Abstract
Purpose
Cataracts can be broadly divided into two types: congenital cataracts and age-related cataracts. ΔG91 is a previously discovered congenital mutation in βA3-crystallin that impairs protein solubility. On the other hand, the deamidation of β-crystallin is a significant feature in aged and cataractous lenses. Several deamidation sites were also identified in βA3-crystallin. The present study is to compare the functional consequence of ΔG91 mutation and the deamidation of βA3-crystallin in terms of folding properties and protein–protein interaction.
Methods
Protein secondary structure and hydrophobic properties were investigated by in silica analysis of the wild type and mutants sequences. Full-length βA3-crystallin was cloned into a mammalian two-hybrid system in order to investigate protein–protein interactions. Deletion and deamidation were introduced by site-directed mutagenesis protocols. Both the Q85 and Q180 deamidation sites were substituted with glutamic acid residues to mimic deamidation. Different combinations of plasmid constructs were transfected in HeLa cells, and changes of protein–protein interactions were analyzed by the luciferase assay.
Results
Bioinformatics prediction suggested that ΔG91 mutation alters both the predicted secondary structure and hydrophobic character of βA3-crystallin, while deamidation only exhibits minimal effects. Mammalian two-hybrid results indicated that both ΔG91 mutation and Q85/Q180 deamidation could significantly decrease the interaction of the βA3-crystallin homodimer.
Conclusion
Our results provided evidence that both mutations involved in congenital cataracts and deamidation in aged lenses commonly altered protein–protein interaction between human lens βA3-crystallins, which may lead to protein insolubilization and contribute to cataracts.
Introduction
According to the World Health Organization, a cataract is defined as “clouding of the lens of the eye which impedes the passage of light” [1]. Worldwide, it is estimated that nearly half of all cases of blindness are caused by cataracts. Cataracts can be broadly divided into two types: congenital cataracts and age-related cataracts. Although most cataract cases are associated with the aging process, congenital cataracts are the leading cause of visual disability in children [2].
Crystallins are major component proteins in intact lens. β-Crystallin is one of the three main lens crystallin components (α-, β-, and γ-crystallin) and is further subdivided into acidic (βA1-, βA2-, βA3-, and βA4-crystallin) and basic (βB1-, βB2-, and βB3-crystallin) groups, which can form homo- or hetero-oligomers in the lens [3]. Previous reports have indicated that mutations in various crystallin genes are deleterious factors contributing to the loss of protein stability and lens transparency [4,5]. For example, since 1998, three kinds of congenital mutation in βA3-crystallin gene have been identified in seven cataract pedigrees. The βA3-crystallin gene, located at 17q11–12, encodes two crystallin proteins (βA3- and βA1-crystallin) from a single mRNA. The βA1-crystallin protein lacks the NH2-terminal 17 AA due to an alternate translation initiation site. These congenital mutations include a three base pair deletion (ΔG91) and splice site mutations (IVS3+1G>C and IVS3+1G>A). Interestingly, although the ethnical backgrounds of patients are diverse, including India, Brazil, China, the UK, Switzerland, and Australia, five of them manifest the ΔG91 mutation, which indicated the functional importance of this site [2,5-9].
On the other hand, proteomics analysis of post-translational modifications in young and aged lenses have identified extensive modification sites in human crystallins [10,11]. Among the several potential post-modifications, deamidation is the most abundant modification in the lens and is significantly increased in aged and cataractous lenses [11]. These post-translational modifications were hypothesized to relate with the age-dependent loss of crystallin solubility. For example, among those previously identified deamidation sites in βA3-crystallin, Takata et al. [12-14] provided evidence that deamidation at Q85 and Q180 destabilizes βA3-crystallin homodimer and disrupts interaction with other β-crystallin subunits.
