Abstract
The UV portion of sunlight is mutagenic and can modify DNA by producing various photoproducts. UV photodamage often occurs at dipyrimidine sites, to give cyclobutane, pyrimidine-(6-4)-pyrimidone (6-4), and pyrimidine-(6-4)-Dewar pyrimidone (Dewar) photoproducts, and at TA and AA sites. There is no reported evidence, however, of UV-photoproduct formation between C or 5-methylC (mC) and A. Irradiation of d(GTATmCATGAGGTGC) with UVB light at physiological pH gives an unexpected photoproduct that undergoes fast thermal deamination but does not revert to its original structure under UVC irradiation. Evidence from nuclease P1 (NP1) digestion coupled with electrospray ionization (ESI)-MS/MS is in accord with product formation between mC and A. HPLC analysis indicates that deamination gives a T<>A photoproduct that coelutes on reverse-phase chromatography with the well-known TA* photoproduct, formed from an initial [2+2] reaction between C5-C6 and C6-C5 of the adjacent thymine and adenine (as shown by Zhao, X., et al. (1996) Nucleic Acids Res. 24, 1554-1560 and Davies, R. J. et al. (2007) Nucleic Acids Res. 35, 1048-1053). Furthermore, the deamination product of the unknown mC<>A photoproduct and the TA* photoproduct undergo nearly identical fragmentation in tandem MS. The evidence, taken together, indicates that the deamination product of the unknown mCA photoproduct has the same chemical structure as the TA* photoproduct. Therefore, the unknown photoproduct is referred to mCA* photoproduct, which, upon deamination, gives the TA* photoproduct.
Keywords: deamination, DNA, enzyme-coupled, HPLC, mass spectrometry, 5-methylC, nuclease P1, photoproduct, TA*, UV
Introduction
Sunlight-induced DNA photo damage is one cause of mutations leading to skin cancer. The toxic, mutagenic and carcinogenic effects of sunlight, however, have been principally attributed to the ultraviolet (UV) portion, which can produce a variety of photoproducts in DNA (1-5). If not repaired, these lesions lead to mutations, increasing the risk for cancer. Until now, the most prevalent UV photoproducts are found at pyrimidine sites, giving cyclobutane dimers, cyclobutane, pyrimidine-(6-4)-pyrimidone (6-4)1, and their pyrimidine-(6-4)-Dewar pyrimidone (Dewar) valence isomers (10). Other UV photoproducts containing adenine such as TA*, which forms from an initial [2+2] cycloaddition reaction between C5-C6 and C6-C5 of the adjacent thymine and adenine (11, 12), A=A and AA* (13-16) are also well known. No UV photoproduct formation between other pairs of bases, however, has been reported.
Mutations due to UV irradiation are predominantly C-to-T transition and CC-to-TT tandem transitions at a dipyrimidine sequence (17, 18). Most C-to-T mutation hotspots occur at methylated CpG sites (19-21). The most common hypothesis is that the C-to-T transition occurs after the deamination of C or 5-methylC (mC), leading to insertion of A instead of a G in the next replication cycle (22). The important deamination reaction of mC-containing photoproducts is affected by flanking bases, pH, and secondary structure (23).
In the course of synthesizing TmC photoproducts of d(GTATmCATGAGGTGC) produced by irradiation with biologically relevant UVB (290-320 nm) light for deamination studies (Scheme 1), we found an unknown photoproduct. To elucidate the structure of the unknown, we employed enzyme digestion, correlation of HPLC retention times of unknown and reference materials, and mass spectrometry (MS). The combined approach allowed us to identify the unknown as a mCA* photoproduct, the name of which indicates covalent linkages between C5-C6 and C6-C5 of mC and A as in TA*. The identification took advantage of its rapid deamination to afford the well-known TA* photoproduct (11, 12, 16, 24, 25). In this communication, we report the structural determination of this unusual and first known photoproduct between mC and A.
Scheme 1.
Known photoproducts that could originate from the designed sequence.
