Abstract
Background
A quadrivalent meningococcal conjugate vaccine (MCV4) was licensed in the United States in 2005; no serogroup B vaccine is available. Neisseria meningitidis changes its capsular phenotype through capsular switching, which has implications for vaccines that do not protect against all serogroups.
Methods
Meningococcal isolates from 10 Active Bacterial Core Surveillance sites from 2000–2005 were analyzed so that changes following MCV4 licensure can be identified. Isolates were characterized by multilocus sequence typing (MLST) and outer membrane protein gene sequencing. Isolates expressing capsular polysaccharide different from that associated with the MLST lineage were considered to demonstrate capsule switching.
Results
Among 1,160 isolates, the most common genetic lineages were the ST-23, ST-32, ST-11, and ST-41/44 clonal complexes. Of serogroup B and Y isolates, 8 (1.5%) and 3 (0.9%), respectively, demonstrated capsular switching, compared to 36 (12.9%) for serogroup C (p <0.0001); most serogroup C switches were from virulent serogroup B and/or serogroup Y lineages.
Conclusions
A limited number of genetic lineages caused the majority of invasive meningococcal infections. A substantial proportion of isolates had evidence of capsular switching. The high prevalence of capsular switching requires surveillance to detect changes in meningococcal population structure that may impact effectiveness of meningococcal vaccines.
Keywords: Neisseria meningitidis; meningococcus, meningococcal; population structure; multilocus sequence typing; DNA sequencing; outer membrane proteins; fetA; porA; porB; molecular epidemiology; capsular switching
INTRODUCTION
Neisseria meningitidis remains a leading cause of meningitis and other serious invasive bacterial infections throughout the world (1, 2). In 2005, a new quadrivalent (A/C/W-135/Y) polysaccharide protein conjugate vaccine (MCV4) was licensed in the United States and is currently approved for persons 2-55 years old. The Advisory Committee on Immunization Practices (ACIP) recommends MCV4 for all U.S. adolescents 11–18 years old and other groups at high risk (3, 4). In addition, it is likely that meningococcal conjugate vaccines will be licensed in the U.S. for infants in the near future (5–8). Serogroup B strains are a major cause of meningococcal disease in the United States and many other parts of the world (1). Although substantial progress is being made towards the development of a vaccine that covers the highly diverse population of serogroup B strains causing endemic disease, this serogroup will most likely not be covered by available vaccines for at least the next several years.
N. meningitidis has a highly plastic genome and multiple genetic mechanisms to alter its antigenic profile. One of the mechanisms is allelic replacement by transformation and homologous recombination of genes involved in capsule biosynthesis (9). These are not just theoretical concerns as both capsule switching or non-capsule antigenic shifts have been observed in the United States and worldwide (9–14). As examples, serogroup C strains with identical genotype to serogroup B clonal strains causing outbreaks have been found (9). Also, a serogroup W-135 clone emerged in 2000 to cause outbreaks of meningococcal disease among Hajj pilgrims and subsequently caused large epidemics in parts of sub-Saharan Africa and case clusters in other parts of the world (11). Genetically, the epidemic clone belonged to the sequence type (ST)-11 clonal complex, which is typically associated with invasive serogroup C meningococcal strains, suggesting that capsular switching had occurred. These data suggest that serogroups not covered by MCV4 could emerge by a similar mechanism. In fact, capsular switching has been observed since licensure of the pediatric hepatavalent pneumococcal polysaccharide protein conjugate vaccine (PCV7). Although PCV7 has been a huge public health success, there has been a marked increase in some non-vaccine pneumococcal serotypes, including apparent capsular switching from vaccine serotypes (15–19). This has prompted the development of new pneumococcal conjugate vaccines that include additional serotypes. Similarly, capsule switching of virulent lineages of N. meningitidis of serogroups A, C, Y, and W-135 that are currently covered by MCV4 could lead to selection of additional virulent serogroup B strains.
Multilocus sequence typing (MLST) of N. meningitidis, based on DNA sequencing of segments of seven housekeeping genes, is a standard molecular subtyping approach for determining the genetic lineage of this organism. MLST is also used to infer capsular switching, which is presumed to have occurred when two meningococcal isolates share the same ST or clonal complex but differ in their polysaccharide capsule. DNA sequencing of genes that encode outer membrane proteins (OMPs) further discriminates among STs and provides additional molecular epidemiologic characterization of meningococcal strains (10, 20). Together, MLST and OMP genotype profile data provide useful epidemiologic information regarding changes in meningococcal population structure over time.
Information about the population structure of N. meningitidis and of the prevalence of capsular switching events is insufficient. The purpose of this study was to determine, during the six year period before MCV4 licensure, the population structure of invasive N. meningitidis in the U.S. and to identify the proportion of meningococcal isolates that demonstrated capsular switching.
METHODS
Study isolates and determination of serogroup
The study isolates were obtained through active, laboratory-based surveillance from January 1, 2000-December 31, 2005, from 10 Active Bacterial Core Surveillance (ABCs) sites. ABCs is a component of the Centers for Disease Control and Prevention (CDC) Emerging Infections Program Network (21). Since MCV4 was licensed in May 2005 and initial vaccine uptake was slow, the entire study period was considered to be the pre-MCV4 era (22).
The following ABCs sites participated: California (three counties in the San Francisco Bay Area), Colorado (5 counties), Connecticut, Georgia, Maryland, Minnesota, New Mexico, New York (15 counties in the Rochester and Albany areas), Oregon, and Tennessee (11 counties). Not all areas participated throughout the study period. According to post-census estimates, the population under surveillance during 2005 was approximately 39.5 million persons, which is approximately 13% of the U.S. population.
The case definition for invasive meningococcal disease is the isolation of N. meningitidis from a normally sterile body fluid from an ABCs site resident (23, 24). Laboratory audits are conducted to identify previously unreported cases.
Laboratory assays
Laboratory work for this study was performed at the CDC and the University of Pittsburgh. Serogrouping was performed as previously described (25). Isolates with discrepant phenotypic serogroup results between the submitting laboratories and CDC or that were non-groupable underwent serogroup specific PCR (SGS-PCR) (26). For isolates with discrepancies between serogrouping and SGS-PCR, the SGS-PCR results were used.
MLST was performed to determine the genetic lineage of each meningococcal isolate and to identify capsular switching. To further define specific meningococcal clones that had undergone capsular switching, OMP gene sequencing of porA VR1 and VR2, porB, and fetA VR was performed as described previously (27–32). DNA sequences were determined using forward and reverse strands.
For clones that had undergone capsular switching, defined below, MLST and SGS-PCR were repeated, using the same template DNA for both tests in the same laboratory at the University of Pittsburgh. For those isolates, the final serogroup result was taken to be the result of the SGS-PCR.
Data analysis
Assembly of MLST sequences was performed using the Staden sequence analysis package or Lasergene 8 (DNAStar, Madison, WI). Sequence Typing Analysis and Retrieval System (STARS) software was used for the determination of alleles (33). The assignment of sequence types (STs), clonal complexes, and porA, porB, and fetA alleles was performed by querying the neisseria.org (neisseria.org/nm/typing/) and PubMLST (pubmlst.org/neisseria/) web sites. STs are considered to belong to the same ST clonal complex when they share alleles at 4 or more of the 7 MLST loci. OMP gene sequence typing results are expressed as porB allele:P1.porA VR1 allele, porA VR2 allele:F.fetA allele (OMP genotype profile) (34, 35).
Capsular switching in a meningococcal isolate was defined as an ST in an isolate of a serogroup not generally associated with that clonal complex and commonly associated with another serogroup.
Bionumerics software version 5.10 (Applied Maths, Austin, Texas) was utilized to create minimum spanning trees (36). The ST with the greatest number of single locus variants (SLVs) was defined as the founder ST.
RESULTS
During the study period, there were 1,301 meningococcal cases that met the ABCs case definition. The study occurred during a time of declining and unusually low meningococcal disease incidence in the United States (1, 37).
Of the 1,301 ABCs cases during the study period, isolates were available for 1,160 (89.2%) (Table 1). The serogroup distribution among the 1,160 isolates was 44.8% serogroup B, 24.1% serogroup C, 2.1% serogroup W-135, and 27.8% serogroup Y. Excluding Oregon, which has an ongoing serogroup B outbreak (38, 39), the serogroup distribution was 35.8% serogroup B, 28.0% serogroup C, 2.2% serogroup W-135, and 32.6% serogroup Y. There were also 2, 1 and 10 serogroup X, A and non-groupable isolates, respectively.
Table 1.