From the functional viewpoint, proper folding and normal protein–protein interactions are two key aspects for ensuring crystallin's cellular function. Therefore, to elucidate the functional consequences of ΔG91 mutation and the deamidation of βA3-crystallin, we predicted the folding characteristics using bioinformatics analysis and further investigated into their homodimer formation by a mammalian two-hybrid system. The two deamidation sites, Q85 and Q180, were used as representative samples in this study because Q85 is located in the NH2-terminal domain and Q180 is located in the COOH-terminal domain. However, both sites are located in the critical interface. Our results indicated that both mutations involved in congenital cataracts and deamidation in aged lenses commonly alter protein–protein interaction in human lens βA3-crystallin, which potentially contributes to decreased protein solubility and formation of cataract.
Methods
Protein secondary structure prediction
The secondary structures of wild type and βA3-crystallin mutants were predicted by the widely used SSpro8 algorithm on the Scratch server [15]. Based on recurrent neural networks and PSI-BLAST-derived profiles, SSpro8 predicts protein secondary structures according to the DSSP classification [16].
Protein hydrophobicity analysis
We used Kyte-Doolittle hydrophobicity plots to detect the potential effects of protein mutants. Hydrophobicity for both wild type and mutants was calculated in a window size of five, which is good for finding hydrophilic regions. Regions with values below zero are hydrophilic in character.
Mammalian two-hybrid system
The mammalian two-hybrid assay kit, obtained from Stratagene (La Jolla, CA), was used in this study. In this assay, a protein of interest (bait) is fused to the DNA-binding domain of the yeast protein GAL4 in the pCMV-BD vector, while another protein (prey) is fused to the transcriptional activation domain of the mouse protein NF-κB in the pCMV-AD vector. The pBD-p53 control plasmid expresses the GAL4 binding domain and amino acids 72–390 of murine p53 as a hybrid protein. The pAD-SV40T control plasmid expresses a hybrid protein that contains the NF-κB transcriptional activation domain fused to amino acids 84–708 of the SV40 large T-antigen. pFR-Luc was used as a reporter vector, which contains a synthetic promoter with five tandem repeats of the yeast GAL4 binding sites that control the expression of a luciferase gene.
Plasmid constructs and site-directed mutagenesis
The βA3-crystallin (CRYBA3) gene was obtained from Fulengen Co., Ltd. (Guangzhou, China). Full-length CRYBA3 was subcloned into pCMV-AD and pCMV-BD vectors with the corresponding forward and reverse primers (Table1). Site-directed mutagenesis was conducted according to DpnI mediated protocol [17]. The primers used for mutagenesis were listed in Table 1. All constructs were verified by sequencing.
Table 1. Primers for sub-cloning of βA3 genes and site-directed mutagenesis.
| Constructs | Forward primer | Reverse primer |
|---|---|---|
|
Sub-cloning | ||
| βA3-crystallin |
TTGGATCCGAGACCCAGGCTGAG |
GCTAAGCTTCTACTGTTGGATTCG |
|
Mutagenesis | ||
| βA3ΔG91 |
GTTTATCCTGGAGAGAGAATACCCTCGCTG |
CAAATAGGACCTCTCTCTTATGGGAGCGAC |
| βA3Q85E |
CTGTGGGCAAGAGTTTATCCTGG |
GACACCCGTTCTCAAATAGGACC |
| βA3Q180E | CGTGGGTATGAGTATATCTTGG | GCACCCATACTCATATAGAACC |
The underlined sequences are BamHI and HindIII restriction enzyme sites for forward and reverse primers, respectively.
Transfection and luciferase assay
HeLa cells were grown in Dulbecco's modified Eagle's medium (Gibco,Carlsbad, CA) supplemented with 10% FBS (Hyclone, Logan, UT) and 100U/ml penicillin/streptomycin under humidified air containing 5% CO2 at 37 °C. Cells were seeded 24 h before transfection in 96-well tissue culture plates. The next day, cells were co-transfected with various constructs with reporter plasmid pFR-Luc and internal control pRL-TK using Lipofectamine 2000 in accordance with the manufacturer’s instructions. Cell lysate was collected 24 h after transfection. Firefly and Renilla luciferase activities were measured using the Promega dual luciferase reporter assay system (Promega, Madison, WI). This system allows the simultaneous expression and measurement of two individual reporter enzymes. The ‘experimental’ reporter (i.e., pFR-luc) is correlated with the effect of protein–protein interaction, while the activity of the co-transfected ‘control’ reporter (i.e., pRL-TK) provides an internal control that minimizes experimental variability caused by protein/cells total amount. The ratios of Firefly to Renilla luciferase activities were then normalized to basal transfection of control vectors (pCMV-AD+ pCMV-BD). All of the assays were performed in triplicate in three independent experiments. We assessed statistical significance using two-tailed Student t test.
