Abstract
Background
Vibrio parahaemolyticus is a leading cause of seafood-related bacterial gastroenteritis and outbreaks worldwide. Sensitive and specific detection methods are needed to better control V. parahaemolyticus infections. This study aimed at developing a highly specific and sensitive loop-mediated isothermal amplification (LAMP) assay for detecting V. parahaemolyticus in oysters. A set of five LAMP primers, two outer, two inner, and one loop were designed based on the published V. parahaemolyticus toxR sequence. Specificity of the assay was evaluated using a panel of 36 V. parahaemolyticus and 39 other strains. The assay sensitivity was determined using serial dilutions of V. parahaemolyticus ATCC 27969 culture ranging from 108 CFU/ml to extinction. The assay was also tested in experimentally inoculated oyster samples.
Results
The toxR-based LAMP assay was able to specifically detect all of the 36 V. parahaemolyticus strains without amplification from 39 other strains. The detection limit was 47-470 cells per reaction in pure culture, up to 100-fold more sensitive than that of toxR-PCR. When applied in spiked oysters, the assay was able to detect 1.1 × 105 V. parahaemolyticus cells per gram of oyster without enrichment, up to 100-fold more sensitive than that of toxR-PCR. Standard curves generated for detecting V. parahaemolyticus in both pure culture and spiked oyster samples showed good linear relationship between cell numbers and the fluorescence or turbidity signals.
Conclusions
The toxR-based LAMP assay developed in this study was sensitive, specific, and quantitative, holding great potential for future field detection of V. parahaemolyticus in raw oysters.
Background
The Gram-negative, halophilic marine bacterium Vibrio parahaemolyticus has emerged as a major cause of seafood-associated outbreaks throughout the world and become a significant concern of seafood safety [1-3]. Shellfish, particularly oysters, has been frequently implicated in V. parahaemolyticus infections [4,5]. Typically within 24 h after eating contaminated seafood, V. parahaemolyticus causes acute, self-limiting gastroenteritis characterized by diarrhea, abdominal cramps, nausea, vomiting, fever, and chills, which lasts for 1-3 days [6]. Two hemolysins, the thermostable direct hemolysin (TDH) and the TDH-related hemolysin (TRH) are well-characterized virulence factors for pathogenic V. parahaemolyticus strains [7]. However, the majority of V. parahaemolyticus strains in the environment and seafood samples lack these two hemolysin genes [8-10], thus the number of total V. parahaemolyticus has been used as an indicator for preventing V. parahaemolyticus infections from seafood consumption [11,12].
Traditional culture-based methods for isolating and enumerating V. parahaemolyticus from seafood samples involve the most probable number (MPN) technique [13]. Although widely used, such methods are labor-intensive and time-consuming (4-7 days). Molecular-based methods such as DNA probe hybridization and PCR assays have been developed for V. parahaemolyticus and yielded rapid and specific results [14-18]. However, the probe hybridization procedure and the gel electrophoresis technique used to analyze PCR amplicons are tedious and time-consuming. Recently, several real-time PCR assays have been developed for the detection of V. parahaemolyticus with increased speed and sensitivity [12,19-21]. Nonetheless, these assays require a dedicated real-time PCR machine, which is rather expensive and not yet widely available.
Loop-mediated isothermal amplification (LAMP), a novel DNA amplification technique invented in 2000 [22], has since been applied in detecting many bacterial and viral agents [23-26]. Because the LAMP assay was carried out under isothermal conditions, a simple heater that maintains a constant temperature (60-65°C) is sufficient. LAMP assays were reported to be highly specific, sensitive, rapid, and cost-effective [23-26]. Very recently, LAMP was adopted to detect V. parahaemolyticus and yielded promising results [11]. However, in this LAMP assay, primers were designed to target the V. parahaemolyticus thermolabile hemolysin gene (tlh), and an increasing number of hemolysin gene sequences with various levels of identity to this gene have been described in other vibrios such as Vibrio harveyi and Vibrio campbellii [27,28]. A search of the GenBank also revealed significant homologies among these hemolysin genes http://www.ncbi.nih.gov/BLAST. Additionally, Croci et al. [29] evaluated several PCR assays for the identification of V. parahaemolyticus by targeting different genes. Among 48 V. parahaemolyticus and 115 other Vibrio spp. strains examined, the two tlh-based PCR protocols [13,14] obtained 100% inclusivity but had 50% and 91% exclusivity, respectively. In contrast, a toxR-based PCR assay [18] simultaneously evaluated in the same study achieved 100% for both inclusivity and exclusivity [29].
The toxR gene was initially described in V. cholerae as the regulatory gene for the cholera toxin and other virulence determinants [30], and was subsequently found in V. parahaemolyticus [31]. Although present in many Vibrio spp., a membrane "tether" region within the coding sequence of toxR possesses significant heterogeneity and could be used to distinguish various Vibrio species [32]. The objective of this study was to develop a highly specific and sensitive toxR-based LAMP assay for the detection of V. parahaemolyticus in raw oyster samples.
Results
Specificity of the LAMP assay
The V. parahaemolyticus toxR-based LAMP assay run on two platforms by using either a real-time PCR machine or a real-time turbidimeter successfully detected 36 V. parahaemolyticus strains while showing negative results for 39 non- V. parahaemolyticus strains (Table 1), indicating that the toxR-based LAMP assay was highly specific. On the real-time PCR platform, mean cycle threshold (Ct; cycles when fluorescence signals reach 30 units) values for the 36 V. parahaemolyticus clinical and environmental strains ranged between 13.58 and 23.95 min, with an average of 17.54 ± 2.27 min. The melting temperatures (Mt) for these strains consistently fell between 81.25 and 82.55°C, with an average Mt of 81.97 ± 0.25°C. For the 39 non- V. parahaemolyticus strains, no Ct value was obtained, with melting curve analysis showing no peaks, suggesting no amplification occurred.
