Abstract
Monocytes recruited into tissues from peripheral blood differentiate into macrophages, which are critical in the pathogenesis of many diseases. There is limited data concerning the global changes in the expression of genes during monocyte to macrophage differentiation, and how the patterns of change identify the mechanism contributing to macrophage differentiation or function. Employing microarray technology, we examined the transcriptional profile of in vitro adherence-induced differentiation of primary human monocytes into macrophages. We found the significant up regulation of genes contributing to the functions of macrophages, including those regulating to immunity and defense; lipid, fatty acid and steroid metabolism; cell adhesion, carbohydrate metabolism; amino acid metabolism and endocytosis. In contrast, the vast majority of transcription factors affected were down regulated during monocyte to macrophage differentiation, suggesting that transcriptional repression may be important for the transition from monocytes to macrophages. However, a limited number of transcription factors were up regulated, among these was C/EBPα, which may contribute to differentiation by regulating down stream genes, which are a characteristic of differentiated macrophages. These observations suggest that examination of the transcriptional profile in monocytes and macrophages in patients may identify relevant therapeutic targets in diseases mediated by macrophages.
Keywords: mononuclear phagocytes, gene regulation, transcription factors
INTRODUCTION
Monocytes originate from bone marrow hematopoietic progenitors, colony-forming units-monocyte (CFU-M) which proliferate and differentiate in the presence of cytokines, including macrophage colony stimulating factor (M-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-3 (IL-3) [1]. Peripheral blood monocytes are capable of differentiating into morphologically and functionally heterogeneous effector cells, including macrophages, myeloid dendritic cells, and osteoclasts [2]. Monocytes leave the bone marrow and travel through peripheral blood vessels. Once they reach the tissue, they differentiate into tissue resident or inflammatory macrophages [2].
In vitro, monocytes adhere to plastic surfaces and, even in the absence of exogenous M-CSF, undergo spontaneous, time-dependent differentiation [3]. The signals required for monocyte to macrophages differentiation in vitro and in vivo are not well characterized, although M-CSF is an important cofactor for the survival and differentiation of monocytes [4, 5], and the M-CSF receptor (CSFR-1), coded by c-fms gene, is required for the differentiation, proliferation, and survival of monocytic phagocytes. Deficiency of c-fms is associated with marked reduction of monocytes and macrophages [6].
In myelomonocytic cells, several transcription factors have been identified that are important for lineage commitment, subsequent differentiation, and cell type-specific gene regulation. These include members of the Ets, C/EBP, and CBF families and several other transcription factors [7, 8]. For example, the c-fms gene is regulated by the coordinated effects of three transcription factors, PU.1, AML1, and CCAAT enhancer-binding proteins (C/EBPs) [1]. In particular the Ets family transcription factor PU.1 is essential for normal monocyte development and macrophage differentiation and for cell type-specific gene expression [6, 9]. The defect of PU.1−/− hematopoietic progenitors may be attributed to their failure to express the receptors for M-CSF, G-CSF and GM-CSF, which are critical for progenitor cell survival and proliferation [1].
Although studies with murine models with specific gene deletions have identified a number of mediators that are critical for monocyte and macrophage differentiation, an understanding of the differences between circulating monocytes and differentiated macrophages is incompletely understood, even though the monocyte and macrophage phenotypes are clearly distinct. Many authors refer to the cells as monocytes/macrophages because many of their functions, such as cytokine and chemokine production, overlap and because there are no unique markers that clearly differentiate the two cell types. These studies were performed to characterize the extent of the differences in the expression of genes between monocytes and macrophages and in an attempt to identify pathways that may be important for the process of monocyte to macrophage differentiation.
METHODS
Cell isolation and culture
Buffy coats (Lifesource, Glenview, IL) were obtained from healthy donors. Mononuclear cells, isolated by Histopaque (Sigma, St. Louis, MO) gradient centrifugation, were separated by countercurrent centrifugal elutriation (JE-6B, Beckman Coulter, Palo Alto, CA) in the presence of 10 µg/ml polymyxin B sulfate, as previously described [10–13]. Isolated monocytes were ≥ 90% pure as determined by morphology, non-specific esterase staining and CD14 expression examined by flow cytometry (data not shown). Monocytes were adhered to plates for 1 hour in RPMI and 1 µg/ml polymyxin B sulfate. Following adherence, monocytes were differentiated into macrophages in vitro for 16 hours or for 7 days in RPMI containing 20% heat-inactivated FBS, 1 µg/ml polymyxin B sulfate, 0.35 mg/ml L-glutamine, 120 Units/ml penicillin and streptomycin (20% FBS/RPMI), without the addition of supplemental growth factors [10–13]. Freshly isolated monocytes, monocytes isolated16 hours following adherence, and 7-day differentiated macrophages from six different normal donors were used to isolate the total RNA. An additional 3 donors were obtained to repeat the qRT PCR data at 0, 16 and 168 hours.
Microarray
Affymetrix HG_U133A chips, which includes approximately 23,000 probe sets, was employed, as previously described [14].
Analysis of microarray data
The microarray data was normalized by dChip software (http://biosun1.harvard.edu/complab/dchip/) and gene ontology (GO) analysis was performed using the online application PANTHER (www.pantherdb.org) [15, 16]. The significance of enrichment of differentially expressed genes for each GO term was determined by the Bonferroni corrected p value as the default setting in PANTHER, whereas the significance of differential expression was determined by nominal p value < 0.05 from the t test and a fold change > 2. Multidimensional Scaling (MDS) analysis was performed to reveal the global changes of gene expression profiles occurring during differentiation.
qRT PCR
Reverse transcription with oligo (dT) primers was performed with the reverse transcription system (Promega, Madison, WI) as previously described employing the ΔΔCt method [14]. The expression value was normalized to GAPDH.
SiRNA transfection
The forced reduction of M-CSF receptor (CSF1R) and CCAAT/enhancer binding protein, alpha (CEBPA) was achieved employing the SMARTpools for CSF1R or CEBPA siRNA, (Dharmacon, Lafayette, CO). A non-specific siRNA was employed as the control. Monocytes differentiated for 2 days were transfected with 100nM siRNAs by lipofectomine methods following the manufacturers directions. After 4 hours, FBS was added to bring the culture medium to 20% FBS and the cells cultured for an additional 5 days.
ELISA
The culture supernatants were collected at the time of 0 (culture medium), 24 and 48hours. M-CSF levels quantified employing a commercially available Human M-CSF ELISA kit (Ray Biotech, Inc, Norcross, GA), according to the manufacturer protocol.
