Abstract
Brome mosaic virus (BMV), a member of the alphavirus-like superfamily of positive-strand RNA viruses, encodes two proteins, 1a and 2a, that interact with each other, with unidentified host proteins, and with host membranes to form the viral RNA replication complex. Yeast expressing 1a and 2a support replication and subgenomic mRNA synthesis by BMV RNA3 derivatives. Using a multistep selection and screening process, we have isolated yeast mutants in multiple complementation groups that inhibit BMV-directed gene expression. Three complementation groups, represented by mutants mab1–1, mab2–1, and mab3–1 (for maintenance of BMV functions), were selected for initial study. Each of these mutants has a single, recessive, chromosomal mutation that inhibits accumulation of positive- and negative-strand RNA3 and subgenomic mRNA. BMV-directed gene expression was inhibited when the RNA replication template was introduced by in vivo transcription from DNA or by transfection of yeast with in vitro transcripts, confirming that cytoplasmic RNA replication steps were defective. mab1–1, mab2–1, and mab3–1 slowed yeast growth to varying degrees and were temperature-sensitive, showing that the affected genes contribute to normal cell growth. In wild-type yeast, expression of the helicase-like 1a protein increased the accumulation of 2a mRNA and the polymerase-like 2a protein, revealing a new level of viral regulation. In association with their other effects, mab1–1 and mab2–1 blocked the ability of 1a to stimulate 2a mRNA and protein accumulation, whereas mab3–1 had elevated 2a protein accumulation. Together, these results show that BMV RNA replication in yeast depends on multiple host genes, some of which directly or indirectly affect the regulated expression and accumulation of 2a.
Upon infection, the genomes of positive-strand RNA viruses are translated to yield a variety of proteins. Some of these direct the assembly of an RNA replication complex, which first synthesizes a negative-strand RNA replication intermediate and then uses this negative strand as a template for producing more positive-strand genomic RNAs. Several lines of evidence suggest that multiple steps in positive-strand RNA virus RNA replication depend on specific host factors. Different host cells show differing levels of permissiveness for various intracellular replication steps (1, 2). The replication complex of each virus assembles on specific membrane sites in the infected cell (3–5), and such association with cell membranes appears particularly important for positive-strand RNA synthesis (6). Partial purification of some positive-strand RNA replication complexes has shown them to be complexes of viral and cellular proteins, and some of the cell proteins in such complexes have been implicated as functional contributors to replication (7, 8).
To facilitate studying the mechanisms of positive-strand RNA virus replication and the nature and function of host proteins involved, we have shown that brome mosaic virus (BMV) RNAs and their derivatives can replicate and direct gene expression in the yeast Saccharomyces cerevisiae, the rapid growth, facile genetics, and completely sequenced genome of which offer potentially useful features for virus replication studies. BMV encodes two RNA replication factors, 1a and 2a, containing three domains conserved throughout the large alphavirus-like superfamily of animal and plant viruses (9). BMV 1a (109 kDa) contains an N-proximal domain implicated in RNA capping and a C-proximal helicase-like domain, whereas 2a (94 kDa) contains a central polymerase-like domain. BMV 1a and 2a interact (10–12) and in vivo colocalize on the endoplasmic reticulum at the sites of BMV RNA synthesis (5). BMV 1a and 2a are encoded by BMV RNA1 and RNA2, respectively. A third genomic RNA, RNA3, encodes the 3a cell-to-cell movement protein and the coat protein, which are required for BMV infection spread in its natural plant hosts but are dispensable for RNA replication (13, 14). The 3′-proximal coat gene is not translatable from RNA3 but only from a subgenomic mRNA, RNA4, synthesized from negative-strand RNA3 (Fig. 1). Host factor involvement in BMV RNA replication is suggested by host-specific replication effects, biochemical studies, and cell biology studies as noted above and by the presence of multiple tRNA-related sequences and functions in the cis-acting replication signals on BMV RNAs (1, 5, 7, 9).
Figure 1.
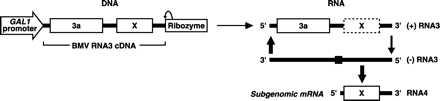
Schematic of a cDNA cassette (Left) for in vivo transcription of wt RNA3 or RNA3 derivatives B3URA3 or B3GUS, and the subsequent replication of these RNAs (Right). 3a, the BMV 3a movement protein ORF; and X, the wt BMV coat gene, URA3 gene, or GUS gene, as appropriate for the relevant RNA3 derivative. The 5′-flanking GAL1 promoter and 3′-flanking hepatitis delta virus ribozyme are also shown. The horizontal arrow (Center) represents in vivo, DNA-dependent synthesis of RNA3 transcripts as an inoculum to initiate replication, whereas the vertical arrows (Right) depict BMV 1a- and 2a-directed, RNA-dependent RNA3 replication and subgenomic mRNA synthesis. The solid box near the center of negative-strand RNA3 represents the subgenomic mRNA promoter. The coat protein gene or its replacements are not translated from their internal position in RNA3 (dotted box marked X, Upper Right), but only from their 5′-proximal position in the subgenomic mRNA, RNA4 (solid box marked X, Lower Right), making their expression dependent on BMV RNA replication and subgenomic mRNA synthesis (15, 16).