Results
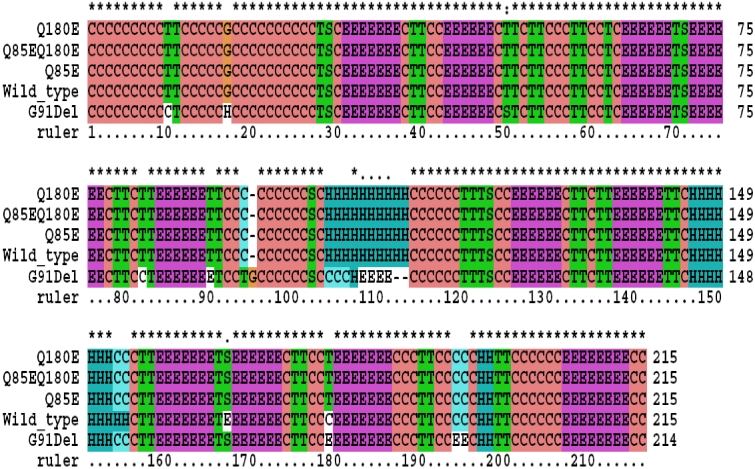
Since the crystal structure of βA3-crystallin has not been resolved experimentally, we used the SSpro8 algorithm to predict the protein secondary structures of wild type βA3-crystallin and mutants based on their sequences. This bioinformatics analysis revealed that the predicted secondary structure of the ΔG91 mutation was significantly different from that of the wild type βA3-crystallin; notably, the mutation completely destroyed the first alpha helix located at position 104–113 of the βA3-crystallin. Furthermore, we predicted that this mutation shortened the second alpha helix at residues 153–155 (Figure 1). However, for both single deamidation mutants and double deamidated mutants, the first alpha helix structure remained unchanged, although the second alpha helix was also found shortened. These results indicated that the secondary structural changes in the deamidated mutants seemed minor, which is consistent with far UV and near UV circular dichroism spectroscopy results [13].
Figure 1.
Predicted secondary structures of the wild type and the mutant βA3-crystallin. The following sequences were used for secondary structures prediction: wild type (Wild_type); glutamine that residues at position 85 is replaced by a glutamic acid residue (Q85E); glutamine that residues at position 180 is replaced by a glutamic acid residue (Q180E); βA3-crystallin double deamidated mutant (Q85EQ180E); or glycine residue at position 91 is removed (G91Del). The secondary structure types are designated as follows (according to DSSP program): H=alpha helix; B=residue in isolated beta-bridge; E=extended strand, participates in beta ladder; G=3-helix (3/10 helix); I=5 helix (pi helix); T=hydrogen bonded turn; S=bend; C=the rest.
Hydrophobic analysis by Kyte-Doolittle plot showed significant changes in the physicochemical properties of the surrounding region in the ΔG9 mutant compared to the wild type. In the mutant form, the hydrophobicity environment has become more hydrophilic with calculated values decreased to −3 (Figure 2A,B). However, compared to wild type, the hydrophobic property of the Q85E/Q180E mutant seemed unchanged (Figure 2A,C). Similar observations were also obtained from single deamidation mutants (data not shown). The above results suggested a ΔG91 mutation, but the Q85E or Q180E deamidation mutants did not alter the hydrophobic properties of βA3-crystallin remarkably.
Figure 2.