RESULTS
Monocyte to macrophage differentiation following adherence
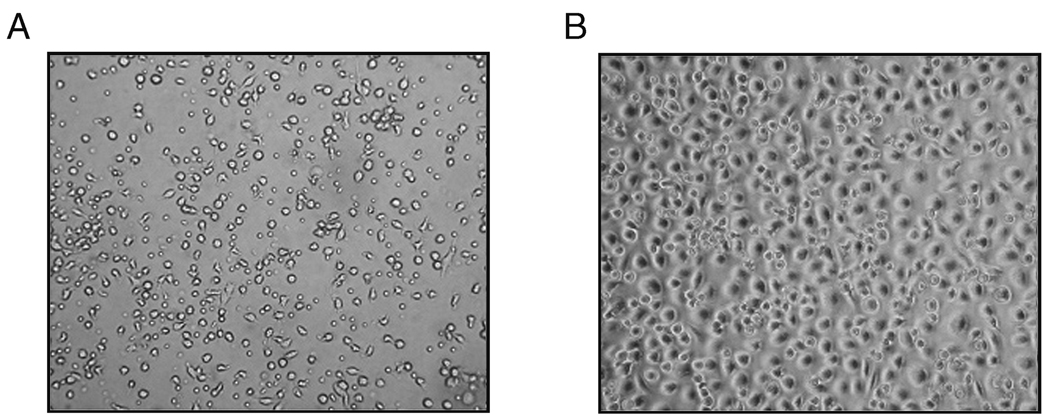
These studies were aimed at characterizing the patterns of gene expression during monocyte to macrophage differentiation, providing only those factors necessary to ensure survival and differentiation. In order to employ an unbiased approach, the culture plates were not coated with extracellular matrix molecules such as collagen or fibronectin, such as might be encountered as monocytes migrate into tissues and with which they may interact during differentiation. Monocytes added to culture plates in the presence of serum, adhered poorly and did not acquire the morphologic appearance of differentiated macrophages (Figure 1A). In contrast, when monocytes were added to the culture dishes in the absence of serum for 1 hour, followed by the addition of serum, they strongly adhered to the plates and after 7 days possessed the morphologic appearance of in vitro differentiated macrophages, appearing egg-shaped, were strongly adherent with a round nucleus appearing above the surface of the surrounding cytoplasm (Figure 1B). The macrophages did not show evidence of activation since multiple activation-induced cytokine genes including TNFα, IL-1β, IL-15 and IL-8 were all reduced in macrophages (168 hours) compared to monocytes at 0 hours (−2.2 to −50.3 fold, p < 0.05). These data indicate that the macrophages had not undergone classical activation [17].
Figure1. The adherence of monocytes to culture plates promotes in vitro monocyte-macrophage differentiation.
Monocytes, isolated from the buffy coats from healthy donors, were added to the culture plates in the presence (panel A) or absence (panel B) of 20% FCS for 1 hour. The cells were incubated for 168 hours in the presence of 20% FCS and then examined.
Multidimensional Scaling (MDS) Analysis
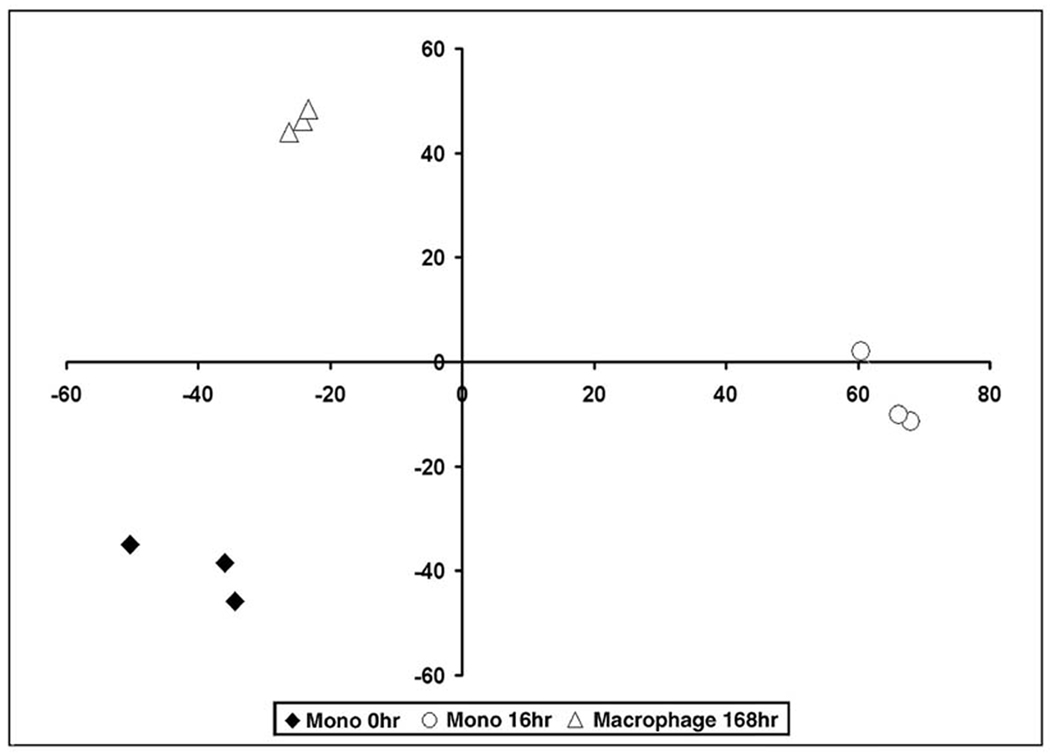
MDS analysis is a multivariate statistical technique which summarizes the high dimensional microarray data for exploring similarities or dissimilarities in data. This technique allows visualization of global expression patterns between experiments. The MDS plot revealed clear and reproducible differences of the gene expression profiles at each time point examined, as monocytes at 0 hours, monocytes at 16 hours and macrophages at 168 hours cluster independently on the graph (Figure 2). This observation demonstrates that the differences in the patterns of genes expressed between monocytes at 0 and 16 hours and macrophages were distinct and not random.
Figure 2. MDS plot reveals distinct clusters during monocyte to macrophage differentiation.
Multidimensional Scaling (MDS) Analysis was performed on data obtained from monocytes (Mono) at 0 hours, monocytes at 16 hours and macrophages at 168 hours.
Differential gene expression between monocytes and macrophages
The extent of the changes in gene expression during monocyte to macrophage differentiation was quite dramatic at each of the time points examined (Table 1). Between 0 and 16 hours, 2384 probe sets had changed (> 2 fold and p < 0.05), and 1621 were different between 16 and 168 hours. This occurred because some genes that initially changed returned to baseline and some that had not changed at 16 hours, were different by 168 hours. Overall, 2322 probe sets were different between monocytes (0 hours) and macrophages (168 hours). Of the probe sets that changed, about 60% were up and 40% down between 0 and 16 and 0 and 168 hours, while those that changed between 16 and 168 hours were equally divided.
Table I.
Number of probe sets changed more than 2 fold and p<0.05 during macrophage differentiation.
