain—a domain found in Cyclase-associated proteins (CAPs; supplementary Fig S1A,B online)—but their amino-acid sequence identity is low outside these domains.
Through a yeast complementation assay, we observed TBCCD1 to be unable to complement yeast TBCC (CIN2) deletion (supplementary Fig S1C online). This suggests that, unlike RP2, TBCCD1 does not overlap functionally with TBCC. This could be due to TBCCD1 lacking the arginine conserved in TBCC and RP2 that is crucial for their GAP activity towards tubulin (Veltel et al, 2008).
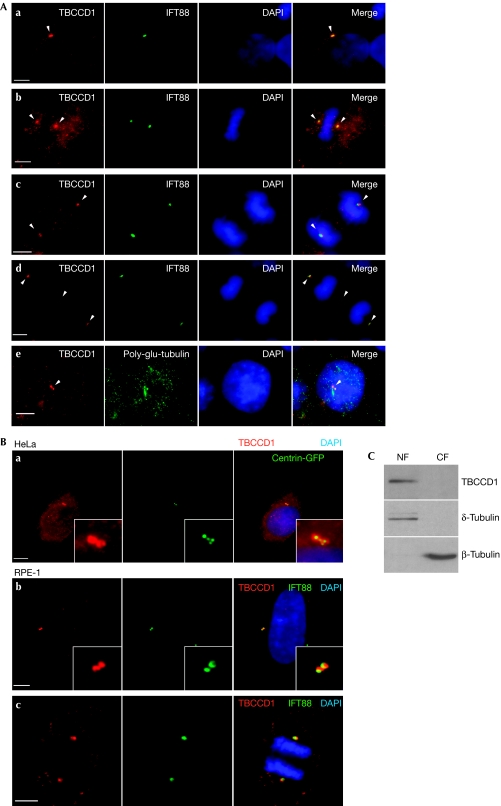
To study the subcellular localization of TBCCD1, a specific mouse polyclonal anti-TBCCD1 serum was raised (supplementary Fig S2A,B online) and used to perform immunofluorescence analysis of human embryonic kidney 293T (HEK293T), HeLa and RPE-1 cells. With this analysis we observed TBCCD1 to be localized at the cytoplasm and at the centrosome throughout the cell cycle (Fig 1A,B). Furthermore, by using HeLa and RPE-1 cells expressing centrin–green fluorescent protein (GFP) we showed that TBCCD1 localizes at the pericentriolar matrix (Fig 1B; supplementary Fig S6A online). TBCCD1 was also observed in the basal body of primary cilia (Fig 1A; supplementary Fig S4 online). Immunolocalization in mouse brain primary cultures showed TBCCD1 also in the basal bodies of motor cilia (supplementary Fig S3A online). The localization data were supported by the expression of fluorescently tagged TBCCD1 (supplementary Fig S4 online). Tagged TBCCD1 accumulated at the centrosome during the cell cycle and also at the spindle midzone and the midbody. However, when using the antiserum, TBCCD1 was barely detected at the latter two sites—unless HEK293T cells were transfected with untagged TBCCD1 (supplementary Fig S2C online)—indicating that the endogenous TBCCD1 levels are too low to be detected or those localizations result from overexpression.
Figure 1.
TBCCD1 sub-cellular localization in human cells. (A) HEK293T cells were immunostained with antibodies against TBCCD1 and either IFT88 or poly-glutamylated tubulin. DNA was stained with DAPI. (a) Interphase cell, (b) cell in metaphase, (c) cell in anaphase, (d) cell in telophase and (e) cell-cycle-arrested cell after serum starvation for 24 h. The arrowheads point to TBCCD1 at the centrosomes (a,d), the constriction zone (d) and the basal body (e). Panels are representative of five independent experiments. (B) Immunostaining of HeLa and RPE-1 interphase cells with anti-TBCCD1 showing pericentriolar matrix localization (a,b). RPE-1 cell in anaphase (c). Scale bars, 5 μm. Panels are representative of five independent experiments. (C) Western blot analysis of nuclear and cytosolic protein fractions (NF and CF) of RPE-1 cells shows that TBCCD1 is mainly present in the nuclear fraction. Results are representative of three independent experiments. DAPI, 4,6-diamidino-2-phenylindole; HEK, human embryonic kidney; TBCCD1, TBCC-domain containing 1.