Characteristics of 1,160 meningococcal disease patients and their meningococcal isolates, 10 ABCs sites, 2000–2005.
| Characteristic | No. Isolates | % |
|---|---|---|
| Site of isolation | ||
| Blood | 852 | 73.5 |
| CSF | 284 | 24.5 |
| Other | 24 | 2.0 |
| Male gender | 612 | 52.8 |
| Median age (range) | 19 (0–95) | N.A. |
| Deceased | 122 | 10.5 |
| ABCs site | ||
| California | 115 | 9.9 |
| Colorado | 54 | 4.7 |
| Connecticut | 83 | 7.2 |
| Georgia | 163 | 14.1 |
| Maryland | 118 | 10.2 |
| Minnesota | 152 | 13.1 |
| New Mexico | 11 | 1.0 |
| New York | 81 | 7.0 |
| Oregon | 310 | 26.7 |
| Tennessee | 73 | 6.3 |
| Serogroup | ||
| A | 1 | 0.1 |
| B | 520 | 44.8 |
| C | 280 | 24.1 |
| W-135 | 24 | 2.1 |
| X | 2 | 0.2 |
| Y | 323 | 27.8 |
| Non-groupable | 10 | 0.9 |
Serogroup B
The 520 serogroup B isolates were heterogeneous by MLST and comprised 16 known clonal complexes (Table 2 and Figure 1). However, the majority of isolates belonged to the ST-32 (259 [49.8%] isolates), ST-41/44 (130 [25.0%]), ST-162 (33 [6.3%]), ST-269 (20 [3.8%]), and ST-35 (20 [3.8%]) clonal complexes, accounting for 88.8% of serogroup B isolates. Removing Oregon isolates, the respective percentages among the remaining 304 serogroup B isolates are 32.2%, 32.2%, 10.9%, 5.6%, and 5.3%.
Table 2.
Outer membrane protein (OMP) genotypes of 1160 meningococcal isolates, by serogroup and clonal complex, 10 ABCs sites, 2000–2005. OMP genotype profile are expressed as porB allele:P1.porA VR1 allele, porA VR2 allele:F.fetA allele.
| Serogroup A | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-5 complex/subgroup III – ST-4789 | 3-219: P1.20,9: F3-1 | 1 |
| Serogroup B | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-32 complex/ET-5 complex Represented by ST-32 (222), ST-3584 (11), ST- 1364 (6), ST-5101 (2) and 18 other STs (1 each) |
3-24: P1.7,16: F3-3 | 126 |
| 3-24: P1.7,16-20: F3-3 | 30 | |
| 3-1: P1.7,16: F3-3 | 22 | |
| 3-36: P1.7,16: F3-3 | 18 | |
| 3-84: P1.7,16: F3-3 | 8 | |
| 3-133: P1.7,16-20: F3-3 | 5 | |
| 3-1: P1.7,16-20: F3-3 | 3 | |
| 3-1: P1.7,16-33: F3-3 | 3 | |
| 3-45: P1.7,16-20: F3-3 | 3 | |
| 3-107: P1.22-1,14: F3-3 | 2 | |
| 3-1: P1.19,15: F5-1 | 2 | |
| 3-24: P1.7,16: F1-7 | 2 | |
| 2-88: P1.5-1,10-4: F3-6 | 1 | |
| 3-135: P1.7,16-20: F3-3 | 1 | |
| 3-14: P1.7,16: F3-3 | 1 | |
| 3-150: P1.7,16-20: F3-3 | 1 | |
| 3-160: P1.7,16: F3-3 | 1 | |
| 3-162: P1.22-1,14: F3-3 | 1 | |
| 3-162: P1.7-2,13-1: F3-3 | 1 | |
| 3-164: P1.7,16-20: F3-3 | 1 | |
| 3-1: P1.12-1,13-1: F5-1 | 1 | |
| 3-1: P1.12-1,13-7: F5-1 | 1 | |
| 3-1: P1.12-1,13: F5-1 | 1 | |
| 3-1: P1.17,10-1: F3-3 | 1 | |
| 3-1: P1.7,16-17: F3-3 | 1 | |
| 3-1: P1.7,16-33: F1-7 | 1 | |
| 3-1: P1.7,16-58: F3-3 | 1 | |
| 3-1: P1.7,16: F1-7 | 1 | |
| 3-221: P1.7-2,13: F3-3 | 1 | |
| 3-226: P1.7,16: F3-3 | 1 | |
| 3-231: P1.7,16: F3-3 | 1 | |
| 3-24: P1.18,13: F3-3 | 1 | |
| 3-24: P1.19,15: F3-3 | 1 | |
| 3-24: P1.5-1,16: F3-3 | 1 | |
| 3-24: P1.5-2,10-2: F3-1 | 1 | |
| 3-24: P1.7,16-16: F3-3 | 1 | |
| 3-24: P1.7,16-20: F3-6 | 1 | |
| 3-24: P1.7,16-72: F3-3 | 1 | |
| 3-24: P1.7,16-86: F3-3 | 1 | |
| 3-24: P1.7-2,13-1: F3-3 | 1 | |
| 3-24: P1.7-2,16-32: F3-3 | 1 | |
| 3-24: P1.7-2,16: F3-3 | 1 | |
| 3-24: P1.7-2,3: F3-3 | 1 | |
| 3-25: P1.7-2,16-20: F3-3 | 1 | |
| 3-31: P1.7,16: F3-3 | 1 | |
| 3-38: P1.7,16: F3-3 | 1 | |
| 3-8: P1.7,16-33: F3-3 | 1 | |
| ST-41/44 complex/Lineage 3 Represented by ST-136 (22), ST-44 (13), ST-170 (10), ST-437 (7), ST-41 (7), ST-154 (6), ST-4682 (5), ST-5111 (3), ST-409 (3), ST-5097 (3), ST-42 (3), ST-40 (2), ST-43 (2), ST-2459 (2),ST-1374 (2), ST-1213 (2), ST-4489 (2), ST-318 (2), ST-5890 (2) and 32 other STs (1 each) |
3-1: P1.7-2,4: F1-5 | 15 |
| 3-71: P1.22-1,14: F5-2 | 10 | |
| 3-107: P1.17,16-3: F5-5 | 8 | |
| 3-138: P1.21,16: F1-5 | 8 | |
| 3-38: P1.7-1,1: F1-7 | 5 | |
| 3-45: P1.7-4,1: F1-7 | 4 | |
| 3-107: P1.17,16-23: F5-5 | 3 | |
| 3-45: P1.21,16: F1-7 | 3 | |
| 3-154: P1.17,16-3: F5-13 | 1 | |
| 3-114: P1.22-1,14: F5-2 | 2 | |
| 3-16: P1.19,15-1: F1-5 | 2 | |
| 3-172: P1.7-2,4: F1-5 | 2 | |
| 3-1: P1.7-2,13-2: F1-5 | 2 | |
| 3-1: P1.7-2,13-9: F1-5 | 2 | |
| 3-1: P1.7-2,4: F1-7 | 2 | |
| 3-45: P1.7-1,1: F5-5 | 2 | |
| 3-82: P1.18-1,34-2: F1-5 | 2 | |
| 2-55: P1.5-1,2-2: F5-8 | 1 | |
| 3-100: P1.22-1,14: F5-2 | 1 | |
| 3-100: P1.7-2,13: F1-5 | 1 | |
| 3-24: P1.7,16: F3-3 | 1 | |
| 3-107: P1.17,16-3: F3-6 | 1 | |
| 3-107: P1.17,16-3: fetA frameshift mutation |
1 | |
| 3-107: P1.5-2,10-1: F1-7 | 1 | |
| 3-107: P1.5-2,10-2: F5-5 | 1 | |
| 3-134: P1.7-1,1: F1-7 | 1 | |
| 3-137: P1.17,16-3: F5-5 | 1 | |
| 3-138: P1.7,16: F1-5 | 1 | |
| 3-139: P1.21,16: F1-7 | 1 | |
| 3-146: P1.12-6,13-4: F1-7 | 1 | |
| 3-14: P1.17,16-91: F3-1 | 1 | |
| 3-151: P1.19,15-1: F1-5 | 1 | |
| 3-152: P1.17,16-3: F5-5 | 1 | |
| 3-154: P1.17,16-49: F5-13 | 1 | |
