Abstract
Purpose
To identify human transient receptor potential cation channel, subfamily M, member 1 (TRPM1) gene mutations in patients with congenital stationary night blindness (CSNB).
Methods
We analyzed four different Japanese patients with complete CSNB in whom previous molecular examination revealed no mutation in either nyctalopin (NYX) or glutamate receptor, metabotropic 6 (GRM6). The ophthalmologic examination included best-corrected visual acuity, refraction, biomicroscopy, ophthalmoscopy, fundus photography, Goldmann kinetic perimetry, color vision tests, and electroretinography (ERG). Exons 2 through 27 and the exon-intron junction regions of human TRPM1 were sequenced.
Results
Five different mutations in human TRPM1 were identified. Mutations were present in three unrelated patients with complete CSNB. All three patients were compound heterozygotes. Fundus examination revealed no abnormalities other than myopic changes, and the single bright-flash, mixed rod-cone ERG showed a “negative-type” configuration with a reduced normal a-wave and a significantly reduced b-wave amplitude. Our biochemical and cell biologic analyses suggest that the two identified IVS mutations lead to abnormal TRPM1 protein production, and imply that the two identified missense mutations lead to the mislocalization of the TRPM1 protein in bipolar cells (BCs).
Conclusions
Human TRPM1 mutations are associated with the complete form of CSNB in Japanese patients, suggesting that TRPM1 plays an essential role in mediating the photoresponse in ON BCs in humans as well as in mice.
Introduction
The complete form of congenital stationary night blindness (CSNB) is a subtype of Schubert-Bornschein CSNB in which the fundus is essentially normal except for myopic changes [1-5]. From early childhood, patients with complete CSNB lack rod function and experience night blindness. Nystagmus and amblyopia sometimes accompany the other symptoms, and the clinical course is stationary. Best-corrected visual acuity is mildly reduced, and high to moderate myopia is usually found. Electroretinogram (ERG) examinations reveal absent depolarization of neuron by light (ON)-responses (b-wave), and previous extensive physiologic studies indicated that the pathology in complete CSNB lies in the dysfunction of the depolarizing ON bipolar cell (BC). There are two hereditary patterns for complete CSNB: X-linked recessive and autosomal recessive [4].
To date, two genes, the leucine-rich proteoglycan nyctalopin gene (NYX)—encoding the glycosylphosphatidylinositol (GPI)-anchored extracellular protein nyctalopin [6,7]—and the GRM6 gene—encoding the metabotropic glutamate receptor mGluR6—have been identified as the mutated gene in X-linked recessive and autosomal recessive complete CSNB [8-10], respectively. Both nyctalopin and mGluR6 proteins are distributed on the postsynaptic ON BCs and are required for the depolarization of the cell. The NYX gene appears to be the major, and possibly only, causative gene for X-linked recessive complete CSNB since NYX gene mutations were identified in the majority of X-linked recessive families with complete CSNB. In contrast, GRM6 gene mutations have been found in only some of the autosomal recessive families with complete CSNB, indicating the existence of other unknown genes for autosomal recessive complete CSNB.
We previously identified a mouse transient receptor potential cation channel, subfamily M, member 1 (Trpm1) homolog of human TRPM1 [11]. Trpm1 is alternatively spliced, resulting in the production of a long form protein (trpm1-L) and a short N-terminal form devoid of transmembrane segments (trpm1-S). Although mouse trpm1-S was previously identified as melastatin [12], mouse trpm1-L has not been identified. Without distinction between trpm1-L or trpm1-S, trpm1 has been reported to be detected in human primary melanocytes [13], poorly metastatic melanoma cell lines [14,15], mouse retinal pigment epithelium (RPE) [16], and subsets of ON and hyperpolarization of neuron by light (OFF) BCs [17,18]. We found that trpm1-L localization is developmentally restricted to the dendritic tips of ON BCs in co-localization with mGluR6 [11,19]. Trpm1 null mutant mice completely lose the photoresponse of ON BCs. Trpm1-L channel activity is negatively regulated by activated Go in the mGluR6 cascade, and we showed that Trpm1-L is a component of the ON BC transduction channel. Interestingly, the trpm1-deficient mice showed similar electrophysiological responses to the responses of those with complete CSNB—significantly reduced b-waves—which led us to examine whether the gene is mutated in human patients with complete CSNB.
Very recently three groups reported the association of TRPM1 mutations with CSNB [20-22]. Independently from these studies, in our present study, we identified five different novel mutations in the human TRPM1 gene: IVS2–3C>G, IVS8+3_6delAAGT, R624C (c.1870C>T), S882X (c.2645C>A), and F1075S (c.3224T>C). In addition, our biochemical and cell biologic analyses suggested that the two intron mutations were likely to result in abnormal protein production by abnormal splicing, and the two missense mutations lead to the mislocalization of the TRPM1 protein in BCs. Fundus examination revealed no abnormalities other than myopic changes, and the single bright-flash, mixed rod-cone ERG showed a “negative-type” configuration with a reduced normal a-wave and a significantly reduced b-wave amplitude.
Methods
Subjects
Four separate patients with complete CSNB (#204, #373, #437, and #484) in whom previous molecular examination revealed no mutation in either NYX or GRM6 among 11 separate patients were analyzed. They were all male, and their ages were 26, 9,19, and 27 years, respectively. They complained of night blindness from early childhood, and otherwise had no health problems. All individuals examined had been under observation at the Department of Ophthalmology of Nagoya University, Nagoya, Japan. The research protocol was designed in compliance with the Declaration of Helsinki and approved by the institutional review board. Written informed consent was obtained from the subjects after an explanation of the purpose of this study was provided. Briefly, the protocol entails molecular analysis of causative genes from blood samples of patients with complete CSNB and in vitro analysis including expression assay and cytological assay using the samples without any disadvantages for the patients.
DNA analysis of patients with complete CSNB
Genomic DNA was extracted from the peripheral blood leukocytes. Exons 2 through 27 of TRPM1 were amplified by polymerase chain reaction (PCR) using the DNA Thermal Cycler 9700 (Applied Biosystems, Foster City, CA). Primers were designed by and purchased from Invitrogen (Carlsbad, CA). The primer sets used in this study are shown in Table 1. For all exons, 100 ng of genomic DNA was amplified in a 50 µl reaction with 0.5 µM of each primer, 0.2 mM of each dNTP, and AmpliTaq Gold polymerase (Applied Biosystems). The PCR conditions were as follows: 5 min at 94 °C; 35 cycles at 94 °C for 30 s, followed by 30 s at 54 °C, and 45 s at 72 °C; and a final extension step at 72 °C for 7 min. The PCR products were directly sequenced using an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems) and BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). To search for polymorphisms, exons 3, 8, 16, 21, and 24 of TRPM1 from 200 alleles (50 men and 50 women) from unrelated, normal Japanese individuals were directly sequenced.