We also observed that TBCCD1 centrosomal localization is not affected by MT depolymerization with nocodazole (supplementary Fig S5A online), indicating it to be an integral centrosome component. Also, its amino-terminal domain (aa 1–328) is involved in centrosome targeting (supplementary Fig S5B online). TBCCD1 was detected, with δ-tubulin, mainly in the nuclear fraction of RPE-1 cells (Fig 1C). As centrosomes are known to fractionate with nuclei, this observation also supports TBCCD1 centrosomal localization. Finally, reverse transcription (RT)–PCR analysis showed that tbccd1 was transcribed in all mouse tissues tested (supplementary Fig S3B online), whereas the protein was detected effectively in the brain and the testis, the two tissues where tubulin was more abundant. These results suggest that TBCCD1 might be regulated post-transcriptionally in the different tissues.
TBCCD1 RNAi affects centrosome and Golgi positioning
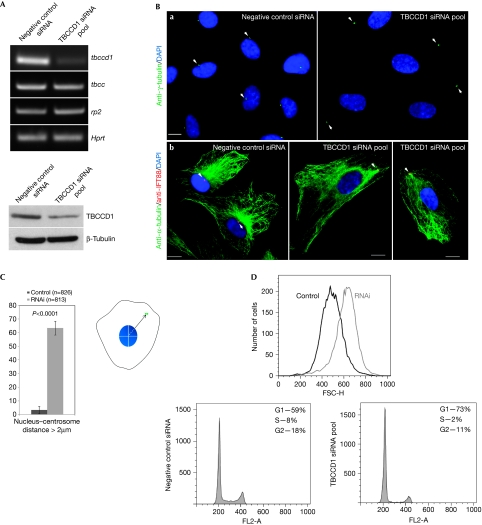
To study the function of TBCCD1, the gene was silenced with a small interfering RNA (siRNA) pool in RPE-1 cells. TBCCD1 silencing was confirmed by RT–PCR, western blotting and immunolocalization analysis, which showed decreased fluorescence intensity at the centrosome and cytoplasm (Fig 2A; supplementary Fig S6A online). Seventy-two hours after transfection, the cells treated with the siRNA pool did not become confluent (supplementary Fig S7A online). Flow cytometry analysis showed that at 48 h after transfection, TBCCD1-silenced cells were larger than control cells (Fig 2D) and presented a 14% increase in the G1 peak and a corresponding decrease in the number of cells in S phase and G2/M phase (Fig 2D; supplementary Fig S7B online), suggesting a cell-cycle delay in G1. By using a γ-tubulin antibody to visualize the centrosome, we observed that TBCCD1 depletion severely affected centrosome localization. Contrary to control cells where the centrosome is located in the cell centre and closely associated with the nucleus, in TBCCD1-depleted cells the centrosome was often located at the cell periphery (Fig 2Ba). Only 3.3±2.7% of the control population had the centrosome more than 2 μm away from the nucleus, whereas in siRNA-treated cells this value rose to 63.5±5.1% (Fig 2C; supplementary Fig S6C online).
Figure 2.