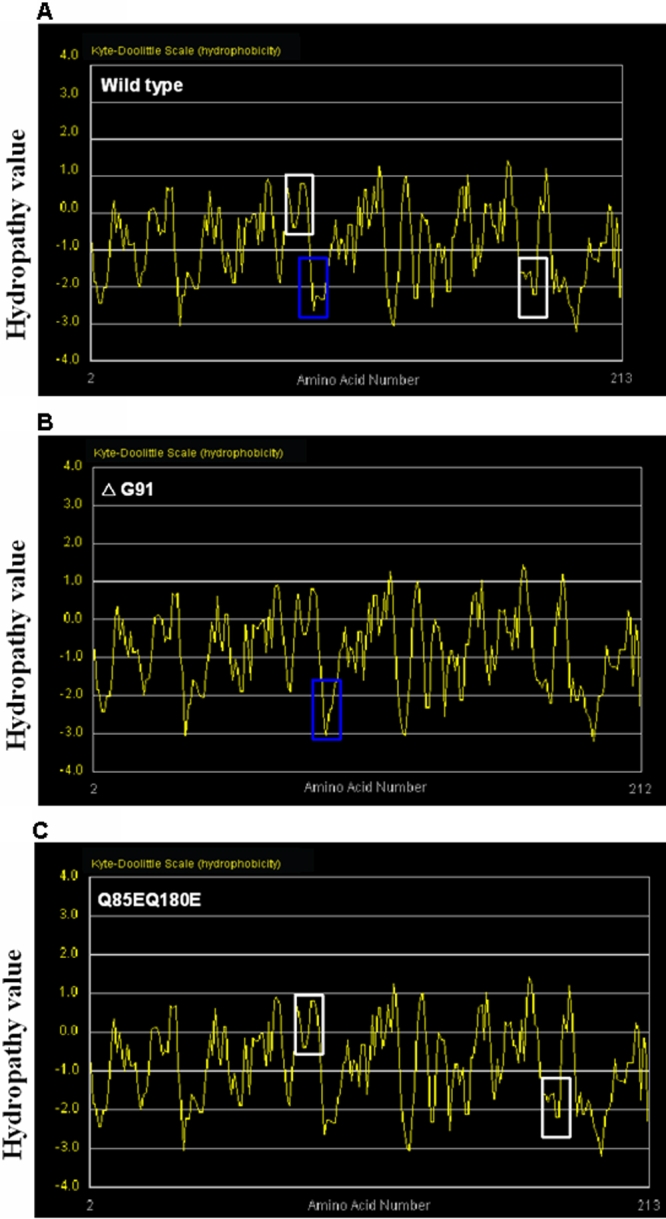
Protein hydrophobicity analysis of the wild type and the mutant βA3-crystallin. Kyte-Doolittle hydrophobicity plot of wild type βA3-crystallin (A), ΔG91 mutant (B), and Q85E/Q180E mutant (C). X-axis represents amino acid position of, and y-axis represents hydropathy value in a window size of 5. The region of interest is marked by blue box (for ΔG91 mutant) or white boxes (Q85E/Q180E mutant).