| 3-154: P1.18-1,3: F5-13 | 1 | |
| 3-154: P1.22,14-6: F5-13 | 1 | |
| 3-157: P1.7-2,4: F1-1 | 1 | |
| 3-165: P1.5-1,10-4: F1-5 | 1 | |
| 3-167: P1.22-1,14: F5-2 | 1 | |
| 3-169: P1.17,16-3: F4-1 | 1 | |
| 3-16: P1.20,23-1: F1-5 | 1 | |
| 3-16: P1.7-1,1: F3-29 | 1 | |
| 3-16: P1.7-1,1: F5-67 | 1 | |
| 3-170: P1.22-1,14: F5-2 | 1 | |
| 3-175: P1.7-1,1: F5-2 | 1 | |
| 3-192: P1.7-2,4: F1-5 | 1 | |
| 3-1: P1.18-1,3: F5-51 | 1 | |
| 3-1: P1.21,16-2: F1-5 | 1 | |
| 3-1: P1.7-1,1: F3-29 | 1 | |
| 3-1: P1.7-1,1: F5-2 | 1 | |
| 3-1: P1.7-2,4-2: F1-5 | 1 | |
| 3-216: P1.7-2,4: F5-8 | 1 | |
| 3-224: P1.22-1,14: F5-2 | 1 | |
| 3-225: P1.7-2,4: F1-5 | 1 | |
| 3-227: P1.17,16-3: F3-6 | 1 | |
| 3-228: P1.7-4,1: F1-7 | 1 | |
| 3-24: P1.7-2,4: F1-5 | 1 | |
| 3-260: P1.7-1,1: F1-7 | 1 | |
| 3-35: P1.21,16: fetA deletion | 1 | |
| 3-38: P1.17,1-7: F3-1 | 1 | |
| 3-38: P1.17,16-3: F3-6 | 1 | |
| 3-39: P1.17,16-3: F5-5 | 1 | |
| 3-44: P1.22-1,2-2: F1-59 | 1 | |
| 3-45: P1.18-1,3: F1-5 | 1 | |
| 3-45: P1.7-1,1: F1-7 | 1 | |
| 3-45: P1.7-1,1: F3-1 | 1 | |
| 3-45: P1.7-1,1: F5-14 | 1 | |
| 3-45: P1.7-4,1: F5-5 | 1 | |
| 3-58: P1.22,14-6: F3-1 | 1 | |
| 3-71: P1.5-1,2-2: F5-2 | 1 | |
| 3-82: P1.7-2,13-31: F2-4 | 1 | |
| 3-84: P1.7-1,1: F1-7 | 1 | |
| 3-84: P1.7-2,4: F1-5 | 1 | |
| 3-90: P1.19,15: F3-6 | 1 | |
| ST-162 complex Represented by ST-162 (29) and 4 other STs (1 each) |
3-73: P1.22,14: F5-9 | 21 |
| 3-115: P1.22,14: F5-9 | 1 | |
| 3-143: P1.22,14-16: F5-9 | 1 | |
| 3-153: P1.22,14: F5-9 | 1 | |
| 3-155: P1.22,14: F5-9 | 1 | |
| 3-173: P1.22,14: F5-9 | 1 | |
| 3-174: P1.22,14: F5-9 | 1 | |
| 3-73: P1.22,14-20: F5-9 | 1 | |
| 3-73: P1.22,14: F5-8 | 1 | |
| 3-73: P1.22,15: F5-9 | 1 | |
| 3-81: P1.7-2,4-21: F5-9 | 1 | |
| 3-81: P1.7-2,4-22: F5-9 | 1 | |
| 3-81: P1.7-2,4: F5-9 | 1 | |
| ST-269 complex Represented by ST-2976 (3), ST-2974 (2), ST-2738 (2) and 13 other STs (1 each) |
3-113: P1.7-2,13-1: F5-7 | 2 |
| 3-25: P1.19-1,15-11: F5-1 | 2 | |
| 3-25: P1.22,9: F5-12 | 2 | |
| 3-100: P1.7-2,13-1: F3-9 | 1 | |
| 3-14: P1.19,15: F1-7 | 1 | |
| 3-16: P1.12-1,23: F1-7 | 1 | |
| 3-16: P1.7-2,13-1: F3-9 | 1 | |
| 3-16: P1.7-2,13-25: F3-9 | 1 | |
| 3-16: P1.7-2,13-2: F3-9 | 1 | |
| 3-16: P1.7-2,13: F3-9 | 1 | |
| 3-19: P1.5-1,10-19: F3-1 | 1 | |
| 3-1: P1.12-1,14-2: F5-8 | 1 | |
| 3-217: P1.7-4,1: F5-1 | 1 | |
| 3-45: P1.5-3,10-2: F5-1 | 1 | |
| 3-55: P1.19,13-2: F3-9 | 1 | |
| 3-55: P1.19,15: F3-9 | 1 | |
| 3-55: P1.31,26: F3-9 | 1 | |
| ST-35 complex Represented by ST-35 (7), ST-457 (3), ST-5885 (2), ST-2612 (2) and 6 other STs (1 each) |
3-39: P1.22-1,14: F4-1 | 9 |
| 3-39: P1.22-1,14: fetA deletion |
3 | |
| 3-171: P1.22-1,14: F4-1 | 1 | |
| 3-1: P1.12-1,16: F1-7 | 1 | |
| 3-25: P1.7-2,13-1: F1-7 | 1 | |
| 3-261: P1.19,15: F4-1 | 1 | |
| 3-35: P1.22-1,14: F4-1 | 1 | |
| 3-39: P1.5-1,2-2: F5-7 | 1 | |
| 3-39: P1.5-2,10-9: F4-1 | 1 | |
| 3-45: P1.22-1,14: F1-7 | 1 | |
| ST-60 complex Represented by ST-60 (6) and 2 other STs (1 each) |
3-8: P1.22-1,14: F3-9 | 2 |
| 2-152: P1.5-1,2-2: F1-7 | 1 | |
| 2-158: P1.21,16: F1-87 | 1 | |
| 2-161: P1.5,2: F1-7 | 1 | |
| 2-37: P1.5,2: F1-7 | 1 | |
| 2-37: P1.5-1,2: F1-7 | 1 | |
| 2-65: P1.5-1,2-2: F1-7 | 1 | |
| ST-213 complex Represented by ST-213 (4) and 3 other STs (1 each) |
3-14: P1.22,14: F5-5 | 3 |
| 3-64: P1.22,14: F5-5 | 2 | |
| 3-152: P1.22,14: F5-5 | 1 | |
| 3-213: P1.19,15: F5-5 | 1 | |
| ST-103 complex Represented by ST-103, ST-3594, ST-5966, ST-6063 (1 each) |
2-160: P1.17,16-3: F5-5 | 1 |
| 2-22: P1.18-1,3: F3-9 | 1 | |
| 2-22: P1.5-2,10: F3-9 | 1 | |
| 2-22: P1.7-1,1: F1-18 | 1 | |
| ST-37 complex Represented by ST-916 (2) and ST-917 (2) |
2-130: P1.7-2,13: F1-5 | 1 |
| 2-132: P1.5-4,2-2: F3-3 | 1 | |
| 2-64: P1.18,2-2: F3-3 | 1 | |
| 2-76: P1.7-2,13-1: F1-5 | 1 | |
| ST-254 complex Represented by ST-254 (2) and ST-3590 (1) |
3-223: P1.19,15: F1-5 | 2 |
| 3-36: P1.22,14: F1-5 | 1 | |
| ST-11 complex/ET-37 complex – ST-11 | 2-2: P1.22,14: F1-30 | 1 |
| 2-2: P1.5,2: F3-6 | 1 | |
| ST-549 complex ST-5887 and ST-5874 |
3-1: P1.21,16: F1-20 | 1 |
| 3-45: P1.21,16-41: F1-20 | 1 | |
| ST-22 complex – ST-5870 | 2-23: P1.18-1,3: F4-1 | 1 |
| ST-334 complex – ST-5873 | 2-134: porA deletion: F1-5 | 1 |
| ST-364 complex – ST-4046 | 2-81: P1.18-7,9: F5-5 | 1 |
| ST-461 complex – ST-461 | 3-14: P1.19-2,13: F5-68 | 1 |
| No clonal complex Represented by ST-2048 (6), ST-2875 (2) and 16 other STs (1 each) |
2-136: porA deletion: F4-1 | 2 |
| 3-154: P1.17,16-3: F5-13 | 1 | |
| 3-16: P1.12-1,16-8: F3-6 | 2 | |
| 2-54: P1.5-1,10-4: F3-29 | 1 | |
| 2-55: P1.22,14: F1-7 | 1 | |
| 3-107: P1.17,16-79: F5-5 | 1 | |
| 3-136: P1.5-1,10-4: F1-5 | 1 | |
| 3-146: P1.22,14: F3-6 | 1 | |
| 3-156: P1.17,16-3: F5-13 | 1 | |
| 3-166: P1.17,9: F1-15 | 1 | |
| 3-168: P1.22-15,28-2: F3-5 | 1 | |
| 3-16: P1.12-1,16-8: F1-62 | 1 | |