TBCCD1 knockdown in RPE-1 cells leads to loss of centrosome–nucleus association and cell-cycle delay. (A) Top: reverse transcription–PCR analysis of tbccd1, tbcc and rp2 expression in control and TBCCD1 siRNA-treated cells. Hprt expression was used as an internal control. Bottom: western blot analysis of TBCCD1 levels in control and TBCCD1 siRNA-treated cells. TBCCD1 levels decreased by about 65% in TBCCD1-silenced cells. β-Tubulin was used as loading control. (B) Control and TBCCD1-silenced cells immunostained with (a) anti-γ-tubulin, (b) anti-α-tubulin and anti-IFT88. Scale bars, 5 μm. DNA was stained with DAPI. The arrowheads point to centrosomes. (C) The nuclear edge–centrosome measurement approach used ImageJ software. The mean percentage of cells (±s.d.) with a centrosome–nucleus distance greater than 2 μm is shown (n=total number of cells scored in three independent experiments). Statistical significance was calculated using a t-test. (D) Flow cytometric analysis of cell cycle and size in control and TBCCD1-depleted cells stained with propidium iodide. All experiments were performed using 72 h post-transfection cells, except for the flow cytometric analysis, which was performed at 48 h. DAPI, 4,6-diamidino-2-phenylindole; HEK, human embryonic kidney; siRNA, small interfering RNA; TBCCD1, TBCC-domain containing 1.
The effect of centrosome displacement in the MT cytoskeleton was analysed by immunofluorescence, which showed that in TBCCD1-silenced cells, MTs were still focused on the centrosome (Fig 2B,b). Next, we analysed MT recovery after depolymerization with nocodazole in TBCCD1-silenced cells. Similarly to control cells, the MT aster was visible 5 min after nocodazole washout (supplementary Fig S8 online) and MTs continued to grow from the centrosome. This showed that misplaced centrosomes were still able to nucleate MTs and that TBCCD1 is either not necessary for this centrosome function, or its remaining levels are sufficient.
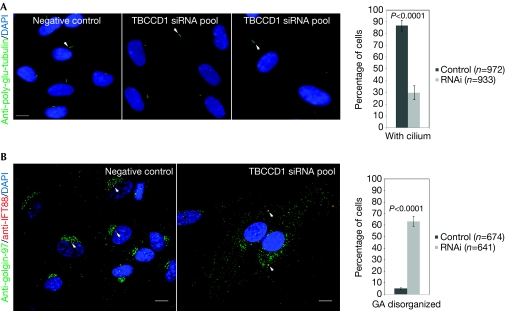
The ability of displaced centrosomes to assemble primary cilia was also tested. As confluent cells are more efficient in assembling primary cilia, more cells were seeded before transfection to obtain a dense monolayer of siRNA-treated cells. The cells were retransfected after 48 h and serum-starved. TBCCD1-depleted cells, under similar density conditions as control cells, were able to assemble primary cilia even when centrosomes were far from the nucleus, but the percentage of cells presenting cilia was lower (30±5.9%) than in control cells (87±4.6%; Fig 3A; supplementary Fig S6D online). Thus, TBCCD1 depletion affects the ability of RPE-1 cells to assemble primary cilia.
Figure 3.
TBCCD1 silencing affects Golgi organization and primary cilia assembly. RPE-1 control and TBCCD1-silenced cells were immunostained 72 h after transfection with antibodies against (A) poly-glutamylated tubulin or (B) IFT88 and golgin-97. The graphs represent the percentage (±s.d.) of cells (A) with cilia and (B) with disorganized GA (n=total number of cells scored in four or three independent experiments, respectively). Statistical significance was calculated using a t-test. Scale bars, 10 μm. DAPI, 4,6-diamidino-2-phenylindole; siRNA, small interfering RNA; TBCCD1, TBCC-domain containing 1.
The MT network is responsible for the internal organization of a cell, being crucial for organizing and positioning the GA close to the nucleus (Rios & Bornens, 2003). As expected, by using an antibody against the centrosome marker IFT88 and GA marker golgin-97, we observed that in control cells the GA has a perinuclear position and is organized around the centrosome (Fig 3B; supplementary Fig S6E online). However, in TBCCD1-silenced cells the GA was disorganized and appeared to follow the centrosome or be fragmented and spread out in the cytoplasm (Fig 3B; supplementary Fig S6E online). A similar GA phenotype was observed during Toxoplasma host invasion, where the centrosome is displaced from its perinuclear position (Coppens et al, 2006).