| Differentiation duration | Total number of changed genes |
Number of genes up- regulated |
Number of genes down-regulated |
|---|---|---|---|
| 0h– 16h | 2384 | 1433 (60%) | 951 (40%) |
| 16h–168h | 1621 | 771 (48%) | 850 (52%) |
| 0h–168h | 2322 | 1444 (62%) | 878 (38%) |
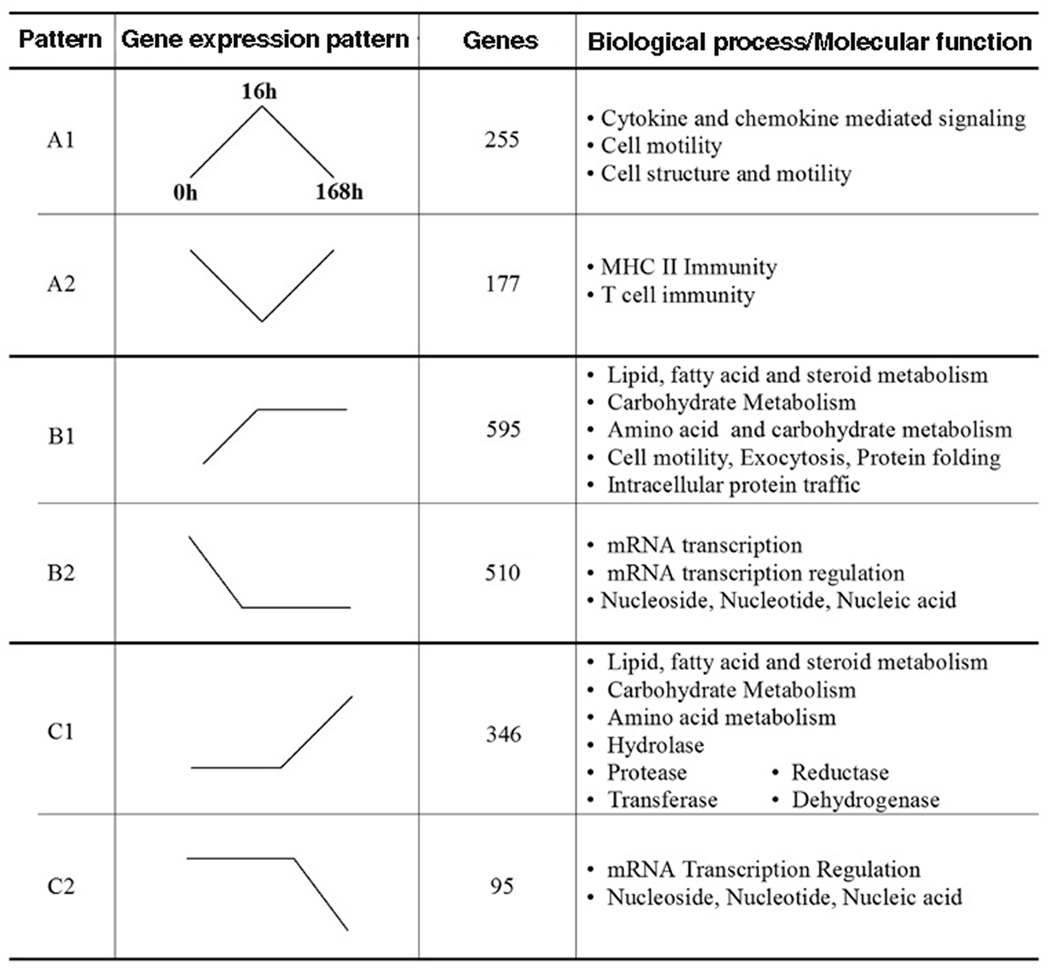
Patterns of Gene Expression
To characterize the potential relevance of the changes, those probe sets identified as > 2 fold change up or down and p < 0.05 were subjected to PANTHER GO analysis by pathway, biological process and molecular function. The results obtained as biological process were the most robust and were primary focus of the analysis. The majority of the changes of the genes expressed identified by biological process, could be assigned to one of 6 patterns. Some changes occurred early (up or down) at 16 hours, but returned to baseline by 168 hours (pattern A1 or A2 respectively) (Figure 3). A second pattern observed was change at 16 hours (up or down), and the changes were maintained or enhanced at 168 hours (pattern B1 or B2). The third pattern of change noted was no change at 16 hours, with significant change (up or down) by 168 hours (pattern C1 or C2). We reasoned that the first pattern (pattern A) may relate to functions that might be important during the process of differentiation, but may not be necessary for macrophage function, that pattern B may represent genes needed during the process of differentiation, but may also be critical for the function of differentiated macrophages, while genes identified by pattern C may be important for the function or maintenance of macrophages.
Figure 3. Patterns of gene expression during monocyte (Mono)-macrophage (MΦ) differentiation.
The patterns of gene expression (represented by the line diagrams) were identified following GO analysis. Pattern A designates categories that were up (A1) or down (A2) regulated at 16 hours, but returned to baseline by 168 hours. Pattern B represents genes up (B1) or down (B2) regulated at 16 hours, and these changes are maintained or enhanced at 168 hours. Pattern C identifies groups with no changes at 16 hours, but which are significant increased (C1) or decreased (C2) by 168 hours. The number of genes included within each pattern is represented in column 3. Potentially important signaling pathways/functions are identified in the column to the right.
Of the 242 biological processes, for pattern A1, only 8 processes demonstrated significant enrichment from those expected by chance. The processes, represented by pattern A1 included the cytokine and chemokine mediated signaling pathway, immunity and defense, granulocyte-mediated immunity, cell motility, signal transduction, and cell surface receptor mediated signal transduction. The genes identified in the cytokine and chemokine pathway were also identified in 6 of the other 8 biological processes. The chemokines included in pattern A1 included CCL2, CCL3, CCL4, CCL5, CCL7, CCL24, CXCL7, as well as CCR1. Additionally, M-CSF and IL-6 receptor [6, 18–20], which may be important in the process of macrophage differentiation, were included in pattern A1.
Pattern A2 identified only 6 of 242 biological processes, which included MHCII-mediated immunity, immunity and defense, T-cell immunity and lipid, fatty acid and steroid metabolism. The genes included in the first 3 categories were highly redundant and included HLA-DR, -DQ and -DP. These observations suggest that early during the process of differentiation, genes important in antigen presentation are decreased but return to the monocyte baseline in differentiated macrophages. Included within the lipid metabolism group were PI3Kδ, arachidonate 5-lipoxygense (ALOX5), and catalase (CAT), but not genes that would be associated with foam cell formation.
Pattern B is represented by genes which changed (up, B1 or down, B2, > 2 fold, and p < 0.05) at 16 hours and this change persisted or was further enhanced at 168 hours (Figure 3). Fourteen biologic processes are represented by pattern B1, and 10 of these processes were only identified as pattern B1. The majority of the processes that unique to pattern B1 related to cell mobility, protein metabolism and trafficking, protein folding and exocytosis. However 4 of the biological processes identified by pattern B1 were also identified in pattern C1 (up, C1 or down, C2, > 2 fold, and p < 0.05), including lipid, fatty acid and steroid metabolism, carbohydrate metabolism, amino acid metabolism, and immunity and defense. Within the immunity and defense category, genes up regulated as pattern B1 or C1 included CSF1R, HSP 70, 90, and 96, macrophage scavenger receptor 1, CD163, and integrin β5.
There were 12 of the 242 biological process represented by pattern B2, down (> 2 fold, p < 0.05) at 16 hours, with the change maintained or enhanced at 168 hours. Seven of the processes were unique to this pattern, including cell communication, B cell and antibody related immunity, cell communication and intracellular signaling cascade. Five of the processes included in B2, were also identified by pattern C2, including mRNA transcription regulation, mRNA transcription, nucleoside, nucleotide and nucleic acid, apoptosis and immunity and defense. There was substantial overlap of genes represented by transcription factors in the first 3 processes. Within the immunity and defense ontogeny, genes represented by pattern B2 or C2 included G-CSF, VEGF, ICAM3, CD86, CD1d, IL-8, and IL-10.
There were several biological processes represented only by pattern C1 including coenzyme and prosthetic group metabolism, vitamin metabolism and electron transport or by pattern C2, cell proliferation and differentiation, ligand-mediated signaling and signal transduction. In contrast to the scarcity of biological processes identified solely by pattern C1, genes identified by molecular function were highly represented (11 of 243 molecular functions) and clearly stood out as genes potentially important for the phagocytic and processing functions of macrophages, including reductase, oxyreductase, hydrolase, deyhydogenase, transferase and aclytransferase. These observations demonstrate the marked diversity in the expression of genes during the process of monocyte to macrophage differentiation.