| 3-16: P1.22,9: F3-6 | 1 | |
| 3-196: P1.12-1,16-8: F3-6 | 1 | |
| 3-1: P1.19-2,15: F1-86 | 1 | |
| 3-212: P1.22,9: F3-6 | 1 | |
| 3-220: P1.5-1,10-4: F5-1 | 1 | |
| 3-38: P1.17,16-3: F5-5 | 1 | |
| 3-38: P1.19,15: F5-28 | 1 | |
| 3-45: P1.21,16: F1-20 | 1 | |
| 3-73: P1.12-1,16-8: F1-5 | 1 | |
| Serogroup C | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-11 complex/ET-37 complex Represented by ST-11 (170), ST-2962 (25), ST- 2961 (6) and 13 other STs (1 each) |
2-2: P1.5,2: F3-6 | 46 |
| 2-2: P1.5,2: F1-30 | 25 | |
| 2-2: P1.22-1,14: F3-6 | 15 | |
| 2-75: P1.5-1,10-4: F3-6 | 15 | |
| 2-2: P1.5-1,10-8: F3-6 | 11 | |
| 2-88: P1.5-1,10-4: F3-6 | 10 | |
| 2-2: P1.5,2: F3-3 | 8 | |
| 2-48: P1.5,2: F1-30 | 6 | |
| 2-2: P1.5-1,2-2: F3-6 | 5 | |
| 2-73: P1.5,2: F3-6 | 5 | |
| 2-2: P1.5,2: F1-5 | 4 | |
| 2-2: P1.5,2: F5-5 | 4 | |
| 2-2: P1.17,16-3: F3-6 | 3 | |
| 2-2: P1.5,2: F4-12 | 3 | |
| 2-2: P1.5-1,2: F5-36 | 3 | |
| 2-85: P1.22-1,14: F3-6 | 3 | |
| 2-2: P1.5,2: F3-1 | 2 | |
| 2-2: P1.5,2: F4-1 | 2 | |
| 2-2: P1.5-2,10-2: F5-36 | 2 | |
| 2-108: P1.22,14: F3-6 | 1 | |
| 2-111: P1.22-1,14: F3-6 | 1 | |
| 2-112: P1.22-1,14: F3-6 | 1 | |
| 2-114: P1.5,2: F3-6 | 1 | |
| 2-115: P1.22-1,14: F3-6 | 1 | |
| 2-118: P1.5,2: F3-6 | 1 | |
| 2-121: P1.22,14: F3-6 | 1 | |
| 2-122: P1.22,14: F5-5 | 1 | |
| 2-123: P1.22-1,14: F3-6 | 1 | |
| 2-124: P1.5,2: F3-6 | 1 | |
| 2-125: P1.5-1,2-2: F3-6 | 1 | |
| 2-126: P1.5,2: F3-6 | 1 | |
| 2-127: P1.5-1,2: F5-36 | 1 | |
| 2-138: P1.5,2-51: F3-6 | 1 | |
| 2-153: P1.5,2: F3-6 | 1 | |
| 2-154: P1.5,2: F3-3 | 1 | |
| 2-2: P1.22,14: F1-30 | 1 | |
| 2-2: P1.5,13-7: F3-1 | 1 | |
| 2-2: P1.5,2-1: F5-5 | 1 | |
| 2-2: P1.5,2-47: F3-6 | 1 | |
| 2-2: P1.5,2-54: F1-30 | 1 | |
| 2-2: P1.5,2-55: F3-6 | 1 | |
| 2-2: P1.5,2: F1-1 | 1 | |
| 2-2: P1.5,2: F1-7 | 1 | |
| 2-2: P1.5-1,10-4: F5-58 | 1 | |
| 2-2: P1.5-1,10-55: F3-6 | 1 | |
| 2-2: P1.5-1,10-8: F4-1 | 1 | |
| 2-2: P1.5-1,10-8: F5-13 | 1 | |
| 2-2: P1.5-1,2-2: F5-9 | 1 | |
| 2-2: P1.5-2,2-2: F3-6 | 1 | |
| 2-2: P1.7-1,1: F1-30 | 1 | |
| 2-2: porA deletion: F3-6 | 1 | |
| 2-39: P1.22-1,14: F3-6 | 1 | |
| 2-48: P1.5,2: F1-33 | 1 | |
| 2-48: P1.5,2: F3-6 | 1 | |
| 2-48: P1.5-1,2: F1-1 | 1 | |
| 2-52: P1.5,2: F3-3 | 1 | |
| 2-53: P1.5-1,10-8: F4-1 | 1 | |
| 2-65: P1.5,2: F1-30 | 1 | |
| 2-78: P1.5-1,2-2: F3-6 | 1 | |
| 2-79: P1.17,16-3: F3-6 | 1 | |
| 2-79: P1.5,2: F1-30 | 1 | |
| ST-103 complex Represented by ST-2006 (17), ST-5837 (2) and ST- 5831 (1) |
2-110: P1.5-1,10-4: F3-9 | 14 |
| 2-22: P1.17,16-3: F1-18 | 2 | |
| 2-110: P1.5,2: F3-9 | 1 | |
| 2-113: P1.5-1,10-4: F3-9 | 1 | |
| 2-116: P1.5-1,10-4: F3-9 | 1 | |
| 2-22: P1.5-1,10-4: F3-9 | 1 | |
| ST-32 complex/ET-5 complex Represented by ST-32 (9), ST-1364 (2) and 2 other STs (1 each) |
3-24: P1.7,16: F3-3 | 7 |
| 3-107: P1.7,16-26: F3-3 | 1 | |
| 3-195: P1.19-7,15: F3-3 | 1 | |
| 3-1: P1.5-2,10: F5-1 | 1 | |
| 3-24: P1.7,16: F5-2 | 1 | |
| 3-24: porA deletion: F3-3 | 1 | |
| 3-36: P1.7,16: F3-3 | 1 | |
| ST-35 complex Represented by ST-278 (5) and 3 other STs (1 each) |
3-194: P1.7-2,13-2: F3-3 | 1 |
| 3-200: P1.7-1,1: F1-7 | 1 | |
| 3-254: P1.7-2,13-1: F1-7 | 1 | |
| 3-36: P1.7-2,13-2: F1-7 | 1 | |
| 3-39: P1.22-1,14: fetA deletion |
1 | |
| 3-45: P1.18-1,3: F1-7 | 1 | |
| 3-45: P1.7-2,13-1: F1-7 | 1 | |
| 3-45: P1.7-2,13-34: F1-7 | 1 | |
| ST-41/44 complex/Lineage 3 Represented by ST-41 (2), ST-437 (2) and 3 other STs (1 each) |
3-1: P1.7-2,4: F1-5 | 2 |
| 3-71: P1.5-1,2-2: F1-28 | 2 | |
| 3-183: P1.21-2,28: F1-5 | 1 | |
| 3-38: P1.7-2,13-2: F1-7 | 1 | |
| 3-71: P1.22-1,14: F5-2 | 1 | |
| ST-269 complex Represented by ST-5840 (2) and 2 other STs (1 each) |
3-197: P1.7-2,13: F3-9 | 2 |
| 3-146: P1.12-1,16-8: F1-5 | 1 | |
| 3-45: P1.5-1,10-4: fetA deletion |
1 | |
| ST-8 complex/Cluster A4 – ST-8 | 2-3: P1.5,2: F5-8 | 2 |
| ST-213 complex – ST-213 | 3-14: P1.22,14: F5-5 | 1 |
| ST-23 complex/Cluster A3 – ST-183 | 3-53: P1.5-2,10-2: F4-1 | 1 |
| ST-461 complex – ST-4027 | 3-14: P1.19-28,13-1: F1-5 | 1 |
| ST-60 complex – ST-60 | 2-99: P1.5-1,10-8: F1-77 | 1 |
| No clonal complex Represented by ST-2048 (5) and 3 other STs (1 each) |
3-16: P1.5,2: F3-6 | 3 |
| 2-100: P1.5-2,10: F1-66 | 1 | |
| 2-54: P1.5-1,10-4: F3-29 | 1 | |
| 3-145: P1.7-2,13-1: F5-5 | 1 | |
| 3-16: P1.19-27,15-1: F3-6 | 1 | |
| 3-16: P1.22-1,2: F3-6 | 1 | |
| Serogroup W135 | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-22 complex Represented by ST-22 (10), ST-1265 (2), ST-1476 (2), ST-1065 (2) and 8 other STs (1 each) |
2-23: P1.18-1,3: F4-1 | 12 |
| 2-109: P1.18-1,3: F4-1 | 2 | |
| 2-109: P1.18-1,3: F3-6 | 1 | |
| 2-109: porA deletion: F1-20 | 1 | |
| 2-119: P1.18-1,3: F4-1 | 1 | |
| 2-120: P1.18-1,3: F4-1 | 1 | |
| 2-156: P1.18-1,3: F4-1 | 1 | |
| 2-22: P1.18-1,3: F1-5 | 1 | |
| 2-23: P1.18-1,3: F1-5 | 1 | |