Table 1.
Bacterial strains used in this study
| Strain group | Strain ID and serotype a | Source and reference |
|---|---|---|
| V. parahaemolyticus | ATCC 17802; O1:K1 | Shirasu food poisoning, Japan |
| (n = 36) | ATCC 27969 | Blue crab, Maryland |
| ATCC 33847 | Gastroenteritis, Maryland | |
| ATCC 49529; O4:K12 | Feces, California | |
| CT-6636; O3:K6 | Clinical, Connecticut | |
| M350A; O5 | Oyster, Washington | |
| NY477; O4:K8 | Oyster, New York | |
| TX-2103; O3:K6 | Clinical, Texas | |
| 8332924; O1:K56 | Oyster, Gulf of Mexico | |
| 83AO8757 | Clinical, feces | |
| 83AO9148 | Clinical, feces | |
| 83AO9756; O4:K12 | Clinical, feces | |
| 84AO1516; O4:K12 | Clinical, feces | |
| 84AO4226 | Clinical, feces | |
| 916i, 916e, 541-0-44c, V68, V69, V154, V155, V166 | Oyster, Gulf, Louisiana [10] | |
| V5, V15, V16, V32, V38, V39, V50, V86, V150, V426, V427, V428, V429, V430 | Oyster, Retail, Louisiana [10] | |
| V. vulnificus | ATCC 27562 | Blood, Florida |
| (n = 18) | ATCC 29306 | Corneal ulcer, Virginia |
| ATCC 33815 | Leg ulcer, Wisconsin | |
| ATCC 33816 | Blood, Alaska | |
| C7184 | Thumb drainage, Texas [39] | |
| 1003 | Wound, Louisiana [40] | |
| 1006 | Blood, Louisiana [40] | |
| WR1 | Sea water, Washington | |
| 515-4C2 | Oyster, California | |
| 541-0-84c | Gulf oyster, Louisiana [10] | |
| V373, V416, V578, V598 | Retail oyster, Louisiana [10] | |
| 132A1, 132T5, 212B6, 342E3 | Gulf oyster, Louisianab | |
| Other Vibrio spp. (n = 10) | ||
| Vibrio alginolyticus | ATCC 17749 | Spoiled horse mackerel, Japan |
| ATCC 33787 | Seawater, Hawaii | |
| Vibrio cholerae | ATCC 14035; O:1 | United Kingdom |
| Vibrio cincinnatiensis | ATCC 35912 | Blood/cerebrospinal fluid, Ohio |
| Vibrio fluvialis | ATCC 33809 | Human feces, Bangladesh |
| Vibrio harveyi | ATCC 14126 | Dead amphipod, Massachusetts |
| ATCC 35084 | Brown shark, Maryland | |
| Vibrio mimicus | ATCC 33653 | Human ear, North Carolina |
| ATCC 33655 | Feces, Tennessee | |
| Vibrio natriegens | ATCC 14048 | Salt marsh mud, Georgia |
| Non-Vibrio spp. (n = 11) | ||
| Campylobacter jejuni | 81-176 | Human |
| Enterobacter aerogenes | ATCC 13048 | Sputum, South Carolina |
| Enterococcus faecalis | ATCC 29212 | Urine |
| Escherichia coli | ATCC 25922 | Human |
| Listeria monocytogenes | ATCC 13932; 4b | Spinal fluid, Germany |
| Pseudomonas aeroginosa | ATCC 27853 | Human blood |
| Salmonella enterica | LT2; Typhimurium | Unknown |
| Shigella flexneri | ATCC 12022; 2b | Unknown |
| Shigella sonnei | ATCC 25931 | Human feces, Panama |
| Staphylococcus aureus | ATCC 29213 | Wound |
| Streptococcus pneumoniae | ATCC 49619; type 59 | Sputum, Arizona |
a ATCC, American Type Culture Collection, Manassas, VA.
b Isolated from three Louisiana coastal locations (designated as 132, 212, and 342) between 2006-2007.
On the real-time turbidimeter platform, time threshold (Tt; time when turbidity values reach 0.1) values for the 36 V. parahaemolyticus clinical and environmental strains ranged from 28.3 to 33.5 min with an average of 31.13 ± 1.67 min. For the 39 non- V. parahaemolyticus strains, no Tt value was obtained, indicating negative results for V. parahaemolyticus toxR-based LAMP assay.
Similarly, no false positive or false negative results for the 75 bacterial strains were observed by PCR using two primer sets, F3/B3 and toxR-PCR primers (Table 2), indicating good specificity.
Table 2.
LAMP and PCR primers used in this study to detect Vibrio parahaemolyticus
| Primer name | Sequence (5'-3') | Positiona | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| F3 | TTGGATTCCACGCGTTAT | 528-545 | Ladder-like bands for LAMP; 183 bp for F3/B3 PCR | This study |
| B3 | CGTTCAATGCACTGCTCA | 693-710 | ||
| FIP | TGAGATTCCGCAGGGTTTGTAA TTATTTTTGGCACTATTACTACCG |
587-608 (F1c) 547-570 (F2) |
||
| BIP | GTTCCGTCAGATTGGTGAGTATC TAGAAGGCAACCAGTTGTT |
609-631(B1c) 673-691(B2) |
||
| Loop | AGAACGTACCAGTGATGACACC | 632-653 | ||
| toxR-F | GTCTTCTGACGCAATCGTTG | 453-472b | 367b | [18] |
| toxR-R | ATACGAGTGGTTGCTGTCATG | 799-819b |
a The positions are numbered based on the coding sequence of V. parahaemolyticus strain AQ3815 toxR gene [GenBank: L11929].
b Differences in primer positions and amplicon size were noted from those originally published after reanalysis of the sequences.