Yeast expressing 1a and 2a from DNA plasmids replicate RNA3 or RNA3 derivatives and synthesize subgenomic mRNAs to express the coat gene or other genes substituted for it. Replicatable RNA3 derivatives can be introduced into yeast by transfection of in vitro transcripts (15) or by in vivo transcription of an RNA3 cDNA flanked 5′ by a DNA-dependent RNA polymerase promoter and 3′ by a self-cleaving ribozyme (Fig. 1) (16). Such cDNA-based RNA3 launching cassettes can be carried on yeast plasmids (16) or, as shown here, integrated into a yeast chromosome. Expression of reporter genes substituted for the coat gene in RNA3 launching cassettes provides colony-selectable or -screenable markers for all forms of BMV RNA-dependent RNA synthesis, since such expression requires 1a-, 2a-directed negative-strand RNA synthesis, and subgenomic mRNA synthesis, and is strongly reduced if RNA-dependent positive-strand RNA amplification is blocked (15, 16).
To identify cellular processes and factors that contribute to BMV replication, we have initiated screens for yeast mutants with defects in supporting BMV RNA replication and gene expression. Here we describe a multistep selection and screening strategy able to identify such mutants, the isolation and characterization of recessive mutants in several distinct complementation groups that suppress the accumulation of BMV RNA synthesis products, a new form of BMV gene regulation, and differential effects of several yeast mutants on the accumulation of a BMV-encoded RNA replication factor. The results show that BMV RNA replication depends on contributions from multiple host genes, some of which directly or indirectly affect the virus-regulated expression and accumulation of the viral polymerase-like protein.
MATERIALS AND METHODS
Plasmids.
BMV 1a and 2a were expressed from pB1CT19 and pB2CT15, yeast 2-μ plasmids with HIS3 and LEU2 selectable markers, respectively (15). BMV RNA3 was expressed in vivo from pB3RQ39 (hereafter pB3), a CEN4 plasmid with the TRP1 marker (16). B3CAT RNA was transcribed in vitro from pB3CA101 (15).
B3GUS expression plasmid pB3MI22 was constructed as follows: pBI101.2 (CLONTECH), a pBI101 (17) derivative with an extra A between the multiple cloning site and Escherichia coli β-glucuronidase (GUS) gene, was cut with SacI, blunted with T4 DNA polymerase, cut with SmaI, and the resulting 1.8-kb GUS gene fragment cloned in the SmaI site of pBluescript II KS(+) (Stratagene) to make pGUSKM2 (K. Mise and P.A., unpublished results). The 1.8-kb SmaI-XbaI fragment from pGUSKM2 was ligated between the XbaI site and Klenow DNA polymerase-treated SalI site of pB3CAT+3(A) (16), replacing the chloramphenicol acetyltransferase (CAT) gene. In the resulting plasmid, pB3MI22, the initiating ATG of the BMV coat gene was followed by TCGAGGGTAGGTCAGTCCCTTATG, where ATG is the GUS initiation codon. To integrate B3GUS into the yeast lys2 locus, pMI-LYS22 was made by cloning the 4.2-kb EcoRI-HindIII yeast DNA fragment containing the LYS2 gene into YIplac211 (18). Then pB3MI24 was constructed by inserting the BamHI-EcoRI fragment of pB3MI22, containing the GAL1 promoter-B3GUS-ribozyme-ADH1 poly(A) signal cassette, between the BamHI-XhoI sites of pMI-LYS22. The EcoRI and XhoI sites were filled in with Klenow DNA polymerase prior to BamHI cleavage, fragment isolation and ligation.
To integrate B3URA3 into the yeast can1 locus, pMI-CAN11 was constructed by cloning the 1.8-kb BamHI-SalI yeast DNA fragment containing the CAN1 gene into pBluescript II KS(+). Then pB3MI27 was constructed by inserting the EcoRI fragment from pB3URA3+3(A) (16), containing the [GAL1 promoter-B3URA3-ribozyme] cassette, into the EcoRI site of pMI-CAN11, oriented so that transcription from the GAL1 promoter is in the same direction as that of the CAN1 locus.
Yeast Strains.
Standard yeast genetic techniques and media were used (19, 20). Except where noted, yeast were grown at 30°C in synthetic defined medium lacking histidine and leucine to select for the 1a and 2a expression plasmids, and containing 2% galactose (gal) or glucose (glc) as indicated. Because high frequency diploid sporulation required pregrowth on rich medium containing histidine and leucine, some spore progeny lost the 1a and/or 2a expression plasmids, which were reintroduced if necessary.