| 2-23: P1.18-1,3: F4-18 | 1 | |
| 2-23: P1.18-1,3: F5-5 | 1 | |
| 3-36: P1.5-2,10-1: F3-6 | 1 | |
| Serogroup X | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-103 complex – ST-5848 | 2-22: P1.18-1,3: F4-1 | 1 |
| ST-175 complex – ST-2980 | 3-100: P1.5-1,10-1: F5-6 | 1 |
| Serogroup Y | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-23 complex/Cluster A3 Represented by ST-23 (254), ST-1625 (13), ST-183 (4), ST-3587 (4), ST-3582 (4), ST-1621 (3),ST- 6315 (2) and 19 other STs (1 each) |
3-36: P1.5-2,10-1: F4-1 | 159 |
| 2-55: P1.5-1,2-2: F5-8 | 84 | |
| 3-36: P1.5,2: F4-1 | 9 | |
| 2-141: P1.5-1,2-2: F5-8 | 4 | |
| 3-36: P1.5-2,10-2: F4-1 | 4 | |
| 3-53: P1.5-2,10-2: F4-1 | 4 | |
| 3-117: P1.5-2,10-1: F4-1 | 2 | |
| 3-36: P1.5-1,2-2: F5-8 | 2 | |
| 3-36: P1.5-2,10-12: F4-1 | 2 | |
| 3-36: P1.5-2,10-29: F4-1 | 2 | |
| 2-131: P1.5-1,2-2: F5-8 | 1 | |
| 2-137: P1.5-1,2-2: F5-8 | 1 | |
| 2-155: P1.5-1,2-2: F5-8 | 1 | |
| 2-157: P1.5-1,2-2: F5-8 | 1 | |
| 2-159: P1.5-1,2-2: F5-8 | 1 | |
| 2-55: P1.12-1,2-2: F5-8 | 1 | |
| 2-55: P1.22,2-2: F5-8 | 1 | |
| 2-55: P1.22-1,2-2: F5-8 | 1 | |
| 2-55: P1.5-1,2-2: F5-1 | 1 | |
| 2-55: P1.5-1,2-30: F5-8 | 1 | |
| 2-55: P1.5-17,2-2: F5-8 | 1 | |
| 2-55: P1.5-2,10-1: F4-1 | 1 | |
| 2-55: P1.5-2,10-9: F5-8 | 1 | |
| 2-55: porA deletion: F5-8 | 1 | |
| 2-68: P1.5-1,2-2: F5-8 | 1 | |
| 2-89: P1.5-1,2-2: F5-8 | 1 | |
| 3-112: P1.5-2,10-1: F4-1 | 1 | |
| 3-198: P1.5-2,10-1: F4-1 | 1 | |
| 3-198: P1.5-2,10-1: F5-8 | 1 | |
| 3-211: P1.5-2,10-1: F4-1 | 1 | |
| 3-214: P1.5-2,10-1: F4-1 | 1 | |
| 3-215: P1.5-2,10-1: F4-1 | 1 | |
| 3-218: P1.5-2,10-29: F4-1 | 1 | |
| 3-230: P1.5-2,10-2: F4-1 | 1 | |
| 3-259: P1.5-2,10-1: F4-1 | 1 | |
| 3-36: P1.5-2,10-1: F5-8 | 1 | |
| 3-36: P1.5-2,10-37: F4-1 | 1 | |
| 3-36: P1.5-2,10-51: F4-1 | 1 | |
| 3-36: P1.5-2,10-63: F4-1 | 1 | |
| 3-53: P1.5,2: F4-1 | 1 | |
| ST-167 complex Represented by ST-1624 (10),ST-167 (2) and 4 other STs (1 each) |
2-55: P1.5-1,10-4: F3-4 | 8 |
| 2-135: P1.5-1,10-4: F3-4 | 1 | |
| 3-222: P1.5-1,10-4: F3-4 | 1 | |
| 3-229: P1.5-1,10-4: F3-6 | 1 | |
| 3-36: P1.5-1,10-8: F1-3 | 1 | |
| 3-45: P1.5-1,10-1: F4-1 | 1 | |
| 3-48: P1.5-1,10-1: F4-1 | 1 | |
| 3-48: P1.5-1,10-4: F4-1 | 1 | |
| 3-71: P1.5-1,10-4: F3-4 | 1 | |
| ST-22 complex – ST-1265 | 2-23: P1.18-1,3: F1-7 | 2 |
| ST-174 complex – ST-1466 | 3-35: P1.21,16: F3-7 | 1 |
| ST-32 complex/ET-5 complex – ST-6065 | 3-24: P1.7,16: F3-3 | 1 |
| Nongroupable | ||
| Clonal Complex | OMP Genotype Profile | No. Isolates |
| ST-60 complex Represented by ST-60 (3) |
2-133: P1.5,2: F1-83 | 1 |
| 2-139: P1.5,2: F1-7 | 1 | |
| 2-65: P1.5,2: F1-7 | 1 | |
| ST-103 complex Represented by ST-103 and ST-5842 |
2-22: P1.18-1,3: F3-9 | 1 |
| 2-22: P1.7-2,13-1: F1-18 | 1 | |
| ST-198 complex Represented by ST-823 and ST-6504 |
3-84: P1.17,9: F5-5 | 2 |
| ST-23 complex/Cluster A3 – ST-23 | 3-36: P1.5-2,10-1: F4-1 | 1 |
| ST-32 complex/ET-5 complex – ST-5138 | 3-24: P1.7,16-20: F3-3 | 1 |
| No clonal complex – ST-6068 | 3-14: P1.18,25: F1-5 | 1 |
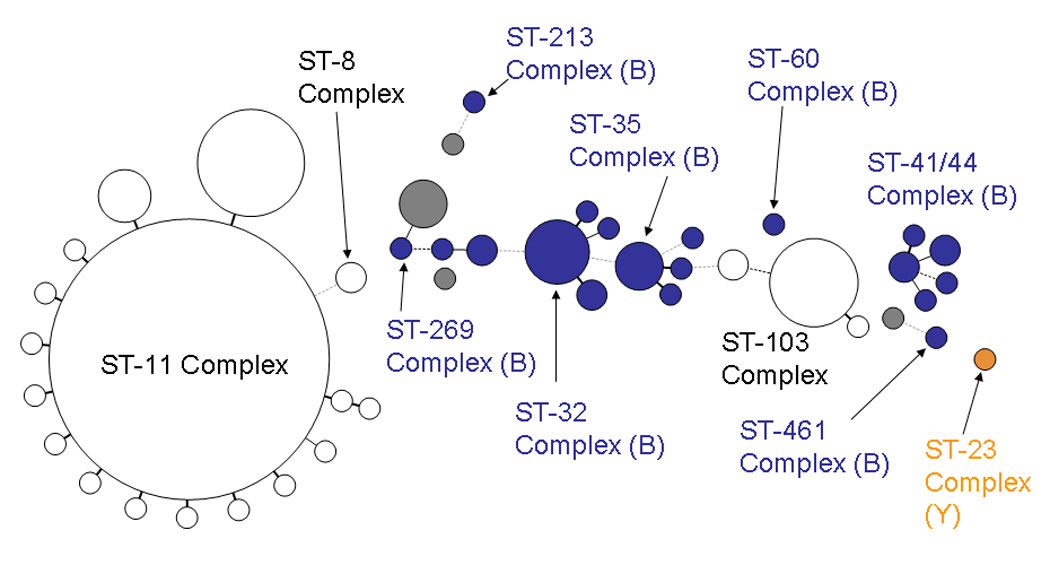
Figure 1.

Figure 1–Figure 4. Minimum spanning tree analysis of ABCs isolates by sequence type, 2000–2005. Figure 1, serogroup B (520 isolates); Figure 2, serogroup C (280 isolates); Figure 3, serogroup Y (323 isolates); Figure 4, serogroup W-135 (24 isolates).
The size of the circles is proportional to the number of isolates represented. Circles in white are isolates with STs that are generally associated with the serogroup represented in the figure (e.g., serogroup B for Figure 1). Circles in color represent isolates with STs generally associated with another serogroup (shown in parentheses), indicating capsular switching. Heavy solid lines represent single locus variants, light solid lines are double locus variants, heavy dotted lines are triple locus variants, light dotted lines are 4-locus variants, grey circles represent STs that are not part of any clonal complex
The ST-32 clonal complex was comprised of 22 STs and two predominant OMP genotype profiles. The clone comprising the largest number of isolates (126 isolates) was 3-24:P1.7,16: F3-3, which is the clone causing the prolonged serogroup B epidemic in Oregon (38, 39). The clone predominated in Oregon but was also present in 7 of the other 9 ABCs sites. The next most common ST-32 OMP genotype profile was 3-24:P1.7,16-20: F3-3, which differed from the Oregon clone in that it is missing 3 amino acids in PorA VR2 (neisseria.org/nm/typing/pora/vr2.shtml). This clone was observed primarily in California and Minnesota, but was present also in 5 other ABCs sites, including Oregon. These two OMP genotype profiles accounted for 156 (60.2%) of the 259 ST-32 clonal complex isolates. There were other relatively uncommon antigenic variants of the ST-32 clonal complex.