Experimental Procedures
Materials
Oligodeoxynucleotides (ODNs) were purchased from Integrated DNA Technologies, Inc. (IDT) (Coralville, Iowa). Nuclease P1 (NP1) from Penicillium citrinum was from Sigma (St. Louis, MO). Milli-Q (18.2 mΩ/ cm) water was prepared by using a Millipore Corporation (Billerica, MA) Milli-Q water purification system. HPLC solvents were from Fisher Scientific (Fair Lawn, NJ).
Instrumentation
UVB irradiation was carried out with a 302 nm transilluminator (Spectroline model TR-302) or a 312 nm lamp with two Spectroline® XX-15B UV 15-watt tubes (peak UV intensity of 1,150 μW/cm2 at 25 cm from the source) from Spectronics Corporation (Westbury, New York). UVC (100-290 nm) irradiation was with a model UVG-254 Mineralight lamp (254 nm, Ultra-Violet Products, Inc., San Gabriel, CA). HPLC separation and analysis were carried out with a System Gold HPLC BioEssential HPLC with binary gradient 125 pumps and a diode array Model 168 detector (Beckman Coulter, Inc., Fullerton, CA). An X-Bridge column (C18, 4.6 ×75 mm, 2.5 μm, 135 Ǻ) from Waters Corporation (Milford, MA) was used for reversed-phase HPLC (RP-HPLC). Mass spectra were obtained with a Thermo Finnigan LTQ-FT mass spectrometer (Thermo Fisher Scientific, Inc., Waltham, MA) or an Applied Biosystems 4700 tandem time-of-flight mass spectrometer (Applied Biosystems, Foster City, CA).
UV irradiation of ODNs
ODNs from IDT were used without further purification. ODN solutions for UV irradiation were prepared as 50 μM in 10 mM tris-(hydroxymethyl)aminomethane HCl (pH 7.6). Samples were then purged with nitrogen gas for 5 min and placed in a polyethylene Ziplock bag. For 302 nm irradiation, the bags were placed on the transilluminator and irradiated for 2.5 h in a cold room (4 °C). For the 312 nm and UVC irradiations, the bags were placed on ice and irradiated for 2.5 h at a distance of approximately 1 cm from the lamp. All samples from irradiation were immediately transferred to Eppendorf tubes and stored in a −80 °C freezer to prevent deamination.
RP-HPLC analysis and purification of products
Basic solvents were used for HPLC to prevent any mC-containing photoproducts from deamination during separation. The flow rate was 1 mL/min with a gradient of 0-20% solvent B in solvent A for 50 min (solvent A: 50 mM triethylammonium acetate, pH 7.8; solvent B: 50% acetonitrile in 50 mM triethylammonium acetate, pH 7.8). Elutes were immediately put on dry ice and then stored in a −80 °C freezer until MS analyses could be conducted.
NP1 digestion
Separated photoproducts were collected accompanying the HPLC separation, put on dry ice, and then stored in a −80 °C freezer. HPLC fractions were thawed on ice before digestion. A 2-5 μL aliquot was mixed with 0.5 μL, 1 unit/μL NP1 and digested on ice for 3-5 min. The digested samples were then submitted to MS analysis.
MS Analysis
Electrospray ionization (ESI) mass spectra were obtained in the negative-ion mode on a LTQ-FTICR instrument. Typically, samples were introduced to the mass spectrometer by injecting in a 10 μL loop. A solution of 50/50 (v/v) methanol/water was used as the spray solvent at a flow rate of 5 μL /min. The spray voltage was 3.5 kV. The capillary voltage and temperature were 46 V and 250 °C, respectively. ESI-MS/MS experiments were done by using collision-activated dissociation with helium as the collision gas. The mass window for precursor-ion selection was 2.5 m/z units, and the collision energy was 30% of the maximum value, producing product ions of good signal-to-noise ratio. Exonuclease digestion coupled MALDI-MS experiments were done on the 4700 MALDI TOF/TOF instrument by using the procedures previously reported (26).