YMI04, the parental strain for mutant isolation, was a derivative of YPH500 (MATα ura3–52 lys2–801 ade2–101 trp1-Δ63 his3-Δ200 leu2-Δ1) (15, 21) with the following modifications: (i) the [GAL1 promoter-B3GUS-ribozyme-ADH1 poly(A) site] cassette was integrated in the lys2 locus by the two-step method (see chapter 19 in ref. 20) with HpaI-linearized pB3MI24. (ii) The [GAL1 promoter-B3URA3-ribozyme] cassette was integrated in the can1 locus by the one-step method (see chapter 19 in ref. 20) with BamHI-, SalI-digested pB3MI27. (iii) pB1CT19 and pB2CT15 were introduced.
YMI06 was a derivative of YPH499 (MATa ura3–52 lys2–801 ade2–101 trp1-Δ63 his3-Δ200 leu2-Δ1) (21) in which lys2–801 was replaced with LYS2+ by two-step gene replacement with HpaI-linearized pMI-LYS22. YMI08 was a derivative of strain 51 (MATa ura3 lys2 ade2 trp1 his3 leu2 cup1Δ; C. F. Lesser and C. Guthrie, personal communication) with the lys2::B3GUS insertion.
GUS Assays.
Filter-lifts of yeast colonies were made with Hybond-N+ membrane (Amersham). Cells were permeabilized by freezing and thawing and assayed for GUS activity by using 5-bromo-4-chloro-3-indolyl β-d-glucuronide (22). To measure GUS activity, yeast cells were grown in liquid media, washed with water, resuspended in 50 mM sodium phosphate (pH 7.0), 10 mM EDTA, 0.1% N-lauroylsarcosine, 0.1% Triton X-100, 10 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and disrupted by vigorous agitation with glass beads. Cell extracts were clarified by centrifugation and assayed for protein concentration (23) and for GUS activity by using 4-methylumbelliferyl β-d-glucuronide (24).
RNA Transfection, CAT, and Luciferase Assays.
Synthesis of capped in vitro transcripts, RNA transfections, CAT assays, and luciferase assays were carried out as described (15, 25).
RNA and Protein Analysis.
Total yeast RNA isolation, Northern blot analysis with strand-specific, 32P-labeled in vitro transcript probes, and primer extension analysis of the RNA3 5′ end were carried out as described (16). Probes for positive- and negative-strand RNA3 and RNA4 were derived from pBCPSN1 or pB3HE1 (16). Probes for 1a and 2a mRNAs were transcripts complementary to RNA1 bases 570–2,926 and to RNA2 bases 31–2,474. Protein was extracted for Western blots as for GUS assays except that extraction buffer was augmented with 10 mM each phenylmethylsulfonyl fluoride and benzamidine and 10 μg/ml each pepstatin A, aprotinin, and leupeptin, and the cell extract clarified by centrifugation through glass wool. Protein electrophoresis and Western blot analyses were as described (5).
RESULTS
Isolation of S. cerevisiae Mutants Inhibiting BMV-Directed Gene Expression.
To facilitate isolation of yeast mutants with reduced ability to support BMV-directed RNA replication and/or gene expression, we constructed yeast strain YMI04. YMI04 was a YPH500 derivative containing constitutive 1a and 2a expression plasmids and, integrated in the chromosomal can1 and lys2 loci, respectively, cDNAs of RNA3 derivatives B3URA3 and B3GUS, which contain yeast uracil biosynthesis gene URA3 and E. coli GUS gene as coat gene replacements (Fig. 1). The B3URA3 and B3GUS cDNAs were linked to the gal-inducible, glc-repressible GAL1 promoter for in vivo transcription. If and only if 1a and 2a were also expressed, the B3URA3 and B3GUS transcripts were replicated and directed synthesis of subgenomic mRNAs necessary to express URA3 and GUS (Fig. 1). Accordingly, YMI04 grew on gal medium lacking uracil but not glc medium lacking uracil. Similarly, when incubated with the GUS substrate 5-bromo-4-chloro-3-indolyl β-d-glucuronide, filter lifts of gal-grown YMI04 colonies became visibly blue in 20–30 min, whereas YMI04 lacking 1a or 2a developed no blue color after 24-h incubation. By using a fluorescence assay with the substrate 4-methylumbelliferyl β-d-glucuronide, extracts of gal-grown YMI04 had an average GUS activity of ≈45 nmol 4-methylumbelliferone/mg total yeast protein per min.
YMI04 yeast on glc plates were mutagenized by ultraviolet irradiation to 70% survival, grown overnight on the same plates in the dark to fix the mutations, pooled, and plated on gal plates containing 0.1% 5-fluoroorotic acid, which selects against cells expressing active URA3 (chapter 18 in ref. 20). After 5–7 days incubation, ≈1% of the cells plated developed into colonies. Two thousand such colonies were streaked on gal plates, grown 3 days, and filter lifts assayed for GUS activity. Thirty four isolates that showed slow or no blue color development were selected and mated with YMI06, a YPH499 derivative lacking any BMV sequences and having the mating type (MATa) opposite to that of YMI04 (MATα). Of the resulting 34 diploids, seven showed restored GUS activity, implying that inhibition of BMV-directed GUS expression in the corresponding YMI04-derived parental haploids was due to recessive yeast chromosomal mutations complemented by the YMI06 genome. Three of these original YMI04-derived Gus− haploid strains were chosen for initial analysis. Based on complementation and recombination data below, these strains are hereafter referred to individually as mab1, mab2, and mab3, where mab refers to their defects in maintenance of BMV functions. The 27 Gus− strains that did not recover GUS activity after mating with YMI06 did so after mating with YMI06 harboring active BMV 1a or 2a expression plasmids, suggesting that the original Gus− haploids carried recessive mutations in 1a or 2a.