The ST-41/44 clonal complex was comprised of 51 STs and many OMP genotype profiles. The OMP genotype profile 3-1:P1.7-2,4:F1-5, accounting for 15 (11.5%) of ST-41/1 44 clonal complex isolatesis consistent with the serogroup B clone causing the longstanding epidemic in New Zealand (40–43). Isolates with this OMP genotype profile were present in 5 ABCs sites over all 6 years.
The ST-162 clonal complex was comprised of 5 STs with ST-162, accounting for 29 (87.9%) of ST-162 clonal complex isolates. The predominant OMP genotype profile for this clonal complex was 3-73: P1.22,14:F5-9, accounting for 21 (63.6%) isolates. Among 20 ST-269 clonal complex isolates, there were 16 different STs with no single ST predominating. There was also no predominant OMP genotype profile: only 3 profiles were present in more than one isolate. ST-35 clonal complex had 20 isolates with 10 STs. The predominant OMP genotype profile for the ST-35 complex isolates was 3-39:P1.22-1,14:F4-1 (9/20) and an additional three isolates shared this profile with the exception of a fetA deletion.
Serogroup C
Of 280 serogroup C isolates, 214 (76.4%) belonged to the ST-11 clonal complex (Table 2 and Figure 2). The serogroup C “early” and “late” clones that were identified in Maryland in the 1990s were the predominant clones, differing only at the fetA locus (2-2:P1.5,2:F1-30 [25 (11.7%)] and 2-2:P1.5,2:F3-6 [46 (21.5%)], respectively) (10). There were 20 ST-103 clonal complex isolates (44). Most of these isolates (13 [65.0%]) were ST-2006 had OMP genotype profile 2-110:P1.5-1,10-4:F3-9.
Figure 2.
Serogroup Y
Of 323 serogroup Y isolates, 303 (93.8%) belonged to the ST-23 clonal complex, 254 (83.8%) of which were ST-23 (Table 2 and Figure 3). Two OMP genotype profiles were highly predominant: 2-55:P1.5-1,2-2:F5-8 (early) and 3-36:P1.5-2,10-1:F4-1 (late) comprising 84 (27.7%) and 159 (52.5%)of the ST-23 complex isolates, respectively (10). The 16 ST-167 complex isolates were comprised of 6 STs, with 10 (62.5%) isolates being ST-1624. OMP 2-55:P1.5-1,10-4:F3-4 was present in 8 (50.0%) of 16 isolates.
Figure 3.

Serogroup W-135
Of serogroup W-135 isolates, all 24 belonged to the ST-22 clonal complex, 10 (41.7%) of which were ST-22 (Table 2 and Figure 4). ST-22 clonal complex isolates were predominantly 2-23:P1.18-1,3:F4-1, accounting for 12 (50.0%) of isolates. There were no ST-11 serogroup W-135 isolates, indicating absence of the clone associated with the world-wide 2000 Hajj-associated outbreak.
Figure 4.

Serogroups A and X
There was one serogroup A isolate, which belonged to ST-4789, part of the ST-5 clonal complex (Table 2). There were two serogroup X isolates. One was ST-2980, which belongs to the ST-175 clonal complex and is usually associated with serogroup W-135 and the other was an ST-103 complex isolate which is usually associated with serogroup C.
Deletion of OMP genes
(Table 2). There were 7 isolates with evidence of porA deletion -2 serogroup C, 3 serogroup B, 1 serogroup W-135, and 1 serogroup Y isolates (10). In addition, there were 4 serogroup B and 2 serogroup C isolates with evidence of fetA deletion (45, 46). One nongroupable ST41/44 isolate had a single nucleotide substitution in the fetA coding region resulting in a frameshift mutation and a protein that is predicted to be non-functional.
Capsular switching
Capsular switching was observed in 8 (1.5%) of serogroup B, 36 (12.9%) of serogroup C, none of serogroup of W-135, both (100%) of the serogroup X, and 3 (0.9%) of serogroup Y isolates (Figure 1–Figure 4). The frequency of capsular switching observed in serogroup C isolates was statistically significantly higher than observed in serogroup B or Y isolates (p value <0.0001).
Of serogroup C isolates, 35 (97.2%) of the 36 isolates demonstrating capsular switching appeared to have arisen from serogroup B clones. Four belonged to the ST-269 clonal complex, 1 belonged to the ST-213 clonal complex, 8 belonged to the ST-35 clonal complex, 13 belonged to the ST-32 clonal complex, 1 belonged to the ST-60 clonal complex, 1 belonged to the ST-461 clonal complex, and 7 belonged to the ST-41/44 clonal complex. There was one serogroup C ST-23 isolate, an ST which is usually associated with serogroup Y. There were 3 (0.9%) serogroup Y isolates that demonstrated capsular switching, 1 belonging to the ST-32 clonal complex (generally associated with serogroup B) and 2 belonging to the ST-22 clonal complex (serogroup W-135). For serogroup B, there were 4 ST-103, 1 ST-334, and 2 ST-11 (all associated with serogroup C); and 1 ST-22 (serogroup W-135) clonal complex isolates.
To determine whether capsular switching could be identified within specific meningococcal clones, as defined by MLST and OMP genotype profile, we further analyzed by OMP genotyping isolates that had demonstrated switching. Eight meningococcal clones that had undergone capsular switching were identified (Table 3). For example, a serogroup C to serogroup B switch occurred within an ST-11 2-2:P1.22,14: F1-30 clone. Similarly, a serogroup B to serogroup C switch was identified within an ST-32 3-24:P1.7,16: F3-3 clone. In many instances, there was overlap by ABCs site and year in the presence of isolates of both serogroups representing capsular switching within a specific clone (Table 3).
Table 3.
Isolates from 10 ABCs sites demonstrating capsular switching that match by outer membrane protein genotyping, 2000–2005. For each group of capsular switches, the serogroup generally associated with the ST is shown at the top (e.g., ST-11 is most commonly associated with serogroup C, ST-32 is associated with serogroup B).
| ST | CC | OMP genotype profile | Serogroup | No. Isolates isolates |
ABCs site/year(s) |
|---|---|---|---|---|---|
| 11 | 11 | 2-2:P1.22,14: F1-30 | C | 1 | GA/2003 |
| B | 1 | MN/2003 | |||
| 11 | 11 | 2-2:P1.5,2: F3-6 | C | 43 | GA/2000-03 TN/2001 CA/2001-03 CO/2000 CT/2001-05 MD/2000-01, 2003 MN/2000, 2003-04 NM/2004 NY/2000-01 OR/2002 |
| B | 1 | GA/2001 | |||
| 32 | 32 | 3-24:P1.7,16: F3-3 | B | 123 | MN/2001-05 OR/2000-05 CA/2000, 2003-04 CO/2003-05 CT/2005 GA/2001 MD/2000 TN/2001 |
| C | 5 | MN/2004-05 OR/2004-05 |
|||
| 32 | 32 | 3-36:P1.7,16: F3-3 | B | 17 | OR/2000, 2001, 2003, 2005 CT/2005 |
| C | 1 | OR/2005 | |||
| 1364 | 32 | 3-24:P1.7,16: F3-3 | B | 2 | CA/2000 OR/2005 |
| C | 2 | OR/2000-01 | |||
| 41 | 41/44 | 3-1:P1.7-2,4: F1-5 | B | 2 | CT/2002, 2004 |
| C | 2 | CA/2003-04 | |||
| 183 | 23 | 3-53:P1.5-2,10-2: F4-1 | Y | 2 | NY/2000, 2005 |
| C | 1 | MD/2001 | |||
| 213 | 213 | 3-14:P1.22,14: F5-5 | B | 1 | OR/2001 |
| C | 1 | MN/2004 |
ST, sequence type; CC, clonal complex; OMP, outer membrane protein
DISCUSSION
This study defines the population structure and OMP genotype profile of invasive N. meningitidis isolates circulating in the United States during the 6 years before licensure of MCV4, as well as isolates that had undergone capsular switching. To our knowledge, this is the first population-based assessment of capsular switching prevalence among invasive meningococcal isolates.
As expected, the population of invasive isolates was primarily composed of a select group of recognized hypervirulent lineages. Clonal complexes ST-11, ST-23, and ST-22 accounted for the majority of isolates belonging to serogroup C, Y, and W-135, respectively. For serogroup B, clonal complexes ST-32 and ST-41/44 predominated. In addition to their presence in the United States, these lineages also have a global distribution (1).