Microtubules are important for GA biogenesis as its precursors are carried by motor proteins towards MT minus ends anchored at the centrosome (Rios & Bornens, 2003). Thus, the observed GA phenotype is probably explained by the displacement of the centrosome and the MT aster. With time this would lead to displacement of the GA to the new centrosomal position. However, the centrosome might not adopt a fixed position after being uncoupled from the nucleus. To test this hypothesis, we analysed non-confluent RPE-1 cells expressing centrin–GFP by live imaging under control and RNA interference (RNAi) conditions. Control cells are highly motile and during their movement the centrosome is in close association with the nucleus without adopting a preferential localization in relation to it (supplementary Fig S9 online and supplementary Movie S1 online). By contrast, TBCCD1-depleted cells migrate more slowly and the centrosome tends to lag behind the nucleus, coming close to it as the cell progresses and the tail retracts (supplementary Fig S9 online and supplementary Movie S2 online). In RNAi cells, although there is no increase in centrosome movement, as observed in p160ROCK silencing (Chevrier et al, 2002), its position relative to the nucleus changes over time, which could result in GA disorganization. However, direct involvement of TBCCD1 in GA organization cannot be excluded, although no apparent TBCCD1 localization at the GA was observed (supplementary Fig S6E online). Furthermore, the GA disorganization could be implicated in the lower efficiency of these cells to assemble primary cilia as GA proteins have been implicated in developmental problems owing to defective cilium assembly (reviewed in Barr, 2009).
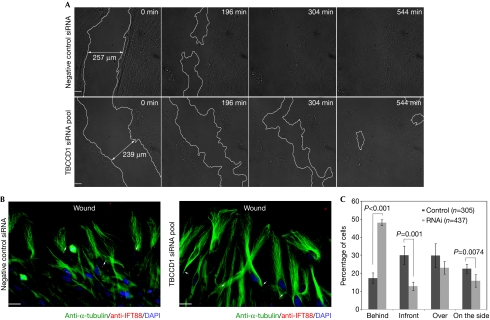
It is well documented that centrosome and GA positioning are important for cell polarization and directed cell migration (Vinogradova et al, 2009). Given the phenotypes described above, we tested whether TBCCD1 depletion caused defects in directed cell migration by performing a wound-healing assay. Thus, control and RNAi cell monolayers, obtained as described for cilia assembly, were wounded to trigger directional migration of cells towards the wound. By performing live imaging of wound closing, we observed that RNAi cells were able to close the wound but were delayed compared with the controls (Fig 4A; supplementary Movies S3, S4 online), which is consistent with the previous observations. The centrosome position relative to the nucleus was also analysed and showed that in control cells there was no preferential orientation of the centrosome (Fig 4B,C). However, in the RNAi population, 48.1±1.7% of the cells had the centrosome behind the nucleus (Fig 4B,C), which agrees with the observations made when using migrating isolated cells.
Figure 4.
TBCCD1 RPE-1-depleted cells are defective in directed cell migration. (A) Control and TBCCD1-silenced cells were grown to confluence, wounded and imaged for 9 h. Frames from 0, 196, 304 and 544 min are shown. Lines were used to limit the wound edges. Scale bars, 50 μm. (B) Wounded control and RNAi cells were allowed to migrate for 1 h and immunostained with α-tubulin and IFT88 antibodies. The arrows point to centrosomes. DNA was stained with DAPI. Scale bars, 20 μm. (C) Graphical representation of centrosome positioning relative to the nucleus in cells at the wound edge (n=total number of cells scored in three independent experiments). The differences between control and RNAi were statistically significant, except for ‘over' (P=0.0075). DAPI, 4,6-diamidino-2-phenylindole; RNAi, RNA interference; siRNA, small interfering RNA; TBCCD1, TBCC-domain containing 1.