Sensitivity of the LAMP assay
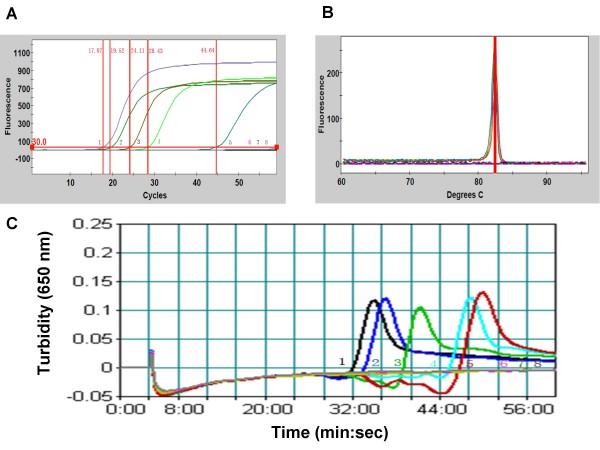
Figure 1 presents sensitivity of the toxR-based LAMP assay when testing 10-fold serial dilutions of V. parahaemolyticus ATCC 27969 DNA templates. A representative optic graph and corresponding melting curve analysis for the real-time PCR platform and a representative turbidity graph for the real-time turbidimeter platform are shown in Figure 1A-1C, respectively. On the real-time PCR platform, for templates ranging in concentration from 4.7 × 105 to 4.7 × 101 CFU per reaction tube, the average Ct values of six repeats ranged from 17.35 to 40.72 min, with melting temperatures consistently falling at around 83°C. No amplification was obtained for the 4.7 CFU and 4.7 × 10-1 CFU templates. Therefore, the detection limit of the toxR-based LAMP assay run in a real-time PCR machine was approximately 47 CFU per reaction. In the real-time turbidimeter platform, the average Tt values fell between 34.43 and 49.07 min for templates ranging from 4.7 × 105 to 4.7 × 102 CFU per reaction tube. In two out of six repeats, amplification of the 4.7 × 101 CFU template occurred (Figure 1C). Therefore, the lower limit of detection for turbidity-based real-time LAMP assay was 47-470 CFU per reaction.
Figure 1.
Sensitivity of the LAMP assay when detecting Vibrio parahaemolyticus ATCC 27969 in pure culture. (A) A representative optic graph generated using the real-time PCR machine; (B) Corresponding melting curve analysis for samples in (A); (C) A representative turbidity graph generated using the real-time turbidimeter. Samples 1-7 corerspond to serial 10-fold dilutions of V. parahaemolyticus ATCC 27969 cells ranging from 4.7 × 105 to 4.7 × 10-1 CFU/reaction; sample 8 is water.
The two PCR assays used to test the same set of V. parahaemolyticus ATCC 27969 templates by using F3/B3 and toxR-PCR primers had the same level of sensitivity, approximately 4.7 × 103 CFU per reaction tube (data not shown), i.e., up to 100-fold less sensitive than the toxR-based LAMP assay.
Quantitative capability of LAMP for detecting V. parahaemolyticus in pure culture
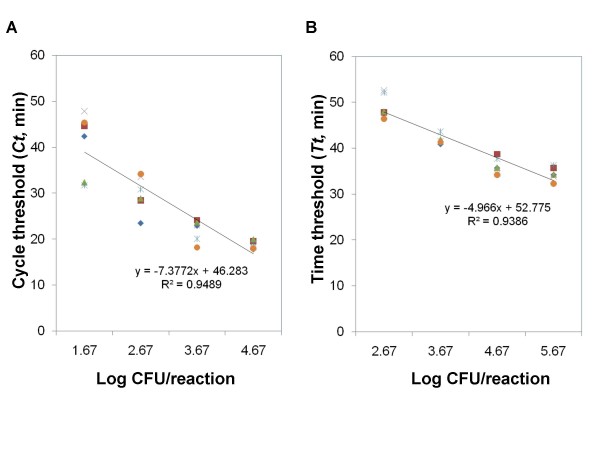
Figure 2 shows the standard curves generated when detecting V. parahaemolyticus ATCC 27969 in pure culture based on six independent repeats in both real-time PCR machine (Figure 2A) and a real-time turbidimeter (Figure 2B). On the real-time PCR platform, the correlation coefficient (r2) was calculated to be 0.95. When run in the real-time turbidimeter platform, the toxR-based LAMP assay had an r2 value of 0.94.
Figure 2.
Standard curves generated when detecting Vibrio parahaemolyticus ATCC 27969 in pure culture. (A) Based on six independent repeats in a real-time PCR machine; (B) Based on six independent repeats in a real-time turbidimeter.