Results and Discussion
HPLC Analysis of the New Photoproduct
After irradiation of d(GTATmCATGAGGTGC) with 302 nm light, RP-HPLC analysis showed four significant photoproducts (PPs): PP1 (~ 18% yield based on HPLC peak areas), PP2, PP3, and PP4 (Figure 1A, upper panel). After incubating the irradiation mixture in a 37 °C water bath for 1 h to cause deamination, we found that the peaks corresponding to PP1 and PP2 decreased, whereas two new peaks, labeled as PP1* and PP2* increased (Figure 1A, lower panel). PP1 deaminates faster than PP2 judging from HPLC peak areas: approximately 60% of PP1 had deaminated whereas only 30% of PP2 had done so. These new peaks may correspond to deamination products of mC photoproducts. When 312 nm light was used, we saw only two significant photoproducts PP1 (~ 22% yield based on HPLC peak areas) and PP4 in the HPLC analysis (Figure 1B, upper panel). Thermal incubation of the irradiation mixture, as described above, caused the peak corresponding to PP1 to decrease and to be replaced by a new peak labeled as PP1* (Figure 1B, lower panel). The sum of peak areas corresponding to PP1 and PP1* is roughly that of the original peak PP1, an observation that is consistent with the assignment that PP1* corresponds to deamination of an mC photoproduct in PP1. Furthermore, PP2* is likely to be the deamination product of PP2, which is also an mC-containing photoproduct.
Figure 1.
RP-HPLC of UVB-induced photoproducts of d(GTATmCATGAGGTGC) at pH 7.6 (upper panel) and of corresponding deamination products (lower panel) with A) 302 nm light and B) 312 nm light. SM: starting material, * indicates photoproduct that has undergone deamination
NP1-coupled ESI-MS and ESI-MS/MS assay
When we treated the photodamaged ODN with nuclease P1, we found a trinucleotide triphosphate, which upon mass spectrometric analysis, gave ions of m/z of 937 for PP1, PP2, PP3 and PP4, and of m/z 938 for PP1* and PP2*. These are the expected digestion products of an ODN containing a photoproduct between adjoining bases (27, 28). The trinucleotide that gives an ion of m/z 937 has a composition of [pd(T<>mCA) - H]− or [pd(mC<>AT) - H]−, and the one of m/z 938 is either [pd(T<>TA) - H]− or [pd(T<>AT) - H]−. To resolve these possibilities, we carried out MS/MS experiments on selected ions of the tri-nucleotides released from the ODN photoproducts in which only the non-damaged base can be lost while the photochemically coupled bases cannot be fragmented. We found the most abundant product ions of m/z 615 and 616 for PP1 and PP1*, respectively, both of which were formed by loss of a 2′-deoxythymidine-5′-phosphate (pdT) (Supplementary Information, Figure S1). These observations allow us to propose that the trinucleotides formed in NP1 digestion of PP1 and PP1* are pd(mC<>AT) and pd(T<>AT), respectively; the latter is a deamination product of the former.
We also observed that the most abundant fragment ions are of m/z of 802 and 803 for PP2, PP2*, respectively; these arise from a loss of 2′-deoxyadenosine-5′-phosphate (pdA) (Supplementary Information, Figure S2). We can now propose that the trinucleotides from PP2 and PP2* are pd(T<>mCA) and pd(T<>TA), respectively; the latter is a deamination product of the former. Similarly, we found that the most abundant product ions are of m/z of 802 for the digestion products of PP3 (Supplementary Information, Figure S3) and PP4 (Supplementary Information, Figure S4), owing to losses of pdA. The trinucleotides liberated in the digestion of PP3 and PP4 are both assigned as pd(T<>mCA), because they can only lose a pdA species.