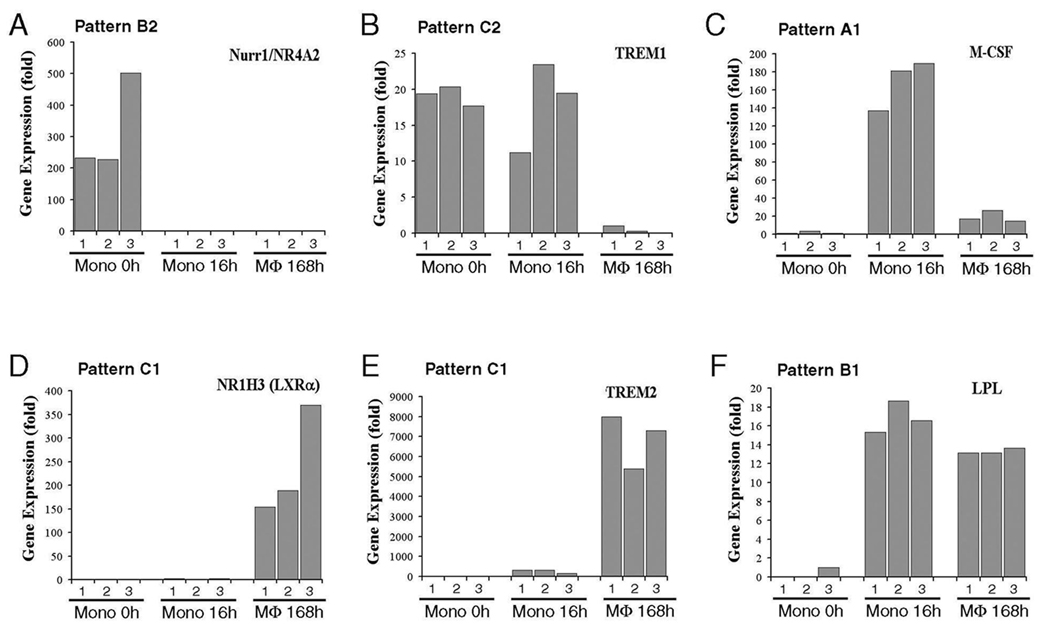
Verification of patterns by qRT-PCR
In order to verify the results from microarray, 6 genes that were represented by pattern A, B and C were employed to perform qRT PCR, initially employing the same RNA that was used to run the Affymetrix microarray. These genes included NR4A2 (Nurr1, nuclear receptor subfamily 4), NR1H3 (LXRα), TREM1, TREM2, LPL, and M-CSF receptor (Figure 4). Our data from qRT PCR matched the patterns identified by microarray. In order to determine if these results were generalizable, monocytes from an additional 3 individuals were isolated and examined at 0, 16 and 168 hours. The results demonstrated patterns similar to those identified in Figure 3 for each of the genes examined. Therefore, the patterns of change during monocyte to macrophage differentiation were consistent.
Figure 4. Verification of gene expression patterns by qRT-PCR.
Six genes (Nurr1, LXRα, TREM1, TREM2, LPL, and M-CSF) representing different expression patterns A, B and C were chosen and examined by qRT-PCR. The triplicate total RNA samples of monocyte 0hr, 16hrs and 168hrs that were used to run the Affymetrix microarray were employed. The gene expression pattern from Figure 3 for each gene is shown on the top of each panel.
Lipid, fatty acid, and steroid process
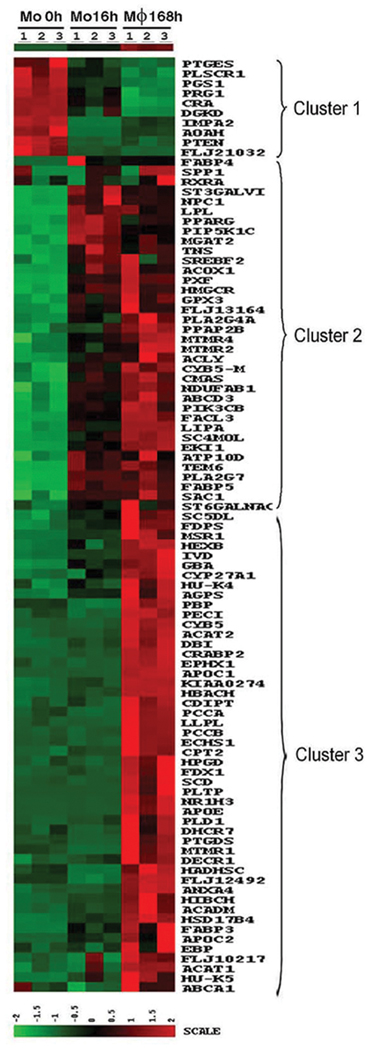
Of the processes that showed marked changes during macrophage differentiation, the Lipid, Fatty Acid and Steroid Metabolism process stood out. Even though abundant data to supports the role of macrophages in LDL oxidation and the pathogenesis of atherosclerosis [21, 22], it is unclear to what extent, the process of differentiation prepares macrophages for these roles, nor is it clear at what point the relevant genes are expressed. To obtain a more complete picture of this process, all genes identified in this biological process as undergoing a significant (> 2 fold, p < 0.05) change during monocyte to macrophage differentiation are presented in a heat map (Figure 5). Three clusters were identified which corresponded to patterns B2 (cluster 1), B1 (cluster 2), and C1 (cluster 3). The small number of genes in cluster 1 included those involved in prostaglandin synthesis such as PGS1 and PTGES, while the majority of the changes demonstrated increased expression, identified in clusters 2 and 3. The majority of the genes identified by this process were up regulated during macrophage differentiation, and they were fairly evenly divided between those that were already up regulated at 16 hours (cluster 2) and those that increased later (cluster 3). It is worthy to mention that Apolipoprotein (APO) C-1 and C-2, Apolipoprotein E, and lipoprotein lipase (LPL) were up regulated dramatically. Additional genes important in lipid metabolism and foam cell formation or function included, cholesterol 27-hydroxylase (CYP27A1), acyl-Coenzyme A: cholesterol acyltransferase 1 and 2 (ACAT1 and 2), fatty acid binding protein (FABP) 3, 4 and 5, 3-hydroxy-3methylglutaryl-Coenzyme A reductase (HMGCR), and lysosomal phospholipase A2 (LLPL). Also up regulated and recognized as potentially important in foam cells was macrophage scavenger receptor 1 (MSR1 or SR-A), scavenger receptor class B member 2 (SCARB2) and class D (CD68, SCARD1), and LXRα (NR1H3). These observations demonstrate that macrophage differentiation, even in an unbiased in vitro environment, prepares macrophages to participate in atherosclerosis.
Figure 5. Heat map of lipid, fatty acid, and steroid metabolism genes differentially regulated during monocyte (Mo) to macrophage differentiation.
The genes included are those identified following GO analysis, as well as additional genes identified in other categories, which are know to be involved in foam cell function. The time points at which the cells were harvested are indicated at the top of the map, and the gene symbol for each gene is to the right of the map.
Transcriptional regulation
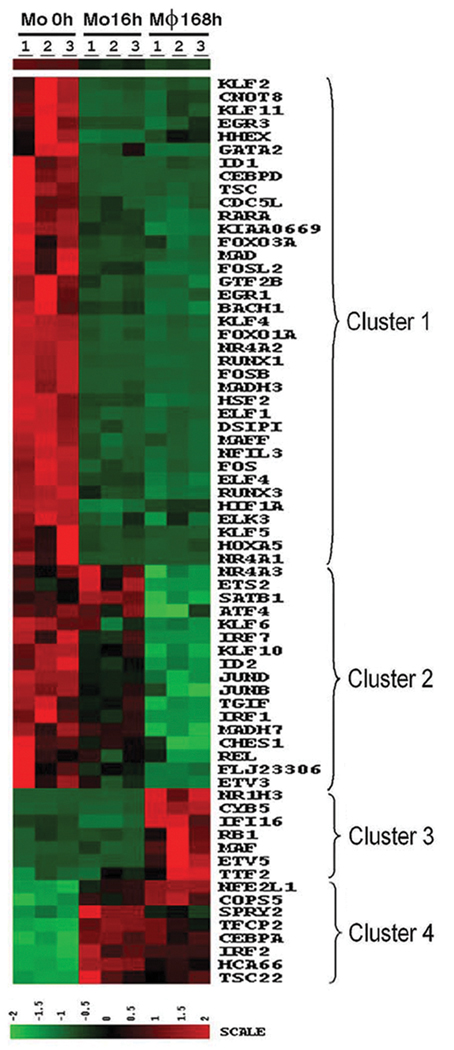
We initially hypothesized that the up regulation of transcription factors would be import for regulating macrophage differentiation. In contrast, the majority (78%) of the transcription factors were down regulated during macrophage differentiation, identified in clusters 1 and 2 (patterns B2 and C2) (Figure 6). Among the genes that were decreased in macrophages FOS, FOSB, JUNB, JUND, CCAAT/enhancer binding protein δ (C/EBPδ), interferon regulatory factor 1 and 7 (IRF1 and 7), MAFF, FOXO3A, FOX01A, SMAD3 and 6, many members of the Kruppel-like factor (KLF) and early growth response (EGR) families, and acute myeloid leukemia 1 and 3 (AML or RUNX1 and 3). A minority of transcription factors identified in clusters 3 and 4 (pattern C1 and B1) were up regulated, including C/EBPα, sprouty homolog 2 (SPRY2), IFR2 and musculoaponeurotic fibrosarcoma oncogene homolog (MAF). These observations identify several candidate genes whose up-regulation may contribute to macrophage differentiation, however, the suppression of genes important for transcriptional regulation appears critical for monocyte to macrophage differentiation.