Detection of V. parahaemolyticus cells in spiked oysters
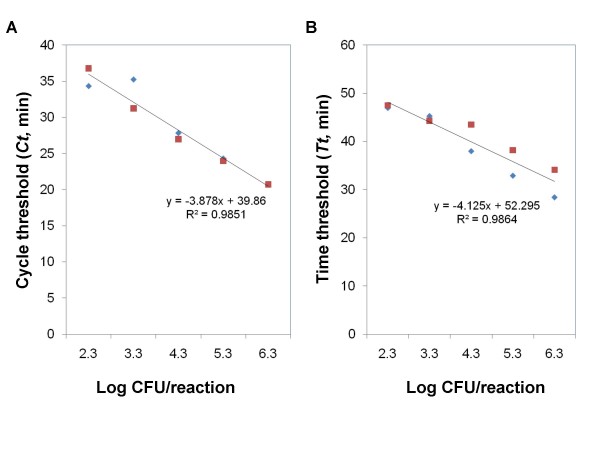
The sensitivity of detecting V. parahaemolyticus ATCC 27969 cells in spiked oyster samples is shown in Table 3. In three independent spiking experiments, the toxR-based LAMP assay using the two platforms consistently detected down to 200 V. parahaemolyticus cells (i.e., 1.1 × 105 CFU/g) in spiked oyster samples without enrichment. However, for the two PCR assays using F3/B3 and toxR-PCR primers, the lowest detection limit achieved was 1.1 × 106 CFU/g and 1.1 × 107 CFU/g, which were up to 100-fold less sensitive than that of the toxR-based LAMP assay. Standard curves (Figure 3) generated for the quantitative detection of V. parahaemolyticus cells in spiked oyster samples had an r2 value of 0.99 for both real-time LAMP platforms.
Table 3.
Comparison of quantitatively detecting Vibrio parahaemolyticus ATCC 27969 in spiked oysters by using the toxR-based LAMP assay in two platforms and PCRa
| Rep. | Levels of spiking (CFU/g) | Amount of cellsb (CFU/rxn) | LAMP | PCR | |||
|---|---|---|---|---|---|---|---|
| Fluorescence-based | Turbidity-based | F3/B3 | toxR | ||||
| Ct (min) | Mt (°C) | Tt (min) | |||||
| 1 | 5.6 × 108 | 1.0 × 106 | 20.61 ± 2.04 | 82.16 ± 0.05 | 31.2 ± 2.97 | + | + |
| 5.6 × 107 | 1.0 × 105 | 22.02 ± 2.04 | 81.36 ± 1.20 | 35.3 ± 1.13 | + | + | |
| 5.6 × 106 | 1.0 × 104 | 25.26 ± 0.56 | 81.87 ± 0.10 | 42.55 ± 2.2 | + | + | |
| 5.6 × 105 | 1.0 × 103 | 34.58 ± 2.25 | 82.45 ± 0.23 | 52.45 ± 2.75 | + | - | |
| 5.6 × 104 | 1.0 × 102 | - | - | - | - | - | |
| 5.6 × 103 | 10 | - | - | - | - | - | |
| 2 | 1.7 × 108 | 3.1 × 105 | 21.78 ± 0.59 | 82.41 ± 0.11 | 29.4 ± 0.85 | + | + |
| 1.7 × 107 | 3.1 × 104 | 23.68 ± 0.16 | 82.25 ± 0.10 | 33.25 ± 0.35 | + | + | |
| 1.7 × 106 | 3.1 × 103 | 29.08 ± 0.45 | 82.60 ± 0.34 | 40.4 ± 4.67 | + | - | |
| 1.7 × 105 | 3.1 × 102 | 31.77 ± 2.23 | 82.50 ± 0.18 | 47.7 ± 1.27 | - | - | |
| 1.7 × 104 | 31 | - | - | - | - | - | |
| 1.7 × 103 | 3.1 | - | - | - | - | - | |
| 3 | 1.1 × 109 | 2.0 × 106 | 20.74 ± 0.03 | 82.48 ± 0.01 | 31.25 ± 4.02 | + | + |
| 1.1 × 108 | 2.0 × 105 | 24.14 ± 0.24 | 82.37 ± 0.05 | 35.55 ± 3.73 | + | + | |
| 1.1 × 107 | 2.0 × 104 | 27.42 ± 0.60 | 82.48 ± 0.11 | 40.75 ± 3.88 | + | + | |
| 1.1 × 106 | 2.0 × 103 | 33.26 ± 2.84 | 82.50 ± 0.26 | 44.8 ± 0.7 | + | - | |
| 1.1 × 105 | 2.0 × 102 | 35.57 ± 1.73 | 82.65 ± 0.09 | 47.25 ± 0.35 | - | - | |
| 1.1 × 104 | 20 | - | - | - | - | - | |
Bolded are detection limits by each assay.
a For each independently prepared template, two times of LAMP reactions were performed and the data presented are means ± standard deviations for the 2 LAMP repeats.
b CFU/reaction was calculated by using CFU/g × 0.09 g/ml × 10 × 2 × 10-3, i.e., CFU/g × 1.8 × 10-3.
Figure 3.
Standard curves generated when testing Vibrio parahaemolyticus ATCC 27969 in spiked oysters. Three sets of independent spiking experimetns were performed, and the LAMP reactions were repeated two times for each inoculation set. The data shown are for the inoculation set 3 ranging from 1.1 × 105 to 1.1 × 109 CFU/g. (A) The assay was run in a real-time PCR machine; (B) The assay was run in a real-time turbidimeter.
Discussion
In this study, we designed a set of five LAMP primers to specifically target the V. parahaemolyticus toxR gene, a gene previously shown to possess better specificity for V. parahaemolyticus detection by PCR than other target genes, such as tlh and gyrB [29]. We also developed real-time LAMP assays using two platforms - a real-time PCR machine and a real-time turbidimeter to quantitatively detect V. parahaemolyticus in pure culture and spiked oyster samples. This is the first report demonstrating the efficacy of a toxR-based LAMP assay for detecting V. parahaemolyticus in oysters.