Stereochemistry of PP2, PP3 and PP4
Given that PP1 and PP2 deaminate, they may be cyclobutane photoproducts (6, 29, 30). It is known that UVC light can revert [2 + 2] cyclobutane photoproducts to their parent ODNs and convert Dewar photoproducts to their (6-4) valence isomers, which are detectable at 325 nm. Deamination does not change the stereochemistry of the photoproducts. We carried out UVC irradiation with 254 nm light on the HPLC-separated PP1*, PP2*, PP3 and PP4. Given that no photoreversion or photoconversion of PP1* occurred, we can exclude PP1 and PP1* as cyclobutane photoproducts. PP2* and PP3, however, reverted to the parent ODN, indicating that they are cyclobutane photoproducts of cis-syn (c,s) or trans-syn (t,s) stereochemistry. We assigned PP2 and PP2* as mC-containing (c,s) photoproduct because PP2 deaminates rapidly. We assigned PP3 as a mC-containing (t,s) photoproduct because it undergoes no significant deamination under the conditions used here, as would be expected on the basis of previous work (6, 10, 29). PP4 undergoes a transition to its (6-4) isomer, suggesting a Dewar photoproduct (Table 1)
Table 1.
Identification of UVB induced photoproducts of d(GTATmCATGAGGTGC) at pH 7.6.
| Photoproduct (PP) | PP1 | PP1* | PP2 | PP2* | PP3 | PP4 |
| Photodimer Assignment | mCA* | TA* | T[c,s]mC | T[c,s]T | T[t,s]mC | T[Dewar]mC |
Comparison of PP1* and the authentic TA* Photoproduct by HPLC and NP1-coupled ESI-MS
Considering that PP1 is an mC<>A photoproduct and that it may also contribute tomC to T mutations via deamination, we were motivated to determine its structure. Interestingly, PP1 deaminates even faster than the T[c,s]mC photoproduct. Given that deamination doesn't change the stereochemistry of a photoproduct, we decided to investigate PP1*, the thermally stable deamination product of PP1. Further, given that the deamination product PP1* contains a T<>A photoproduct but is not a cyclobutane adduct, we suspected that it has the structure of the well known TA* photoproduct (11, 12). Thus, we compared the RP-HPLC retention time of the T<>A photoproduct formed from deamination of PP1* to that of the authentic TA* photoproduct formed within the same sequence context.
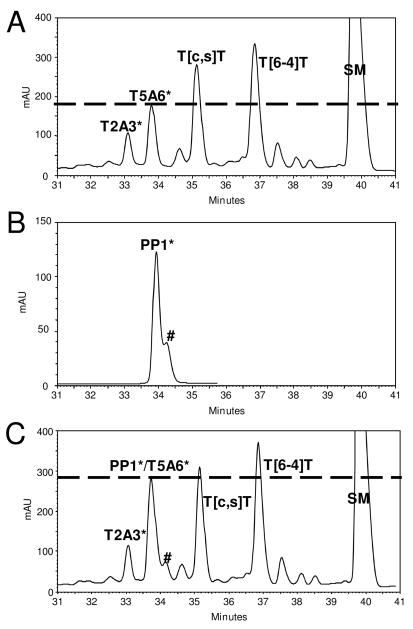
The authentic TA* photoproduct was obtained by UVC (254 nm) of d(GTATTATGAGGTGC). We assigned the reference photoproducts on the basis of photoconversion, photoreversion, and NP1-coupled ESI-MS/MS as described above (Figure 2). Particularly, we can distinguish T5A6* from T2A3* with bovine spleen phosphodiesterase-coupled ladder sequencing by MALDI-MS (data not shown) (26, 31, 32), taking advantage of exonuclease digestion that goes from 5′ to 3′ and terminates just before T5. The approach was to inject onto the HPLC the same amount of sample individually and then to co-inject them. Co-elution of PP1* and the T5A6*-containing photoproduct 14-mer strongly suggests that the two photoproducts have the same structure (Figure 2). This suggestion was then confirmed by NP1-coupled ESI-MSn analysis, whereby both MS2 and MS3 of the NP1 digested PP1* and authentic T5<>A6 photoproduct 14-mer show the same fragmentation pattern (Figure 3).
Figure 2.