Figure 6. Heat map of transcriptional regulation genes.
The genes include those identified following GO analysis, as well as additional genes identified in other categories, which are know to be involved in transcriptional regulation, such a nuclear receptors. The time points at which the cells were harvested are indicated at the top of the map, and the gene symbol for each gene is to the right of the map.
Modulation of genes during differentiation
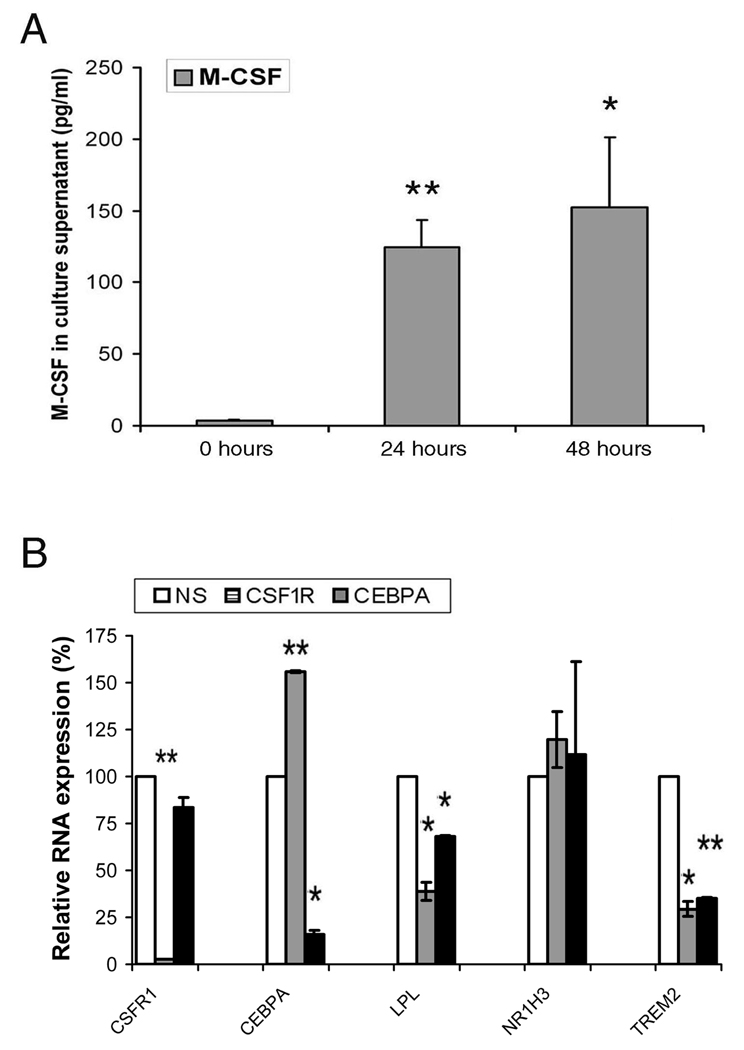
Further studies were performed to determine if genes differentially regulated might effect the expression of other genes during the process of differentiation. Since M-CSF was up regulated at 16 hours, culture supernatants were examined to determine if it was secreted. At 24 and 48 hours M-CSF was readily detected into the macrophage culture supernatants (Figure 7A). Additional studies employed siRNA for the M-CSF receptor (CSF1R) and C/EBPα, which were up regulated at 16 hours and remained elevated at 168 hours, to determine if they contributed to the expression of other genes that were differentially regulated. The siRNA were added to differentiating macrophages at 48 hours, a time at which the expression each of these genes was already increased. To determine the effect of suppression of these genes, 3 genes expressed as pattern B1, lipoprotein lipase (LPL), or pattern C1, LXRα (NR1H3) and triggering receptor expressed on myeloid cells 2 (TREM2), were examined by qRT-PCR. The knock down of the expression of CSF1R and C/EBPα with their respective siRNAs was highly effective (Figure 7B). There was no change in the morphology of the macrophages at day 7 (data not shown). Unexpectedly, the forced reduction of CSF1R resulted in the increased expression of C/EBPα. Further, the knock down of either CSF1R or C/EBPα resulted in significant (p < 0.05 to 0.01) reduction of LPL and TREM2, but had no effect on LXRα. These observations demonstrate the potential relevance of secretion of M-CSF and the expression of C/EBPα during the macrophage differentiation.
Figure 7. Modulation of genes during differentiation.
A. Monocytes secrete M-CSF into the culture supernatant during differentiation. `The culture supernatants were collected at 0, 24 and 48 hours. The results represent the mean ± 1SE of 3 independent experiments. B. The forced reduction of CSF1R and C/EBPα affected the expression of other genes during the process of differentiation. CSF1R and CEBPA or non-specific siRNA (100 nM) were added at 48 hours and the cells harvested at 168 hours and RNA employed for qRT-PCR. The results represent the mean ± 1SE of 2 independent experiments. * and ** represent p<0.05 and p<0.01 versus 0 hours (panel A) or the NS siRNA control (panel B).
DISCUSSION
These studies document a diverse and extensive difference in the patterns of gene expression between monocytes and macrophages. Greater that 10% of the probe sets employed were differentially expressed between monocytes and macrophages. We employed an unbiased approach, since there are well-recognized differences between resident or inflammatory monocytes and macrophages, depending on the tissue and the environment [2, 23]. The changes observed were not random, and the differences identified were related to characteristic functions of macrophages. Of particular note, this study documented extensive up regulation of genes related to lipid, fatty acid and steroid metabolism, and the reduction of genes contributing to transcriptional regulation.
M-CSF promotes the differentiation of macrophages from circulating monocytes [5], but factors in serum, in addition to M-CSF, are required for optimal differentiation [24]. M-CSF was highly expressed at 16 hours, and was detected in the culture supernatants at 24 and 48 hours, consistent with earlier observations [25]. Therefore, early in the process of differentiation M-CSF is expressed, and it is capable of enhancing differentiation. These observations identify a positive feedback loop where by the process of differentiation promotes the local induction of M-CSF and its receptor CSF1R. For technical reasons we were not able to knock down genes prior to 48 hours. Mice deficient in c-fms (CSF1R) or M-CSF (op/op) demonstrate deficient development of monocytes [6, 7, 26]. Nonetheless, our data clearly document the importance of M-CSF and CSF1R in some, but not all, aspects of macrophage differentiation. It is also possible the differentiation was promoted through the firbonectin receptor [27], since the fibronectin gene was highly induced in macrophages, and fibronectin is secreted by macrophages [27].
Additionally, αv, which may serve as a vitronectin or fibronectin receptor, was strongly induced at 16 and 168 hours, while its binding partner integrin β3 was enhanced at 16 hours. Additionally, integrins α5 and β1, which also bind fibronectin, were highly expressed in monocytes, and both increased significantly, but less than 2 fold, in macrophages. Therefore, following the initial adherence of monocytes, the process of differentiation may have been promoted by the expression of both M-CSF [4, 20] and fibronectin [27].