The LAMP primers were selected from regions of the V. parahaemolyticus toxR gene coding sequence that are highly specific to V. parahaemolyticus [18,32]. The five primers (F3, B3, FIP, BIP, and loop) targeted seven regions of V. parahaemolyticus toxR (Table 2), providing additional levels of specificity compared to PCR primers (targeting two regions). Among a total of 36 V. parahaemolyticus and 39 non-V. parahaemolyticus strains tested, the toxR-based LAMP assay run on both real-time platforms obtained 100% inclusivity and 100% exclusivity. This level of specificity was the same as that of two toxR-based PCR assays evaluated simultaneously in this study and that of a tlh-based LAMP assay developed by Yamazaki et al. [11]. Future pairwise comparison of the two LAMP assays (toxR-based and tlh-based) using an extensive collection of Vibrio strains as done previously for PCR [29] would be desired to further evaluate the performance of the two LAMP assays on both inclusivity and exclusivity.
When comparing the sensitivity of LAMP with PCR, the toxR-LAMP assays were able to detect 47-470 V. parahaemolyticus cells per reaction tube, in contrast to 4.7 × 103 cells for toxR-PCR. Similarly, the tlh-based LAMP assay for V. parahaemolyticus was reported to be 10-fold more sensitive than PCR, with a detection limit of 2 CFU per reaction for LAMP [11]. In a recent report on the detection of pathogenic V. parahaemolyticus by targeting the tdh gene, both LAMP and PCR were capable of detecting less than 1 CFU of TDH-producing V. parahaemolyticus in a reaction tube, although for different serotypes tested, slight difference in terms of sensitivity was observed [33]. Additionally, several studies on the detection of other Vibrio spp. also found LAMP to be 10-fold more sensitive than PCR [23,34,35].
Running the toxR-LAMP assay in a real-time PCR machine consistently achieved a lower limit of detection of 47 cells per reaction, whereas in a real-time turbidimeter, a detection limit of 47 cells was only occasionally achieved (2 out of 6 attempts). In addition, the average time to positive results as indicated by Ct (17.54 min) for the real-time PCR platform was markedly shorter than that of the real-time turbidimeter platform as indicated by Tt (31.13 min), suggesting that the real-time LAMP assay based on fluorescence was faster and slightly more sensitive than that based on turbidity. This finding agrees with a previous study which reported that a fluorescent intercalation dye (YO-PRO-1)-based real-time LAMP was 10-fold more sensitive and faster than a turbidimetry real-time LAMP [36]. However, in that report, the fluorescence-based LAMP assay was found to generate anomalous and irreproducible results in low-concentration templates (less than 1 × 103 copies), which could be due to the effect of the intercalating dye on DNA amplification efficiency [36].
In this study, we chose SYTO-9 as the intercalating dye for the real-time PCR platform instead of the commonly used real-time PCR dye SYBR Green I. Based on a previous study [37] comparing the use of these two dyes in real-time PCR, SYTO-9 was found to generate highly reproducible DNA melting curves over a broader range of dye concentrations than SYBR Green I and was far less inhibitory. We also evaluated the use of EvaGreen (Biotium, Hayward, CA) as the intercalating dye on the real-time PCR platform for LAMP, but found it to be inhibitory for LAMP amplifications (data not shown).
The strong linear correlation (r2 = 0.94-0.99) between the number of V. parahaemolyticus cells in the LAMP reaction and the associated Ct or Tt values over a dynamic range of template concentrations (101 to 106 cells) illustrates the quantitative capability of the toxR-based real-time LAMP assays when detecting this organism in both pure culture and spiked oysters. Very few reports have examined the quantitative ability of LAMP. One study monitoring ammonia-oxidizing bacteria using LAMP also reported it to possess good quantitative capability between 1 × 104 and 1 × 1010 DNA copies [36].
In spiked oyster samples, we found the detection limit of the toxR-based LAMP assay to be 200 V. parahaemolyticus cells per reaction, which translates to 1.1 × 105 cells per gram of oyster sample. In contrast, the detection limit of the tlh-based LAMP in spike shrimp samples was reported to be 5.3 × 102 CFU/g (2 CFU/reaction) [11]. The U.S. Food and Drug Administration requires that all postharvest-processed oysters have lower than 30 MPN/g of either V. vulnificus or V. parahaemolyticus [38]. This indicates that without enrichment, DNA amplification assays such as LAMP, although potentially quantitative, lack the needed sensitivity when applied to food samples [23]. Therefore, combining MPN overnight enrichment [19] or pre-enrichment for 6 h [33] with LAMP or other DNA amplification assays is a desirable approach to achieve the needed sensitivity.
When testing spiked oyster samples, we observed the time to positive samples (Ct for the real-time PCR platform and Tt for the real-time turbidimeter) was delayed several minutes compared to pure culture samples and the detection limit was higher (200 V. parahaemolyticus cells in oyster samples vs. 47 cells in pure culture). Nonetheless, no extensive sample preparation other than homogenization and two simple centrifugation steps was required. This significantly reduced the total assay time. Combined with less than 1 h for the real-time LAMP assay, the complete LAMP detection system was markedly faster than either PCR or conventional methods.
Conclusions
The toxR-based real-time LAMP assay developed in this study was a highly specific, sensitive, and rapid method for the detection of V. parahaemolyticus in oysters. Future testing with natural or commercial oyster samples is desired to further evaluate the efficacy of the assay in detecting V. parahaemolyticus in oysters in a field setting.
Methods
Bacterial strains and DNA templates preparation
Strains used in this study (Table 1) were maintained in Luria-Bertani broth (BD Diagnostic Systems, Sparks, MD) containing 30% glycerol at -80°C. V. parahaemolyticus ATCC 27969, originally isolated from blue crab hemolymph was used for sensitivity testing. Additional 35 V. parahaemolyticus clinical and environmental strains and 39 non- V. parahaemolyticus strains were used to evaluate assay specificity. All Vibrio strains were routinely cultured using trypticase soy agar or broth (TSA or TSB; BD Diagnostic Systems) supplemented with 2% NaCl at 35°C overnight. Non-Vibrio strains were grown on Luria-Bertani agar or blood agar (BD Diagnostic Systems).