RP-HPLC assay of A) ~ 5 nmol UVC irradiation mixture of d(GTATTATGAGGTGC), B) ~ 360 pmol of deamination product of HPLC fraction of d(GTATmCATGAGGTGC), and C) a coinjected sample of A and B. # is the contamination peak of PP3 from HPLC separation (Figure 1A, lower panel)
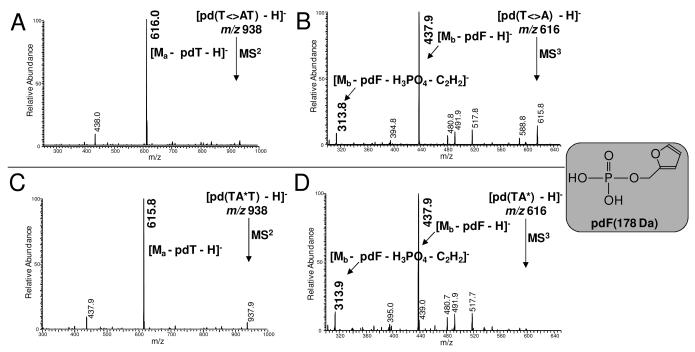
Figure 3.
NP1 coupled ESI- MS2 and MS3 assays of mC<>A photoproduct of d(GTATmCATGAGGTGC) following deamination to T<>A (A and B, respectively), and TA* photoproduct of T5A6 of d(GTATTATGAGGTGC) (C and D, respectively).
As shown in the product-ion spectra, both compounds undergo the expected major neutral loss of pdT yielding a most abundant product ion of m/z 616. An MS3 experiment shows that the m/z 616 ion formed from the precursor ion gives the same major product ion of m/z 438. The fragmentation to give the m/z 438 ion is a loss of the 2-deoxyribose ring with the attached phosphate group (pdF). The other distinctive product ion is of m/z 314, which is most abundant in an MS4 experiment whereby the m/z 438 ion is probed as formed by the sequence m/z 938 → m/z 616 ion → m/z 438 ion. The ion of m/z 314 likely arises by the loss of phosphoric acid (H3PO4), possibly via an ion-dipole complex, accompanied by the elimination of acetylene (C2H2). These fragments are consistent with a photoproduct structure made up of a stable tricyclic center that loses groups on the periphery. More importantly, they add support that the newly discovered mC<>A photoproduct is an analog of the well-known TA* photoproduct (Scheme 2).
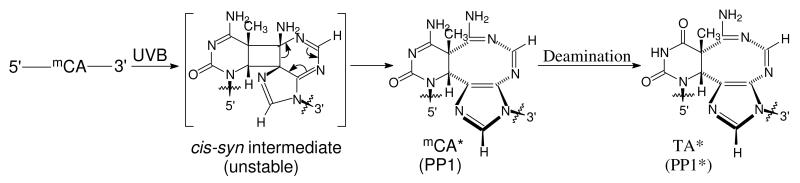
Scheme 2.
Proposed structure of the mC<>A photoproduct and its mechanism of formation and deamination.
Conclusion
We discovered a new photoproduct of d(GTATmCATGAGGTGC) involving reaction of mC and A. Deamination gave the well-known TA* photoproduct (11, 12, 16, 24, 25) whose structure was previously completely established by NMR (11), MS (16), and X-ray crystallography (12), and its mutation spectrum determined in bacteria (33). Comparisons of HPLC retention times and tandem MS show that the properties of the mC<>A deamination product are nearly identical to those of the authentic TA* photoproduct, substantiating their identical structures. Interestingly, TA* forms under UVC whereas mCA arises under UVB. This is consistent with the UV absorbance of thymine (λ max = 267 nm) has greater overlap with UVC whereas mC (λ max = 278 nm) has greater overlap with the UVB lamp.
The mC residues in DNA play an important role in regulation of gene expression and chromatin structure (34, 35), and UV-induced damage of mC could be mutagenic. Although methylation of C predominantly occurs at d(CpG) sites in mammals (35, 36), it is conceivable that an inter-strand type mCA* could form in DNA that is folded to bring mC and A proximate to each other (26, 37-39). Non-CG methylation is present primarily in viral or stably integrated plasmid DNA sequences (40) and has also been detected in significant abundance in human embryonic stem cells and induced pluripotent stem cells (41, 42), although photochemical damage should not be an issue for them, and as well as in plants (43-45). Further photochemistry studies could be conducted targeting a series of DNA duplexes containing a mCA site and folded DNA having mC and A in close proximity to explore the possibility of intra-and inter-strand type mCA* formation, respectively. Future studies will also required to define better the biological consequences of mCA* photoproduct formation.