A recent study also examined monocyte to macrophage differentiation, however, they employed the addition of exogenous M-CSF and coated the culture dishes with FCS, rather than using an initial adherence [28]. Nonetheless, the extent of the changes, and many of the patterns observed in the two studies were similar. For example both studies noted an increase of genes related to lipid and fatty acid metabolism in macrophages and the reduction of genes induced during classical activation of macrophages activation, such as CXCL8, TNFα, IL-1β, and CD69. Additionally, in both studies, macrophage scavenger receptor 1 (MSR1) and the mannose receptor (MRC1) were increased in unactivated macrophage, as well as in alternatively activated M2 macrophages [28]. There were differences noted between the two methods of differentiation. In both studies the transcriptional expression of chemokine receptors CCR2, CCR7 and CX3CR1 was increased, although changes in CCR5 were different between the two studies. Further there were differences in genes related to regulation of prostaglandins. Overall, these observations demonstrate marked and consistent differences in the transcriptional profile between circulating monocytes and in vitro differentiated macrophages, employing both methods of differentiation.
A unique characteristic noted early in the process of differentiation was the expression of chemokine genes, including CCL2, CCL3, CCL4, CCL7, CCL24, and CXCL5 which are capable of attracting leukocytes, particularly monocytes [29]. CCR1, but not other chemokine receptors, was also transiently up regulated. These changes returned to baseline by 168 hours, and suggest that as monocytes begin to differentiate, they signal for the recruitment of additional leukocytes. Since both CD14+CD16+ and CD14+CD16− cells were included, we can not determine whether inflammatory or resident monocytes [2] may respond differently. Additionally, at this early time point, MHC class II molecules, DR, DQ and DP, were down regulated. The reduction of MHC class II molecules at this time suggests that early during differentiation the ability to present antigen is diminished.
The categories of genes regulated in patterns B and C demonstrated unique and overlapping categories that provide insight into the functional diversity that occurs during the process of differentiation. The genes up regulated as identified by patterns B1 and C1 relate to known functions of macrophages including metabolic pathways for lipid, fatty acid and steroid, carbohydrate, and amino acid metabolism, while those down regulated include those related to the mRNA transcription and apoptosis. Additionally, pattern C1 was also represented by genes identified in the molecular function ontogeny, including oxyreductase, reductase, hydrolase, dehydrogenase, transferase, glycosidase, lyase, protease, and acyltransferase. Many of the genes in the hydrolase molecular function, together with others identified by Medline search, were enriched in genes critical for osteoclast differentiation and function, including tartrate resistant acid phosphatase 5, cathepsins D, K, L and L2, and a large number of ATPase, H+ transporting lysosomal proteins. However, the cells were not osteoclasts, since they did not express calcitonin receptor.
Of the biologic processes identified by patterns B1 and C1, the lipid, steroid and fatty acid metabolism pathway was among the most prominent. Macrophage differentiation in the absence of an additional signal, such as the addition of oxidized LDL, resulted in an extensive modulation of genes that are important in the development of atherosclerosis. Proteins important in lipoprotein homeostasis such as APOE, APOC1 and 2 were highly induced (8 to 20 fold) in macrophages, consistent with another study that employed the serial analysis of gene expression method, which utilized short sequence tag oligonucleotides, and M-CSF [30]. Receptors important for the binding of modified lipoproteins such as MSR1, SCARB2, and CD68 were up regulated during macrophage differentiation, while others such as CD36 (SCARB3), LDLR and lectin-like oxidized receptor (LOX-1), LDL related protein 1(LRP1, APOER) were already highly expressed in monocytes and were not significantly increased in macrophages.
Additionally, genes important in the metabolism and transport of long chain fatty acids including FABP 3, 4, and 5, cholesterol 27-hydroxylase, which regulates the export of cholesterol [31], lipase A (LIPA) which promotes chylomicron formation and atherosclerosis [32], and HMGCR, the target of statins [33] were all up regulated in macrophages. LPL, which may contribute to foam cell formation and hydrolyzes triglycerides in chylomicrons, VLDL and HDL and lysosomal phospholipase A2, which is an independent risk factor of coronary heart disease [34] were also increased during macrophage differentiation. Further, both ACAT1 and ACAT2, which esterify cholesterol, promoting lipid droplet formation [35], were increased during macrophage differentiation. However, some genes important in the formation and function of foam cells such as the ABC transport proteins ABCA1 and ABCG1, important for the export of cholesterol and oxidized phospholipids [36], were highly expressed in monocytes and changed less than 2 fold in macrophages. However, ABCA1 was up regulated in classically, compared to alternatively activated, macrophages [28]. Both ABCA1 and ABCG1 may also be induced by oxysterols mediated through the LXRα [37]. Overall, these observations demonstrate that macrophage differentiation, even in the absence of exogenous M-CSF or oxidized LDLs, directs macrophages toward foam cell development.
Transcriptional repression in eukaryotes has been less well studied than transcriptional activation, partially due to the notion that it is more economical to turn on genes than to keep them off [19]. Our data demonstrate a marked reduction of the expression of transcription factors during macrophage differentiation. Of the classical transcription factors and the nuclear receptors that regulate transcription, almost 80% of the genes identified were down regulated in macrophages. Among the genes down regulated were three members of nuclear receptor subfamily 4 (NR4A1–3), AML1 and AML3 (RUNX1 and 3), C/EBPδ, ETS2, IRF1, FOSB, FOS, JUNB, JUND, MAFF, HIF1A, FOX03A and FOX01A. While many of these genes had been considered important for macrophage development, our observations suggest that these genes may contribute to the maintenance of monocytes, and that down-regulation allowed the monocytes to differentiate. Supporting this concept, both AML-1 and AML-3, which may function by recruiting co-repressors and histone deacetylases, are involved in the transcriptional repression and gene silencing, such as silencing CD4 during T cell development [38, 39]. Supporting a role of AML-1 in maintaining the monocyte phenotype, the forced reduction of AML-1 by antisense oligonucleotides, or over expression of dominant negative form of AML-1 induced the expression of non-specific esterase, a marker of differentiated macrophages [40].
It is of note that many nuclear receptors were down regulated during the differentiation, especially the NR4A1–3 (Nurr77, Nurr1, and NOR-1). They have been identified as immediate-early genes induced in vascular smooth muscle and endothelial cells by growth factors such as platelet derived growth factor and vascular endothelial cell growth factors, and they are found in atherosclerotic lesions [41]. Therefore, NR4A receptors may contribute to maintaining the monocyte phenotype or for differentiation in the context of a pathologic environment. Cooperation between p57kip2 and NR4A2 contributes to the differentiation of dopaminergic neural cells from stem cells [42]. LXRα (NR1H3) is the only nuclear factor, which was up regulated during monocyte to macrophage differentiation. This observation is consistent with previous finding that LXRα was expressed during GM-CSF-induced monocyte to macrophage differentiation [43]. LXRα is important for macrophage survival, suppression of macrophage activation and the export of cholesterol by regulating the expression of ABCA1 and ABCG1 [44, 45]. Given the extent of the repression of the transcription factors, it was surprising that Kruppel-like factors (KLF 2, 4, 5, 6, 10 and 11) were all reduced, since KLFs may serve as transcriptional repressors of certain genes [46–48]. It is possible that they serve as transcriptional repressors in monocytes. The observation that ERG1 and 3 are repressed during macrophage differentiation appears at odds with earlier studies that suggested ERG1 was essential for macrophage differentiation [49, 50]. However, a recent study demonstrated normal macrophage differentiation in Erg1, 2 or 3 deficient mice [51]. Therefore, the differential regulation of nuclear receptor genes and the repression of KLF and ERG family members may be important for macrophage differentiation and function.