Table 1. The primer sets used in this study.
| Exon | Direction | Primer sequence (5′-3′) |
|---|---|---|
| 2 |
Forward |
GCTTGGGAATGTATTGGTCC |
| 2 |
Reverse |
TTCTGGACTCCTCTTTCTGC |
| 3–1 |
Forward |
TTGCTAATGAAGGGGAAAGG |
| 3–1 |
Reverse |
TGAAAGCTGTGATCCTAGTG |
| 4–2 |
Forward |
TACCCAACAGATTCCTATGG |
| 4–2 |
Reverse |
TTGTGGATTCAGAGCTTTGG |
| 4 |
Forward |
TGCTTGGACATGAGCAATAG |
| 4 |
Reverse |
TATTAGGAAGCTCTTTGGGG |
| 5 |
Forward |
GATCTGTAGGGAATGTAGGG |
| 5 |
Reverse |
CAACTGCATCTGAATCAACG |
| 6 |
Forward |
TATTGTGCTTGCTTCCCTTG |
| 6 |
Reverse |
GCAATCAATCTCCTTCTCAG |
| 7 |
Forward |
TACTCTTCCCCATACTTCAG |
| 7 |
Reverse |
CGTTTGGAATGTCTAAGCAG |
| 8 |
Forward |
CCACAGCAAAGTCTCAAATC |
| 8 |
Reverse |
AATAACCCTGTCATGCACAC |
| 9 |
Forward |
CAGTCTCTGTGGTCATTTTG |
| 9 |
Reverse |
TTCATGCTGAGACTGCTCAAG |
| 10 |
Forward |
AAGCCAGCAATGCTTAGTTC |
| 10 |
Reverse |
TGTCACAAGGGAGAACATAC |
| 11 |
Forward |
ATGTGTGCACATACAGAGGT |
| 11 |
Reverse |
GAAGCAAGGACAAGAACCAT |
| 12 |
Forward |
ACCCTTTCCTCTGTGTTTCC |
| 12 |
Reverse |
GCACAATATGCACCAGTGAC |
| 13 |
Forward |
TACTGACCTCAAGTGATCTG |
| 13 |
Reverse |
TTCATTATCTCCTGTGCCTG |
| 14 |
Forward |
CTGTTACATGAACTCCCAAG |
| 14 |
Reverse |
TTGTTGAAAAGCAAGCGAGG |
| 15 |
Forward |
CCATTCGTATGTCCACTATC |
| 15 |
Reverse |
GAGTAGTATCGTATATTCGC |
| 16 |
Forward |
AGTTCTGGAACTCTTTGTTG |
| 16 |
Reverse |
AGAGACAAGTACTCAGGATG |
| 17 |
Forward |
GGTGGATATGCCTGTCTAAG |
| 17 |
Reverse |
AAAGTGCTCAGTTCAGCCTG |
| 18 |
Forward |
GGCTCCTAGGTTATTGAATA |
| 18 |
Reverse |
GTGGTACAAGAAATCTCAAC |
| 19 |
Forward |
TTCTAATGGTGCCTGTGTGC |
| 19 |
Reverse |
CAATGTTAGCCAGAGATCTC |
| 20 |
Forward |
GTGTCAACCTGAAAGAAAAC |
| 20 |
Reverse |
TTTCAGTAAGGAGCCATATC |
| 21 |
Forward |
ATGACCTGGAATGTTCCATC |
| 21 |
Reverse |
GCCTGTTCTTATGTCCTAAC |
| 22 |
Forward |
TATAATGAGCCAGGGTGAAC |
| 22 |
Reverse |
TTTGGAACTGATCATCTGCC |
| 23 |
Forward |
CTTTTCTCTCAACCATCATC |
| 23 |
Reverse |
ACAGTGAATTTGTGGTTCTC |
| 24 |
Forward |
AGCTGGGGCTACAGAGTTTA |
| 24 |
Reverse |
TATCTTGGGAGCGTTCTGAG |
| 25 |
Forward |
GTAACTTTGACTGCTCTGGG |
| 25 |
Reverse |
CGAGCAAGTAGTTGAGTGAG |
| 26 |
Forward |
GGGCTGTGAAATTCTCATTG |
| 26 |
Reverse |
AACTCTGGCATCCCCCATAG |
| 27–1 |
Forward |
CATGTTAGGGGATTGTGAAG |
| 27–1 |
Reverse |
ATACGTTGCCTCACATTCAG |
| 27–2 |
Forward |
CAGAATGGTGAATGCTCTTG |
| 27–2 |
Reverse |
GAAAGGTGAAGACTGTTCTG |
| 27–3 |
Forward |
AAAAAAACCTGTTCCTTCCG |
| 27–3 |
Reverse |
TTTGTCGTTTCCACTGTTAG |
| 27–4 |
Forward |
GAAGAAACTATTTCCCCAAG |
| 27–4 |
Reverse |
GAATATCTGTGCTATGAGAG |
| 27–5 |
Forward |
TAGAGTACAGTTCAATCACG |
| 27–5 |
Reverse |
ATGGATTTCACATTCCTAGG |
| 27–6 |
Forward |
TCTGTGAAGCCAGATCAAAC |
| 27–6 | Reverse | GTTTAGATGGCCAAGATGAC |
TRPM1 splicing minigene constructs
DNA fragments of TRPM1 exon 2 with its downstream splice donor region (nucleotide position in GenBank # NC_000015: 24738–25055; exon 2+SD) and exon 8 with its downstream splice donor region (39020–39392; exon 8+SD) were PCR-amplified using human genomic DNA as a PCR template; SacI and KpnI sites were appended to the 5′ and 3′ ends, respectively. A partial fragment of β-globin (HBB) intron 2 (614–1166 in GenBank # NC_000011) was PCR-amplified; KpnI and SalI sites were appended to the 5′ and 3′ ends, respectively. DNA fragments of TRPM1 exon 3 with its upstream splice acceptor region (31325–31691; SA+exon 3) and exon 9 with its splice acceptor region (39976–40280; SA+exon 9) were PCR-amplified using human genomic DNA as a PCR template; SalI and BamHI sites were appended to the 5′ and 3′ ends, respectively. The exon 2+SD, HBB intron, and SA+exon 3 fragments (exon 2+3 construct), or exon 8+SD, HBB intron, and SA+exon 9 (exon 8+9 construct) were ligated and subcloned into the SacI and BamHI sites of pBluescript KS+ (Stratagene), containing three repeats of the Flag-tag between the BamHI and NotI sites. The generated exon 2+3 construct and exon 8+9 construct were digested with SacI and NotI and subcloned into SacI and BamHI digested pEGFP-C3 and pEGFP-C2 (Clontech, Palo Alto, CA), respectively, each with a synthesized BglII-NotI linker.
Transfection and western blots analysis
HEK293T cells were cultured in D-MEM containing 10% fetal bovine serum (FBS; Nissui, Tokyo, Japan). These cells were grown under 5% carbon dioxide at 37 °C. Transfections were performed using the calcium phosphate method. Transfected cells were incubated at 37 °C for 48 h, and harvested for further analysis. Cell extract proteins were separated by SDS–PAGE, and transferred to a polyvinylidene difluoride membrane (ATTO, Tokyo Japan). The membrane was incubated with a mouse anti-Flag antibody (1:1,000; Sigma, St Louis, MO), a rabbit anti-β−gal antibody (1:10,000; Chemicon, Temecula, CA), or a mouse anti-β−actin antibody (1:2,000; Sigma), and incubated with a horseradish peroxidase-conjugated goat anti-mouse or -rabbit IgG (1:10,000; Zymed Laboratories, San Francisco, CA). The bands were visually developed using Chemi-Lumi One L (Nacalai Tesque, Kyoto, Japan).