Here, we describe a new human pericentriolar matrix component, TBCCD1, and show that its depletion in RPE-1 cells severely affects the centrosome position relative to the nucleus, with profound consequences on GA organization, cell shape and cell migration.
Many of the proteins that have been implicated in centrosome perinuclear positioning, such as Zyg-12, Emerin and Samp1, are nuclear envelope proteins (Malone et al, 2003; Salpingidou et al, 2007; Buch et al, 2009) constituting physical links between the nucleoskeleton and the centrosome and cytoskeleton. TBCCD1 cellular localization suggests that its involvement in centrosome nuclear association will be through a different mechanism. As TBCCD1 contains a TBCC domain, we considered that it could be a GAP for tubulin. This was not confirmed as it does not complement the yeast CIN2 deletion. However, the possibility that TBCCD1 is a GAP cannot be excluded. As observed for the p160ROCK Rho-associated kinase (Chevrier et al, 2002) and the Polo/Gwl mitotic kinases (Archambault et al, 2007), it is conceivable that this TBCCD1 putative regulatory activity might be responsible for the described phenotypes. TBCCD1 also possesses a CARP domain—a characteristic of CAP proteins, which regulate actin polymerization. Thus, an attractive hypothesis is that TBCCD1 could promote crosstalk between the centrosome and MT and actin cytoskeletons that are required for centrosome positioning.
Finally, during review of this paper, Feldman & Marshall (2009) reported that Chlamydomonas reinhardtii TBCCD1 localizes in centrioles and in the region between the two nucleus–centriole connectors (rhizoplasts). A TBCCD1 insertion mutant showed an altered number of flagella and affected centriole linkage and positioning, which leads to defects in spindle orientation. In spite of the differences between the two models, both studies show TBCCD1 to be a centrosomal protein important for centrosome–nucleus connection.
Methods
Cell lines and primary cultures. HEK293T and HeLa-centrin–GFP cells were grown in Dulbecco's modified Eagle's medium (DMEM), whereas hTERT-RPE-1 and hTERT-RPE-1-centrin–GFP were grown in DMEM/F12, both supplemented with 10% fetal bovine serum. Mixed primary cell cultures were obtained from 2-day-old murine dissociated cerebella.
Plasmid construction and transfection. Human tbccd1 entire or truncated coding sequences were amplified by PCR from testis cDNA and cloned in mammalian expression vectors (pIC111, pIC112 and pIC113); yeast expression vector pRS413 (also used to clone tbcc) and bacterial expression vector pGEX-4T-1 (GE Healthcare, Pittsburgh, PA, USA). The human tbcc coding sequence was amplified from brain cDNA and cloned in pRS413. Plasmid transfections used Lipofectamine-2000 (Invitrogen, Carlsbad, CA, USA).
RNA interference. RPE-1 cells (2 × 104 cells seeded in 12-well plates) were transfected with 100 nM of a pool of four siRNAs directed to TBCCD1 obtained from Dharmacon (ON-TARGETplus Duplex; Lafayette, CO, USA) and Ambion (Silencer Select siRNAs; Austin, TX, USA), using Oligofectamine (Invitrogen). As negative controls, Silencer Select Negative Control 2 siRNA (Ambion) or a non-target fluorescent control siRNA (siGlo RISC-free siRNA; Dharmacon) was used.
For MT regrowth assays, MTs were depolymerized with 30 μM of nocodazole for 40 min, then the cells were washed thoroughly with culture medium and placed in medium at 37°C to enable MT repolymerization.
To induce primary cilia assembly, the cells were seeded at a 2.5-fold higher density. Forty-eight hours after transfection the cells were retransfected and serum-starved (0.25% serum) for 24 h. A similar strategy was used in wound-healing assays, except for serum starvation. Twenty-four hours after the second transfection, the cell monolayer was wounded with a micropipette tip. The cells were either live-imaged or processed for immunofluorescence.