While the vast majority of transcription factors that changed were down regulated, a number were up regulated including MAF, CYP5 and C/EBPα. Our data demonstrates the potential importance of C/EBPα, since the forced reduction of C/EBPα suppressed the induction of TREM2 and LPL, which were induced after 16 hours. Even though C/EBPα may be important in some aspects of macrophage differentiation, reduction of C/EBPα failed to reduce the expression of NR1H3 (LXRα) or to modify the morphology of the differentiating macrophages. Supporting the importance of C/EBPα, fetal livers deficient in C/EBPα demonstrated an impaired ability to generate macrophages [52]. Additionally, committed T cell progenitors were reprogrammed to macrophages by the ectopic expression of C/EBPα [53]. Together, these observations suggest that although repression of the transcriptional program may contribute to monocyte to macrophage differentiation, C/EBPα may be important for this process.
ACKNOWLEDGEMENTS
This work was supported awards from the National Institute of Health (AR049217 and AR048269). All authors read and approved the final manuscript. We thank Dr. Nadereh Jafari from Northwestern University Microarray Core Facility, Center of Genetic Medicine for conducting the Affymetrix microarray chips.
REFERENCES
- 1.Gangenahalli GU, Gupta P, Saluja D, Verma YK, Kishore V, Chandra R, Sharma RK, Ravindranath T. Stem cell fate specification: role of master regulatory switch transcription factor PU.1 in differential hematopoiesis. Stem Cells Dev. 2005;14:140–152. doi: 10.1089/scd.2005.14.140. [DOI] [PubMed] [Google Scholar]
- 2.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 3.Kaplan G, Gaudernack G. In vitro differentiation of human monocytes. Differences in monocyte phenotypes induced by cultivation on glass or on collagen. J Exp Med. 1982;156:1101–1114. doi: 10.1084/jem.156.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker S, Warren MK, Haskill S. Colony-stimulating factor-induced monocyte survival and differentiation into macrophages in serum-free cultures. J Immunol. 1987;139:3703–3709. [PubMed] [Google Scholar]
- 5.Brugger W, Kreutz M, Andreesen R. Macrophage colony-stimulating factor is required for human monocyte survival and acts as a cofactor for their terminal differentiation to macrophages in vitro. J Leukoc Biol. 1991;49:483–488. doi: 10.1002/jlb.49.5.483. [DOI] [PubMed] [Google Scholar]
- 6.Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 2002;99:111–120. doi: 10.1182/blood.v99.1.111. [DOI] [PubMed] [Google Scholar]
- 7.Wiktor-Jedrzejczak W, Bartocci A, Ferrante AW, Jr, Ahmed-Ansari A, Sell KW, Pollard JW, Stanley ER. Total absence of colony-stimulating factor 1 in the macrophage-deficient osteopetrotic (op/op) mouse. Proc Natl Acad Sci U S A. 1990;87:4828–4832. doi: 10.1073/pnas.87.12.4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang B, Berger J, Hu E, Szalkowski D, White-Carrington S, Spiegelman BM, Moller DE. Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha. Mol Endocrinol. 1996;10:1457–1466. doi: 10.1210/mend.10.11.8923470. [DOI] [PubMed] [Google Scholar]
- 9.Valledor AF, Borras FE, Cullell-Young M, Celada A. Transcription factors that regulate monocyte/macrophage differentiation. J Leukoc Biol. 1998;63:405–417. doi: 10.1002/jlb.63.4.405. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Ma Y, Cole SM, Zander C, Chen KH, Karras J, Pope RM. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- 11.Liu H, Perlman H, Pagliari LJ, Pope RM. Constitutively activated Akt-1 is vital for the survival of human monocyte-differentiated macrophages. Role of Mcl-1, independent of nuclear factor (NF)-kappaB, Bad, or caspase activation. J Exp Med. 2001;194:113–126. doi: 10.1084/jem.194.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pagliari LJ, Perlman H, Liu H, Pope RM. Macrophages require constitutive NF-kappaB activation to maintain A1 expression and mitochondrial homeostasis. Mol Cell Biol. 2000;20:8855–8865. doi: 10.1128/mcb.20.23.8855-8865.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perlman H, Pagliari LJ, Georganas C, Mano T, Walsh K, Pope RM. FLICE-inhibitory protein expression during macrophage differentiation confers resistance to fas-mediated apoptosis. J Exp Med. 1999;190:1679–1688. doi: 10.1084/jem.190.11.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Huang Q, Shi B, Eksarko P, Temkin V, Pope R. Regulation of Mcl-1 expression in rheumatoid synovial macrophages. Arthritis Rheum. 2006;54:3174–3181. doi: 10.1002/art.22132. [DOI] [PubMed] [Google Scholar]
- 15.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, Guo N, Muruganujan A, Doremieux O, Campbell MJ, Kitano H, Thomas PD. The PANTHER database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33:D284–D288. doi: 10.1093/nar/gki078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, Ladunga I, Ulitsky-Lazareva B, Muruganujan A, Rabkin S, Vandergriff JA, Doremieux O. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Pope RM. Phagocytes: mechanisms of inflammation and tissue destruction. Rheum Dis Clin North Am. 2004;30:19–39. doi: 10.1016/S0889-857X(03)00107-8. v. [DOI] [PubMed] [Google Scholar]
- 18.Chomarat P, Banchereau J, Davoust J, Palucka AK. IL-6 switches the differentiation of monocytes from dendritic cells to macrophages. Nat Immunol. 2000;1:510–514. doi: 10.1038/82763. [DOI] [PubMed] [Google Scholar]
- 19.Graeber TG, Shuai K. Rapid gene repression triggered by interleukin-6 at the onset of monocyte differentiation. Biochem Biophys Res Commun. 2000;267:863–869. doi: 10.1006/bbrc.1999.2041. [DOI] [PubMed] [Google Scholar]
- 20.Pixley FJ, Stanley ER. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 2004;14:628–638. doi: 10.1016/j.tcb.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 21.Jessup W, Kritharides L. Metabolism of oxidized LDL by macrophages. Curr Opin Lipidol. 2000;11:473–481. doi: 10.1097/00041433-200010000-00005. [DOI] [PubMed] [Google Scholar]
- 22.Jessup W, Wilson P, Gaus K, Kritharides L. Oxidized lipoproteins and macrophages. Vascul Pharmacol. 2002;38:239–248. doi: 10.1016/s1537-1891(02)00174-x. [DOI] [PubMed] [Google Scholar]
- 23.Gordon S. Macrophage heterogeneity and tissue lipids. J Clin Invest. 2007;117:89–93. doi: 10.1172/JCI30992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreutz M, Krause SW, Hennemann B, Rehm A, Andreesen R. Macrophage heterogeneity and differentiation: defined serum-free culture conditions induce different types of macrophages in vitro. Res Immunol. 1992;143:107–115. doi: 10.1016/0923-2494(92)80087-2. [DOI] [PubMed] [Google Scholar]
- 25.Scheibenbogen C, Andreesen R. Developmental regulation of the cytokine repertoire in human macrophages: IL-1, IL-6, TNF-alpha, and M-CSF. J Leukoc Biol. 1991;50:35–42. doi: 10.1002/jlb.50.1.35. [DOI] [PubMed] [Google Scholar]
- 26.Cecchini MG, Dominguez MG, Mocci S, Wetterwald A, Felix R, Fleisch H, Chisholm O, Hofstetter W, Pollard JW, Stanley ER. Role of colony stimulating factor-1 in the establishment and regulation of tissue macrophages during postnatal development of the mouse. Development. 1994;120:1357–1372. doi: 10.1242/dev.120.6.1357. [DOI] [PubMed] [Google Scholar]