In vivo electroporation and section immunohistochemistry
In vivo electroporation was performed at postnatal day 0 (P0) as described previously [23]. Approximately 0.3 μl of DNA solution (5 μg/μl) was injected into the subretinal space of P0 mouse pups, and square electric pulses (80 V; five 50 ms pulses with 950 ms intervals) were applied with electrodes. Wild-type (WT) and mutant forms of TRPM1 (R624C and F1075S) containing 3× Flag at the C-terminus were expressed from the mouse mGluR6 promoter (mGluR6-TRPM1–3×Flag). To generate this construct, the chicken β-actin gene (CAG) promoter region in CAG-TRPM1 was replaced with the 9.5 kb fragment of mGluR6 5′ upstream genomic sequence [24]. mGluR6-TRPM1–3×Flag (4 μg/μl) and CAG-GFP (1 μg/μl) were co-electroporated and harvested at P14. Immunohistochemistry to observe the subcellular localization of TRPM1–3×Flag expressed in the electroporated ON BCs was performed on sections as previously described [23]. Mouse anti-Flag antibody (1:500), rabbit anti-PKCα antibody (1:20,000; Sigma), and rat anti-GFP antibody (1:1,000; Nacalai Tesque) were used. The signal intensity of Flag immunostaining at the dendritic tips and somas of ON BCs was measured using AxioVision software (Carl Zeiss, Oberkochen, Germany). The ON BCs that showed more intensity in the dendritic tips than in the somas were counted. One hundred electroporated ON BCs were counted from three retinas independently prepared for each construct.
Clinical evaluations in patients associated with TRPM1 mutations
The ophthalmologic examination included best-corrected visual acuity, refraction, biomicroscopy, ophthalmoscopy, fundus photography, Goldmann kinetic perimetry, color vision testing, and electroretinography [25]. Standardized full-field ERGs were elicited by Ganzfeld stimuli after pupillary dilation with 0.5% tropicamide and 0.5% phenylephrine hydrochloride and 20 min of dark-adaptation. The rod (scotopic) ERGs were elicited by a blue stimulus with a luminance of 5.2×10−3 cd/sec•m2. The mixed rod-cone, single flash (bright white) ERGs were elicited by a white stimulus of 44.2 cd/sec•m2. The photopic (cones) single-flash ERGs and 30 Hz flicker ERGs were elicited by white stimuli at a luminance of 4 cd/sec•m2 and 0.9 cd/sec•m2, respectively, on a white background of 68 cd/m2. The single flash bright white ERGs were recorded again after 17 h of dark adaptation.
ERG recordings in the trpm1 null mutant (trpm1–/–) mice
We generated trpm1–/– mice as described previously [11]. Our methods for recording the ERGs from the mice have been described in detail [26]. In brief, the mice were dark-adapted overnight, and then anesthetized with an intramuscular injection of 70 mg/kg ketamine and 14 mg/kg xylazine. ERGs were recorded with a gold-wire loop electrode placed on the anesthetized cornea. The mice were placed in a Ganzfeld bowl and stimulated with stroboscopic stimuli of 1.0 log cd-s/m2 (photopic units) maximum intensity. Four levels of stimulus intensity ranging from −5.0 to 1.0 log cd-s/m2 were used for the dark-adapted ERG recordings, and four levels of stimulus intensity ranging from −0.5 to 1.0 log cd-s/m2 were used for the light-adapted ERGs. The light-adapted ERGs were recorded on a rod-suppressing white background of 1.3 log cd-s/m2 (photopic units). The amplitude of the a-wave was measured from the baseline to a set time of 8 msec after the initial negative wave for the dark-adapted ERG, and 11 msec for the light-adapted ERG. The b-wave amplitude was measured from the negative trough of the a-wave to the maximal positive peak.
Animal care
All procedures conformed to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research, and these procedures were approved by the Institutional Safety Committee on Recombinant DNA Experiments and the Animal Research Committee of Osaka Bioscience Institute. The mice were housed in a temperature-controlled room with a 12h:12h light-dark cycle. Fresh water and rodent diet were available at all times.
Results
DNA analysis of the complete CSNB patients
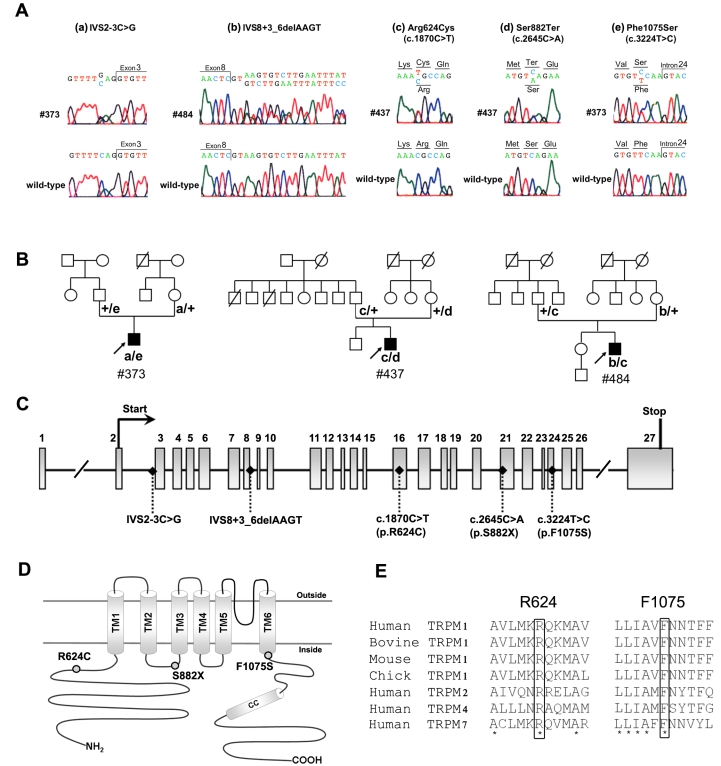
Five different mutations in TRPM1 were identified: IVS2–3C>G, IVS8+3_6delAAGT, R624C (c.1870C>T), S882X (c.2645C>A), and F1075S (c.3224T>C). Mutations were present in three unrelated patients with complete CSNB (#373, #437, and #484). All three patients were compound heterozygotes. Their pedigrees and the DNA sequence chromatogram for the patients harboring these mutations are presented in Figure 1A,B.
Figure 1.
Compound heterozygous TRPM1 mutations identified in patients #373, #437, and #484. A: Sequence chromatograms showing the mutations: IVS2–3C>G in patient #373 (a), IVS8+3_6delAAGT in patient #484 (b), Arg624Cys (c. 1870C>T) in patients #437 and #484 (c), Ser882Ter (c. 2645C>A) in patient #437 (d), and Phe1075Ser (c. 3224T>C) in patient #373(e). B: Complete CSNB pedigrees of Japanese patients #373, #437, and #484. These three patients are compound heterozygotes of TRPM1 mutations. C: Exon structures of human TRPM1. The first methionine (Start) and a stop codon (Stop) of the TRPM1 open reading frame are indicated. All mutations found in this study are shown. D: Putative topology of the human TRPM1. All mutations found in this study are illustrated. The six transmembrane domains are indicated as TM1-TM6. E: Alignment of R624 and F1075 in TRPM proteins. Sequence alignment of TRPM1 from human, bovine, mouse, chick, and TRPM2, TRPM4, and TRPM7 from human. Amino acid residues R624 and F1075 are boxed. The asterisks indicate completely conserved residues.
The two intron changes, IVS2–3C>G and IVS8+3_6delAAGT (Figure 1C), were interpreted to be pathogenic because no RNA splicing activities were detected when each of the two changes was present (see splicing assay section). They were also predicted to affect RNA splicing, as assessed by an online splice-site prediction tool. The donor site prediction score from the software for the exon 8 donor site in TRPM1 was 0.99 for the WT sequence, whereas it was under 0.01 for the corresponding mutated sequence with the IVS8+3_6delAAGT sequence change. The acceptor site prediction score for the exon 3 acceptor site in TRPM1 was 0.98 for the WT sequence, whereas it was 0.44 for the mutated sequence with the IVS2–3C>G sequence change. The two mutations were not found in an analysis of 100 Japanese normal controls.