RT–PCR and western blot analysis. Total RNA samples from mouse tissues and cell lines were prepared by using the RNeasy Mini kit (Qiagen, Germantown, MD, USA) followed by cDNA synthesis by using Superscript II RT (Invitrogen).
Total protein extracts were prepared by using a lysis buffer containing 50 mM 4-(2-hydroxyethyl)-1-piperazineethane sulphonic acid (HEPES; pH 8), 200 mM NaCl, 5 mM EDTA, 0.5% NP-40, with protease inhibitors. The cytoplasmic and nuclear extracts were prepared by lysing the cells with 50 mM HEPES (pH 8), 10 mM NaCl, 2 mM ethylenediamine tetraacetic acid (EDTA), 250 ml sucrose and 0.1% NP-40, with protease inhibitors, followed by centrifugation at 3,000g (10 min at 4°C). The supernatant was collected as the cytosolic fraction. Nuclei were washed with buffer without NP-40 and resuspended in 50 mM HEPES (pH 8), 400 mM NaCl, 2 mM EDTA and 20% glycerol (v/v) with protease inhibitors. After incubation on ice for 20 min the samples were centrifuged at 10,000g (10 min at 4°C). The supernatant was collected as the nuclear fraction. Equal amounts of nuclear and cytosolic extracts were analysed.
Antibody generation and immunofluorescence. Human TBCCD1 was expressed as a glutathione-S-transferase fusion in BL21 Rosetta DE3 cells and purified from inclusion bodies using a standard protocol. The purified protein was used to immunize Balb/c mice.
For immunofluorescence, the cells were fixed with cold methanol (10 min at −20°C), blocked with 3% bovine serum albumin (20 min), incubated with the primary antibodies (1 h), washed and incubated with secondary antibodies (1 h). The primary and secondary antibodies are listed in the supplementary information online. The cells were analysed by using a Deltavision System or with Leica DMRA2 and Zeiss LSM510 confocal microscopes. Images were analysed with the ImageJ software.
Flow cytometric analysis. Control and TBCCD1-silenced cells were collected, fixed with ethanol and stained with propidium iodide. Flow cytometry analysis was performed using a FACSCalibur system.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Mónica Bettencourt-Dias (Instituto Gulbenkian Ciência; IGC) and Leonor Saúde (Instituto Medicina Molecular) for critically reviewing the paper, and Lars Jansen and Cláudia Florindo at IGC for help with live imaging. hTERT-RPE-1-centrin–GFP cells were provided by A. Khodjakov, and mammalian expression vectors pIC111, pIC112 and pIC113 were provided by I. Cheeseman. This work was partly supported by PTDC/CVT/71630/2006 (Fundação para a Ciência e Tecnologia), Consolider-Ingenio SMES (Centrosome-3D and BFU2007-64882) and IFIMAV grants. Fellowships were given to J.G. (SFRH/BD/24532/2005) and S.N. (SFRH/BPD/20681/2004).
Footnotes
The authors declare that they have no conflict of interest.