- 27.Laouar A, Collart FR, Chubb CB, Xie B, Huberman E. Interaction between alpha 5 beta 1 integrin and secreted fibronectin is involved in macrophage differentiation of human HL-60 myeloid leukemia cells. J Immunol. 1999;162:407–414. [PubMed] [Google Scholar]
- 28.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 29.Mantovani A, Bonecchi R, Locati M. Tuning inflammation and immunity by chemokine sequestration: decoys and more. Nat Rev Immunol. 2006;6:907–918. doi: 10.1038/nri1964. [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto S, Suzuki T, Dong HY, Nagai S, Yamazaki N, Matsushima K. Serial analysis of gene expression in human monocyte-derived dendritic cells. Blood. 1999;94:845–852. [PubMed] [Google Scholar]
- 31.Hansson M, Ellis E, Hunt MC, Schmitz G, Babiker A. Marked induction of sterol 27-hydroxylase activity and mRNA levels during differentiation of human cultured monocytes into macrophages. Biochim Biophys Acta. 2003;1593:283–289. doi: 10.1016/s0167-4889(02)00398-1. [DOI] [PubMed] [Google Scholar]
- 32.Kodvawala A, Ghering AB, Davidson WS, Hui DY. Carboxyl ester lipase expression in macrophages increases cholesteryl ester accumulation and promotes atherosclerosis. J Biol Chem. 2005;280:38592–38598. doi: 10.1074/jbc.M502266200. [DOI] [PubMed] [Google Scholar]
- 33.Moghadasian MH. Clinical pharmacology of 3-hydroxy-3-methylglutaryl coenzyme A reductase inhibitors. Life Sci. 1999;65:1329–1337. doi: 10.1016/s0024-3205(99)00199-x. [DOI] [PubMed] [Google Scholar]
- 34.Packard CJ, O'Reilly DS, Caslake MJ, McMahon AD, Ford I, Cooney J, Macphee CH, Suckling KE, Krishna M, Wilkinson FE, Rumley A, Lowe GD West of Scotland Coronary Prevention Study Group. Lipoprotein-associated phospholipase A2 as an independent predictor of coronary heart disease. N Engl J Med. 2000;343:1148–1155. doi: 10.1056/NEJM200010193431603. [DOI] [PubMed] [Google Scholar]
- 35.Rudel LL, Shelness GS. Cholesterol esters and atherosclerosis-a game of ACAT and mouse. Nat Med. 2000;6:1313–1314. doi: 10.1038/82110. [DOI] [PubMed] [Google Scholar]
- 36.Beyea MM, Heslop CL, Sawyez CG, Edwards JY, Markle JG, Hegele RA, Huff MW. Selective up-regulation of LXR-regulated genes ABCA1, ABCG1, and APOE in macrophages through increased endogenous synthesis of 24(S),25-epoxycholesterol. J Biol Chem. 2007;282:5207–5216. doi: 10.1074/jbc.M611063200. [DOI] [PubMed] [Google Scholar]
- 37.Pennings M, Meurs I, Ye D, Out R, Hoekstra M, Van Berkel TJ, Van Eck M. Regulation of cholesterol homeostasis in macrophages and consequences for atherosclerotic lesion development. FEBS Lett. 2006;580:5588–5596. doi: 10.1016/j.febslet.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 38.Durst KL, Hiebert SW. Role of RUNX family members in transcriptional repression and gene silencing. Oncogene. 2004;23:4220–4224. doi: 10.1038/sj.onc.1207122. [DOI] [PubMed] [Google Scholar]
- 39.Lutterbach B, Hiebert SW. Role of the transcription factor AML-1 in acute leukemia and hematopoietic differentiation. Gene. 2000;245:223–235. doi: 10.1016/s0378-1119(00)00014-7. [DOI] [PubMed] [Google Scholar]
- 40.Sakakura C, Yamaguchi-Iwai Y, Satake M, Bae SC, Takahashi A, Ogawa E, Hagiwara A, Takahashi T, Murakami A, Makino K, et al. Growth inhibition and induction of differentiation of t(8;21) acute myeloid leukemia cells by the DNA-binding domain of PEBP2 and the AML1/MTG8(ETO)-specific antisense oligonucleotide. Proc Natl Acad Sci U S A. 1994;91:11723–11727. doi: 10.1073/pnas.91.24.11723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martinez-Gonzalez J, Badimon L. The NR4A subfamily of nuclear receptors: new early genes regulated by growth factors in vascular cells. Cardiovasc Res. 2005;65:609–618. doi: 10.1016/j.cardiores.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Joseph B, Wallen-Mackenzie A, Benoit G, Murata T, Joodmardi E, Okret S, Perlmann T. p57(Kip2) cooperates with Nurr1 in developing dopamine cells. Proc Natl Acad Sci U S A. 2003;100:15619–15624. doi: 10.1073/pnas.2635658100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kohro T, Nakajima T, Wada Y, Sugiyama A, Ishii M, Tsutsumi S, Aburatani H, Imoto I, Inazawa J, Hamakubo T, Kodama T, Emi M. Genomic structure and mapping of human orphan receptor LXR alpha: upregulation of LXRa mRNA during monocyte to macrophage differentiation. J Atheroscler Thromb. 2000;7:145–151. doi: 10.5551/jat1994.7.145. [DOI] [PubMed] [Google Scholar]
- 44.Joseph SB, Bradley MN, Castrillo A, Bruhn KW, Mak PA, Pei L, Hogenesch J, O'Connell RM, Cheng G, Saez E, Miller JF, Tontonoz P. LXR-dependent gene expression is important for macrophage survival and the innate immune response. Cell. 2004;119:299–309. doi: 10.1016/j.cell.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 45.Zelcer N, Tontonoz P. Liver X receptors as integrators of metabolic and inflammatory signaling. J Clin Invest. 2006;116:607–614. doi: 10.1172/JCI27883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen ZY, Shie JL, Tseng CC. Gut-enriched Kruppel-like factor represses ornithine decarboxylase gene expression and functions as checkpoint regulator in colonic cancer cells. J Biol Chem. 2002;277:46831–46839. doi: 10.1074/jbc.M204816200. [DOI] [PubMed] [Google Scholar]
- 47.Shie JL, Chen ZY, Fu M, Pestell RG, Tseng CC. Gut-enriched Kruppel-like factor represses cyclin D1 promoter activity through Sp1 motif. Nucleic Acids Res. 2000;28:2969–2976. doi: 10.1093/nar/28.15.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shie JL, Chen ZY, O'Brien MJ, Pestell RG, Lee ME, Tseng CC. Role of gut-enriched Kruppel-like factor in colonic cell growth and differentiation. Am J Physiol Gastrointest Liver Physiol. 2000;279:G806–G814. doi: 10.1152/ajpgi.2000.279.4.G806. [DOI] [PubMed] [Google Scholar]
- 49.Krishnaraju K, Hoffman B, Liebermann DA. The zinc finger transcription factor Egr-1 activates macrophage differentiation in M1 myeloblastic leukemia cells. Blood. 1998;92:1957–1966. [PubMed] [Google Scholar]
- 50.Nguyen HQ, Hoffman-Liebermann B, Liebermann DA. The zinc finger transcription factor Egr-1 is essential for and restricts differentiation along the macrophage lineage. Cell. 1993;72:197–209. doi: 10.1016/0092-8674(93)90660-i. [DOI] [PubMed] [Google Scholar]
- 51.Carter JH, Tourtellotte WG. Early growth response transcriptional regulators are dispensable for macrophage differentiation. J Immunol. 2007;178:3038–3047. doi: 10.4049/jimmunol.178.5.3038. [DOI] [PubMed] [Google Scholar]
- 52.Heath V, Suh HC, Holman M, Renn K, Gooya JM, Parkin S, Klarmann KD, Ortiz M, Johnson P, Keller J. C/EBPalpha deficiency results in hyperproliferation of hematopoietic progenitor cells and disrupts macrophage development in vitro and in vivo. Blood. 2004;104:1639–1647. doi: 10.1182/blood-2003-11-3963. [DOI] [PubMed] [Google Scholar]
- 53.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]