The S882X mutation was considered to be a pathogenic null mutation since mRNA containing the predicted early stop codon would most likely be subject to nonsense mediated decay without being translated into a protein [27]. Even if the translated protein was produced, it would lack the C-terminus region of TRPM1, including transmembrane domains 4–6, which is very likely to lead to a loss of function as a cation channel (Figure 1D). This change was not found among the 100 healthy control individuals we examined. The two missense changes in the TRPM1 gene, R624C and F1075S, were located in the N-terminal region and in the C-terminal end of the transmembrane domain, respectively (Figure 1D). They were interpreted as likely pathogenic because the arginine at position 624 and the phenylalanine at 1075 are well conserved among the TRPM subfamily, suggesting the importance of these amino acid residues for TRPM function (Figure 1E). The two mutations were not found in an analysis of 100 Japanese normal controls. None of the five mutations mentioned above have been reported in a SNP database.
Analysis of protein expression from TRPM1 mutant minigenes
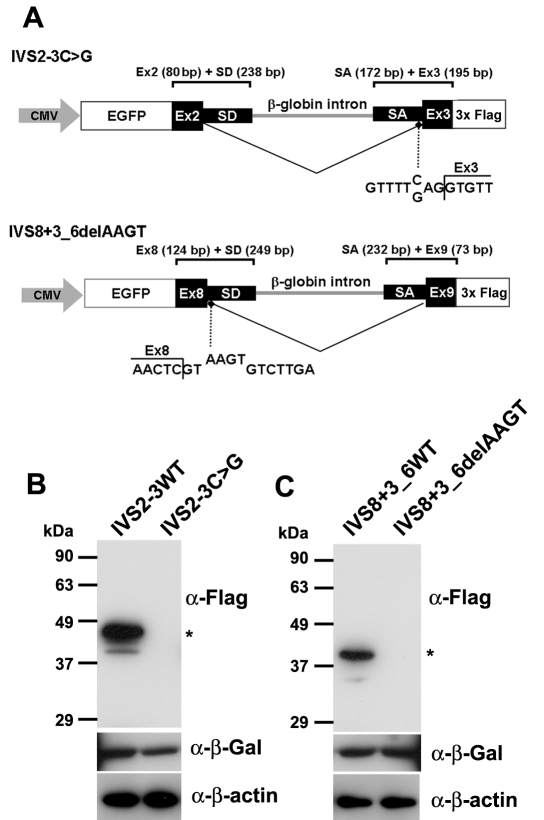
Our previous expression analysis of mouse TRPM1 showed that the long form (TRPM1-L), which functions as a cation channel, is predominantly expressed in the retina [11], suggesting that human patient samples for directly analyzing splicing are not readily available. To overcome this limitation, minigenes for expression in tissue culture cells were designed; these minigenes included the upstream exons (exon 2 or 8) fused with EGFP, downstream exons (exon 3 or 9) fused with 3×Flag-tags, and a HBB intron, which is often used in splicing assays (an exon 2+3 minigene and an exon 8+9 minigene; Figure 2A). Since the signals required for constitutive splicing are usually within ~100 nucleotides of splice junctions [11], we took ~200 nucleotide regions for both splicing donors and acceptors to ensure the splicing assay. We also generated mutated minigenes containing the IVS2–3C>G mutation in the exon 2+3 minigene or the IVS8+3_6delAAGT mutation in the exon 8+9 minigene. Minigene splicing was analyzed in HEK293T cells since they have been used extensively to study constitutive RNA splicing. HEK293T cells were transfected with minigene DNA and a lacZ expression internal control vector, and western blotting was performed on whole cell extracts isolated 48 h later. We then analyzed protein products of the EGFP-exon 2+3 fusion and the EGFP-exon 8+9 fusion proteins by western blot analysis of whole cell extracts transfected with the WT and mutated minigenes. We detected an expected 45 kDa or 39 kDa band with the anti-Flag antibody in HEK293T cell extracts transfected with the WT exon 2+3 minigene or the exon 8+9 minigene (Figure 2B,C). In contrast, we did not detect any bands in either of the cell extracts transfected with the mutated minigenes. We detected the β-gal bands of the transfection internal control, and the β-actin bands of the cell extract control in both the WT and mutant minigene transfected cells. These results showed that both the IVS2–3C>G and IVS8+3_6delAAGT mutations abrogate normal splicing and lead to abnormal protein production, suggesting that these trpm1 alleles are loss-of-function alleles.
Figure 2.
Analysis of protein expression from TRPM1 mutant minigenes. A: Schematic of TRPM1 minigenes. The minigene IVS2–3C>G contains an EGFP gene, exon 2, the exon 2 splice donor region, a portion of the β-globin (HBB) intron 2, the exon 3 splice acceptor region, and exon 3, and uses the CMV promoter and SV40 polyA site from the pEGFP-C3 vector. The minigene IVS8+3_6delAAGT contains an EGFP gene, exon 8, the exon 8 splice donor region, a portion of the HBB intron 2, the exon 9 splice acceptor region, and exon 9, and uses the CMV promoter and SV40 polyA site from the pEGFP-C2 vector. Mutations of IVS2–3C>G and IVS8+3_6delAAGT are shown. B-C: western blots of HEK293 cell extracts transfected with minigenes IVS2–3C>G (B) or IVS8+3_6delAAGT (C). WT represents protein from a wild-type minigene transfection, and asterisks represent spliced EGFP detected by an anti-Flag antibody. β-Gal and β-actin were used for transfection control and loading control, respectively.
In vivo analysis of intracellular localization of TRPM1 protein carrying missense mutations
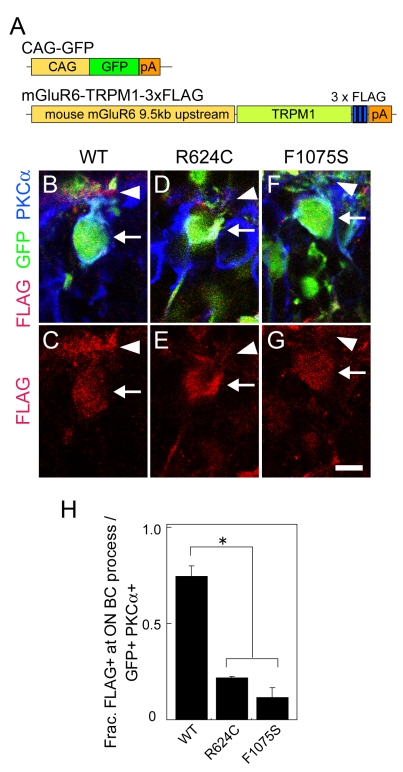
The TRPM1 missense mutations R624C and F1075S are located in the N-terminal and the C-terminal end of the six-transmembrane domains (6TM), respectively (Figure 1D). Mutations in these regions often cause misfolding/mislocalization of the protein, and/or a defect in channel activity [28-32]. To determine the functional relevance of R624C and F1075S TRPM1 residues, we mutagenized the TRPM1 cDNA to introduce R624C and F1075S mutations. We first prepared expression vectors encoding R624C or F1075S mutant proteins attached with a Flag-tag at the C-terminus, and expressed them both, as well as WT vectors, in HEK293T cells. We did not observe a significant difference in the protein level between the mutants and WT by western blotting, suggesting that the protein stability of R624C and F1075S was not affected (data not shown). We then prepared expression vectors of R624C- and F1075S-TRPM1 cDNAs fused with the mouse mGluR6 9.5 kb promoter that drives ON BC-specific expression (Figure 3A). We electroporated each of them as well as the WT vector into the P0 mouse retinas. We examined the localization of the expressed TRPM1 mutant proteins in ON BCs at P14 when the endogenous mouse TRPM1 is preferentially localized to the dendritic tips [11,19]. The electroporated human TRPM1 was visualized by an anti-Flag tag antibody. TRPM1 WT was localized at the soma (arrow) and dendritic tips (arrowheads) of the ON BCs (Figure 3B,C). On the other hand, the immunostaining signals of both the R624C and the F1075S TRPM1 proteins were significantly weaker than those of the WT protein (Figure 3D-G). We quantified the number of electroporated ON BCs in which the WT TRPM1 protein was localized more in the dendritic tips than somas. About 70% of ON BCs that electroporated with WT TRPM1 showed a brighter TRPM1 signal at the dendritic tips, and the signals on the dendritic tips were dramatically reduced in both mutant proteins (Figure 3H). These results may imply that R624C and F1075S mutations lead to mislocalization of TRPM1 protein and are responsible for the protein trafficking of TRPM1 channel proteins. However, it should be noted that there is also a possibility that the transfected TRPM1 proteins are misfolded due to the addition of the 3xFlag tag, leading to the undetectability of Flag-tagged mutant proteins specifically in the dendrites. Future analysis is required to obtain a conclusive result on this point.