To prepare DNA template, a single bacterial colony grown on appropriate agar plates was suspended in 500 μl of TE buffer (10 mM Tris, pH 8.0; 1 mM EDTA; Sigma-Aldrich, St. Louis, MO) and heated at 95°C for 10 min in a dry heating block. The crude cell lysate was centrifuged at 12,000 g for 2 min and the supernatant was stored at -20°C until use.
LAMP primers and reaction conditions
The V. parahaemolyticus toxR gene [GenBank: L11929] was used as the target for LAMP primer design. Five primers, two outer (F3 and B3), two inner (FIP and BIP), and one loop (Loop) which recognized seven distinct regions of the target sequence were designed using the PrimerExplorer software version 4 (Fujitsu Limited, Japan; http://primerexplorer.jp/e. Oligonucleotide sequences and locations of the primers are shown in Table 2. The primers were synthesized by Invitrogen (Carlsbad, CA).
The LAMP reaction mix in a 25 μl total volume consisted of the following: 1 × Thermo buffer, 6 mM of MgSO4, 0.8 M of betaine (Sigma-Aldrich), 1.4 mM of deoxynucleotide triphosphate (dNTP), 0.2 μM of each outer primer (F3 and B3), 1.6 μM of each inner primer (FIP and BIP), 0.8 μM of the loop primer, 8 U of Bst DNA polymerase (New England Biolabs, Ipswich, MA), and 2 μl of DNA template. Additionally, 0.4 μM of SYTO-9 green fluorescent dye (Invitrogen) was added when the LAMP reaction was carried out in a real-time PCR machine as described below.
Two platforms were used to run the LAMP reactions. On the first platform, a real-time PCR machine (SmartCycler II System; Cepheid, Sunnyvale, CA) was used and the SYTO-9 green fluorescent dye was added. The assay was conducted at 63°C for 1 h. Fluorescence readings were acquired every 60 s using the FAM channel (excitation at 450-495 nm and detection at 510-527 nm), followed by melting curve analysis from 63°C to 96°C with 0.2°C increment per second. The fluorescence threshold unit was set to be 30. On the second platform, the LAMP reaction was carried out in a Loopamp real-time turbidimeter (LA-320C; Teramecs, Kyoto, Japan) at 63°C for 1 h and terminated at 80°C for 5 min. Turbidity readings at 650 nm were obtained real-time and a turbidity threshold value of 0.1 was used. A negative control was included for each LAMP run.
PCR
As a comparison, two sets of PCR reactions were performed, one using LAMP outer primers (F3 and B3) and the other one using the toxR-PCR primers (Table 2) published previously [18]. Each PCR mix in a 25 μl total volume contained 1 × PCR buffer, 0.2 mM of each dNTP, 1.5 mM of MgCl2, 0.5 μM of each forward and reverse primer, 0.625 U of GoTaq Hot Start Polymerase (Promega, Madison, WI), and 2 μl of DNA template. The PCR reactions were conducted using initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 94°C for 1 min, primer annealing at 60°C (50°C for F3/B3 primers) for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 7 min in a Bio-Rad C1000 Thermal Cycler (Hercules, CA). Aliquots (10 μl) of PCR products were analyzed by electrophoresis on 1.5% agarose gel containing ethidium bromide, and visualized under UV light. Gel images were documented by a Gel Doc XR system (Bio-Rad).
LAMP specificity and sensitivity
Seventy-five bacterial strains (Table 1) were used to determine the LAMP specificity. DNA templates were made from fresh overnight bacterial cultures and aliquots (2 μl) were subjected to both LAMP and PCR amplifications. Specificity tests were repeated twice.
To determine LAMP sensitivity, serial 10-fold dilutions (ca. 108 CFU/ml to extinction) of a mid-log phase V. parahaemolyticus ATCC 27969 culture grown in TSB were prepared in phosphate buffered saline (PBS; BD Diagnostic Systems) and quantified using the standard plating method. DNA templates were prepared from each dilution by the boiling method described above and aliquots (2 μl) were subjected to both LAMP and PCR amplifications. Sensitivity tests were repeated six times and the lower limits of detection (CFU/reaction) were reported. Standard curves were generated by plotting Ct (cycle threshold; for the real-time PCR platform) or Tt (time threshold; for the real-time turbidimeter platform) values against log CFU/reaction and the linear regression was calculated using the Microsoft Excel Software (Seattle, WA).
LAMP testing in experimentally inoculated oyster samples
Oyster samples were obtained from local seafood restaurants and determined to be V. parahaemolyticus-negative as described previously [10]. Oyster samples were processed following a previous study with slight modifications [11]. Briefly, 25 g of oyster sample was mixed with 225 ml of alkaline peptone water (APW; BD Diagnostic Systems) and homogenized in a food stomacher (Model 400; Tekmar Company, Cincinnati, OH) for 90 s to generate 1:10 oyster in APW homogenate. Serial 10-fold dilutions of a mid-log phase V. parahaemolyticus ATCC 27969 culture were prepared in PBS as described above. Aliquots (100 μl) of each dilution were inoculated into 900 μl of the 1:10 oyster in APW homogenate. The spiked oyster samples were mixed well and centrifuged at 900 g for 1 min to remove oyster tissues. The supernatants were transferred to a fresh tube and centrifuged at 10,000 g for 5 min to pellet bacterial cells. After removing the supernatants, pellets were resuspended in 100 μl of TE and boiled for templates as described above. Aliquots (2 μl) of the supernatant were used for both LAMP and PCR amplifications. The spiked oyster sensitivity tests were repeated three times and the lower limits of detection (CFU/g) were reported. Standard curves were generated similarly as in pure culture sensitivity testing.