A substantial proportion of invasive serogroup C, Y, and B isolates demonstrated capsular switching, indicating that this is a common natural phenomenon in Neisseria meningitidis. These isolates retain their invasiveness; however, during the period of this study, the isolates that arose through capsular switching caused less disease than the isolates from the same genetic lineage of the original serogroup. For example, there was only one serogroup B, ST-11 isolate, whereas ST-11 isolates made up the bulk of serogroup C isolates. Whether this change in type of capsular polysaccharide expression in a different genotype affects transmission, carriage, virulence, or some other aspects of meningococcal biology is unknown. The timing of the capsular switches that we identified is also unknown. In the case of the Hajj serogroup W-135 outbreak, the serogroup C capsular switch appears to have occurred long before the onset of the 2000 outbreak in Saudi Arabia, but may have been selected for by serogroup A/C polysaccharide vaccine pressure in Hajj pilgrims (11). In contrast, Vogel et al demonstrated rapid capsule switching in the case of a teenage girl who died of ST-32 serogroup B meningococcal disease and her boyfriend who had pharyngeal carriage with an otherwise indistinguishable serogroup C strain (12). Although it is generally assumed that the serogroup of the progenitor in a capsular switch is the one most commonly associated with the genetic lineage, in reality the direction of the switch is unknown.
For serogroups B and Y isolates, fewer than 2% demonstrated capsular switching. However, almost 13% of serogroup C isolates belonged to genetic lineages associated with other serogroups, mostly serogroup B. This suggests fewer barriers for the acquisition of the serogroup C capsular genes. A gene conversion event to change the specificity of the capsule polymerase from (α2→8)-linked polysialic acid] (serogroup B) to (α2→9)-linked polysialic acid] (serogroup C) may be facilitated by the close similarity of the DNA sequence of the gene between these serogroups. Also there is evidence for selection processes that favor or restrict transformation events, such as the restriction modification system differences noted between ST-11 and ST-32 isolates (47). These hypotheses can be tested using in vitro experiments of horizontal gene transfer of meningococcal isolates of different serogroups and genetic lineages.
The Oregon serogroup B clone had previously been observed to have undergone capsular switching to serogroup C (9). We identified 5 additional serogroup C isolates that were indistinguishable from the Oregon clone by MLST and OMP genotyping. We also identified a serogroup Y isolate in Colorado that belonged to the ST-32 complex (ST 6065) and had the same OMP genotype profile as the Oregon clone. We also observed the Oregon serogroup B clone causing disease in 7 of the 9 other ABCs sites. However, circulation of the Oregon clone has not resulted in hyperendemic serogroup B disease in these other sites as it has done in Oregon (38). The reasons for this are not clear but continued surveillance is required.
In summary, we have defined the population and antigenic structure of invasive meningococcal isolates at ABCs sites throughout the U.S. Capsular switching was common, particularly among serogroup C isolates. Capsular switching from virulent serogroup C and Y lineages to serogroup B with clonal expansion is one mechanism by which replacement serogroup B disease could occur after meningococcal conjugate vaccine introduction. Although this has not occurred following introduction of monovalent serogroup C conjugate vaccines in the U.K. (48), the broader serogroup coverage of MCV4 could conceivably have a larger impact. Continuing to monitor for these events will be an important component of N. meningitidis surveillance in the setting of new meningococcal vaccines in the United States.
ACKNOWLEDGMENTS
This publication made use of the Neisseria Multi Locus Sequence Typing website (pubmlst.org/neisseria/) developed by Keith Jolley and Man-Suen Chan and located at the University of Oxford (34). The development of this site has been funded by the Wellcome Trust and European Union.
We thank the following Emerging Infections Program (EIP)/Active Bacterial Core surveillance (ABCs) site investigators and staff: Art Reingold, Pam Daily, Joelle Nadle, and Gretchen Rothrock (California); Ken Gershman and Steve Burnite (Colorado); Matt Carter and Susan Petit (Connecticut); Kathryn Arnold, Monica Farley, Wendy Baughman, and Paul Malpiedi (Georgia); Rosemary Hollick and Terresa Carter (Maryland), Ruth Lynfield, Brenda Jewell, and Jean Rainbow (Minnesota); Joan Baumbach and Joseph Bareta (New Mexico); Nancy Bennett and Nancy Spina (New York); Mark Schmidt and Ann Thomas (Oregon); William Schaffner and Brenda Barnes (Tennessee); and Chris Van Beneden, Carolyn Wright, Emily Weston, and Karrie-Ann Toews (CDC ABCs program). We also thank participating microbiology laboratory personnel and hospital infection preventionists in ABCs site hospitals and laboratories for identifying the N. meningitidis cases and submitting the bacterial isolates and Melina Lenser for performing molecular subtyping.
STUDY FUNDING
This study was supported by the Centers for Diseases Control and Prevention and by a grant from Sanofi Pasteur.
Footnotes
FINANCIAL DISCLOSURES
Dr. Harrison receives funding from the Centers for Disease Control and Prevention and the National Institute of Allergy and Infectious Diseases. He receives research support and lecture fees from Sanofi Pasteur; lecture fees from Novartis Vaccines; and has served as a consultant to GlaxoSmithKline, Novartis Vaccines, Sanofi Pasteur, and Wyeth. Ms. Shutt and Dr. Marsh receive research support from Sanofi Pasteur. Dr. Stephens has research funding from the National Institute of Allergy and Infectious Diseases, the Department of Veterans Affairs and the Georgia Research Alliance. Other authors: no disclosures.
Literature cited
- 1.Harrison LH, Trotter CL, Ramsay ME. Global epidemiology of meningococcal disease. Vaccine. 2009;27 Suppl 2:B51–B63. doi: 10.1016/j.vaccine.2009.04.063. [DOI] [PubMed] [Google Scholar]
- 2.Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease. N Engl J Med. 2001;344:1378–1388. doi: 10.1056/NEJM200105033441807. [DOI] [PubMed] [Google Scholar]
- 3.Bilukha OO, Rosenstein N. Prevention and control of meningococcal disease. Recommendations of the Advisory Committee on Immunization Practices (ACIP) MMWR Recomm Rep. 2005;54:1–21. [PubMed] [Google Scholar]
- 4.Revised recommendations of the Advisory Committee on Immunization Practices to Vaccinate all Persons Aged 11–18 Years with Meningococcal Conjugate Vaccine. MMWR Morb Mortal Wkly Rep. 2007;56:794–795. [PubMed] [Google Scholar]
- 5.Snape MD, Perrett KP, Ford KJ, et al. Immunogenicity of a tetravalent meningococcal glycoconjugate vaccine in infants: a randomized controlled trial. JAMA. 2008;299:173–184. doi: 10.1001/jama.2007.29-c. [DOI] [PubMed] [Google Scholar]
- 6.Harrison LH. A multivalent conjugate vaccine for prevention of meningococcal disease in infants. JAMA. 2008;299:217–219. doi: 10.1001/jama.2007.57-c. [DOI] [PubMed] [Google Scholar]
- 7.Nolan T, Lambert S, Roberton D, et al. A novel combined Haemophilus influenzae type b-Neisseria meningitidis serogroups C and Y-tetanus-toxoid conjugate vaccine is immunogenic and induces immune memory when co-administered with DTPa-HBV-IPV and conjugate pneumococcal vaccines in infants. Vaccine. 2007;25:8487–8499. doi: 10.1016/j.vaccine.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Keyserling H, Pina M, Hou V, Bassily E, Reinhardt A. Immunogenicity of a quadrivalent meningococcal polysaccharide diphtheria toxoid conjugate vaccine against serogroups A, C, Y, and W-135 (MCV-4) in 9 to 15-month olds. Infectious Diseases Society of America Annual Meeting; October 12-15, 2006; Toronto, Canada. Abstract 640. [Google Scholar]
- 9.Swartley JS, Marfin AA, Edupuganti S, et al. Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci U S A. 1997;94:271–276. doi: 10.1073/pnas.94.1.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison LH, Jolley KA, Shutt KA, et al. Antigenic shift and increased incidence of meningococcal disease. J Infect Dis. 2006;193:1266–1274. doi: 10.1086/501371. [DOI] [PubMed] [Google Scholar]