References
- Archambault V, Zhao X, White-Cooper H, Carpenter ATC, Glover DM (2007) Mutations in Drosophila Greatwall/Scant reveal its roles in mitosis and meiosis and interdependence with Polo kinase. PLoS Genet 3: e200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr FA (2009) Membrane traffic: Golgi stumbles over cilia. Curr Biol 19: R253–R255 [DOI] [PubMed] [Google Scholar]
- Bartolini F, Bhamidipati A, Thomas S, Schwahn U, Lewis SA, Cowan NJ (2002) Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J Biol Chem 277: 14629–14634 [DOI] [PubMed] [Google Scholar]
- Bartolini F, Tian G, Piehl M, Cassimeris L, Lewis SA, Cowan NJ (2005) Identification of a novel tubulin-destabilizing protein related to the chaperone cofactor E. J Cell Sci 118: 1197–1207 [DOI] [PubMed] [Google Scholar]
- Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E (2009) An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci 122: 2100–2107 [DOI] [PubMed] [Google Scholar]
- Burakov A, Nadezhdina E, Slepchenko B, Rodionov V (2003) Centrosome positioning in interphase cells. J Cell Biol 162: 963–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevrier V, Piel M, Collomb N, Saoudi Y, Frank R, Paintrand M, Narumiya S, Bornens M, Job D (2002) The Rho-associated protein kinase p160ROCK is required for centrosome positioning. J Cell Biol 157: 807–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens I, Dunn JD, Romano JD, Pypaert M, Zhang H, Boothroyd JC, Joiner KA (2006) Toxoplasma gondii sequesters lysosomes from mammalian hosts in the vacuolar space. Cell 125: 261–274 [DOI] [PubMed] [Google Scholar]
- Cunningham LA, Kahn RA (2008) Cofactor D functions as a centrosomal protein and is required for the recruitment of the γ-tubulin ring complex at centrosomes and organization of the mitotic spindle. J Biol Chem 283: 7155–7165 [DOI] [PubMed] [Google Scholar]
- de Anda FC, Pollarolo G, da Silva JS, Camoletto PG, Feiguin F, Dotti CG (2005) Centrosome localization determines neuronal polarity. Nature 436: 704–708 [DOI] [PubMed] [Google Scholar]
- Feldman JL, Marshall WF (2009) ASQ2 encodes a TBCC-like protein required for mother–daughter centriole linkage and mitotic spindle orientation. Curr Biol 19: 1238–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontalba A, Paciucci R, Avila J, Zabala JC (1993) Incorporation of tubulin subunits into dimers requires GTP hydrolysis. J Cell Sci 106: 627–632 [DOI] [PubMed] [Google Scholar]
- Higginbotham HR, Gleeson JG (2007) The centrosome in neuronal development. Trends Neurosci 30: 276–283 [DOI] [PubMed] [Google Scholar]
- Lüders J, Stearns T (2007) Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol 8: 161–167 [DOI] [PubMed] [Google Scholar]
- Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. (2003) The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell 115: 825–836 [DOI] [PubMed] [Google Scholar]
- Pouthas F, Girard P, Lecaudey V, Ly TB, Gilmour D, Boulin C, Pepperkok R, Reynaud EG (2008) In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum. J Cell Sci 121: 2406–2414 [DOI] [PubMed] [Google Scholar]
- Rios RM, Bornens M (2003) The Golgi apparatus at the cell centre. Curr Opin Cell Biol 15: 60–66 [DOI] [PubMed] [Google Scholar]
- Salpingidou G, Smertenko A, Hausmanowa-Petrucewicz I, Hussey PJ, Hutchison CJ (2007) A novel role for the nuclear membrane protein emerin in association of the centrosome to the outer nuclear membrane. J Cell Biol 178: 897–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephan A, Vaughan S, Shaw MK, Gull K, McKean PG (2007) An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic 10: 1323–1330 [DOI] [PubMed] [Google Scholar]
- Tian G, Bhamidipati A, Cowan NJ, Lewis SA (1999) Tubulin folding cofactors as GTPase-activating proteins. GTP hydrolysis and the assembly of the α/β-tubulin heterodimer. J Biol Chem 274: 24054–24058 [DOI] [PubMed] [Google Scholar]
- Veltel S, Gasper R, Eisenacher E, Wittinghofer A (2008) The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol 15: 373–380 [DOI] [PubMed] [Google Scholar]
- Vinogradova T, Miller PM, Kaverina I (2009) Microtubule network asymmetry in motile cells: role of Golgi-derived array. Cell Cycle 8: 2168–2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM (2001) Cell motility: can Rho GTPases and microtubules point the way? J Cell Sci 114: 3795–3803 [DOI] [PubMed] [Google Scholar]
- Yvon AM, Walker JW, Danowski B, Fagerstrom C, Khodjakov A, Wadsworth P (2002) Centrosome reorientation in wound-edge cells is cell type specific. Mol Biol Cell 13: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.