Authors' contributions
SC carried out the LAMP and PCR assays, conducted data analysis, and drafted the manuscript; SC and BG conceived of the study and participated in its design. BG coordinated the study and helped to finalize the manuscript. Both authors read and approved the final manuscript.
Contributor Information
Siyi Chen, Email: csiyi1@lsu.edu.
Beilei Ge, Email: bge@lsu.edu.
Acknowledgements
We thank Feifei Han for technical support and helpful discussions. This study was supported in part by funding from the Louisiana Sea Grant Office under a Program Developmental Project R/PMO-20-PD.
References
- Butt AA, Aldridge KE, Sanders CV. Infections related to the ingestion of seafood Part I: Viral and bacterial infections. Lancet Infect Dis. 2004;4(4):201–212. doi: 10.1016/S1473-3099(04)00969-7. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Preliminary FoodNet Data on the incidence of infection with pathogens transmitted commonly through food--10 States, 2008. MMWR Morb Mortal Wkly Rep. 2009;58(13):333–337. [PubMed] [Google Scholar]
- Su YC, Liu C. Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 2007;24(6):549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- Altekruse SF, Bishop RD, Baldy LM, Thompson SG, Wilson SA, Ray BJ, Griffin PM. Vibrio gastroenteritis in the US Gulf of Mexico region: the role of raw oysters. Epidemiol Infect. 2000;124(3):489–495. doi: 10.1017/S0950268899003714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePaola A, Kaysner CA, Bowers J, Cook DW. Environmental investigations of Vibrio parahaemolyticus in oysters after outbreaks in Washington, Texas, and New York (1997 and 1998) Appl Environ Microbiol. 2000;66(11):4649–4654. doi: 10.1128/AEM.66.11.4649-4654.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Vibrio parahaemolyticus infections associated with consumption of raw shellfish--three states, 2006. MMWR Morb Mortal Wkly Rep. 2006;55(31):854–856. [PubMed] [Google Scholar]
- Iida T, Park K, Honda T. In: The biology of vibrios. Thompson FL, Austin B, Swings J, editor. Washington, DC: ASM Press; 2006. Vibrio parahaemolyticus; pp. 341–348. [Google Scholar]
- Cook DW, Oleary P, Hunsucker JC, Sloan EM, Bowers JC, Blodgett RJ, Depaola A. Vibrio vulnificus and Vibrio parahaemolyticus in U.S. retail shell oysters: a national survey from June 1998 to July 1999. J Food Prot. 2002;65(1):79–87. doi: 10.4315/0362-028x-65.1.79. [DOI] [PubMed] [Google Scholar]
- DePaola A, Nordstrom JL, Bowers JC, Wells JG, Cook DW. Seasonal abundance of total and pathogenic Vibrio parahaemolyticus in Alabama oysters. Appl Environ Microbiol. 2003;69(3):1521–1526. doi: 10.1128/AEM.69.3.1521-1526.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Walker RD, Janes ME, Prinyawiwatkul W, Ge B. Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl Environ Microbiol. 2007;73(21):7096–7098. doi: 10.1128/AEM.01116-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki W, Ishibashi M, Kawahara R, Inoue K. Development of a loop-mediated isothermal amplification assay for sensitive and rapid detection of Vibrio parahaemolyticus. BMC Microbiol. 2008;8:163. doi: 10.1186/1471-2180-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Iwade Y, Konuma H, Hara-Kudo Y. Development of a quantitative real-time PCR method for estimation of the total number of Vibrio parahaemolyticus in contaminated shellfish and seawater. J Food Prot. 2005;68(5):1083–1088. doi: 10.4315/0362-028x-68.5.1083. [DOI] [PubMed] [Google Scholar]
- Kaysner CA, DePaola A. Bacteriological Analytical Manual online. 9. Chapter 9. U.S Food and Drug Administration, Rockville, MD; 2001. Vibrio. [Google Scholar]
- Bej AK, Patterson DP, Brasher CW, Vickery MC, Jones DD, Kaysner CA. Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods. 1999;36(3):215–225. doi: 10.1016/S0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- Gooch JA, DePaola A, Kaysner CA, Marshall DL. Evaluation of two direct plating methods using nonradioactive probes for enumeration of Vibrio parahaemolyticus in oysters. Appl Environ Microbiol. 2001;67(2):721–724. doi: 10.1128/AEM.67.2.721-724.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Pan SF, Chen CH. Sequence of a cloned pR72H fragment and its use for detection of Vibrio parahaemolyticus in shellfish with the PCR. Appl Environ Microbiol. 1995;61(4):1311–1317. doi: 10.1128/aem.61.4.1311-1317.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswaran K, Dohmoto N, Harayama S. Cloning and nucleotide sequence of the gyrB gene of Vibrio parahaemolyticus and its application in detection of this pathogen in shrimp. Appl Environ Microbiol. 1998;64(2):681–687. doi: 10.1128/aem.64.2.681-687.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol. 1999;37(4):1173–1177. doi: 10.1128/jcm.37.4.1173-1177.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordstrom JL, Vickery MC, Blackstone GM, Murray SL, DePaola A. Development of a multiplex real-time PCR assay with an internal amplification control for the detection of total and pathogenic Vibrio parahaemolyticus bacteria in oysters. Appl Environ Microbiol. 2007;73(18):5840–5847. doi: 10.1128/AEM.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Lee GG, Kim J, Kwon JY, Kwon ST. The development of rapid real-time PCR detection system for Vibrio parahaemolyticus in raw oyster. Lett Appl Microbiol. 2008;46(6):649–654. doi: 10.1111/j.1472-765X.2008.02368.x. [DOI] [PubMed] [Google Scholar]