Figure 3.
In vivo electroporation of WT, R624C, and F1075S TRPM1 expression vectors fused with the mGluR6 promoter. A: DNA constructs used for electroporation. CAG-GFP was co-electroporated to identify electroporated regions. WT and mutant forms of TRPM1 fused with 3×Flag were expressed under the mouse mGluR6 9.5 kb promoter. B-G: Section immunohistochemistry of the P14 retinas electroporated in vivo at P0 with CAG-GFP and mGluR6-TRPM1–3×Flag. The sections were immunostained with PKCα (blue), GFP (green), and Flag (red) antibodies (B, D, and F). Immunostaining with Flag was visualized separately (C, E and G). Arrows represent the soma and arrowheads represent the dendritic tips of ON BC. Scale bar: 10 μm. H. Composition of electroporated ON BC (GFP+ PKCα+), where bright Flag signal was observed in the dendrites. The error bars represent standard deviation (SD). Asterisks show that the differences are statistically significant (n=3) (Student’s t-test, p<0.05).
Clinical characteristics of the complete CSNB patients associated with TRPM1 mutations
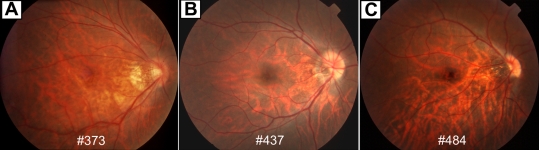
The clinical characteristics of the three complete CSNB patients with TRPM1 mutations are summarized in Table 2. All the patients showed the typical clinical features of complete CSNB. They complained of night blindness from their early childhood, and the best-corrected visual acuities were normal in two patients (#373 and #437) and mildly reduced in one (#484). The refractive errors were highly or moderately myopic, and astigmatism was present in all of the patients (Table 2). Mild nystagmus and esotropia was observed in the patient with reduced visual acuity (#484). Slit lamp examination revealed no abnormality, and all fundus examinations revealed no abnormalities in the posterior pole other than myopic changes, including the tilted discs and temporal pallor of the optic discs (Figure 4). Goldmann kinetic visual fields and color vision tests with the Ishihara plates, the AO H-R-R pseudoisochromatic plates, and Farnsworth D-15 panel test were conducted in two patients (#437 and #484) with normal results. No family history was obtained from the patients (Figure 1B).
Table 2. Clinical Characteristics in complete CSNB with TRPM1 mutations.
| Mutation | Corrected visual acuity | Refraction | Color vision | Fundus appearance | Visual field | Night blindness | Nystagmus | Strabismus |
|---|---|---|---|---|---|---|---|---|
|
patient #373 (Male, 9 years old) | ||||||||
| IVS3–3C>G, F1075S |
1.0 (OD) |
−8.25 −2.75 X 40 (OD) |
ND |
Myopic |
ND |
+ |
− |
Orthophoria |
| 1.0 (OS) |
−8.25 −0.50 X 180 (OS) |
|||||||
|
patient #437 (Male, 19 years old) | ||||||||
| R624C, S882X |
1.0 (OD) |
−4.50 −2.25 X 120 (OD) |
Normal |
Myopic |
Normal |
+ |
− |
Orthophoria |
| 1.0 (OS) |
−4.50 −2.75 X 50 (OS) |
|||||||
|
patient #484 (Male, 27 years old) | ||||||||
| IVS9+3_6delAAGT, R624C | 0.5 (OD) | −13.00 −3.00 X 90 (OD) | Normal | Myopic | Normal | + | + | Esotropia |
Figure 4.
Fundus photographs of patients with mutations of TRPM1. A-C: Fundus images of patient #373 (A), patient #437 (B), and patient #484 (C). All fundus examinations revealed no abnormalities in the posterior pole other than myopic changes, including the tilted discs and temporal pallor of the optic discs. The patient’s number is indicated in each photograph.
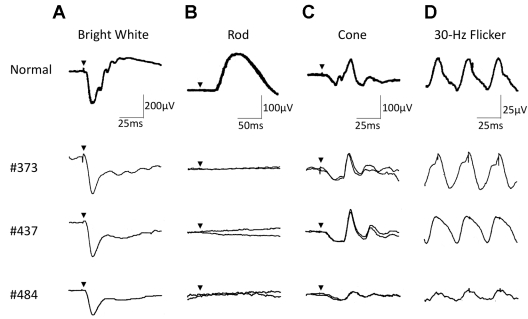
The full-field scotopic (rod) ERGs elicited by a blue light were non-recordable after 20 min of dark-adaptation (Figure 5B). The photopic ERG showed an a-wave with apparently normal implicit time and a b-wave with delayed implicit time (Figure 5C). The amplitudes of the 30 Hz flicker ERG were within normal range (Figure 5D). The single bright-flash, mixed rod-cone ERG elicited by a white stimulus had a “negative-type” configuration with a reduced normal a-wave and with a significantly reduced b-wave amplitude (Figure 5A). The oscillatory potentials were reduced.
Figure 5.
Full-field ERGs of patients with mutations of the TRPM1 gene. A-D: Full-field ERGs recorded in a normal subject and three affected individuals. The single bright-flash, mixed rod-cone ERGs showed a “negative-type” configuration with a reduced normal a-wave and a significantly reduced b-wave amplitude (A). The scotopic ERGs showed no response after 20 min of dark-adaptation (B). The photopic ERGs showed an apparent a-wave with normal implicit time and a b-wave with delayed implicit time (C). The amplitudes of the 30 Hz flicker ERGs were within normal range (D). The oscillatory potentials were reduced. Arrowheads: stimulus onset. Each patient’s number is noted at left.