GUS activity per mg of total protein in extracts of gal-grown mab1, mab2, and mab3 yeast averaged 4%, 9%, and 18%, respectively, of that for wild type (wt) YMI04 (Fig. 2A). To test whether inhibition of BMV-directed gene expression could be due to interference with the initial, DNA-directed transcription or nucleocytoplasmic transport of RNA3 derivatives such as B3GUS, we bypassed these nuclear steps by transfecting wt, mab1, mab2, and mab3 yeast with in vitro transcripts of B3CAT (15), an RNA3 derivative with the coat gene replaced by the bacterial CAT gene (Fig. 2B). For mab2, average CAT expression relative to wt was almost identical to that for GUS. For mab1 and mab3, average CAT expression relative to wt was higher than for GUS, but the increase was not significantly different within the statistical variations of both measurements. Thus, cytoplasmic steps of BMV RNA synthesis must be inhibited in mab1, mab2, and mab3 yeast.
Figure 2.
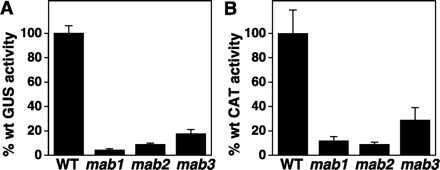
(A) BMV-directed GUS expression in 1a- and 2a-expressing yeast containing a chromosomally integrated B3GUS expression cassette. Parental strain YMI04 and mutant strains mab1, mab2 and mab3 derived from it were grown in gal-containing liquid medium for 48 h, protein was extracted, and GUS activity per mg of total protein was measured. Averages and standard deviations from four independent cultures of each strain are shown. (B) BMV-directed CAT expression in 1a- and 2a-expressing yeast transfected with B3CAT in vitro transcripts. Parental strain YMI04 and mab1, mab2 and mab3 mutant strains were cotransfected with in vitro transcripts of B3CAT and luciferase mRNA (transcribed from pGEM-luc, Promega), incubated 21 h in glc media, and assayed for CAT and luciferase activity. Luciferase mRNA was included as an internal standard since luciferase was translated directly from the transfected mRNA, requiring no BMV-directed RNA synthesis, and showed no strain-specific variation between YMI04 and mab mutant strains. Thus, in each sample, CAT activity was normalized to luciferase activity to control for any variations in transfection efficiency. Averages and SD from three independent transfections of each strain are shown.
mab1, mab2, and mab3 Are Recessive, Chromosomal Mutations in Distinct Complementation Groups.
To test whether reduced BMV-directed GUS expression was due to a single mutation each in mab1, mab2, and mab3 yeast, these mutants were crossed with wt MATa strain YMI08, the diploid progeny were sporulated, and tetrad segregation of GUS expression per mg of total yeast protein was analyzed. YMI08 was used for these crosses because it gave higher sporulation and spore germination frequencies than YMI06. Tetrads were analyzed for crosses with mab1 (24 tetrads), mab2 (28 tetrads), and mab3 (20 tetrads). For all tetrads from crosses of mab1 or mab2 yeast with YMI08, two spores showed high GUS activity and wt growth, whereas the other two showed low GUS activity. The average difference between high and low GUS expression was ≈10-fold for mab1 and 20-fold for mab2. For mab1 crosses with YMI08, low GUS expression cosegregated with temperature-sensitive inhibition of growth at 36°C, a characteristic of the original mab1 isolate (Fig. 3). Similarly, for mab2 crosses with YMI08, low GUS expression cosegregated with slow growth at 30°C (Fig. 3). mab2 strains also showed a temperature-sensitive growth defect at 36°C, though this effect was weaker than for mab1. The 2:2 segregation of GUS activity and growth phenotype implied that inhibition of BMV-directed GUS expression in mab1 and mab2 yeast was due in each strain to a single mutant gene also responsible for altered growth. Superimposed on the 2:2 segregation of this major shift in GUS activity induced by mab1 and mab2 was a smaller, independent effect introduced by YMI08 (i.e., in the absence of mab mutations, BMV-directed GUS expression in YMI08 was ≈50% of that in YMI04). In tetrad analysis, this YMI08-specific reduction segregated as a single modifier gene difference between YMI08 and YMI04, unlinked to and phenotypically additive with the mab mutations.
Figure 3.