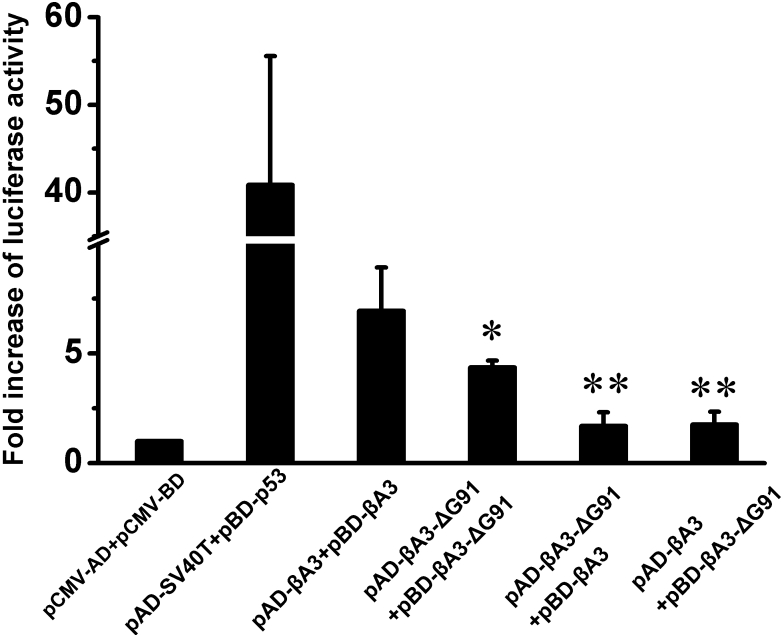
Previously, a mammalian two-hybrid system has been introduced to demonstrate the presence of protein–protein interactions among lens crystallins and to investigate the effects of mutations in αA-crystallin (R116C), αB-crystallin (R120G), and γC-crystallin (T5P) [18-20]. Here we used a mammalian two-hybrid system to investigate whether the ΔG91 mutation and Q85E/Q180E deamidation altered the endogenous interaction of the βA3-crystallin homodimer, which has not been previously addressed. In the absence of fusion protein, the activation domain of pCMV-AD is physically separated from the DNA-binding domain of pCMV-BD; thus, luciferase expression is low and the signal represents baseline level. Since expressed proteins of the pBD-p53 and pAD-SV40T control plasmids interact to significantly induce luciferase activity, the luciferase signal can be used to verify the induction of the luciferase reporter gene. Compared to these positive controls, we observed about 7 fold increase of luciferase activity with cotranfection of pBD-βA3 and pAD-βA3 constructs, indicative of considerable self-association between wild type βA3-crystallins.
Relative to the interaction between the wild type βA3-crystallin, there was an obvious decrease in the interaction between the ΔG91 mutant and the wild type βA3-crystallin. Interestingly, interactions between the mutant and wild type βA3-crystallin decreased to a greater extent than the interactions between the two ΔG91 mutant βA3-crystallins when both were compared with wild type interactions. It should be noted that the decreases in interaction were also observed when proteins were cloned in reverse order (i.e., pAD-βA3-ΔG91+pBD-βA3 versus pAD-βA3 +pBD-βA3-ΔG91), indicating that the decreased interactions were not vector-specific (Figure 3).
Figure 3.
Luciferase activities for detection of protein–protein interactions involving various ΔG91 mutants. Luciferase activity values are expressed as fold activation relative to the basal control (pCMV-AD+pCMV-BD). Various plasmid constructs were co-transfected as labeled. Data represent the mean±SEM of results in three independent experiments. Group differences were all compared with wild type homodimer interaction (pAD-βA3+pBD-βA3). The asterisk indicates a p<0.05 and the double asterisk indicates a p<0.01.
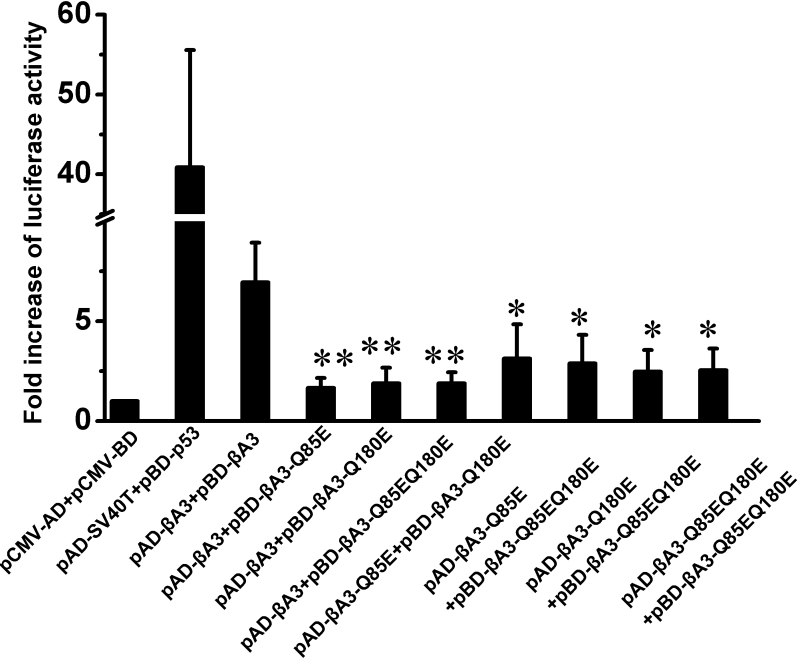
To mimic the deamidation effect, we constructed reporter plasmids; the glutamine residues at position 85 in the NH2-terminal domain and glutamine residues at position 180 in the COOH-terminal domain were substituted with glutamic acid residues by site-directed mutagenesis. We found that single site deamidation at Q85 or Q180 significantly decreased the protein–protein interactions between the wild type and mutant βA3-crystallins. Similarly, the luciferase signal, when co-transfected with double Q85E/Q180E mutant and wild type βA3-crystallin, also decreased several folds compared with the wild type interaction. The interactions among various βA3-crystallin mutants increased slightly compared with interaction between mutant and wild type βA3-crystallin, but were still significantly weaker than that of wild type (Figure 4).