- Ward LN, Bej AK. Detection of Vibrio parahaemolyticus in shellfish by use of multiplexed real-time PCR with TaqMan fluorescent probes. Appl Environ Microbiol. 2006;72(3):2031–2042. doi: 10.1128/AEM.72.3.2031-2042.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Ge B. Evaluation of a loop-mediated isothermal amplification assay for detecting Vibrio vulnificus in raw oysters. Foodborne Pathog Dis. 2008;5(3):311–320. doi: 10.1089/fpd.2008.0084. [DOI] [PubMed] [Google Scholar]
- Hara-Kudo Y, Yoshino M, Kojima T, Ikedo M. Loop-mediated isothermal amplification for the rapid detection of Salmonella. FEMS Microbiol Lett. 2005;253(1):155–161. doi: 10.1016/j.femsle.2005.09.032. [DOI] [PubMed] [Google Scholar]
- Parida M, Posadas G, Inoue S, Hasebe F, Morita K. Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of West Nile virus. J Clin Microbiol. 2004;42(1):257–263. doi: 10.1128/JCM.42.1.257-263.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh HY, Shoemaker CA, Klesius PH. Sensitive and rapid detection of Flavobacterium columnare in channel catfish Ictalurus punctatus by a loop-mediated isothermal amplification method. J Appl Microbiol. 2006;100(5):919–925. doi: 10.1111/j.1365-2672.2006.02853.x. [DOI] [PubMed] [Google Scholar]
- San BB Luis, Hedreyda CT. Analysis of a gene (vch) encoding hemolysin isolated and sequenced from Vibrio campbellii. J Gen Appl Microbiol. 2006;52(6):303–313. doi: 10.2323/jgam.52.303. [DOI] [PubMed] [Google Scholar]
- Zhang XH, Meaden PG, Austin B. Duplication of hemolysin genes in a virulent isolate of Vibrio harveyi. Appl Environ Microbiol. 2001;67(7):3161–3167. doi: 10.1128/AEM.67.7.3161-3167.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croci L, Suffredini E, Cozzi L, Paniconi M, Ciccaglioni G, Colombo MM. Evaluation of different polymerase chain reaction methods for the identification of Vibrio parahaemolyticus strains isolated by cultural methods. J AOAC Int. 2007;90(6):1588–1597. [PubMed] [Google Scholar]
- Miller VL, Taylor RK, Mekalanos JJ. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell. 1987;48(2):271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- Lin Z, Kumagai K, Baba K, Mekalanos JJ, Nishibuchi M. Vibrio parahaemolyticus has a homolog of the Vibrio cholerae toxRS operon that mediates environmentally induced regulation of the thermostable direct hemolysin gene. J Bacteriol. 1993;175(12):3844–3855. doi: 10.1128/jb.175.12.3844-3855.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio CR, Klose KE. A region of the transmembrane regulatory protein ToxR that tethers the transcriptional activation domain to the cytoplasmic membrane displays wide divergence among Vibrio species. J Bacteriol. 2000;182(2):526–528. doi: 10.1128/JB.182.2.526-528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto J, Sugawara C, Akahane K, Hashimoto K, Kojima T, Ikedo M, Konuma H, Hara-Kudo Y. Rapid and specific detection of the thermostable direct hemolysin gene in Vibrio parahaemolyticus by loop-mediated isothermal amplification. J Food Prot. 2009;72(4):748–754. doi: 10.4315/0362-028x-72.4.748. [DOI] [PubMed] [Google Scholar]
- Fall J, Chakraborty G, Kono T, Maeda M, Itami T, Sakai M. Establishment of loop-mediated isothermal amplification method (LAMP) for the detection of Vibrio nigripulchritudo in shrimp. FEMS Microbiol Lett. 2008;288(2):171–177. doi: 10.1111/j.1574-6968.2008.01332.x. [DOI] [PubMed] [Google Scholar]
- Yamazaki W, Seto K, Taguchi M, Ishibashi M, Inoue K. Sensitive and rapid detection of cholera toxin-producing Vibrio cholerae using a loop-mediated isothermal amplification. BMC Microbiol. 2008;8:94. doi: 10.1186/1471-2180-8-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoi Y, Hosogai M, Tsuneda S. Real-time quantitative LAMP (loop-mediated isothermal amplification of DNA) as a simple method for monitoring ammonia-oxidizing bacteria. J Biotechnol. 2006;125(4):484–491. doi: 10.1016/j.jbiotec.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Monis PT, Giglio S, Saint CP. Comparison of SYTO9 and SYBR Green I for real-time polymerase chain reaction and investigation of the effect of dye concentration on amplification and DNA melting curve analysis. Anal Biochem. 2005;340(1):24–34. doi: 10.1016/j.ab.2005.01.046. [DOI] [PubMed] [Google Scholar]
- National shellfish sanitation program guide for the control of molluscan shellfish 2007. http://www.fda.gov/Food/FoodSafety/Product-SpecificInformation/Seafood/FederalStatePrograms/NationalShellfishSanitationProgram/ucm046353.htm
- Oliver JD, Warner RA, Cleland DR. Distribution and ecology of Vibrio vulnificus and other lactose-fermenting marine vibrios in coastal waters of the southeastern United States. Appl Environ Microbiol. 1982;44(6):1404–1414. doi: 10.1128/aem.44.6.1404-1414.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Siebeling RJ. Identification of Vibrio vulnificus O serovars with antilipopolysaccharide monoclonal antibody. J Clin Microbiol. 1991;29(8):1684–1688. doi: 10.1128/jcm.29.8.1684-1688.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]