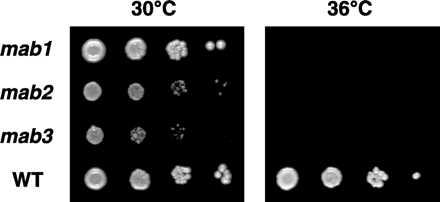
mab1, mab2 and mab3 yeast growth phenotypes, which cosegregate with inhibition of BMV RNA replication and gene expression (see Results). The indicated yeast strains were grown in glc-containing medium and diluted in sterile water to A600 of 0.2, 0.025, 0.003, and 0.0004 (8-fold serial dilutions). Two microliters of each dilution was spotted on glc plates and incubated at 30°C or 36°C for 3 days. For each strain, the dilution series corresponds to plating ≈10 cells in the most dilute spot. At 30°C, the growth of mab1 yeast was similar to that of parental strain YMI04, whereas mab3 and especially mab2 yeast grew more slowly. After prolonged incubation at 36°C, mab1 and mab3 yeast showed no growth, whereas mab2 yeast growth was weakly detectable (Right and results not shown).
For all tetrads from crosses of mab3 yeast with YMI08, two spores showed wt growth and two showed slow growth at 30°C, like the original mab3 strain (Fig. 3). Where tested, slowly growing mab3 progeny also were temperature sensitive for growth at 36°C, like the original mab3 strain. For each tetrad, the two spores with mab3-like growth always supported BMV-directed GUS expression to <20% of the level in YMI04. For the two spores with wt growth, GUS expression varied from 20–100% of YMI04. Much of this variation could be accounted for by the YMI08-derived modifier gene described above. For example, in ≈75% of mab3-derived tetrads, the four spores showed segregation of GUS activity consistent with one of the three patterns—tetratype, parental ditype, and nonparental ditype (see chapter 3 in ref. 20)—expected for the independently segregating effects of a 4- to 5-fold reduction in GUS expression due to mab3 and a 2-fold reduction in GUS expression due to the unlinked modifier gene from YMI08. However, there appeared to be an excess of tetratypes beyond the expected 4:1:1 ratio of tetratypes to parental and nonparental ditypes and, in 10–20% of tetrads, GUS expression in one of the two wt-growing spores was lower than expected from the YMI08 modifier gene alone. These results suggest that most of the reduction in BMV-directed expression in mab3 yeast was due to a single mutation also restricting growth, but that a secondary effect on GUS expression was also present. Such secondary effects might include a second mutation in the original mab3 yeast isolate, an additional gene difference between YMI08 and YMI04 that affects BMV in mab3 but not mab1 or mab2 yeast, or other effects.
To allow complementation tests, mab1, mab2, and mab3 strains with the opposite, MATa mating type were isolated by dissecting tetrads from the original crosses of each mutant with YMI06 (see above) and determining BMV-directed GUS activity levels and mating types in the individual haploids. These MATa mab strains and wt, MATa YMI06 were crossed with the original MATα mab strains and their MATα progenitor, YMI04, in all combinations, and the GUS activity of the diploid progeny was measured (Fig. 4). When MATa and MATα strains bearing the same mab mutation were crossed, the resulting mab-homozygous diploids supported BMV-directed GUS expression at only 5–10% of the level in diploids generated by crossing either of the same mab parents to wt YMI04 or YMI06. When strains with any two independent mab mutations were crossed, the diploid progeny supported BMV-directed GUS expression at 60–130% of the level in diploids generated by crossing either of the same mab parents to wt YMI04 or YMI06. Thus, the BMV-inhibiting mutations in the mab1, mab2, and mab3 strains are in separate complementation groups. To rule out intragenic complementation between mutations in the same gene, further genetic linkage analysis established that the BMV-inhibiting mutations in the mab1, mab2, and mab3 strains were nonallelic and thus represented three independent genes (J.D. and M.I., unpublished results).
Figure 4.
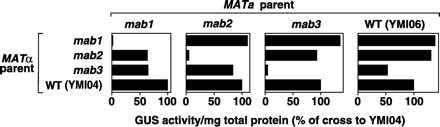
Complementation tests of mab1, mab2, and mab3. Diploid yeast strains expressing 1a and 2a and containing the lys2-integrated B3GUS expression cassette were generated by the indicated crosses among mab1, mab2, and mab3 strains and strains with wt MAB loci (YMI04, YMI06). The diploids were cultured in gal medium for 48 h and GUS activity per mg of total protein measured as in Fig. 2A. Each set of diploids with a common MATa parent was processed in parallel and GUS activity expressed as a % of the activity in the diploid derived by crossing to MATα YMI04, which has wt MAB loci.
mab Mutants Reduce RNA3 and Subgenomic mRNA Accumulation.