Figure 4.
Luciferase activities for detection of protein–protein interactions involving various βA3-crystallin deamidation mutants. Luciferase activity values are expressed as fold activation relative to the basal control (pCMV-AD+pCMV-BD). Various plasmid constructs were co-transfected as labeled. Data represent the mean±SEM of results in three independent experiments. Group differences were all compared with wild type homodimer interaction (pAD-βA3+pBD-βA3). The asterisk indicates a p<0.05 and the double asterisk indicates a p<0.01.
Discussion
ΔG91 is the most common mutation found in βA3-crystallin. This glycine is highly conserved among a wide range of phylogenetic taxa, highlighting its importance [9]. However, the mechanistic understanding of this mutation is still limited; so far, there was only one in vitro report suggesting that removal of the glycine residue destroys the solubility of βA3-crystallin protein [2]. All vertebrate lens β-crystallins consist of two domains and each one folds into two similar ‘Greek key’ motifs. Each ‘Greek key’ motif is comprised of four consecutive anti-parallel β-strands [21]. Since this deletion is located at the boundary of the β-sheet of the second ‘Greek key’, this mutation is expected to destroy the inter-strand hydrogen bonds between β-strands. Importantly, our bioinformatics analysis found it could potentially disrupt the nearby helical region located in residue 104–113, which is also located in the second ‘Greek key’ motif. Therefore, this deletion mutation has the potential to reduce the stability of the second ‘Greek key’ motif. Furthermore, the predicted hydrophobic nature became more hydrophilic in this region, probably causing partial unfolding of the hydrophobic packaging in the mutant protein. Although computer-based analysis indicated that the ‘Greek key’ motif form inter-domain associations, the function of the ‘Greek key’ motif is still a matter of debate [21]. Using a two-hybrid system assay, we demonstrated that the ΔG91 βA3-crystallin mutant exhibited decreased interactions with the wild type βA3-crystallin, or with another ΔG91 βA3-crystallin mutant. This finding provided evidence that the ‘Greek key’ motif is crucial to molecular interactions between domains such as monomer–monomer βA3-crystallin association.
During the aging process, βA3-crystallin undergoes an unusually large number of modifications with deamidation being a major factor. Previous investigation indicated deamidation at Q85 and Q180 destabilizes the βA3-crystallin homodimer; however, the dimer formation was not disrupted [13]. In a mammalian hybrid system, we found that deamidation at these sites could partially destroy protein–protein interaction of βA3-crystallin, although the secondary structure and hydrophobic environment were predicted to be unchanged. We speculated that the differences between the two studies may result from the different experimental conditions and measurement sensitivities. The association of deamidated βA3-crystallin mutants was indirectly inferred from molecular masses and light scattering data while we investigated the interaction in a mammalian hybrid system. This system could detect weak and transient interactions and is considered physiologically relevant [18]. Our result highlighted the importance of deamidation at the critical interface. Indeed, a recent report indicated that the same two sites alter the interactions of βA3-crystallin with βB1- and βB2-crystallin [14].