ERG analysis of the trpm1 null mutant (trpm1–/–) mice
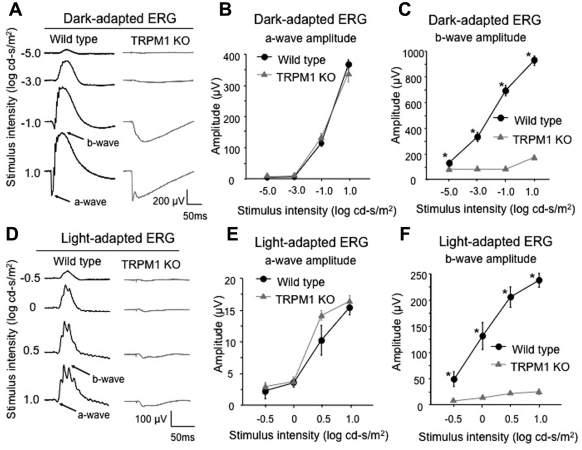
To address a possible role of TRPM1 in ON BC function, we previously generated trpm1 null mutant (trpm1–/–) mice by targeted gene disruption [11]. Next, to compare the physiologic retinal function of trpm1–/– between humans and mice, we recorded mouse ERGs in 8-week-old mice (Figure 6). The dark-adapted (rod) ERGs elicited by different stimulus intensities from WT and trpm1–/– mice are shown in Figure 6A. The amplitudes of the a-wave, originating from the photoreceptors, were approximately equal for both types of mice (Figure 6B). In contrast, the dark-adapted ERG b-wave, originating mainly from the ON BCs, in trpm1–/– mice was not present at the lower intensities, and only a very small positive inflection was seen at the highest stimulus intensity of 1.0 log cd-s/m2 (Figure 6C). The ERGs elicited by different stimulus intensities from light-adapted WT and trpm1–/– mice are shown in Figure 6D. The b-wave was severely depressed or absent leaving only the a-wave in the trpm1–/– mouse (Figure 6E,F). These ERG results are similar to those obtained by other groups [19,33], and suggested that the function of both rod and cone ON BCs, and the signal transmission from rod and cone photoreceptors to BCs were severely impaired in trpm1–/– mice. In contrast, the presence of a normal a-wave indicated that the rod and cone photoreceptors were functioning normally in this mutant mouse.
Figure 6.
ERG of WT and trpm1–/– mice. A-F: ERGs recorded from 8-week-old mice. Dark-adapted (A) and light-adapted (D) ERGs were elicited by four different stimulus intensities in both WT and trpm1–/– mice (n=5). Amplitudes of dark-adapted (B) and light-adapted (E) ERG a-waves as a function of the stimulus intensity are shown. Amplitudes of dark-adapted (C) and light-adapted (F) ERG b-waves as a function of the stimulus intensity are shown. The bars represent the standard error of the mean (SEM). Asterisks show that the differences are statistically significant (Mann–Whitney test, p<0.05). The trpm1–/– mouse had a normal a-wave, but a severely depressed b-wave for both dark- and light-adapted ERGs.
Discussion
An essential step in intricate visual processing is the segregation of visual signals into ON and OFF pathways by retinal BCs [34]. The release of glutamate from photoreceptors modulates the photoresponse of ON BCs [35] via metabotropic glutamate receptor 6 (mGluR6) and the G-protein (Go) that regulates a cation channel [36-38]. We recently reported that we identified a mouse trpm1 long form (trpm1-L) and found that TRPM1-L localization is developmentally restricted to the dendritic tips of ON BCs in co-localization with mGluR6 [11]. Trpm1 null mutant mice completely lose the photoresponse of ON BCs. TRPM1-L channel activity is negatively regulated by activated Go in the mGluR6 cascade. These results suggest that TRPM1-L is a component of the ON BC transduction channel. These observations led us to examine whether the gene is mutated in human patients with complete CSNB in the current study.
To identify human TRPM1 gene mutations in CSNB patients, we analyzed four separate Japanese patients with complete CSNB in whom previous molecular examination revealed no mutation in either the NYX or GRM6 genes. In the current study, we identified five different mutations in TRPM1: IVS2–3C>G, IVS8+3_6delAAGT, R624C (c.1870C>T), S882X (c.2645C>A), and F1075S (c.3224T>C). Mutations were present in three unrelated patients with complete CSNB (#373, #437, and #484). Using a minigene expression assay, we showed that two intron mutations, IVS2–3C> and IVS8+3_6delAAGT, can cause splicing abnormalities leading to defects in protein production. Since these two mutations are located in the N-terminus region of TRPM1, these mutations are likely to produce a loss-of-function allele of TRPM1. The nonsense mutation S882X (c.2645C>A) is located between TM2 and TM3. Thus, the truncated TRPM1 protein is likely to be non-functional as a channel. Regarding the two missense mutations, R624C (c.1870C>T) and F1075S (c.3224T>C), we observed failure of the transportation of the missense mutant channels to the dendritic tips. However, ~20% of ON BCs could transport the mutant forms of TRPM1, suggesting that these ON BCs are still active. Considering that human patients carrying these mutations still have cone ERG responses, this fraction might be cone ON BCs converging inputs from cone photoreceptors. Since these two amino acid residues are evolutionarily conserved among the TRPM subfamily, these amino acid residues are indispensable for the physiologic functions of TRPM channels. For example, R671Q mutation in the Drosophila TRPM1 homolog (trp14) resulted in a significant reduction of ERG response [39]. This mutation is located between the 6TM and the TRP domain [40], which is close to the F1075 residue of human TRPM1. Although this region is not in the functional domains, it still could be responsible for the physiologic function of the TRPM channel.
The scotopic ERG b-waves recorded using Ganzfeld stimuli were nonrecordable, and the photopic ERG showed normal a-wave amplitude, but the amplitude of the b-wave is reduced or absent in the three affected human individuals. There seems to be no apparent genotype-phenotype correlation in our patients with TRPM1 mutations. Full-field ERG results showed no detectable post-receptoral ON-pathway function for the three patients. The ERG amplitudes of patient #484 were smaller than those of the other two patients, but the reductions were considered to be due to high myopia (−12 to −13 D) [41] or other unknown reasons. The function of the inner retina was further analyzed using L-M cone ERGs elicited by rectangular, 100–125 msec duration light stimuli under light-adapted conditions in one patient, and as a result, the amplitude of the b-wave at light onset (ON-response) was significantly reduced, but the d-wave amplitude (OFF-response) at light offset was unaffected with normal OPs on the OFF-responses (data not shown). These electrophysiological results indicated that the pathology in complete CSNB with TRPM1 mutations lay in the dysfunction of the depolarizing ON BCs because it is generally considered that the positive ON-response (b-wave) reflects the depolarizing ON BCs. The impairment in the ON-response was also observed in the trpm1–/– mice in which b-wave amplitude for both scotopic and photopic conditions were completely defective. These results in human and mice strongly indicated that TRPM1 is essential for the function of ON BCs in the visual pathway.
The clinical features of patients with complete CSNB are similar even when the causative gene is different. Most patients complain of night blindness while showing normal fundi accompanied by highly myopic refractive errors [4,5]. The best-corrected visual acuity is mildly reduced, or sometimes normal. We could not clarify any difference in ERG responses that were recorded according to ISCEV protocol among those with different gene defects. To date, the only phenotypic difference among complete CSNB patients with different gene mutations has been detected using a scotopic 15 Hz-flicker ERG with increasing intensities [42]. It was reported that patients with TRPM1 mutations showed 15 Hz-flicker ERG responses similar to those with NYX mutations, but different from those with GRM6 mutations or normal control subjects, suggesting some difference in the rod pathway [21].