Northern blot analysis of RNA from gal-grown mab1, mab2, and mab3 yeast showed that accumulation of B3URA3 RNA and subgenomic URA3 mRNA were reduced relative to YMI04 (results not shown). Quantitative comparisons, however, were difficult because subgenomic URA3 mRNA levels in the mutants were reduced to background level. Moreover, even in wt YMI04, accumulation of B3GUS RNA and its subgenomic mRNA was less than for B3URA3. Accordingly, to compare the accumulation of BMV RNA synthesis products in YMI04 and mutant yeast, we used plasmid pB3 (16) to introduce GAL1 promoter-driven wt RNA3 transcripts, which in wt yeast are replicated to higher levels than B3URA3 or B3GUS RNA. To simplify Northern blot analysis and avoid possible interference effects, mab1, mab2, and mab3 mutant strains lacking the can1-, lys2-integrated B3URA3- and B3GUS-expression cassettes were first obtained by dissecting tetrads from crosses of each mutant with YMI06 (CAN1 LYS2; see above) and screening for CAN1 LYS2 mutant strains. These strains were then transformed with pB3 and grown in gal prior to RNA extraction and Northern blot hybridization. For all three mutants, accumulation of RNA3 and subgenomic RNA4 positive strands (Fig. 5A) and negative strands (Fig. 5B) were reduced from wt. Comparing equal amounts of total yeast RNA from three independent cultures for each strain, positive-strand RNA3 levels in mab1, mab2, and mab3 yeast averaged 14%, 12%, and 16% of wt, subgenomic RNA4 levels averaged 4%, 7%, and 6% of wt, and negative-strand RNA3 levels averaged 7%, 33%, and 27% of wt, respectively.
Figure 5.
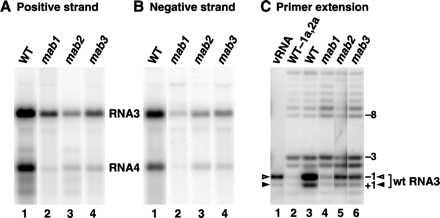
Analysis of RNA3 and RNA4 species in parental strain YPH500 and mab1, mab2, and mab3 yeast expressing 1a, 2a and wt RNA3 after 48 h growth in liquid gal medium. (A) Northern blot analysis of positive-strand RNA3 and RNA4. Total RNA was extracted, glyoxylated, electrophoresed in 1% agarose, transferred to nylon membrane, and probed with a strand-specific, 32P-labeled RNA probe from the BMV coat gene. (B) Northern blot analysis of negative-strand RNA3 and RNA4, as in A except that a strand-specific coat gene probe of opposite polarity was used. (C) Reverse transcriptase primer extension analysis of the 5′ ends of RNA3 species by using a primer complementary to bases 30–44 of RNA3. vRNA, a reaction using BMV virion RNA as template; wt-1a,2a, YPH500 expressing RNA3 but lacking 1a and 2a expression plasmids. Numbers at the right indicate nucleotide position relative to the wt RNA3 5′ end. The solid and open arrowheads indicate the positions of the two bands arising from natural RNA3 (see Results).
Whereas the subgenomic RNA4 and negative-strand BMV RNAs in Fig. 5 A and B were derived from RNA-dependent RNA synthesis, the positive-strand RNA3 bands in Fig. 5A include RNA3 species produced by DNA-dependent transcription of pB3 as well as by RNA-dependent RNA replication. To distinguish DNA-derived and RNA-derived RNA3, the 5′ ends of RNA3 species in pB3-containing wt and mab yeast strains were examined by primer extension with reverse transcriptase (Fig. 5C). Plant-derived BMV virion RNA (lane 1) yielded two bands, a lower, weaker band corresponding to the 5′ end of RNA3 and an upper, intense band from cap-dependent incorporation of an additional nucleotide (16). Gal-induced, wt YPH500 yeast containing pB3 but lacking 1a and 2a contain GAL1 promoter-derived RNA3 transcripts with multiple 5′ ends, with the most prominent initiated at positions −2/−3 and −8/−9 relative to the RNA3 5′ end (lane 2; see also ref. 16). YPH500 yeast containing pB3 and expressing 1a and 2a contained the same DNA-derived RNA3 transcripts plus intense bands at +1/−1, corresponding to the 5′ end of natural BMV RNA3 produced by BMV RNA-dependent RNA replication (lane 3). mab1, mab2, and mab3 mutant strains containing pB3 and expressing 1a and 2a contained the usual pB3-derived RNA3 transcripts, showing that the mutants are not defective in GAL1-promoted transcription of RNA3. However, the ratio of authentic RNA3 5′ ends relative to DNA-derived transcripts was reduced 5- to 10-fold compared with wt (as determined by PhosphorImager analysis of lanes 4–6 and similar reactions).
1a and 2a mRNA and Protein Accumulation.