Can deamidation lead to cataracts? One proteomics study showed that deamidations are the most extensive post-translational modification in aged and cataractous lenses and that the extent of deamidation was significantly increased in the water-insoluble fractions [11]. However, a recent report also suggested there was no apparent increase of deamidation upon the onset of a nuclear cataract [10]. Since deamidation is accompanied with both aged and cataractous lenses, it could be interpreted as an associated rather than a causal factor. Nevertheless, we provided evidence that deamidation at the critical interface of βA3-crystallin may decrease the homodimer interaction. In support of this finding, the same two deamidation sites were found to be involved in the interactions of βA3-crystallin with other β-crystallins [14]. However, the exact mechanistic understanding of deamidation-mediated protein insolubilization needs further investigations.
Perhaps the most interesting aspect of the current investigation is that our data demonstrated the common mechanisms for two distinct protein damages of βA3-crystallin. ΔG91 mutation altered predicted both the secondary structure and hydrophobic character of βA3-crystallin, while deamidation only exhibited a slight effect. It is important to recognize that these apparently distinct protein damages are linked by a common underlying mechanism, decreasing the homodimer interaction. It should be noted that both of the two deamidation sites are at the critical interface. This factor may explain why a minor modification such as deamidation could greatly decrease interaction even though the protein structure is not disturbed. This example also emphasized the crucial role of appropriate crystallin interactions for lens transparency and visual acuity [18]. Crystallins are known to be involved in various homogeneous or heterogeneous interactions and any subtle perturbation to a single crystallin molecule may amplify the deleterious effects at homo- and hetero-oligomer levels.
In conclusion, we elucidated the functional consequences of ΔG91 mutation and the deamidation of βA3-crystallin in terms of the folding properties and homodimer interactions. We provided evidence that both congenial mutations and age-related deamidations could commonly alter protein–protein interaction in human lens βA3-crystallin. Since β-crystallins are known to form both homo- and hetero-oligomers, current investigation can be extended forward to explore the mechanism of ΔG91 mutation and deamidation of βA3-crystallin on its interaction with other β-crystallin subunits.
Acknowledgments
Xiaorong Tan gratefully acknowledges the funding of the science foundation of the education department of Henan province (Grant No. 2006210002). We would like to thank the reviewers for very constructive comments. We would also like to thank Mr. Deyin Yang for his assistance on graphic preparation.
References
- 1.Resnikoff S, Pascolini D, Etya'ale D, Kocur I, Pararajasegaram R, Pokharel GP, Mariotti SP. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82:844–51. [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy MA, Bateman OA, Chakarova C, Ferris J, Berry V, Lomas E, Sarra R, Smith MA, Moore AT, Bhattacharya SS, Slingsby C. Characterization of the G91del CRYBA1/3-crystallin protein: a cause of human inherited cataract. Hum Mol Genet. 2004;13:945–53. doi: 10.1093/hmg/ddh110. [DOI] [PubMed] [Google Scholar]
- 3.Slingsby C, Bateman OA. Quaternary interactions in eye lens beta-crystallins: basic and acidic subunits of beta-crystallins favor heterologous association. Biochemistry. 1990;29:6592–9. doi: 10.1021/bi00480a007. [DOI] [PubMed] [Google Scholar]
- 4.Ajaz MS, Ma Z, Smith DL, Smith JB. Size of human lens beta-crystallin aggregates are distinguished by N-terminal truncation of betaB1. J Biol Chem. 1997;272:11250–5. doi: 10.1074/jbc.272.17.11250. [DOI] [PubMed] [Google Scholar]