Supplementary Material
Acknowledgments
This research was supported by the Washington University NIH Mass Spectrometry Research Resource (Grant No. P41 RR000954) and by NIH (Grant CA40463).
Footnotes
Supporting Information Available: Available are additional mass spectra for photoproducts and their deamination products of d(GTATmCATGAGGTGC), free of charge via the internet at http://pubs.acs.org.
Abbreviations: [c,s], cis,syn; Dewar, pyrimidine-(6-4)-Dewar pyrimidone; ESI, electrospray ionization; mC, 5-methylC; mCA*, photoproduct formed between 5-methylcytosine and adenine that deaminates to give the TA* product (see below); MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; MS3, tandem mass spectrometry in which a first-generation product ion is fragmented further; NP1, Nuclease P1; ODN, oligodeoxynucleotide; PP, photoproduct; (6-4), pyrimidine-(6-4)-pyrimidone; RP-HPLC, reversed phase high performance liquid chromatography; TA*, photoproduct, formed from an initial [2+2] photoreaction between C5-C6 and C6-C5 of thymine and adenine; [t,s], trans,syn; UVB, 290-320 nm; UVC, 100-290 nm.
References
- 1.Tyrrell RM. The molecular and cellular pathology of solar ultraviolet radiation. Mol. Aspects Med. 1994;15:1–77. [PubMed] [Google Scholar]
- 2.Masamitsu Ichihashi MU, Budiyanto Arief, Araki Keishi, Funasaka Yoko. Sunlight and Skin Cancer. Environmental Sciences. 2000:203–227. [Google Scholar]
- 3.Hemminki K, Xu G, Le Curieux F. Ultraviolet radiation-induced photoproducts in human skin DNA as biomarkers of damage and its repair. IARC Sci. Publ. 2001;154:69–79. [PubMed] [Google Scholar]
- 4.Cleaver JE, Crowley E. UV damage, DNA repair and skin carcinogenesis. Front Biosci. 2002;7:d1024–1043. doi: 10.2741/A829. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat. Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 6.Shetlar MD, Basus VJ, Falick AM, Mujeeb A. The cyclobutane dimers of 5-methylcytosine and their deamination products. Photochem. Photobiol. Sci. 2004;3:968–979. doi: 10.1039/b404271a. [DOI] [PubMed] [Google Scholar]
- 7.Taylor JS. In: DNA Damage Recognition. Siede W, Kow YW, Doetsch PW, editors. Taylor and Francis Group; New York: 2006. pp. 67–94. [Google Scholar]
- 8.Pfeifer GP. Formation and processing of UV photoproducts: effects of DNA sequence and chromatin environment. Photochem. Photobiol. 1997;65:270–283. doi: 10.1111/j.1751-1097.1997.tb08560.x. [DOI] [PubMed] [Google Scholar]
- 9.Celewicz L, Mayer M, Shetlar MD. The photochemistry of thymidylyl-(3′-5′)-5-methyl-2′-deoxycytidine in aqueous solution. Photochem. Photobiol. 2005;81:404–418. doi: 10.1562/2004-06-15-ra-201.1. [DOI] [PubMed] [Google Scholar]
- 10.Douki T, Cadet J. Formation of cyclobutane dimers and (6-4) photoproducts upon far-UV photolysis of 5-methylcytosine-containing dinucleotide monophosphates. Biochemistry. 1994;33:11942–11950. doi: 10.1021/bi00205a033. [DOI] [PubMed] [Google Scholar]
- 11.Zhao X, Nadji S, Kao JL, Taylor JS. The structure of d(TpA), the major photoproduct of thymidylyl-(3′5′)-deoxyadenosine. Nucleic Acids Res. 1996;24:1554–1560. doi: 10.1093/nar/24.8.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davies RJ, Malone JF, Gan Y, Cardin CJ, Lee MP, Neidle S. High-resolution crystal structure of the intramolecular d(TpA) thymine-adenine photoadduct and its mechanistic implications. Nucleic Acids Res. 2007;35:1048–1053. doi: 10.1093/nar/gkl1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumar S, Sharma ND, Davies RJ, Phillipson DW, McCloskey JA. The isolation and characterisation of a new type of dimeric adenine photoproduct in UV-irradiated deoxyadenylates. Nucleic Acids Res. 1987;15:1199–1216. doi: 10.1093/nar/15.