Next, we compared 1a and 2a mRNA and protein levels in wt YMI04 and mab1, mab2, and mab3 yeast in multiple experiments. As in the experiments above, 1a and 2a were expressed from separate yeast 2-μ DNA plasmids, by using the same ADH1 promoter and polyadenylation site (15). BMV 1a mRNA and protein accumulation were similar in wt, mab1, mab2, or mab3 yeast in the presence or absence of 2a (Fig. 6A). However, 2a mRNA and protein accumulation varied significantly in several respects. First, in wt yeast, 1a expression increased 2a mRNA and protein accumulation approximately 5-fold (Fig. 6B, lanes 1–2; E. Smirnyagina, J. Chen, and P.A., unpublished results). Second, in mab1 and mab2 yeast, 2a mRNA and protein accumulation were altered in two ways. In the absence of 1a, 2a mRNA and protein levels were reduced 2- to 3-fold relative to wt (Fig. 6B, lanes 3 and 5). The cause of this reduced basal 2a mRNA accumulation is unclear. It appears unlikely to reflect reduced DNA-dependent transcription, since accumulation of 1a mRNA from the same ADH1 promoter and polyadenylation site appeared unchanged from wt (Fig. 6A). Furthermore, similar ratios of 2a mRNA and protein in wt, mab1 and mab2 yeast suggest that there was no defect in nucleocytoplasmic transport or translation of 2a mRNA. In a more striking contrast to wt, expression of 1a in mab1 and mab2 yeast induced no change in 2a mRNA and protein levels (Fig. 6B, lanes 4–6). Thus, mab1 and mab2 blocked the ability of 1a to stimulate 2a mRNA and protein accumulation. Third, in mab3 yeast, 2a mRNA accumulation in the presence of 1a was slightly lower than wt, but 2a protein accumulation, which was stimulated by 1a, was significantly higher than wt in both the presence and absence of 1a (Fig. 6B, lanes 7–8).
Figure 6.
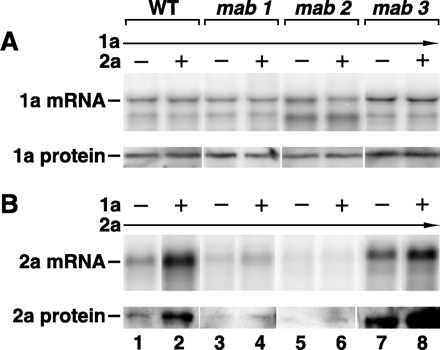
Accumulation of (A) 1a mRNA and protein and (B) 2a mRNA and protein in parental strain YMI04 (wt) and mab1, mab2, and mab3 yeast expressing 1a and 2a alone or together, as indicated. Equal amounts of total yeast RNA or protein were loaded in each lane prior to electrophoresis and Northern blot or immunoblot analysis, respectively, as described in Materials and Methods.
DISCUSSION
Whereas prior evidence for host factors in RNA virus replication has been largely circumstantial, the experiments described here show that BMV RNA replication and gene expression in yeast depend on multiple host functions. In particular, we have isolated and characterized yeast mutants bearing BMV-inhibiting mutations in three complementation groups. The causal mutations, hereafter designated mab1–1, mab2–1, and mab3–1, inhibit BMV RNA-dependent RNA replication and subgenomic mRNA transcription. BMV-directed gene expression decreased similarly whether RNA3-derived replication templates were transcribed in vivo from nuclear DNA or introduced by transfecting in vitro transcripts, bypassing the nucleus. Thus, these yeast mutations must interfere with cytoplasmic steps of BMV RNA synthesis. All three mutants were recessive, showing that the corresponding wt yeast gene contributes an active function supporting BMV replication. For each mutant, the reduction in subgenomic mRNA accumulation was sufficient to account for the reduction in BMV-directed reporter gene expression without appealing to defective translation of subgenomic mRNA. Each mutation also made yeast growth temperature sensitive, implying that the affected genes contribute to normal cell processes, as expected. Below we discuss the possible defects of the mutants and their effects on BMV 2a expression.
Recently, our laboratory found that 1a expression could increase accumulation of the polymerase-like 2a protein (E. Smirnyagina, R. Quadt, and P.A., unpublished results). In the course of this work, we found that 1a increased 2a mRNA accumulation (Fig. 6B). Thus, though translated independently, 2a expression and accumulation are regulated by 1a. Such regulation may be functionally related to the translational readthrough or frameshift mechanisms used by Sindbis virus and other single component positive-strand RNA viruses to regulate the ratio of 1a-like and 2a-like proteins, and by retroviruses to regulate their pol genes (26–28). In keeping with 1a’s cytoplasmic localization (ref. 5; M.R.-H. and P.A., unpublished results), preliminary results suggest that 1a influences 2a mRNA and protein levels through posttranscriptional effects, which are under further study (M. Janda, J. Chen, M. Sullivan, and P.A., unpublished results).
The mab1–1 and mab2–1 mutations interfered with the ability of 1a to stimulate 2a mRNA and protein accumulation (Fig. 6B). However, reduced RNA3 replication and RNA4 transcription do not appear to result solely from reduced 2a protein accumulation, since preliminary studies show that using an alternate promoter to restore 2a protein accumulation to wt levels does not restore wt BMV-directed RNA synthesis in mab1 and mab2 strains (J.D., M.I., M. Janda and P.A., unpublished results). Thus, in mab1 and mab2 yeast, 1a and/or 2a appear unable to function normally in directing RNA3 replication and RNA4 transcription. In particular, BMV RNA replication may be suppressed in mab1 and mab2 yeast due to inability of 1a to function properly, as indicated by its failure to stimulate 2a mRNA and protein accumulation.