- 5.Lu S, Zhao C, Jiao H, Kere J, Tang X, Zhao F, Zhang X, Zhao K, Larsson C. Two Chinese families with pulverulent congenital cataracts and deltaG91 CRYBA1 mutations. Mol Vis. 2007;13:1154–60. [PubMed] [Google Scholar]
- 6.Bateman JB, Geyer DD, Flodman P, Johannes M, Sikela J, Walter N, Moreira AT, Clancy K, Spence MA. A new betaA1-crystallin splice junction mutation in autosomal dominant cataract. Invest Ophthalmol Vis Sci. 2000;41:3278–85. [PubMed] [Google Scholar]
- 7.Ferrini W, Schorderet DF, Othenin-Girard P, Uffer S, Heon E, Munier FL. CRYBA3/A1 gene mutation associated with suture-sparing autosomal dominant congenital nuclear cataract: a novel phenotype. Invest Ophthalmol Vis Sci. 2004;45:1436–41. doi: 10.1167/iovs.03-0760. [DOI] [PubMed] [Google Scholar]
- 8.Kannabiran C, Rogan PK, Olmos L, Basti S, Rao GN, Kaiser-Kupfer M, Hejtmancik JF. Autosomal dominant zonular cataract with sutural opacities is associated with a splice mutation in the betaA3/A1-crystallin gene. Mol Vis. 1998;4:21. [PubMed] [Google Scholar]
- 9.Qi Y, Jia H, Huang S, Lin H, Gu J, Su H, Zhang T, Gao Y, Qu L, Li D, Li Y. A deletion mutation in the betaA1/A3 crystallin gene (CRYBA1/A3) is associated with autosomal dominant congenital nuclear cataract in a Chinese family. Hum Genet. 2004;114:192–7. doi: 10.1007/s00439-003-1049-7. [DOI] [PubMed] [Google Scholar]
- 10.Hains PG, Truscott RJ. Post-translational modifications in the nuclear region of young, aged, and cataract human lenses. J Proteome Res. 2007;6:3935–43. doi: 10.1021/pr070138h. [DOI] [PubMed] [Google Scholar]
- 11.Wilmarth PA, Tanner S, Dasari S, Nagalla SR, Riviere MA, Bafna V, Pevzner PA, David LL. Age-related changes in human crystallins determined from comparative analysis of post-translational modifications in young and aged lens: does deamidation contribute to crystallin insolubility? J Proteome Res. 2006;5:2554–66. doi: 10.1021/pr050473a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lampi KJ, Ma Z, Hanson SR, Azuma M, Shih M, Shearer TR, Smith DL, Smith JB, David LL. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp Eye Res. 1998;67:31–43. doi: 10.1006/exer.1998.0481. [DOI] [PubMed] [Google Scholar]
- 13.Takata T, Oxford JT, Brandon TR, Lampi KJ. Deamidation alters the structure and decreases the stability of human lens betaA3-crystallin. Biochemistry. 2007;46:8861–71. doi: 10.1021/bi700487q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takata T, Woodbury LG, Lampi KJ. Deamidation alters interactions of beta-crystallins in hetero-oligomers. Mol Vis. 2009;15:241–9. [PMC free article] [PubMed] [Google Scholar]
- 15.Pollastri G, Przybylski D, Rost B, Baldi P. Improving the prediction of protein secondary structure in three and eight classes using recurrent neural networks and profiles. Proteins. 2002;47:228–35. doi: 10.1002/prot.10082. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 17.Fisher CL, Pei GK. Modification of a PCR-based site-directed mutagenesis method. Biotechniques. 1997;23:570–1. doi: 10.2144/97234bm01. [DOI] [PubMed] [Google Scholar]
- 18.Fu L, Liang JJ. Detection of protein-protein interactions among lens crystallins in a mammalian two-hybrid system assay. J Biol Chem. 2002;277:4255–60. doi: 10.1074/jbc.M110027200. [DOI] [PubMed] [Google Scholar]
- 19.Fu L, Liang JJ. Alteration of protein-protein interactions of congenital cataract crystallin mutants. Invest Ophthalmol Vis Sci. 2003;44:1155–9. doi: 10.1167/iovs.02-0950. [DOI] [PubMed] [Google Scholar]
- 20.Liu BF, Liang JJ. Protein-protein interactions among human lens acidic and basic beta-crystallins. FEBS Lett. 2007;581:3936–42. doi: 10.1016/j.febslet.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graw J. Genetics of crystallins: cataract and beyond. Exp Eye Res. 2009;88:173–89. doi: 10.1016/j.exer.2008.10.011. [DOI] [PubMed] [Google Scholar]