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma ND, Davies RJ. Extent of formation of a dimeric adenine photoproduct in polynucleotides and DNA. J. Photochem. Photobiol. B. 1989;3:247–258. doi: 10.1016/1011-1344(89)80066-1. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, Joshi PC, Sharma ND, Bose SN, Jeremy R, Davies H, Takeda N, McCloskey JA. Adenine photodimerization in deoxyadenylate sequences: elucidation of the mechanism through structural studies of a major d(ApA) photoproduct. Nucleic Acids Res. 1991;19:2841–2847. doi: 10.1093/nar/19.11.2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Taylor JS, Gross ML. Isolation and mass spectrometric characterization of dimeric adenine photoproducts in oligodeoxynucleotides. Chem. Res. Toxicol. 2001;14:738–745. doi: 10.1021/tx010009u. [DOI] [PubMed] [Google Scholar]
- 17.Horsfall MJ, Borden A, Lawrence CW. Mutagenic properties of the T-C cyclobutane dimer. J. Bacteriol. 1997;179:2835–2839. doi: 10.1128/jb.179.9.2835-2839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brash DE, Rudolph JA, Simon JA, Lin A, McKenna GJ, Baden HP, Halperin AJ, Ponten J. A role for sunlight in skin cancer: UV-induced p53 mutations in squamous cell carcinoma. Proc. Natl. Acad. Sci. USA. 1991;88:10124–10128. doi: 10.1073/pnas.88.22.10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.You YH, Li C, Pfeifer GP. Involvement of 5-methylcytosine in sunlight-induced mutagenesis. J. Mol. Biol. 1999;293:493–503. doi: 10.1006/jmbi.1999.3174. [DOI] [PubMed] [Google Scholar]
- 20.Mitchell DL. Effects of cytosine methylation on pyrimidine dimer formation in DNA. Photochem. Photobiol. 2000;71:162–165. doi: 10.1562/0031-8655(2000)071<0162:eocmop>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 21.Tommasi S, Denissenko MF, Pfeifer GP. Sunlight induces pyrimidine dimers preferentially at 5-methylcytosine bases. Cancer Res. 1997;57:4727–4730. [PubMed] [Google Scholar]
- 22.Lee DH, Pfeifer GP. Deamination of 5-methylcytosines within cyclobutane pyrimidine dimers is an important component of UVB mutagenesis. J. Biol. Chem. 2003;278:10314–10321. doi: 10.1074/jbc.M212696200. [DOI] [PubMed] [Google Scholar]
- 23.Cannistraro VJ, Taylor JS. Acceleration of 5-methylcytosine deamination in cyclobutane dimers by G and its implications for UV-induced C-to-T mutation hotspots. J. Mol. Biol. 2009;392:1145–1157. doi: 10.1016/j.jmb.2009.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bose SN, Davies RJ, Sethi SK, McCloskey JA. Formation of an adeninethymine photoadduct in the deoxydinucleoside monophosphate d(TpA) and in DNA. Science. 1983;220:723–725. doi: 10.1126/science.6836308. [DOI] [PubMed] [Google Scholar]
- 25.Bose SN, Kumar S, Davies RJ, Sethi SK, McCloskey JA. The photochemistry of d(T-A) in aqueous solution and in ice. Nucleic Acids Res. 1984;12:7929–7947. doi: 10.1093/nar/12.20.7929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su DG, Kao JL, Gross ML, Taylor JS. Structure determination of an interstrand-type cis-anti cyclobutane thymine dimer produced in high yield by UVB light in an oligodeoxynucleotide at acidic pH. J. Am. Chem. Soc. 2008;130:11328–11337. doi: 10.1021/ja8010836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Taylor JS, Gross ML. Nuclease P1 digestion combined with tandem mass spectrometry for the structure determination of DNA photoproducts. Chem. Res. Toxicol. 1999;12:1077–1082. doi: 10.1021/tx9900831. [DOI] [PubMed] [Google Scholar]