By contrast, in mab3 yeast, 2a protein accumulation from the ADH1 promoter was stimulated by 1a and was higher than in the parental YMI04 strain in the presence and absence of 1a (Fig. 6B). Since the mab3–1 mutation was recessive and did not elevate 2a mRNA levels, this suggests that the wt MAB3 gene simultaneously contributes to supporting BMV RNA replication and to down-regulating the BMV 2a RNA replication factor. Thus, proper regulation of 2a may be important for efficient BMV RNA replication.
The wt MAB1, MAB2, and MAB3 gene products may be directly involved in BMV replication or may affect other host proteins involved in replication. Possible effects of these mutations include defects in the function, localization, or synthesis of a host factor required for RNA replication, or defects in 1a or 2a mRNA or protein localization or modification. Recently, approaches similar to those described here have been used to isolate additional yeast complementation groups required for normal levels of BMV RNA replication and gene expression (M.I., A. Noueiry, W. M. Lee and P.A., unpublished results), showing that MAB1, MAB2, and MAB3 are not the only host loci involved in supporting these processes. These results suggest that yeast should provide a useful genetic basis for further analysis of the components and mechanisms of BMV RNA replication.
Acknowledgments
We thank K. Mise for pGUSKM2, E. Smirnyagina, J. Chen, and M. Janda for unpublished results; J. Chen and M. Janda for assistance with RNA and protein analysis; C. Yu for excellent technical assistance; D. Ursic for generous advice on yeast techniques; and M. Culbertson for helpful comments on the manuscript. This research was supported by National Institutes of Health Grants GM51301 and GM35072. P.A. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- BMV
brome mosaic virus
- CAT
chloramphenicol acetyltransferase
- gal
galactose
- glc
glucose
- GUS
β-glucuronidase
- wt
wild type
References
- 1.De Jong W, Ahlquist P. J Virol. 1995;69:1485–1492. doi: 10.1128/jvi.69.3.1485-1492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamarnik A V, Andino R. EMBO J. 1996;15:5988–5998. [PMC free article] [PubMed] [Google Scholar]
- 3.Froshauer S, Kartenbeck J, Helenius A. J Cell Biol. 1988;107:2075–2086. doi: 10.1083/jcb.107.6.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienz K, Egger D, Pfister T, Troxler M. J Virol. 1992;66:2740–2747. doi: 10.1128/jvi.66.5.2740-2747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Restrepo-Hartwig M, Ahlquist P. J Virol. 1996;70:8908–8916. doi: 10.1128/jvi.70.12.8908-8916.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu S X, Ahlquist P, Kaesberg P. Proc Natl Acad Sci USA. 1992;89:11136–11140. doi: 10.1073/pnas.89.23.11136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quadt R, Kao C C, Browning K S, Hershberger R P, Ahlquist P. Proc Natl Acad Sci USA. 1993;90:1498–1502. doi: 10.1073/pnas.90.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Osman T A M, Buck K W. J Virol. 1997;71:6075–6082. doi: 10.1128/jvi.71.8.6075-6082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahlquist P. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 10.Kao C C, Quadt R, Hershberger R P, Ahlquist P. J Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kao C C, Ahlquist P. J Virol. 1992;66:7293–7302. doi: 10.1128/jvi.66.12.7293-7302.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smirnyagina E, Lin N S, Ahlquist P. J Virol. 1996;70:4729–4736. doi: 10.1128/jvi.70.7.4729-4736.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Allison R, Thompson C, Ahlquist P. Proc Natl Acad Sci USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mise K, Ahlquist P. Virology. 1995;206:276–286. doi: 10.1016/s0042-6822(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 15.Janda M, Ahlquist P. Cell. 1993;72:961–970. doi: 10.1016/0092-8674(93)90584-d. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa M, Janda M, Krol M A, Ahlquist P. J Virol. 1997;71:7781–7790. doi: 10.1128/jvi.71.10.7781-7790.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gietz R D, Sugino A. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 19.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1987. [Google Scholar]
- 20.Guthrie, C. & Fink, G. R., eds. (1991) Methods Enzymol. 194.
- 21.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hirt H. Curr Genet. 1991;20:437–439. doi: 10.1007/BF00317075. [DOI] [PubMed] [Google Scholar]
- 23.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 24.Schmitz U K, Lonsdale D M, Jefferson R A. Curr Genet. 1990;17:261–264. doi: 10.1007/BF00312618. [DOI] [PubMed] [Google Scholar]
- 25.Russell P J, Hambidge S J, Kirkegaard K. Nucleic Acids Res. 1991;19:4949–4953. doi: 10.1093/nar/19.18.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishikawa M, Meshi T, Motoyoshi F, Takamatsu N, Okada Y. Nucleic Acids Res. 1986;14:8291–8305. doi: 10.1093/nar/14.21.8291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Rice C M. J Virol. 1989;63:1326–1337. doi: 10.1128/jvi.63.3.1326-1337.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farabaugh P J. Annu Rev Genet. 1996;30:507–528. doi: 10.1146/annurev.genet.30.1.507. [DOI] [PubMed] [Google Scholar]