- 11.Mayer LW, Reeves MW, Al-Hamdan N, et al. Outbreak of W135 meningococcal disease in 2000: not emergence of a new W135 strain but clonal expansion within the electophoretic type-37 complex. J Infect Dis. 2002;185:1596–1605. doi: 10.1086/340414. [DOI] [PubMed] [Google Scholar]
- 12.Vogel U, Claus H, Frosch M. Rapid serogroup switching in Neisseria meningitidis. N Engl J Med. 2000;342:219–220. doi: 10.1056/NEJM200001203420319. [DOI] [PubMed] [Google Scholar]
- 13.Linz B, Schenker M, Zhu P, Achtman M. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis. Mol Microbiol. 2000;36:1049–1058. doi: 10.1046/j.1365-2958.2000.01932.x. [DOI] [PubMed] [Google Scholar]
- 14.Tsang RS, Law DK, Tyler SD, Stephens GS, Bigham M, Zollinger WD. Potential capsule switching from serogroup Y to B: The characterization of three such Neisseria meningitidis isolates causing invasive meningococcal disease in Canada. Can J Infect Dis Med Microbiol. 2005;16:171–174. doi: 10.1155/2005/216369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks LA, Harrison LH, Flannery B, et al. Incidence of pneumococcal disease due to non-pneumococcal conjugate vaccine (PCV7) serotypes in the United States during the era of widespread PCV7 vaccination, 1998–2004. J Infect Dis. 2007;196:1346–1354. doi: 10.1086/521626. [DOI] [PubMed] [Google Scholar]
- 16.Moore MR, Gertz RE, Jr., Woodbury RL, et al. Population snapshot of emergent Streptococcus pneumoniae serotype 19A in the United States, 2005. J Infect Dis. 2008;197:1016–1027. doi: 10.1086/528996. [DOI] [PubMed] [Google Scholar]
- 17.Pai R, Moore MR, Pilishvili T, Gertz RE, Whitney CG, Beall B. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J Infect Dis. 2005;192:1988–1995. doi: 10.1086/498043. [DOI] [PubMed] [Google Scholar]
- 18.Scott DA, Komjathy SF, Hu BT, et al. Phase 1 trial of a 13-valent pneumococcal conjugate vaccine in healthy adults. Vaccine. 2007;25:6164–6166. doi: 10.1016/j.vaccine.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Hsu HE, Shutt KA, Moore MR, et al. Effect of pneumococcal conjugate vaccine on pneumococcal meningitis. N Engl J Med. 2009;360:244–256. doi: 10.1056/NEJMoa0800836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison LH. Prospects for vaccine prevention of meningococcal infection. Clin Microbiol Rev. 2006;19:142–164. doi: 10.1128/CMR.19.1.142-164.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuchat A, Hilger T, Zell E, et al. Active bacterial core surveillance of the emerging infections program network. Emerg Infect Dis. 2001;7:92–99. doi: 10.3201/eid0701.010114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National vaccination coverage among adolescents aged 13-17 years--United States, 2006. MMWR Morb Mortal Wkly Rep. 2007;56:885–888. [PubMed] [Google Scholar]
- 23.Harrison LH, Dwyer DM, Maples CT, Billmann L. Risk of meningococcal infection in college students. Jama. 1999;281:1906–1910. doi: 10.1001/jama.281.20.1906. [DOI] [PubMed] [Google Scholar]
- 24.Harrison LH, Pass MA, Mendelsohn AB, et al. Invasive meningococcal disease in adolescents and young adults. Jama. 2001;286:694–699. doi: 10.1001/jama.286.6.694. [DOI] [PubMed] [Google Scholar]
- 25.Wedege E, Hoiby EA, Rosenqvist E, Froholm LO. Serotyping and subtyping of Neisseria meningitidis isolates by co-agglutination, dot-blotting and ELISA. J Med Microbiol. 1990;31:195–201. doi: 10.1099/00222615-31-3-195. [DOI] [PubMed] [Google Scholar]
- 26.Mothershed EA, Sacchi CT, Whitney AM, et al. Use of real-time PCR to resolve slide agglutination discrepancies in serogroup identification of Neisseria meningitidis. J Clin Microbiol. 2004;42:320–328. doi: 10.1128/JCM.42.1.320-328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holmes EC, Urwin R, Maiden MC. The influence of recombination on the population structure and evolution of the human pathogen Neisseria meningitidis. Mol Biol Evol. 1999;16:741–749. doi: 10.1093/oxfordjournals.molbev.a026159. [DOI] [PubMed] [Google Scholar]
- 28.Maiden MC, Bygraves JA, Feil E, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urwin R, Feavers IM, Jones DM, Maiden MC, Fox AJ. Molecular variation of meningococcal serotype 4 antigen genes. Epidemiol Infect. 1998;121:95–101. doi: 10.1017/s0950268898008942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thompson EA, Feavers IM, Maiden MC. Antigenic diversity of meningococcal enterobactin receptor FetA, a vaccine component. Microbiology. 2003;149:1849–1858. doi: 10.1099/mic.0.26131-0. [DOI] [PubMed] [Google Scholar]
- 31.Russell JE, Jolley KA, Feavers IM, Maiden MCJ, Suker J. PorA variable regions of Neisseria meningitidis. Emerging Infectious Diseases. 2004;10:674–678. doi: 10.3201/eid1004.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feavers IM, Gray SJ, Urwin R, et al. Multilocus sequence typing and antigen gene sequencing in the investigation of a meningococcal disease outbreak. J Clin Microbiol. 1999;37:3883–3887. doi: 10.1128/jcm.37.12.3883-3887.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 34.Jolley KA, Chan MS, Maiden MC. mlstdbNet - distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics. 2004;5:86. doi: 10.1186/1471-2105-5-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Russell JE, Jolley KA, Feavers IM, Maiden MC, Suker J. PorA variable regions of Neisseria meningitidis. Emerg Infect Dis. 2004;10:674–678. doi: 10.3201/eid1004.030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–1530. doi: 10.1128/JB.186.5.1518-1530.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granoff DM, Harrison LH, Borrow R. Meningococcal vaccines. In: Plotkin S, Orenstein WA, Offit PA, editors. Vaccines. Fifth ed. Philadelphia: Saunders Elsevier; 2008. pp. 399–434. [Google Scholar]
- 38.Diermayer M, Hedberg K, Hoesly F, et al. Epidemic serogroup B meningococcal disease in Oregon: the evolving epidemiology of the ET-5 strain. Jama. 1999;281:1493–1497. doi: 10.1001/jama.281.16.1493. [DOI] [PubMed] [Google Scholar]
- 39.Haley CE, Hedberg K, Harrison LH, Cieslak P. Evolution of the Oregon serogroup B meningococcal epidemic — persistence of the B:15:P1.7,16 clone. Submitted for publication. [Google Scholar]
- 40.Martin DR, Walker SJ, Baker MG, Lennon DR. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J Infect Dis. 1998;177:497–500. doi: 10.1086/517385. [DOI] [PubMed] [Google Scholar]
- 41.Dyet K, Devoy A, McDowell R, Martin D. New Zealand's epidemic of meningococcal disease described using molecular analysis: implications for vaccine delivery. Vaccine. 2005;23:2228–2230. doi: 10.1016/j.vaccine.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 42.Baker MG, Martin DR, Kieft CE, Lennon D. A 10-year serogroup B meningococcal disease epidemic in New Zealand: descriptive epidemiology, 1991–2000. J Paediatr Child Health. 2001;37:S13–S19. doi: 10.1046/j.1440-1754.2001.00722.x. [DOI] [PubMed] [Google Scholar]
- 43.Dyet KH, Martin DR. Clonal analysis of the serogroup B meningococci causing New Zealand's epidemic. Epidemiol Infect. 2006;134:377–383. doi: 10.1017/S0950268805004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Lemos AP, Yara TY, Gorla MC, et al. Clonal distribution of invasive Neisseria meningitidis serogroup C strains circulating from 1976 to 2005 in greater São Paulo, Brazil. J Clin Microbiol. 2007;45:1266–1273. doi: 10.1128/JCM.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marsh JW, O'Leary MM, Shutt KA, Harrison LH. Deletion of fetA gene sequences in serogroup B and C Neisseria meningitidis isolates. J Clin Microbiol. 2007;45:1333–1335. doi: 10.1128/JCM.02422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Claus H, Elias J, Meinhardt C, Frosch M, Vogel U. Deletion of the meningococcal fetA gene used for antigen sequence typing of invasive and commensal isolates from Germany: frequencies and mechanisms. J Clin Microbiol. 2007;45:2960–2964. doi: 10.1128/JCM.00696-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claus H, Friedrich A, Frosch M, Vogel U. Differential distribution of novel restriction-modification systems in clonal lineages of Neisseria meningitidis. J Bacteriol. 2000;182:1296–1303. doi: 10.1128/jb.182.5.1296-1303.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trotter CL, Ramsay ME, Gray S, Fox A, Kaczmarski E. No evidence for capsule replacement following mass immunisation with meningococcal serogroup C conjugate vaccines in England and Wales. Lancet Infect Dis. 2006;6:616–617. doi: 10.1016/S1473-3099(06)70584-9. author reply 617–618. [DOI] [PubMed] [Google Scholar]