- 28.Vu B, Cannistraro VJ, Sun L, Taylor JS. DNA synthesis past a 5-methylC-containing cis-syn-cyclobutane pyrimidine dimer by yeast pol eta is highly nonmutagenic. Biochemistry. 2006;45:9327–9335. doi: 10.1021/bi0602009. [DOI] [PubMed] [Google Scholar]
- 29.Lemaire DG, Ruzsicska BP. Kinetic analysis of the deamination reactions of cyclobutane dimers of thymidylyl-3′,5′-2′-deoxycytidine and 2′-deoxycytidylyl-3′,5′-thymidine. Biochemistry. 1993;32:2525–2533. doi: 10.1021/bi00061a009. [DOI] [PubMed] [Google Scholar]
- 30.Mosbaugh DW, Bennett SE. Uracil-excision DNA repair. Prog. Nucleic Acid Res. Mol. Biol. 1994;48:315–370. doi: 10.1016/s0079-6603(08)60859-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhang LK, Rempel D, Gross ML. Matrix-assisted laser desorption/ionization mass spectrometry for locating abasic sites and determining the rates of enzymatic hydrolysis of model oligodeoxynucleotides. Anal. Chem. 2001;73:3263–3273. doi: 10.1021/ac010042l. [DOI] [PubMed] [Google Scholar]
- 32.Zhang LK, Ren Y, Rempel D, Taylor JS, Gross ML. Determination of photomodified oligodeoxynucleotides by exonuclease digestion, matrix-assisted laser desorption/ionization and post-source decay mass spectrometry. J. Am. Soc. Mass Spectrom. 2001;12:1127–1135. doi: 10.1016/S1044-0305(01)00291-4. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Taylor JS. Mutation spectra of TA*, the major photoproduct of thymidylyl-(3′5′)-deoxyadenosine, in Escherichia coli under SOS conditions. Nucleic Acids Res. 1996;24:1561–1565. doi: 10.1093/nar/24.8.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Antequera F, Bird A. CpG islands. EXS. 1993;64:169–185. doi: 10.1007/978-3-0348-9118-9_8. [DOI] [PubMed] [Google Scholar]
- 35.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 36.Illingworth RS, Bird AP. CpG islands--‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 37.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 38.Douki T, Laporte G, Cadet J. Inter-strand photoproducts are produced in high yield within A-DNA exposed to UVC radiation. Nucleic Acids Res. 2003;31:3134–3142. doi: 10.1093/nar/gkg408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su DG, Fang H, Gross ML, Taylor JS. Photocrosslinking of human telomeric G-quadruplex loops by anti cyclobutane thymine dimer formation. Proc. Natl. Acad. Sci. USA. 2009;106:12861–12866. doi: 10.1073/pnas.0902386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorincz MC, Groudine M. C(m)C(a/t)GG methylation: a new epigenetic mark in mammalian DNA? Proc. Natl. Acad. Sci. USA. 2001;98:10034–10036. doi: 10.1073/pnas.201392598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Costello JF, Krzywinski M, Marra MA. A first look at entire human methylomes. Nat. Biotechnol. 2009;27:1130–1132. doi: 10.1038/nbt1209-1130. [DOI] [PubMed] [Google Scholar]
- 43.Finnegan EJ, Genger RK, Peacock WJ, Dennis ES. DNA Methylation in Plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:223–247. doi: 10.1146/annurev.arplant.49.1.223. [DOI] [PubMed] [Google Scholar]
- 44.Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 45.Vanyushin BF. DNA methylation in plants. Curr. Top. Microbiol. Immunol. 2006;301:67–122. doi: 10.1007/3-540-31390-7_4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.