Very recent studies reported that TRPM1 gene mutations were the major cause of AR complete CSNB in patients with Caucasian ancestors [20-22]. We previously identified NYX mutations in five families and GRM6 mutations in two families among 11 Japanese families with complete CSNB [43,44], and in the current study, we found TRPM1 mutations in three of the remaining four families. From these results we confirmed that the gene was responsible for patients with complete CSNB. It is likely that one of the three genes that is localized at the dendritic terminals of ON BCs and contributing the cell activity, TRPM1, GRM6, or NYX, would responsible for most patients with complete CSNB.
In conclusion, we identified five different mutations in the TRPM1 gene in three unrelated Japanese patients with complete CSNB. TRPM1 is essential for the depolarizing ON BC function in humans as well as in mice.
Acknowledgments
We thank S. Nakanishi for providing us the MG6-Z vector. We also thank M. Joukan, A. Tani, T. Tsujii, A. Ishimaru, Y. Saioka, K. Sone and S. Kennedy for technical assistance. This work was supported by CREST from Japan Science and Technology Agency, and a Grant for Molecular Brain Science, a Grant-in-Aid for Scientific Research (B) from Ministry of Education, Culture, Sports, Science and Technology, and The Takeda Science Foundation, The Uehara Memorial Foundation, Novartis Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, The Naito Foundation.
References
- 1.Schubert G, Bornschein H. Analysis of the human electroretinogram. Ophthalmologica. 1952;123:396–413. doi: 10.1159/000301211. [DOI] [PubMed] [Google Scholar]
- 2.Carr RE. Congenital stationary nightblindness. Trans Am Ophthalmol Soc. 1974;72:448–87. [PMC free article] [PubMed] [Google Scholar]
- 3.Bech-Hansen NT, Naylor MJ, Maybaum TA, Pearce WG, Koop B, Fishman GA, Mets M, Musarella MA, Boycott KM. Loss-of-function mutations in a calcium-channel alpha1-subunit gene in Xp11.23 cause incomplete X-linked congenital stationary night blindness. Nat Genet. 1998;19:264–7. doi: 10.1038/947. [DOI] [PubMed] [Google Scholar]
- 4.Miyake Y, Yagasaki K, Horiguchi M, Kawase Y, Kanda T. Congenital stationary night blindness with negative electroretinogram. A new classification. Arch Ophthalmol. 1986;104:1013–20. doi: 10.1001/archopht.1986.01050190071042. [DOI] [PubMed] [Google Scholar]
- 5.Ruether K, Apfelstedt-Sylla E, Zrenner E. Clinical findings in patients with congenital stationary night blindness of the Schubert-Bornschein type. Ger J Ophthalmol. 1993;2:429–35. [PubMed] [Google Scholar]
- 6.Bech-Hansen NT, Naylor MJ, Maybaum TA, Sparkes RL, Koop B, Birch DG, Bergen AA, Prinsen CF, Polomeno RC, Gal A, Drack AV, Musarella MA, Jacobson SG, Young RS, Weleber RG. Mutations in NYX, encoding the leucine-rich proteoglycan nyctalopin, cause X-linked complete congenital stationary night blindness. Nat Genet. 2000;26:319–23. doi: 10.1038/81619. [DOI] [PubMed] [Google Scholar]
- 7.Pusch CM, Zeitz C, Brandau O, Pesch K, Achatz H, Feil S, Scharfe C, Maurer J, Jacobi FK, Pinckers A, Andreasson S, Hardcastle A, Wissinger B, Berger W, Meindl A. The complete form of X-linked congenital stationary night blindness is caused by mutations in a gene encoding a leucine-rich repeat protein. Nat Genet. 2000;26:324–7. doi: 10.1038/81627. [DOI] [PubMed] [Google Scholar]
- 8.Dryja TP, McGee TL, Berson EL, Fishman GA, Sandberg MA, Alexander KR, Derlacki DJ, Rajagopalan AS. Night blindness and abnormal cone electroretinogram ON responses in patients with mutations in the GRM6 gene encoding mGluR6. Proc Natl Acad Sci USA. 2005;102:4884–9. doi: 10.1073/pnas.0501233102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeitz C, van Genderen M, Neidhardt J, Luhmann UF, Hoeben F, Forster U, Wycisk K, Mátyás G, Hoyng CB, Riemslag F, Meire F, Cremers FP, Berger W. Mutations in GRM6 cause autosomal recessive congenital stationary night blindness with a distinctive scotopic 15-Hz flicker electroretinogram. Invest Ophthalmol Vis Sci. 2005;46:4328–35. doi: 10.1167/iovs.05-0526. [DOI] [PubMed] [Google Scholar]
- 10.Zeitz C, Forster U, Neidhardt J, Feil S, Kälin S, Leifert D, Flor PJ, Berger W. Night blindness-associated mutations in the ligand-binding, cysteine-rich, and intracellular domains of the metabotropic glutamate receptor 6 abolish protein trafficking. Hum Mutat. 2007;28:771–80. doi: 10.1002/humu.20499. [DOI] [PubMed] [Google Scholar]
- 11.Koike C, Obara T, Uriu Y, Numata T, Sanuki R, Miyata K, Koyasu T, Ueno S, Funabiki K, Tani A, Ueda H, Kondo M, Mori Y, Tachibana M, Furukawa T. TRPM1 is a component of the retinal ON bipolar cell transduction channel in the mGluR6 cascade. Proc Natl Acad Sci USA. 2010;107:332–7. doi: 10.1073/pnas.0912730107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duncan LM, Deeds J, Hunter J, Shao J, Holmgren LM, Woolf EA, Tepper RI, Shyjan AW. Down-regulation of the novel gene melastatin correlates with potential for melanoma metastasis. Cancer Res. 1998;58:1515–20. [PubMed] [Google Scholar]
- 13.Deeds J, Cronin F, Duncan LM. Patterns of melastatin mRNA expression in melanocytic tumors. Hum Pathol. 2000;31:1346–56. [PubMed] [Google Scholar]
- 14.Duncan LM, Deeds J, Cronin FE, Donovan M, Sober AJ, Kauffman M, McCarthy JJ. Melastatin expression and prognosis in cutaneous malignant melanoma. J Clin Oncol. 2001;19:568–76. doi: 10.1200/JCO.2001.19.2.568. [DOI] [PubMed] [Google Scholar]
- 15.Fang D, Setaluri V. Expression and Up-regulation of alternatively spliced transcripts of melastatin, a melanoma metastasis-related gene, in human melanoma cells. Biochem Biophys Res Commun. 2000;279:53–61. doi: 10.1006/bbrc.2000.3894. [DOI] [PubMed] [Google Scholar]
- 16.Miller AJ, Du J, Rowan S, Hershey CL, Widlund HR, Fisher DE. Transcriptional regulation of the melanoma prognostic marker melastatin (TRPM1) by MITF in melanocytes and melanoma. Cancer Res. 2004;64:509–16. doi: 10.1158/0008-5472.can-03-2440. [DOI] [PubMed] [Google Scholar]
- 17.Kim DS, Ross SE, Trimarchi JM, Aach J, Greenberg ME, Cepko CL. Identification of molecular markers of bipolar cells in the murine retina. J Comp Neurol. 2008;507:1795–810. doi: 10.1002/cne.21639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koike C, Sanuki R, Miyata K, Koyasu T, Miyoshi T, Sawai H, Kondo M, Usukura J, Furukawa T. The functional analysis of TRPM1 in retinal bipolar cells. 30th Annual meeting Japan Neuroscience Society 2007; 58(Supplement 1):S41. [Google Scholar]
- 19.Morgans CW, Zhang J, Jeffrey BG, Nelson SM, Burke NS, Duvoisin RM, Brown RL. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc Natl Acad Sci USA. 2009;106:19174–8. doi: 10.1073/pnas.0908711106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Audo I, Kohl S, Leroy BP, Munier FL, Guillonneau X, Mohand-Saïd S, Bujakowska K, Nandrot EF, Lorenz B, Preising M, Kellner U, Renner AB, Bernd A, Antonio A, Moskova-Doumanova V, Lancelot ME, Poloschek CM, Drumare I, Defoort-Dhellemmes S, Wissinger B, Léveillard T, Hamel CP, Schorderet DF, De Baere E, Berger W, Jacobson SG, Zrenner E, Sahel JA, Bhattacharya SS, Zeitz C. TRPM1 is mutated in patients with autosomal-recessive complete congenital stationary night blindness. Am J Hum Genet. 2009;85:720–9. doi: 10.1016/j.ajhg.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Genderen MM, Bijveld MM, Claassen YB, Florijn RJ, Pearring JN, Meire FM, McCall MA, Riemslag FC, Gregg RG, Bergen AA, Kamermans M. Mutations in TRPM1 are a common cause of complete congenital stationary night blindness. Am J Hum Genet. 2009;85:730–6. doi: 10.1016/j.ajhg.2009.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Sergouniotis PI, Michaelides M, Mackay DS, Wright GA, Devery S, Moore AT, Holder GE, Robson AG, Webster AR. Recessive mutations of the gene TRPM1 abrogate ON bipolar cell function and cause complete congenital stationary night blindness in humans. Am J Hum Genet. 2009;85:711–9. doi: 10.1016/j.ajhg.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Onishi A, Peng GH, Hsu C, Alexis U, Chen S, Blackshaw S. Pias3-dependent SUMOylation directs rod photoreceptor development. Neuron. 2009;61:234–46. doi: 10.1016/j.neuron.2008.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueda Y, Iwakabe H, Masu M, Suzuki M, Nakanishi S. The mGluR6 5′ Upstream Transgene Sequence Directs a Cell-Specific and Developmentally Regulated Expression in Retinal Rod and ON-Type Cone Bipolar Cells. J Neurosci. 1997;17:3014–23. doi: 10.1523/JNEUROSCI.17-09-03014.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakamura M, Ito S, Terasaki H, Miyake Y. Novel CACNA1F Mutations in Japanese Patients with Incomplete Congenital Stationary Night Blindness. Invest Ophthalmol Vis Sci. 2001;42:1610–6. [PubMed] [Google Scholar]
- 26.Chen S, Kadomatsu K, Kondo M, Toyama Y, Toshimori K, Ueno S, Miyake Y, Muramatsu T. Effects of flanking genes on the phenotypes of mice deficient in basigin/CD147. Biochem Biophys Res Commun. 2004;324:147–53. doi: 10.1016/j.bbrc.2004.08.232. [DOI] [PubMed] [Google Scholar]
- 27.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 28.Yang K, Fang K, Fromondi L, Chan KW. Low temperature completely rescues the function of two misfolded K ATP channel disease-mutants. FEBS Lett. 2005;579:4113–8. doi: 10.1016/j.febslet.2005.06.039. [DOI] [PubMed] [Google Scholar]
- 29.Smit LS, Strong TV, Wilkinson DJ, Macek M, Jr, Mansoura MK, Wood DL, Cole JL, Cutting GR, Cohn JA, Dawson DC, Collins FS. Missense mutation (G480C) in the CFTR gene associated with protein mislocalization but normal chloride channel activity. Hum Mol Genet. 1995;4:269–73. doi: 10.1093/hmg/4.2.269. [DOI] [PubMed] [Google Scholar]
- 30.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–48. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
- 31.Mukerji N, Damodaran TV, Winn MP. TRPC6 and FSGS: the latest TRP channelopathy. Biochim Biophys Acta. 2007;1772:859–68. doi: 10.1016/j.bbadis.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Montell C. TRP channels in Drosophila photoreceptor cells. J Physiol. 2005;567:45–51. doi: 10.1113/jphysiol.2005.092551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shen Y, Heimel JA, Kamermans M, Peachey NS, Gregg RG, Nawy S. A transient receptor potential-like channel mediates synaptic transmission in rod bipolar cells. J Neurosci. 2009;29:6088–93. doi: 10.1523/JNEUROSCI.0132-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Werblin FS, Dowling JE. Organization of the retina of the mudpuppy, Necturus maculosus. II. Intracellular recording. J Neurophysiol. 1969;32:339–55. doi: 10.1152/jn.1969.32.3.339. [DOI] [PubMed] [Google Scholar]
- 35.Ayoub GS, Copenhagen DR. Application of a fluorometric method to measure glutamate release from single retinal photoreceptors. J Neurosci Methods. 1991;37:7–14. doi: 10.1016/0165-0270(91)90016-s. [DOI] [PubMed] [Google Scholar]
- 36.Masu M, Iwakabe H, Tagawa Y, Miyoshi T, Yamashita M, Fukuda Y, Sasaki H, Hiroi K, Nakamura Y, Shigemoto R, Takada M, Nakamura K, Nakao K, Katsuki M, Nakanishi S. Specific deficit of the ON response in visual transmission by targeted disruption of the mGluR6 gene. Cell. 1995;80:757–65. doi: 10.1016/0092-8674(95)90354-2. [DOI] [PubMed] [Google Scholar]
- 37.Nawy S. The metabotropic receptor mGluR6 may signal through G(o), but not phosphodiesterase, in retinal bipolar cells. J Neurosci. 1999;19:2938–44. doi: 10.1523/JNEUROSCI.19-08-02938.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhingra A, Lyubarsky A, Jiang M, Pugh EN, Jr, Birnbaumer L, Sterling P, Vardi N. The light response of ON bipolar neurons requires G[alpha]o. J Neurosci. 2000;20:9053–8. doi: 10.1523/JNEUROSCI.20-24-09053.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang T, Jiao Y, Montell C. Dissecting independent channel and scaffolding roles of the Drosophila transient receptor potential channel. J Cell Biol. 2005;171:685–94. doi: 10.1083/jcb.200508030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishida M, Hara Y, Yoshida T, Inoue R, Mori Y. TRP channels: molecular diversity and physiological function. Microcirculation. 2006;13:535–50. doi: 10.1080/10739680600885111. [DOI] [PubMed] [Google Scholar]
- 41.Westall CA, Dhaliwal HS, Panton CM, Sigesmun D, Levin AV, Nischal KK, Héon E. Values of electroretinogram responses according to axial length. Doc Ophthalmol. 2001;102:115–30. doi: 10.1023/a:1017535207481. [DOI] [PubMed] [Google Scholar]
- 42.Scholl HP, Langrova H, Pusch CM, Wissinger B, Zrenner E, Apfelstedt-Sylla E. Slow and fast rod ERG pathways in patients with X-linked complete stationary night blindness carrying mutations in the NYX gene. Invest Ophthalmol Vis Sci. 2001;42:2728–36. [PubMed] [Google Scholar]
- 43.Nakamura M, Lin K, Ito S, Terasaki H, Miyake Y. Novel NYX Mutations and Clinical Phenotype in Japanese Patients with Complete Congenital Stationary Night Blindness. ARVO Annual Meeting; 2003 May 4–9; Fort Lauderdale (FL). [Google Scholar]
- 44.Nakamura M, Miyake Y. Molecular genetic study of congenital stationary night blindness. Nippon Ganka Gakkai Zasshi. 2004;108:665–73. [PubMed] [Google Scholar]